ABSTRACT
Amyloids represent protein fibrils that have highly ordered structure with unique physical and chemical properties. Amyloids have long been considered lethal pathogens that cause dozens of incurable diseases in humans and animals. Recent data show that amyloids may not only possess pathogenic properties but are also implicated in the essential biological processes in a variety of prokaryotes and eukaryotes. Functional amyloids have been identified in archaea, bacteria, fungi, and animals, including humans. Plants are one of the most poorly studied groups of organisms in the field of amyloid biology. Although amyloid properties have not been shown under native conditions for any plant protein, studies demonstrating amyloid properties for a set of plant proteins in vitro or in heterologous systems in vivo have been published in recent years. In this review, we systematize the data on the amyloidogenic proteins of plants and their functions and discuss the perspectives of identifying novel amyloids using bioinformatic and proteomic approaches.
KEYWORDS: Amyloid, prion, LD, WALTZ, SARP, PSIA, AFP, plant, yeast, A. thaliana, S. cerevisiae
INTRODUCTION
Amyloids are protein fibrils with a characteristic structure called “cross-β”. This structure results from the formation of intermolecular β-sheets perpendicular to the axis of the amyloid fibrils1 and is detected using two-dimensional X-ray diffraction.2 The stacking of monomers in the fibrils results from the formation of numerous hydrogen bonds between neighboring β-chains.3 Such spatial organization makes amyloids one of the most stable biogenic particles and endows these macromolecules with unique physico-chemical properties: amyloids resist treatment with different detergents (like SDS and sarkosyl), proteinases, high temperatures, and several acids,2,4-7 and can persist in the environment for years.8 In addition, the unique spatial structure of amyloids can be specifically detected with several compounds. The binding of amyloids with Congo red diazo dye leads to apple-green birefringence in the polarized light,9 while the interaction of amyloids with the fluorescent dye Thioflavin-T enhances the emission of its fluorescence.10,11
Amyloids have long been considered as pathogens for humans and animals causing incurable diseases called amyloidoses. The first pathanatomic descriptions of amyloidoses in the liver and spleen were obtained in the XVII or XVIII centuries, while the protein nature of pathological amyloid deposits was revealed only in 1858.12 In 1854, Rudolf Virchow proposed the term “amyloid” (starch-like),13 since amyloid deposits were stained a blue color using iodine similarly to starch granules. It has since been confirmed that the blue staining reflects the complex organization of amyloid deposits comprising, in addition to protein fibrils, proteoglycans and glycosaminoglycans.14 To date, more than 30 proteins forming pathological or pathology-associated amyloids have been identified in humans.15 Infectious neurodegenerative amyloidoses represent a special group of diseases caused by the amyloid state of prion (an acronym from proteinaceous infectious particle) protein (PrP), which was called PrPSc (PrP Scrapie, from scrapie disease of sheep and goats).7,16 Presently, the term “prion” is attributed to all infectious amyloids. Nevertheless, there are prions that are unlikely to form amyloids.17,18 In a broader sense, prions are proteins that persist under the same conditions in two or more conformations, of which at least one conformation possesses infectious properties.19 Thus, amyloids and prions represent partially overlapping sets.
Since 2000, the paradigm of amyloids as pathogens has rapidly shifted. The formation of the amyloid structure not only leads to lethal diseases but is also necessary for essential biological functions. In different organisms, from archaea and bacteria to humans, more than 20 functional amyloids have been described, and their number is constantly growing.20-22 In prokaryotes, at least eight groups of proteins capable of forming functional amyloids were identified. These amyloids participate in the biofilm formation,23 overcoming the surface tension,24 storage of toxins,25 formation of pores in phagolysosome membrane26 or hypersensitive response activation in plants.27 In animals, amyloids are involved in long-term memory formation,28,29 melanin polymerization,30 hormone storage,31 tooth enamel biomineralization,32 programmed necrosis regulation33 and the matrix formation in spermatozoid acrosome.34
Prions of ascomycetes represent a specific group of infectious amyloids. There are approximately ten prions in yeast Saccharomyces cerevisiae.35 In contrast to non-infectious amyloids, the aggregates of prions are constantly fragmented by chaperone proteins,36,37 providing the infectious properties and efficient transmission of prions during cell division.38,39 The prionization of proteins may lead to heritable changes in their functional activities that cause different phenotypic manifestations. This phenomenon is referred to as protein inheritance,40,41 i.e., an inheritance that is not determined by changes in the primary structure of genes, but rather the conformation of particular proteins. Most yeast prions have no significant effect on viability; however, some of them may be harmful under certain conditions,42 and in contrast, some prions may increase cell survival.43,44
Thus, numerous amyloids involved in pathogenesis or implicated in a variety of functions have been identified in different organisms. At the same time, plants remain one of the most poorly studied group of organisms in the field of amyloid biology reflecting, on the one hand, the high complexity of working with plants as objects of research, and, on the other hand, the lack of methods for detecting amyloids at the proteomic level. In this review, we systematize the data on amyloidogenic proteins of plants, analyze their potential biological functions, and discuss the perspectives for the identification of novel amyloids in plants.
