ABSTRACT
RNA interference-based technologies have emerged as an attractive and effective therapeutic option with potential application in diverse human diseases. These tools rely on the development of efficient strategies to obtain homogeneous non-coding RNA samples with adequate integrity and purity, thus avoiding non-targeted gene-silencing and related side-effects that impair their application onto pre-clinical practice. These RNAs have been preferentially obtained by in vitro transcription using DNA templates or via chemical synthesis. As an alternative to overcome the limitations presented by these methods, in vivo recombinant production of RNA biomolecules has become the focus in RNA synthesis research. Therefore, using pre-miR-29b as a model, here it is evaluated the time-course profile of Escherichia coli and Rhodovolum sulfidophilum microfactories to produce this microRNA. As the presence of major host contaminants arising from the biosynthesis process may have important implications in the subsequent downstream processing, it is also evaluated the production of genomic DNA and host total proteins. Considering the rapidly growing interest on these innovative biopharmaceuticals, novel, more cost-effective, simple and easily scaled-up technologies are highly desirable. As microRNA recombinant expression fulfills those requirements, it may take the leading edge in the methodologies currently available to obtain microRNAs for clinical or structural studies.
KEYWORDS: Escherichia coli, fermentation, miRNA, non-coding RNA, recombinant RNA, RNAi technology, Rhodovolum sulfidophilum
RNA interference technology and pre-miR-29b in Alzheimer disease
The discovery of functional small noncoding RNAs such as microRNAs (miRNA or miR), small interfering RNAs and piwi-interacting RNAs as novel agents for regulating gene expression in different eukaryotic cellular processes has opened new opportunities to develop RNA-based therapies for diverse diseases.1,2 This process is termed RNA interference (RNAi) and involves many small RNAs that act as regulators of mRNA expression for therapeutic knockdown of disease-causing genes.1,3 The discovery of RNAi has enabled researchers to exploit various small RNA tools to control target gene expression, to delineate gene function, and investigate cellular signaling and networks.2 In particular, increasing evidences in literature suggest that miR-29 expression is involved in several regulatory actions, namely in tumor-suppressing and immune-modulating processes, as well as in the regulation of pathways of neurodegenerative diseases.4-6 Previously, our research group reported an integrated strategy for the biosynthesis,7 purification8 and delivery9 of recombinant pre-miR-29b, demonstrating in vitro that pre-miR-29b preparations downregulate ß-site amyloid precursor protein cleaving enzyme 1 (BACE1) and Aß42 levels.10 Together, these results showed that recombinant pre-miR-29b improved the outcome regarding the currently available methodologies of microRNA-based therapeutics in Alzheimer disease.10
Advantages and drawbacks of different sources of RNA biopharmaceuticals
Systematic studies of the structure and function of RNA and the establishment of RNA-based therapeutics usually require large quantities of the target RNA with adequate integrity, purity and biological activity.2 The required RNA for biomolecular applications has mostly been obtained by two distinct methods11: the longer oligoribonucleotides by in vitro transcription from DNA templates using T7 RNA polymerase12 whereas short oligoribonucleotides are preferentially obtained via chemical synthesis by phosphoamidite chemistry.13 Despite recent improvements, these sources of agents for RNA research remain costly, laborious and have their limitations with respect to sequence requirements, variations in yield, non-templated nucleotide additions and/or the maximum length of the oligonucleotide.11,14 Also, although these methods are efficient in miRNA production, several purification protocols need to be used to remove process-associated impurities (plasmid DNA, incomplete transcripts, enzymes, salts and others) once they can lead to non-targeted gene-silencing. Actually, this fact still restricts the application of these RNAs onto pre-clinical or clinical trials.15,16 In particular, beyond the fact that large-scale chemical synthesis of RNA remains technically difficult and expensive, these RNAs are traditionally purified by preparative electrophoresis followed by gel extraction, a procedure that is time-consuming and leads to acrylamide-associated impurities within the RNA.17
As an alternative to overcome these limitations, huge efforts have been made in recent years to develop recombinant RNA techniques to cost-effectively produce biologic RNA agents in vivo that can better retain the structure, function and safety properties of natural RNAs.2 In fact, the success of this strategy relies on the fact that these recombinant RNAs are recognized by cellular machinery, being precisely processed, post-transcriptionally modified and not subjected to 3′ polyadenylation, which triggers RNA degradation.18