I. Amyloids Involved in the Adhesion of Green Algae
Life forms of green algae are diverse, and many of these organisms form biofilms attached to different surfaces. The adhesion of the spores of green algae includes two stages: initial and constant adhesion.45 At the initial stage, preceded by a complex mechanism involving the recognition of a proper substrate, spores attach to the surface using a sticky secretion present on their shells. Subsequently, the spores actively produce special gel-like extracellular polymeric substances (EPS) comprising different proteins, glycoproteins, and proteoglycans. EPS bind Thioflavin-T and exhibit apple-green birefringence upon staining with Congo red.46,47 Moreover, Raman spectroscopy revealed EPS peaks similar to those observed for known amyloids.46 These data suggest that the EPS of green algae, including micro- (Coccomyxa sp. or Glaphyrella trebouxiodes)46 and macrophytes (Prasiola linearis),47 may contain protein (or proteins) in an amyloid state. These proteins might be the main determinants of the high mechanical resistance of EPS. Amyloids are involved in the biofilm formation and adhesion of different species of bacteria.22,23 Thus, adhesive proteins represent an example of cross-kingdom conservation of biological functions of amyloids. Nevertheless, the particular amyloid-forming proteins of EPS were not identified, and a complete understanding of the role of amyloid formation for green algae adhesion requires further investigation.
Notably, amyloids are important structural elements for different biological substances. The egg envelopes (chorions) of “annual fish” Austrofundulus limnaeus, which lives in the annually drying reservoirs of South America, contain amyloid proteins. The amount of amyloids in these envelopes increases with dehydration, which increases the survival of embryos.48 The matrix of mice spermatozoid acrosome contains a range of proteins forming detergent-resistant aggregates with amyloid properties.49 The chorion of silk moth eggs bears at least two classes of amyloid-forming proteins.50 Thus, similar to the aforementioned structures, the formation of EPS might implicate ensembles of various amyloid-forming proteins likely to co-aggregate as a result of their structural similarity.
II. Prion-like Protein Luminidependens of Arabidopsis thaliana
Flowering time in Arabidopsis thaliana is controlled by a complex molecular mechanism involving several cascades of transcriptional factors,51 one of which is so-called Autonomous Flowering Pathway (AFP). This regulatory pathway is independent of the photoperiod and vernalization, i.e., induction of flowering as a result of a prolonged influence of low, typically positive temperatures.52
Recently, a paper by S. Chakrabortee and co-authors, from the laboratory of S. Lindquist demonstrated that Luminidependens (LD), an AFP protein in A. thaliana, has prion properties in the heterologous system of the yeast Saccharomyces cerevisiae.53 Most known yeast prion proteins are rich in asparagine (N) and glutamine (Q).54,55 LD was predicted as a potentially prion-forming protein using a recently developed bioinformatic algorithm based on a hidden Markov model,56 as the prion-forming region of A. thaliana protein was closely similar to those of yeast prion proteins.53 Notably, the fraction of proteins acting as flowering regulators was statistically significantly increased in a set of approximately 500 A. thaliana proteins detected using this algorithm.53 Further experimental analysis demonstrated that LD and two other AFP proteins, Flowering Locus PA (FPA) and Flowering Locus CA (FCA), form detergent-resistant oligomers in yeast cells. Moreover, LD fused with the reporter sequences demonstrated properties typical for yeast prions: dominance, cytoplasmic infectivity, and dependence on the level of Hsp70 chaperone production.53
Thus, LD forms a chimeric prion in yeast cells. Does this finding suggest that LD has prion properties in A. thaliana cells? Indeed, drawing this conclusion would be at least premature. The fact that LD forms detergent-resistant oligomers in a heterologous system at an increased level of production confirms neither amyloid fibril formation nor prion-like properties under native conditions in A. thaliana. Nevertheless, the role of epigenetic regulation in flowering induction in A. thaliana has been demonstrated in a number of studies. Vernalization results in the silencing of the FLOWERING LOCUS C (FLC), the product of which is a protein that represses flowering.52 The FLC gene is also a target for LD with a nuclear localization57 and inhibits the expression of FLC in meristems via histone modification58 and the negative regulation of the SUF4 activator of FLC expression.59 Prionization typically entails the functional inactivation of prion-forming protein;60 therefore, according to the hypothesis of Y. Chernoff, the most likely phenotype of LD prionization would be the delay of the flowering.61 Notably, temperature is not only a factor controlling vernalization in plants but also affects both the induction62 and stability of prions.63 Thus, the influence of temperature on flowering could also be modulated through changes in the induction frequency or the effectiveness of LD prion propagation.
III. Plant Proteins with Amyloid Properties in Vitro
Several plant proteins possess amyloid properties in vitro. The monellin protein of the tropical fruit Dioscoreophyllum cumminsii irreversibly denatures when boiled at 85°C in acidic (pH 2.5) buffer. The consequent addition of 100 mM sodium chloride induces the formation of monellin fibrils that bind Congo red.64 Unfortunately, birefringence upon binding of Congo red with monellin fibrils was not analyzed, therefore it is impossible to draw a conclusion concerning the amyloid nature of this protein, even in vitro.
Maize transglutaminase (TGZ) forms insoluble inclusions when overproduced under the control of the strong psbA promoter in vivo in the transplastomic tobacco plants.65 These inclusions contain fibrillar aggregates of TGZ resistant to SDS treatment.65 In vitro, the TGZ protein forms detergent-resistant aggregates of a fibrillar structure showing birefringence upon binding with Congo red. The fibrils of the short C-terminal (aa 466–477) region of TGZ have similar properties in vitro.65 Therefore, maize TGZ forms amyloid-like fibrils in vitro and in the transplastomic tobacco plants.