Recombinant RNA biosynthesis: General considerations about the hosts and the RNA design
The underlying principle of recombinant expression of RNA is rather straightforward: the target RNA coding sequence is introduced into a vector and the resulting plasmid is inserted into host cells grown in appropriate conditions.19 Then, the host transcription machinery will synthesize the RNA of interest that should be accumulated in cytosol and finally, at the end of culture, cells are pelleted, lysed and the RNA is purified by standard chromatographic techniques.20
Recombinant RNA expression has been mostly achieved using Escherichia coli (E. coli) as the host, once it can be grown easily and economically and a large number of plasmids and strains are available, allowing the construction of a rational strategy.20 On the other hand, the production of artificial RNAs in the marine phototrophic bacterium Rhodovolum sulfidophilum (R. sulfidophilum) was first achieved in 2010 by Suzuki and colleagues.21 In particular, as R. sulfidophilum does not accumulate any RNases in the culture medium, its ability to produce extracellular nucleic acids21 opens new perspectives in the RNA recombinant technology once their stability is greatly enhanced. To achieve successful recombinant RNA expression in E. coli, researchers usually use a strategy of “camouflage” of the RNA of interest within a stable RNA scaffold22 that may be achieved using tRNA11,17,22 or rRNA.23 This strategy relies on expressing the RNA of interest inserted in the place of the anticodon stem of the tRNA and as it is hidden within a standard tRNA structure, the recombinant RNA escapes ribonucleases and is processed by the standard E. coli tRNA processing enzymes.20 Then, although for some studies, using the entire RNA chimera might be acceptable, in other cases it might be preferable to induce the cleavage of the desired RNA off the scaffold, what may be achieved using ribozyme, DNAzyme or by cleavage with RNase H using a pair of guide DNA oligonucleotides - the method that seems to work better.22 More recently, motivated by the concept of “prodrug” and the idea to deploy biologic RNAs to perform RNA actions, Chen and collaborators developed a novel optimal non-coding RNA scaffold (OnRS)-based strategy to achieve a consistent high-yield production of chimeric RNAs in E. coli.14 Indeed, they found that using a fusion tRNA/pre-miRNA isolated from bacteria, the pre-miRNA is selectively processed to biologically active miRNA in human cells while the tRNA scaffold is degraded to tRNA fragments.14 On the other hand, using R. sulfidophilum as the expression host, a similar strategy to the first used by Suzuki and colleagues in 201021 was also applied by our research group7 where the target RNA is flanked by two hammerhead ribozyme sequences that possess self-cleavage activities leading to the releasing of the mature recombinant target RNA. Another important issue for evaluating the therapeutic effect of miRNAs is the target sequence cloned into the plasmid. Indeed, to evaluate the biologic effect of miR-29, the production of the pre-miRNA was chosen instead of the mature miRNA since not only its recognition and processing within the cell is more efficient but also because its structural characteristics can facilitate the purification of the target miRNAs.7,24
Choosing the appropriate recombinant microrna microfactory: Escherichia coli and Rhodovolum sulfidophilum
Previously, our group successfully reported the production of recombinant human pre-miR-29b both intra and extracellularly in R. sulfidophilum.7 Indeed, higher extracellular pre-miR-29b levels were obtained after 40 h of bacterial growth reaching a concentration near 182 µg/L while total intracellular pre-miR-29b was of about 358 µg/L at 32 h.7 Moreover, almost 87% cells were viable at the end of the fermentation, guaranteeing that the extracellular medium is not contaminated with intracellular RNases and endotoxins, thus ensuring the integrity of the target RNA.7 Following these results and to evaluate which host can be more suitable for pre-miR-29b recombinant biosynthesis, chemically competent E. coli DH5α cells were transformed with pBHSR1-RM-pre-miR-29b by heat-shock. Specifically, E. coli fermentation experiments were performed in 500 mL shake-flasks containing 125 mL of TB medium (12 g/L tryptone, 24 g/L yeast extract, 4 mL/L glycerol, 0.017 M KH2PO4, 0.072 M K2HPO4) supplemented with 30 µg/mL kanamycin and incubated at 37°C and 250 rpm. A typical growth profile of E. coli DH5α harboring the recombinant plasmid pBHSR1-RM-pre-miR-29b is shown in Fig. 1. According to these results, cell growth was suspended at the beginning of logarithmic decline phase (OD600 ≈5.4 after 8 h) and as it was expected, E. coli growth kinetics was much faster than R. sulfidophilum, allowing to perform a fermentation experiment in just one day, compared with a 4-day period required by R. sulfidophilum.