Prohevein protein is the precursor of hevein, a key component of Hevea brasiliensis latex, and has an evolutionarily conserved C-terminal region, which, in contrast to the full-length protein, forms fibrils in vitro.66 These fibrils exhibit a pattern typical for amyloids in two-dimensional X-ray diffraction and apple-green birefringence upon binding with Congo red.66 Notably, the C-terminal region of progevein is also present in the latex of the hevea and exhibits agglutination.67 Thus, the amyloid properties of the C-terminal region of progevein could enhance its stability in vivo and the agglutination of pathogenic microorganisms and fungi, facilitating their destruction by the enzymes of the lutoid fraction of latex.66
Several defense peptides of plants were recently demonstrated as exhibiting amyloid properties in vitro. For example, the antimicrobial peptide Cn-AMP2 from coconut (Cocos nucifera) liquid endosperm contains bioinformatically predicted amyloidogenic regions and demonstrates amyloid properties in vitro, including the formation of fibrils detectable using electronic microscopy and the enhancement of the fluorescence of Tioflavin-T dye.68 Another example is defensins, which are short plant proteins involved in protection from various pathogens. The RsAFP-19 peptide is a C-terminal fragment (19 residues) of RsAFP1 and RsAFP2 defensins of the radish Raphanus sativus. This peptide exhibits fungicidal activity. Freeze-thaw cycles induce RsAFP-19 aggregation in vitro. These aggregates have a fibrillar structure and exhibit a cross-β pattern in X-ray diffraction typical for amyloids.69 It is not known whether the full-length defensins RsAFP1 and RsAFP2 form amyloid aggregates in vivo, but the fungicidal activity of RsAFP-19 is negatively correlated with the level of its aggregation.69 A similar effect has been reported for the amyloids of bacterial microcin E492.25 Thus, one of the potential biological roles of amyloid formation by plant defense proteins might be the sequestration of the toxic intermediates of these proteins into the functionally inactive fibrils.25,70
Taken together, the studies conducted to date show that several plant proteins or their fragments have amyloid properties in vitro or in heterologous systems (Table 1). Whether these molecules have amyloid properties in the native conditions remains unclear and should be analyzed in additional studies. Further, is it likely that plants have other amyloid-forming proteins?
TABLE 1.
Amyloidogenic proteins in plants.
| Amyloid properties were shown:* |
|||||
|---|---|---|---|---|---|
| Protein | Function | Species | in vivo | in vitro | Reference |
| Proteins of algae extracellular polymeric substances (EPS)** | Attachment to surface | Coccomyxa sp., Glaphyrella trebouxiodes и Prasiola linearis (algae) | + | − | 46, 47 |
| Luminidependens (LD) | Autonomous Flowering Pathway (AFP) component | Arabidopsis thaliana (rockcress), Saccharomyces cerevisiae (yeast)*** | ± | − | 53 |
| Flowering Locus PA (FPA) | AFP component | Arabidopsis thaliana (rockcress), Saccharomyces cerevisiae (yeast)*** | ± | − | 53 |
| Flowering Locus CA (FCA) | AFP component | Arabidopsis thaliana (rockcress), Saccharomyces cerevisiae (yeast)*** | ± | − | 53 |
| Pro-hevein С-terminal domain | Latex component, protective function | Hevea brasiliensis (rubber tree) | − | + | 66 |
| Monellin | Unknown, has a sweet taste for human | Dioscoreophyllum cumminsii | − | ± | 64 |
| Maize transglutaminase (TGZ) | Posttranslational modification of proteins | Zea mays (corn), Nicotiana tabacum (tobacco)**** | ± | + | 65 |
| RsAFP-19 peptide | RsAFP1 and RsAFP2 defensins fragment, fungicide | Raphanus sativus (radish) | − | + | 69 |
| Cn-AMP2 peptide | Antimicrobial peptide of liquid endosperm | Cocos nucifera (coconut tree) | − | + | 68 |
| Peptides resulting after the limited proteolysis of the seed storage proteins** | Nutrient storage | Glycine max (soybean), Pisum sativum (pea), Triticum aestivum (wheat) | ± | − | 91–93 |
«+» Amyloid properties were completely validated; «±» at least one of the amyloid properties was shown (formation of detergent-resistant aggregates in vivo, fibril formation in vitro, apple-green birefringence upon Congo red binding, or increase in Thioflavin-T fluorescence); «-» amyloid properties were not shown.
Particular proteins were not identified.
Amyloid properties of these A. thaliana proteins were investigated in the heterologous S. cerevisiae system.
Amyloid properties of Z. mays TGZ protein were shown under overexpression in transplastomic N. tabacum plants.
IV. Perspectives for the Identification of Novel Amyloids and Prions in Plants
The identification of novel prions and amyloids remains arduous and time-consuming. Significant progress in this field of biology can be provided using novel bioinformatic and proteomic methods. The methodology of proteomic analysis of amyloids is currently at the very dawn of its development. The first attempts to apply proteomics for studying amyloids71-73 were associated with the identification of proteins sequestered by pathological amyloids that form large deposits and can be isolated from fixed tissue by laser capture microdissection (LCM).74 A proteome-wide method for the identification of candidates for novel amyloid-forming proteins, called TAPI (Technique for Amyloid Purification and Identification), was proposed in 2013.75 TAPI uses polyacrylamide gel electrophoresis for the ultrafiltration of detergent-treated amyloid-rich protein fractions, followed by HPLC combined with mass spectrometry to identify candidate proteins.75 In 2014, another method called PSIA (Proteomic Screening and Identification of Amyloids) was developed.76 This method uses differential ultracentrifugation combined with treatment using ionic detergents77 to obtain protein fractions rich in amyloid proteins. These fractions are solubilized, and the proteins (peptides) are separated either by two-dimensional gel electrophoresis76 or HPLC78,79 coupled with mass spectrometry, which provides high resolution sufficient to identify the most minor amyloid-forming proteins.79 Nevertheless, to date, proteomic methods have not been applied for the identification of amyloid proteins in plant species. Is it possible to assess the amyloid properties of plant proteins by other methods?
The primary structure of proteins is the key determinant of their amyloidogenic properties. In 1997, the polyglutamine tracts capable of forming amyloids involved in the development of neurodegenerative diseases in humans were described.80 Subsequently, it was noted that the common feature of different structural proteins of infectious yeast amyloids (prions) is the presence of regions rich in Q or N residues.81 Homopeptides formed by several other amino acids, particularly E, also form amyloids.82 Such amyloidogenic regions (type I amyloidogenic regions) are rich in one or similar (such as Q and N) residues. To identify these regions, several bioinformatic algorithms were developed.54,83 Notably, the relative content of the biased residues is more important for the formation of type I amyloidogenic regions than the positions of the particular residues.84
QN-rich proteins are widely spread throughout plant proteomes. Thus, in Arabidopsis thaliana, approximately 200 such proteins were detected,54 three of which, the transcriptional regulators LD, FPA, and FCA discussed above (see Section II, Table 1), showed some properties of amyloids.53 Enrichment in Q and N is a characteristic feature of transcription factors, and of other DNA- and RNA-binding proteins of various organisms.54 Moreover, polyasparagine and polyglutamine tracts display transcriptional activity.85,86 Transcription factors are abundant in the actively dividing cells of plant meristems. These tissues are promising for identifying novel plant amyloids likely to be involved in regulatory processes and having prion-like properties.
The enrichment in Q and E is a common feature of many plant seed storage proteins. For example, seed storage proteins of maize (Zea mays), rye (Secale sereale), wheat (Triticum aestivum), oat (Avena sativa), and other important crops are rich in Q and P,87 and the storage proteins of legumes are rich in E and D.88 The ability to form fibrils, which are widely used in biotechnology, is well known for maize prolamins (zeins).89 In addition, studies on the structure of zein fibrils did not demonstrate characteristic signals in X-ray diffraction inherent in amyloids.90 The fractions enriched with seed storage proteins of pea (Pisum sativum)91 and soy (Glycine max)92 were subjected to the limited proteolysis at a temperature of more than 80°C and pH = 2, and the resulting peptides formed fibrils with enhanced fluorescence of Thioflavin-T dye, suggesting an amyloid structure.92 Similar results were obtained with peptide mixtures formed after trypsin digestion of the wheat gliadin and gluten storage proteins.93 Unfortunately, the amyloid properties of the full-length seed storage proteins were never tested, and specific peptides capable of forming amyloids were not identified.91-93 Notably, a significant portion of proteins contain bioinformatically predicted amyloidogenic peptides, which are capable of forming amyloid-like fibrils at high concentrations and under special conditions,94 but this fact does not indicate that the corresponding full-length proteins have amyloid properties.
Notably, natural dehydration occurs during maturation of plant seeds. This process is accompanied by the compaction of genetic material, a decrease in metabolic rate, and a change in the structure of proteins.95,96 This process is similar to the dehydration of the eggs of the “annual fish” Austrofundulus limnaeus leading to the amyloidogenesis of the proteins in its envelopes, which increases the survival of embryos.48 It can be assumed that seed storage proteins during dehydration also form amyloids, which increase their stability, and subsequently, under favorable conditions, these proteins revert to a monomeric or oligomeric state that can easily be metabolized by the growing embryo.
QN-rich proteins represent approximately one-third of the currently known amyloids.97 For other amyloid-forming proteins, strict patterns of the primary structure have not been identified. However, various amino acids have different amyloidogenic propensities. For example, hydrophobic aromatic (W, F) and some aliphatic monoamino monocarboxylic (I, V, L) amino acids have the highest amyloidogenic propensity (i.e., the ability to induce amyloid formation).98 The amyloidogenic regions formed by these amino acids (type II amyloidogenic regions), unlike type I regions, do not have one prevalent amino acid. The specific position of the corresponding amino acids is of particular importance for the formation of type II regions. Removing or changing the position of only one amino acid in such amyloidogenic regions can lead to a complete loss of amyloid properties.99 Approximately ten different bioinformatic algorithms have been developed for the prediction of type II amyloidogenic regions.98
Defense proteins and peptides of plants are structurally and functionally heterogeneous groups that are not characterized by the enrichment of Q and N; however, many of these molecules are hydrophobic100 and thus may contain type II amyloidogenic regions. Plants produce a wide range of various defense peptides and proteins.101,102 As previously discussed, the amyloid properties for some of these molecules are shown, but are still not entirely clear (Table 1, Fig. 1). Since amyloids represent one of the most stable variants of the quaternary structure of a protein, we propose that adopting an amyloid state by defense proteins could not only promote the inactivation of toxic intermediates but also increases the effectiveness of these proteins against various pathogens.