Figure 1.

Growth profile of E. coli DH5α harboring the plasmid pHBSR1-RM-pre-miR-29b cultivated in TB medium at 37°C and 250 rpm (red line/primary scale). Time-course analysis of intracellular pre-miR-29b production in E. coli DH5α cultures (black line/secondary scale) measured by quantitative RT-PCR using a specific probe and as described previously9 Error bars indicate standard deviations calculated from 3 independent samples.
Samples were recovered every hour either to assess cell growth or the levels of pre-miR-29b, genomic DNA and total host proteins, according to the experimental protocols described previously.7 For intracellular fractions, the phenol-chloroform method using TripleXtractor reagent (GRISP-Research Solutions, Porto, Portugal) was applied to extract RNA, followed by the synthesis of cDNA and the pre-miR-29b levels were determined using RT-qPCR using a specific probe.7 Initially, the extracted RNA samples obtained during the course of fermentation were analyzed by polyacrylamide urea gel electrophoresis to evaluate their integrity and quality. As shown in Figure 2-A, it is possible to observe that E. coli-derived pre-miR-29b was biosynthesized in a high quality and intact form. Moreover, the same electrophoretic analysis was performed in intracellular R. sulfidophilum fractions (Fig. 2-B) and it was also confirmed the high stability and integrity of pre-miR-29b samples.
Figure 2.
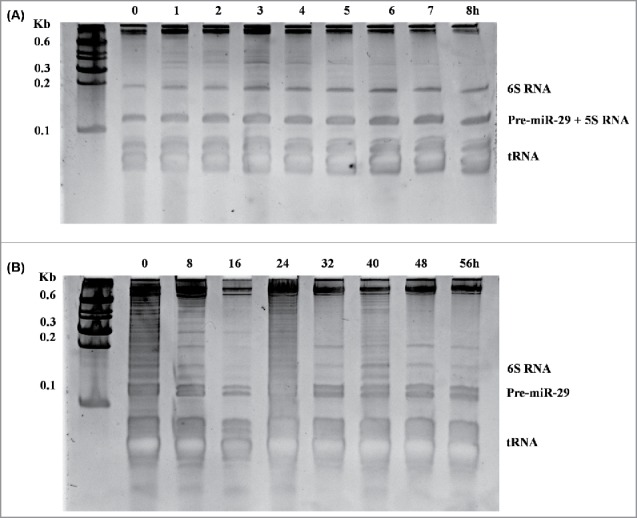
Electrophoretic analysis of nucleic acids from intracellular fractions of E. coli DH5α (A) and R. sulfidophilum (B), both containing the plasmid pHBSR1-RM-pre-miR-29b. Samples were recovered from cell suspensions at different periods of fermentation and the total RNA fractions were obtained by the phenol-chloroform method using TripleXtractor reagent (GRISP-Research Solutions, Porto, Portugal). Lanes, cultivation periods. Lane M, molecular weight marker.