FIGURE 1.
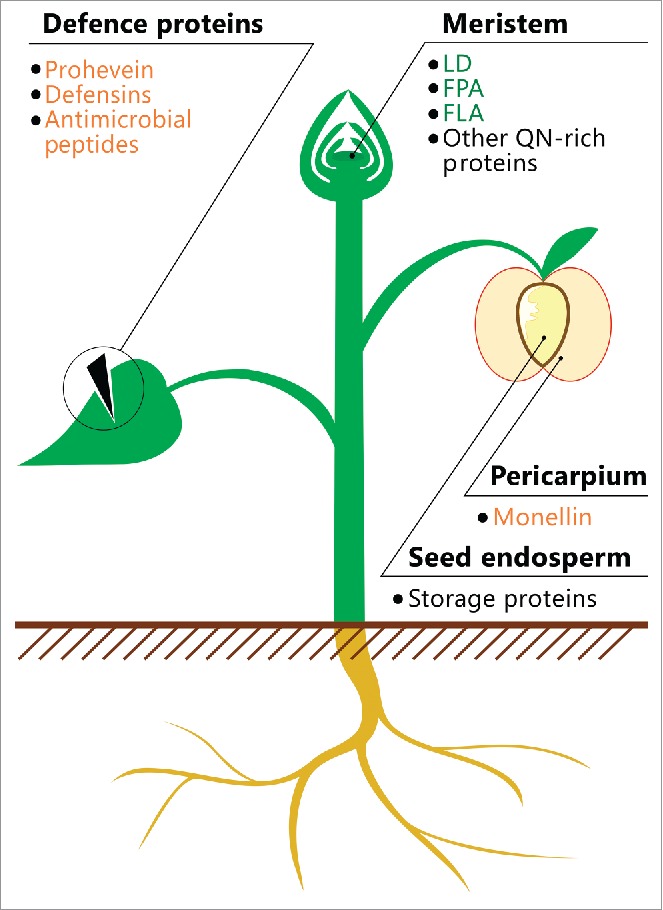
Location of potentially amyloidogenic proteins in plants. A schematic illustration of the plant is shown. The names of proteins or peptides, whose amyloid properties were partially characterized in vivo (green) and in vitro (orange), and proteins, whose fragments have amyloidogenic properties (black), are indicated. The data on these proteins and peptides are summarized in Table 1.
CONCLUSION
To date, amyloid properties were shown for several plant proteins or their fragments in vitro or in heterologous systems in vivo. However, no plant proteins were shown to form amyloids under native conditions. Based on the data obtained in previous studies (Fig. 1, Table 1), we distinguished four groups of plant proteins promising for the identification of novel amyloids: (i) QN-rich proteins, particularly those capable of binding nucleic acids; (ii) defense proteins and peptides containing hydrophobic regions; (iii) seed storage proteins containing Q- and E-rich regions; and (iv) proteins involved in the adhesion of algal cells to the surfaces. Based on the methodology developed to date, we can propose two main strategies to reveal candidates for functional amyloids in the proteomes of plants: (i) bioinformatic prediction of potentially amyloidogenic proteins and (ii) proteomic screenings of proteins resistant to treatment with ionic detergents. In both cases, the amyloid properties of candidate proteins should be verified by analyzing their ability to form fibrils and adopt cross-β structure. Further studies on the bioinformatic prediction and experimental identification of amyloid proteins in the proteomes of different species are required to elucidate the biological roles and functions of plant amyloids.
ABBREVIATIONS
- AFP
Autonomous Flowering Pathway
- EPS
Extracellular Polymeric Substances
- HPLC
High Performance Liquid Chromatography
- LPS
Lowest Probability Subsequences
- PSIA
Proteomic Screening and Identification of Amyloids
- SDS
Sodium Dodecyl Sulfate
- Sarkosyl
Sodium lauroyl sarcosinate
- SARP
Sequence Analysis Based on the Ranking of Probabilities
- TAPI
Technique for Amyloid Purification and Identification; The standard single-letter amino acid code is used
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
Authors declare no potential conflicts of interest.
FUNDING
This work was financially supported by a grant from the Russian Science Foundation (Project No 17-16-01100).