In what concerns to E. coli intracellular fraction, pre-miR-29b production is almost constant from 1 to 4 h of cultivation, reaching a peak in late-log phase – 6 h – with 1640 µg/µL*unit of OD600. These pre-miR-29b production levels are much higher than those previously obtained intra (182 µg/L) and extracellularly (358 µg/L) from R. sulfidophilum.7 Ponchon and collaborators,17 using the tRNA scaffold method in E. coli with subsequent cleavage with RNAse H, reported yields from 5 to 23 mg of RNA per liter of culture for RNA oligonucleotide lengths between 91 and 376 bases, respectively. Despite it is difficult to perform a proper and direct comparison as the results were not presented in the same units and the experimental set-up is different – target RNA, plasmid and promoter, RNA construction design -, the experimental approach here presented can be advantageous in the sense that it isn't necessary to perform any enzyme-mediated cleavage step post-production. As a matter of fact, the bacterial origin of these enzymes may eventually raise safety concerns regarding the employment of the target RNA onto clinical applications. On the other hand, the results obtained with R. sulfidophilum are far from being unsatisfactory, particularly for the extracellular RNA that is directly recovered from the culture medium after the application of a desalting and concentration step, avoiding the use of hazardous chemicals, while simplifying the RNA downstream processing.
The type of the host cells has important effects on downstream processing when the product is expressed intracellularly, since in this case the cellular components are the major impurities.25 Therefore, it was also evaluated the concentration of major host contaminants during the whole fermentation period. Host proteins content in the intracellular fraction recovered from E. coli is depicted in Fig. 3 and ranged from 150 to 250 ng/µL, being 197 ng/µL at 6 hours of fermentation, the period where higher target RNA concentrations were obtained. The R. sulfidophilum protein content was distinct, since the extracellular fraction contained the lowest protein levels with approximately 20 ng/µL at the time-point where RNA production was maximized - 40 h.7 According to Fig. 4, in R. sulfidophilum intracellular fraction, the protein values were in the interval of 159 to 398 ng/µL, but at 32 h of fermentation, the highest protein levels were achieved, matching the period where more pre-miR-29b was obtained.
Figure 3.

Production of intracellular host cell proteins (black line - circles/primary scale), gDNA - assessed by RT-PCR in the extracted RNA samples (red line - triangles/secondary scale) - and gDNA – assessed by RT-PCR after gDNA direct extraction from cells using the Wizard® Genomic DNA Purification Kit Protocol (red line-squares/secondary scale) by E. coli DH5α bearing the pHBSR1-RM-pre-miR-29b plasmid at different periods of fermentation. Each point represents the average of 3 independent experiments whereas the bars indicate standard deviations.
Figure 4.

Levels of intracellular host cell proteins (black line - circles/ primary scale), gDNA – assessed by RT-PCR in the extracted RNA samples (red line - triangle/secondary scale) – and gDNA – assessed by RT-PCR after gDNA direct extraction from cells using the Wizard® Genomic DNA Purification Kit Protocol (red line – squares/secondary scale) produced at different periods of fermentation by R. sulfidophilum bearing the pHBSR1-RM-pre-miR-29b plasmid. Each point represents the average of 3 independent experiments whereas the bars indicate standard deviations.
The concentration of another major host impurity - genomic DNA (gDNA) - was also assessed in each RNA extracted fraction or directly from E. coli (Fig. 3) or R. sulfidophilum cells (Fig. 4). The lowest levels were obtained for R. sulfidophilum extracellular fraction,7 indicating that a minor percentage of the cell population is lysed and, consequently, the release of intracellular gDNA to the culture medium is not significant. As shown in Fig. 4, the R. sulfidophilum intracellular fraction presented gDNA levels in the interval 20 to 40 ng/µL, reaching a peak of 40 ng/µL at 8 h of production. Also, we evaluated gDNA levels directly from R. sulfidophilum cells and higher values were obtained, indicating that the RNA extraction method used within this work is highly efficient and allows the removal of a high percentage of gDNA.
On the other hand, the E. coli gDNA profile depicted in Fig. 3 was quite different once higher levels – from 99 to 105 ng/µL – were obtained from 0 to 2 h, and then the values were considerably reduced. This result was somehow unexpected but the analysis of gDNA directly extracted from E. coli also followed this tendency.