REFERENCES
- 1.Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem. 1968;16:673-7. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 2.Sipe JD, Cohen AS. Review: history of the amyloid fibril. J Struct Biol. 2000;130:88-98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 3.Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729-39. doi: 10.1006/jmbi.1997.1348. PMID:9356260. [DOI] [PubMed] [Google Scholar]
- 4.Hazeki N, Tukamoto T, Goto J, Kanazawa I. Formic Acid Dissolves Aggregates of an N-Terminal Huntingtin Fragment Containing an Expanded Polyglutamine Tract: Applying to Quantification of Protein Components of the Aggregates. Biochem Biophys Res Commun. 2000;277:386-93. doi: 10.1006/bbrc.2000.3682. PMID:11032734. [DOI] [PubMed] [Google Scholar]
- 5.Selkoe DJ, Ihara Y, Salazar FJ. Alzheimer's disease: insolubility of partially purified paired helical filaments in sodium dodecyl sulfate and urea. Science. 1982;215:1243-5. doi: 10.1126/science.6120571. PMID:6120571. [DOI] [PubMed] [Google Scholar]
- 6.Selkoe DJ, Abraham CR, Podlisny MB, Duffy LK. Isolation of Low‐Molecular‐Weight Proteins from Amyloid Plaque Fibers in Alzheimer's Disease. J Neurochem. 1986;46:1820-34. doi: 10.1111/j.1471-4159.1986.tb08501.x. PMID:3517233. [DOI] [PubMed] [Google Scholar]
- 7.Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science 1982;218:1309-11. doi: 10.1126/science.6815801. PMID:6815801. [DOI] [PubMed] [Google Scholar]
- 8.Wiggins RC. Prion Stability and infectivity in the environment. Neurochem Res. 2009;34:158-68. doi: 10.1007/s11064-008-9741-6. PMID:18483857. [DOI] [PubMed] [Google Scholar]
- 9.Puchtler H, Sweat F, Levine M. On the binding of Congo red by amyloid. J Histochem Cytochem. 1962;10:355-64. doi: 10.1177/10.3.355. [DOI] [Google Scholar]
- 10.Vassar PS, Culling CF. Fluorescent stains, with special reference to amyloid and connective tissues. Arch Pathol. 1959;68:487-98. PMID:13841452. [PubMed] [Google Scholar]
- 11.LeVine 3rd H. Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzym. 1999;309:274-84. doi: 10.1016/S0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 12.Friedreich N, Kekule FA. Zur Amyloidfrage. Virchows Arch Path Anat Physiol 1859;16:50-65. doi: 10.1007/BF01945246. [DOI] [Google Scholar]
- 13.Virchow R. Ueber eine im Gehirn und Ruckenmark des Menschen aufgefunde Substanz mit der chemishen Reaction der Cellulose. Virchows Arch Path Anat Physiol. 1854;6:135-8. doi: 10.1007/BF01930815. [DOI] [Google Scholar]
- 14.Kyle RA. Amyloidosis: a convoluted story. Br J Haematol. 2001;114:529-38. doi: 10.1046/j.1365-2141.2001.02999.x. PMID:11552976. [DOI] [PubMed] [Google Scholar]
- 15.Sipe JD, Benson MD, Buxbaum JN, Ikeda S-I, Merlini G, Saraiva MJM, Westermark P. Amyloid fibril proteins and amyloidosis: chemical identification and clinical classification International Society of Amyloidosis 2016 Nomenclature Guidelines. Amyloid. 2016;23:1-5. doi: 10.1080/13506129.2016.1257986. PMID:26646718. [DOI] [PubMed] [Google Scholar]
- 16.Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 1983;35:349-58. doi: 10.1016/0092-8674(83)90168-X. PMID:6418385. [DOI] [PubMed] [Google Scholar]
- 17.Roberts BT, Wickner RB. Heritable activity: a prion that propagates by covalent autoactivation. Genes Dev. 2003;17:2083-7. doi: 10.1101/gad.1115803. PMID:12923060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown JC, Lindquist S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 2009;23:2320-32. doi: 10.1101/gad.1839109. PMID:19797769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146-58. doi: 10.1016/j.cell.2009.02.044. PMID:19345193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pham CLL, Kwan AH, Sunde M. Functional amyloid: widespread in Nature, diverse in purpose. Essays Biochem. 2014;56:207-19. doi: 10.1042/bse0560207. PMID:25131597. [DOI] [PubMed] [Google Scholar]
- 21.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid – from bacteria to humans. Trends Biochem. Sci. 2007;32:217-24. doi: 10.1016/j.tibs.2007.03.003. PMID:17412596. [DOI] [PubMed] [Google Scholar]
- 22.Syed AK, Boles BR. Fold modulating function: Bacterial toxins to functional amyloids. Front. Microbiol. 2014;5. doi: 10.3389/fmicb.2014.00401. PMID:25136340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851-5. doi: 10.1126/science.1067484. PMID:11823641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claessen D, Rink R, De Jong W Siebring J, De Vreugd P Boersma FGH, Dijkhuizen L, Wosten HAB. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003;17:1714-26. doi: 10.1101/gad.264303. PMID:12832396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bieler S, Estrada L, Lagos R, Baeza M, Castilla J, Soto C. Amyloid formation modulates the biological activity of a bacterial protein. J Biol Chem. 2005;280:26880-5. doi: 10.1074/jbc.M502031200. PMID:15917245. [DOI] [PubMed] [Google Scholar]
- 26.Bavdek A, Kostanjsek R, Antonini V, Lakey JH, Dalla Serra M, Gilbert RJC, Anderluh G. PH dependence of listeriolysin O aggregation and pore-forming ability. FEBS J. 2012;279:126-41. doi: 10.1111/j.1742-4658.2011.08405.x. PMID:22023160. [DOI] [PubMed] [Google Scholar]
- 27.Oh J, Kim J-G, Jeon E, Yoo C-H, Moon JS, Rhee S, Hwang I. Amyloidogenesis of type III-dependent harpins from plant pathogenic bacteria. J Biol Chem. 2007;282:13601-9. doi: 10.1074/jbc.M602576200. PMID:17314101. [DOI] [PubMed] [Google Scholar]
- 28.Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A Neuronal Isoform of CPEB Regulates Local Protein Synthesis and Stabilizes Synapse-Specific Long-Term Facilitation in Aplysia. Cell. 2003;115:893-904. doi: 10.1016/S0092-8674(03)01021-3. PMID:14697206. [DOI] [PubMed] [Google Scholar]
- 29.Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EML, Kannan K, Guo F, et al.. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148:515-29. doi: 10.1016/j.cell.2012.01.004. PMID:22284910. [DOI] [PubMed] [Google Scholar]
- 30.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. PMID:16300414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KP, Simon R, Schubert D, et al.. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328-32. doi: 10.1126/science.1173155. PMID:19541956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carneiro KMM, Zhai H, Zhu L, Horst JA, Sitlin M, Nguyen M, Wagner M, Simpliciano C, Milder M, Chen C-L, et al.. Amyloid-like ribbons of amelogenins in enamel mineralization. Sci Rep. 2016;6:23105. doi: 10.1038/srep23105. PMID:27009419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al.. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339-50. doi: 10.1016/j.cell.2012.06.019. PMID:22817896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guyonnet B, Egge N, Cornwall GA. Functional amyloids in the mouse sperm acrosome. Mol Cell Biol. 2014;34:2624-34. doi: 10.1128/MCB.00073-14. PMID:24797071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickner RB. Yeast and Fungal Prions. Cold Spring Harb Perspect Biol. 2016;8:a023531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chernova TA, Wilkinson KD, Chernoff YO. Prions, chaperones, and proteostasis in yeast. Cold Spring Harb Perspect Biol. 2017;9:a023663. doi: 10.1101/cshperspect.a023663. PMID:27815300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chernoff Y, Lindquist S, Ono B, Inge-Vechtomov S, Liebman S. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995;268:880-4. doi: 10.1126/science.7754373. PMID:7754373. [DOI] [PubMed] [Google Scholar]
- 38.Wickner RB, Shewmaker FP, Bateman DA, Edskes HK, Gorkovskiy A, Dayani Y, Bezsonov EE. Yeast prions: structure, biology, and prion-handling systems. Microbiol Mol Biol Rev. 2015;79:1-17. doi: 10.1128/MMBR.00041-14. PMID:25631286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taguchi H, Kawai-Noma S. Amyloid oligomers: Diffuse oligomer-based transmission of yeast prions. FEBS J.2010;277:1359-68. doi: 10.1111/j.1742-4658.2010.07569.x. PMID:20148963. [DOI] [PubMed] [Google Scholar]
- 40.Wickner RB, Taylor KL, Edskes HK, Maddelein ML, Moriyama H, Roberts BT. Prions in Saccharomyces and Podospora spp.: protein-based inheritance. Microbiol Mol Biol Rev 1999;63:844-861. PMID:10585968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inge-Vechtomov SG. Yeast prions as a model of neurodegenerative infectious amyloidoses in humans. Russ J Dev Biol. 2011;42:293-300. doi: 10.1134/S1062360411020068. [DOI] [PubMed] [Google Scholar]
- 42.McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci U S A. 2011;108:5337-41. doi: 10.1073/pnas.1102762108. PMID:21402947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355-9. doi: 10.1126/science.1219491. PMID:22517861. [DOI] [PubMed] [Google Scholar]
- 44.Holmes DL, Lancaster AK, Lindquist S, Halfmann R. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell. 2013;153:153-65. doi: 10.1016/j.cell.2013.02.026. PMID:23540696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fletcher RL, Callow ME. The settlement, attachment and establishment of marine algal spores. Br Phycol J 1992;27:303-29. doi: 10.1080/00071619200650281. [DOI] [Google Scholar]
- 46.Mostaert AS, Giordani C, Crockett R, Karsten U, Schumann R, Jarvis SP. Characterisation of Amyloid Nanostructures in the Natural Adhesive of Unicellular Subaerial Algae. The J of Adhesion. 2009;85:465-483. doi: 10.1080/00218460902996366. [DOI] [Google Scholar]
- 47.Mostaert AS, Higgins MJ, Fukuma T, Rindi F, Jarvis SP. Nanoscale mechanical characterisation of amyloid fibrils discovered in a natural adhesive. J Biol Phys. 2006;32:393-401. doi: 10.1007/s10867-006-9023-y. PMID:19669445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Podrabsky JE, Carpenter JF, Hand SC. Survival of water stress in annual fish embryos: dehydration avoidance and egg envelope amyloid fibers. Am J Physiol Regul Integr Comp Physiol. 2001;280:R123-31. PMID:11124142. [DOI] [PubMed] [Google Scholar]