Actually, although there are no specific regulations for the use of RNA products onto pre-clinical and clinical applications, some authors follow the criteria defined from the regulatory agencies for the use of DNA samples. Therefore, we should keep in mind that although the downstream processing must be able to remove the majority of the contaminants present in the initial sample, the production conditions greatly influence the levels of contaminants and can eventually influence the final purity of the product. In particular, host cell components may also be found as impurities in secreted products, since cell lysis always occurs to some extent during fermentation.25 Nevertheless, in the culture conditions that we set-up, this problem is minimized, allowing to recover pre-miR-29b from R. sulfidophilum culture medium with fewer contaminants.
Intracellular microRNAs downstream processing and assessment of RNA integrity and stability
In recombinant RNA technology, along with the target heterologous RNA, other RNA species are also produced, emphasizing the need to develop accurate and effective purification protocols to isolate the RNA of interest from other contaminants, thus avoiding non-targeted-gene silencing and immunologic responses.16 For intracellular RNA, prior the purification step, it is necessary to perform the extraction of total soluble RNA20 by direct phenol extraction of the cell suspension, based on the original tRNA purification protocol by Zubay.26
Typically, the isolation of the target recombinant RNA is achieved using a chromatographic step. Dual-step chromatographic strategies applying gel-filtration and anion-exchange chromatographic matrices have been used at which a third hydrophobic-interaction chromatography step may be added if the purity with the first two steps is not the desired.17,20 More recently, the downstream strategies based on affinity chromatography evolved and actually represent a highly efficient option for RNA purification to increase not only the yields but also the selectivity in a single chromatographic step.16 In particular, boronate, RNA affinity tags and amino-acid based affinity chromatography have been extensively used for miRNA purification and as the later relies on naturally-occurring biologic interactions established within the cell, it seems particularly promising in RNA purification.16
After extraction, total soluble RNA is ethanol precipitated and can be analyzed by polyacrylamide-urea gel electrophoresis,20 useful to evaluate the integrity of the RNA. On the other hand, quantification of RNA has become increasingly important and is an essential step before RNA-based assays, gene expression analysis and RNAi applications.27 Actually, diverse ways are available for miRNA detection including conventional techniques such as Northern blotting, microarray and RT-qPCR but also biosensor techniques that include electrochemical-based detection or optical-based detection.28 On the other hand, RNA quantification is mainly achieved by ultraviolet absorbance, microcapillary electrophoresis and fluorescence-based quantification.27 In particular, RT-qPCR assays may offer a highly precise and specific way to detect and quantify microRNAs.29
Conclusions
The development of RNAi technologies opened new avenues for treatment options in diverse human diseases. An increasing number of experimental protocols were developed to obtain small RNA molecules for clinical and structural applications. These molecules have been mostly obtained by chemical or enzymatic methods but recent developments on RNA recombinant technology may be shifting the paradigm in RNA synthesis research. Indeed, the lower cost coupled to the higher efficiency and the possibility of growing large-scale cultures turn the attention to the production of RNAs using recombinant sources. In principle, RNA recombinant production involves a host and a plasmid bearing the desired DNA sequence, and appropriate culture conditions need to be set up. Therefore, as understanding the strengths and pitfalls of different hosts can aid in their effective use, here it was evaluated and compared the ability of two distinct hosts to produce pre-miR-29b - the well-characterized E. coli and the marine phototrophic bacterium R. sulfidophilum. Globally, we found that higher pre-miR-29b concentrations were obtained from E. coli intracellular fractions with shorter fermentation periods. Moreover, as the presence of impurities arising from the upstream stage can influence the final purity obtained after their downstream processing, it was also assessed the production of host cell proteins and genomic DNA. In this way, the E. coli fraction seems to be more contaminated, particularly regarding the proteins content, while R. sulfidophilum extracellular fraction presented fewer impurities. Therefore, we recommend working with E. coli when the final aim is to maximize RNA titers. On the other hand, direct recovery of RNA from the culture medium of R. sulfidophilum avoids time-consuming and laborious extraction methods involving hazardous chemicals while simultaneously ensures the target RNA integrity since no host RNases are secreted.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Prof. Yo Kikuchi (Division of Life Science and Biotechnology, Department of Ecological Engineering, Toyohashi University of Technology) for kindly providing the pBHSR1-RM plasmid.
Funding
This work was financially supported by the Portuguese Foundation for Science and Technology (FCT) through the following projects: EXPL/BBB-BIO/1056/2012 and Pest-OE/SAU/UI0709/2014. Patrícia Pereira and Augusto Pedro acknowledge the fellowships SFRH/BD/81914/2011 and SFRH/BD/81222/2011, respectively, from FCT. The authors also acknowledge the program COMPETE (FCOMP-01–0124FEDER-041068—EXPL/QEQ-MED/1068/2013) and the program Fundo Europeu de Desenvolvimento Regional (FEDER) [COMPETE (FCOMP-01–0124-FEDER-027560)].
References
- [1].Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391(6669):806-11; PMID:9486653; http://dx.doi.org/ 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- [2].Py Ho, Yu AM. Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip Rev RNA 2016; 7(2):186-97; PMID:26763749; http://dx.doi.org/ 10.1002/wrna.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ramachandran PV, Ignacimuthu S. RNA interference - a silent but an efficient therapeutic tool. Appl Biochem Biotechnol 2013; 169(6):1774-89; PMID:23340870; http://dx.doi.org/ 10.1007/s12010-013-0098-1 [DOI] [PubMed] [Google Scholar]
- [4].Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol 2009, 6 (6): 419-29; PMID 19434076; http://dx. 420 doi.org/ 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- [5].Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol 2008; 18 (1): 130-8; PMID: 18226108; http://dx.doi.org/ 21278200 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Du L, Pertsemlidis A. Cancer and neurodegenerative disorders: pathogenic convergence through microRNA regulation. J Mol Cell Biol 2011; 3:176-180; PMID:21278200; http://dx.doi.org/ 10.1093/jmcb/mjq058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pereira P, Pedro AQ, Tomás J, Maia CJ, Queiroz JA, Figueiras A, Sousa F. Advances in time course extracellular production of human pre-miR-29b from Rhodovolum sulfidophilum. Appl Microbiol Biotechnol 2016; 100(8):3723-34; PMID:26860940; http://dx.doi.org/ 10.1007/s00253-016-7350-x [DOI] [PubMed] [Google Scholar]
- [8].Pereira P, Sousa A, Queiroz J, Correia I, Figueiras A, Sousa F. Purification of pre-miR-29 by arginine-affinity chromatography. J Chromatogr Analyt Technol Biomed Life Sci 2014; 951–952:16-23; http://dx.doi.org/ 10.1016/j.jchromb.2014.01.020 [DOI] [PubMed] [Google Scholar]
- [9].Pereira P, Jorge AF, Martins R, Pais AA, Sousa F, Figueiras A. Characterization of polyplexes involving small RNA. J Colloid Interface Sci 2012; 387(1):84-94; PMID:22980740; http://dx.doi.org/ 10.1016/j.jcis.2012.07.088 [DOI] [PubMed] [Google Scholar]
- [10].Pereira PA, Tomás JF, Queiroz JA, Figueiras AR, Sousa F. Recombinant pre-miR-29b for Alzheimer's disease therapeutics. Sci Rep 2016; 6:19946; PMID:26818210; http://dx.doi.org/ 10.1038/srep19946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nelissen FH, Leunissen EH, Van de Laar L, Tessari M, Heus HA, Wijmenga SS. Fast production of homogeneous recombinant RNA - towards large-scale production of RNA. Nucleic Acids Res 2012; 40(13):e102; PMID:22457065; http://dx.doi.org/ 10.1093/nar/gks292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Beckert B, Masquida B. Synthesis of RNA by in vitro transcription. Methods Mol Biol 2011; 703:29-41; PMID:21125481 [DOI] [PubMed] [Google Scholar]
- [13].Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 2013; 12(11):847-65; PMID:24172333; http://dx.doi.org/ 10.1038/nrd4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen Q, Wang W, Zeng S, Urayama S, Yu A. A general approach to high-yield biosynthesis of chimeric RNAs bearing various types of functional small RNAs for broad applications. Nucleic Acids Res 2015; 43(7):3857-3869; PMID:25800741; http://dx.doi.org/ 10.1093/nar/gkv228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Martins R, Queiroz JA, Sousa F. Ribonucleic-acid purification. J Chromatogr A 2014; 1355:1-14; PMID:24951289; http://dx.doi.org/ 10.1016/j.chroma.2014.05.075 [DOI] [PubMed] [Google Scholar]
- [16].Pereira Queiroz JA, Figueiras A, Sousa F. Affinity approaches in RNAi-based therapeutics purification. J Chromtogr Analyt Technol Biomed Life Sci 2016; 1021:45-56; http://dx.doi.org/ 10.1016/j.jchromb.2016.01.022 [DOI] [PubMed] [Google Scholar]
- [17].Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F. A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat Protoc 2009; 4(6):947-59; PMID:19478810; http://dx.doi.org/ 10.1038/nprot.2009.67 [DOI] [PubMed] [Google Scholar]
- [18].Ponchon L, Catala M, Seijo B, El Khouri M, Dardel F, Nonin-Lecomte S, Tisné C. Co-expression of RNA-protein complexes in Escherichia coli and applications to RNA biology. Nucleic Acids Res 2013; 41(15):e150; PMID:23804766; http://dx.doi.org/ 10.1093/nar/gkt576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Duan Z, Yu AM. Bioengineered non-coding RNA agent (BERA) in action. Bioengineered 2016; 14:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ponchon L, Dardel F. Large-scale expression and purification of recombinant RNA in Escherichia coli. Methods 2011; 54(2):267-73; PMID:21320602; http://dx.doi.org/ 10.1016/j.ymeth.2011.02.007 [DOI] [PubMed] [Google Scholar]
- [21].Suzuki H, Ando T, Umekage S, Tanaka T, Kikuchi Y. Extracellular production of an RNA aptamer by ribonuclease-free marine bacteria harboring engineered plasmids: a proposal for industrial RNA drug production. Appl Environ Microbiol 2010; 76(3):786-93; PMID:19966026; http://dx.doi.org/ 10.1128/AEM.01971-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat Methods 2007; 4:571-76; PMID:17558412; http://dx.doi.org/ 10.1038/nmeth1058 [DOI] [PubMed] [Google Scholar]
- [23].Liu Y, Stepanov VG, Strych U, Willson RC, Jackson GW, Fox GE. DNAzyme mediated recovery of small recombinant RNAs from a 5S rRNA-derived chimera expressed in Escherichia coli. BMC Biotechnol 2010; 10:85; PMID:21134283; http://dx.doi.org/ 10.1186/1472-6750-10-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol 2011; 18(10):1153-58; PMID:21926993; http://dx.doi.org/ 10.1038/nsmb.2125 [DOI] [PubMed] [Google Scholar]
- [25].Carta G, Jungbauer A. Chromatography Media, in Protein Chromatography: Process development and Scale-up. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2010. [Google Scholar]
- [26].Zubay G. The isolation and fractionation of soluble ribonucleic acid. J Mol Biol 1962; 4(5):347-56; http://dx.doi.org/ 10.1016/S0022-2836(62)80015-1 [DOI] [Google Scholar]
- [27].Aranda R, Dineen SM, Craig RL, Guerrieri RA, Robertson JM. Comparison and evaluation of RNA quantification methods using viral, prokaryotic, and eukaryotic RNA over a 104 concentration range. Anal Biochem 2009; 387(1):122-7; PMID:19454255; http://dx.doi.org/ 10.1016/j.ab.2009.01.003 [DOI] [PubMed] [Google Scholar]
- [28].Hunt EA, Broyles D, Head T, Deo SK. MicroRNA detection: Current technology and research strategies. Annu Rev Anal Chem 2015; 8:217-37; http://dx.doi.org/ 10.1146/annurev-anchem-071114-040343 [DOI] [PubMed] [Google Scholar]
- [29].Niu Y, Zhang L, Qiu H, Wu Y, Wang Z, Zai Y, Liu L, Qu J, Kang K, Gou D. An improved method for detecting circulating microRNAs with S-poly(T) plus real-time PCR. Sci Rep 2015; 5:15100; PMID:26459910; http://dx.doi.org/ 10.1038/srep15100 [DOI] [PMC free article] [PubMed] [Google Scholar]