- 49.Guyonnet B, Egge N, Cornwall GA. Functional amyloids in the mouse sperm acrosome. Mol Cell Biol. 2014;34:2624-34. doi: 10.1128/MCB.00073-14. PMID:24797071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iconomidou VA, Vriend G, Hamodrakas SJ. Amyloids protect the silkmoth oocyte and embryo. FEBS Lett. 2000;479:141-5. doi: 10.1016/S0014-5793(00)01888-3. PMID:10981723. [DOI] [PubMed] [Google Scholar]
- 51.Simpson GG. The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr Opin Plant Biol. 2004;7:570-4. doi: 10.1016/j.pbi.2004.07.002. PMID:15337100. [DOI] [PubMed] [Google Scholar]
- 52.Abou-Elwafa SF, Buttner B, Chia T, Schulze-Buxloh G, Hohmann U, Mutasa-Gottgens E, Jung C, Muller AE. Conservation and divergence of autonomous pathway genes in the flowering regulatory network of Beta vulgaris. J Exp Bot. 2011;62:3359-74. doi: 10.1093/jxb/erq321. PMID:20974738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakrabortee S, Kayatekin C, Newby GA, Mendillo ML, Lancaster A, Lindquist S. Luminidependens (LD) is an Arabidopsis protein with prion behavior. Proc Natl Acad Sci U S A. 2016;113:6065-70. doi: 10.1073/pnas.1604478113. PMID:27114519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. PMID:12801414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910-5. doi: 10.1073/pnas.97.22.11910. PMID:11050225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146-58. doi: 10.1016/j.cell.2009.02.044. PMID:19345193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aukerman MJ, Lee I, Weigel D, Amasino RM. The Arabidopsis flowering-time gene LUMINIDEPENDENS is expressed primarily in regions of cell proliferation and encodes a nuclear protein that regulates LEAFY expression. Plant J. 1999;18:195-203. doi: 10.1046/j.1365-313X.1999.00442.x. PMID:10363371. [DOI] [PubMed] [Google Scholar]
- 58.Domagalska MA, Schomburg FM, Amasino RM, Vierstra RD, Nagy F, Davis SJ. Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development. 2007;134:2841-50. doi: 10.1242/dev.02866. PMID:17611230. [DOI] [PubMed] [Google Scholar]
- 59.Kim S, Choi K, Park C, Hwang H-J, Lee I. SUPPRESSOR OF FRIGIDA4, encoding a C2H2-Type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell. 2006;18:2985-98. doi: 10.1105/tpc.106.045179. PMID:17138694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 1994;264:566-9. doi: 10.1126/science.7909170. PMID:7909170. [DOI] [PubMed] [Google Scholar]
- 61.Chernoff YO. Are there prions in plants? Proc Natl Acad Sci U S A. 2016;113:6097-6099. doi: 10.1073/pnas.1605671113. PMID:27217577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chernova TA, Wilkinson KD, Chernoff YO. Physiological and environmental control of yeast prions. FEMS Microbiol. Rev. 2014;38:326-44. doi: 10.1111/1574-6976.12053. PMID:24236638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hines JK, Li X, Du Z, Higurashi T, Li L, Craig EA. [SWI+], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in hsp70 chaperone system activity. PLoS Genet. 2011;7. doi: 10.1371/annotation/65a80750-95f9-40a1-a509-64ee5febbaa3. PMID:21379326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Konno T, Murata K, Nagayama K. Amyloid-like aggregates of a plant protein: a case of a sweet-tasting protein, monellin. FEBS Lett 1999;454:122-6. doi: 10.1016/S0014-5793(99)00789-9. PMID:10413108. [DOI] [PubMed] [Google Scholar]
- 65.Villar-Piqué A, Sabaté R, Lopera O, Gibert J, Torne JM, Santos M, Ventura S. Amyloid-Like Protein Inclusions in Tobacco Transgenic Plants. PLoS One. 2010;5:e13625. doi: 10.1371/journal.pone.0013625. PMID:21049018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berthelot K, Lecomte S, Coulary-Salin B, Bentaleb A, Peruch F. Hevea brasiliensis prohevein possesses a conserved C-terminal domain with amyloid-like properties in vitro. Biochim Biophys Acta. 2016;1864:388-99. doi: 10.1016/j.bbapap.2016.01.006. PMID:26805576. [DOI] [PubMed] [Google Scholar]
- 67.Soedjanaatmadja UMS, Subroto T, Beintema JJ. Processed products of the hevein precursor in the latex of the rubber tree (Hevea brasiliensis). FEBS Lett 1995;363:211-3. doi: 10.1016/0014-5793(95)00309-W. PMID:7737403. [DOI] [PubMed] [Google Scholar]
- 68.Gour S, Kaushik V, Kumar V, Bhat P, Yadav SC, Yadav JK. Antimicrobial peptide (Cn-AMP2) from liquid endosperm of Cocos nucifera forms amyloid-like fibrillar structure. J Pept Sci. 2016;22:201-7. doi: 10.1002/psc.2860. PMID:27028204. [DOI] [PubMed] [Google Scholar]
- 69.Garvey M, Meehan S, Gras SL, Schirra HJ, Craik DJ, Van der Weerden NL, Anderson MA, Gerrard JA, Carver JA. A radish seed antifungal peptide with a high amyloid fibril-forming propensity. Biochim Biophys Acta – Proteins Proteomics. 2013;1834:1615-23. doi: 10.1016/j.bbapap.2013.04.030. [DOI] [PubMed] [Google Scholar]
- 70.Caughey B, Lansbury PT. Protofibrils, Pores, Fibrils, and Neurodegeneration: Separating the Responsible Protein Aggregates from The Innocent Bystanders. Annu Rev Neurosci. 2003;26:267-98. doi: 10.1146/annurev.neuro.26.010302.081142. PMID:12704221. [DOI] [PubMed] [Google Scholar]
- 71.Schonberger SJ, Edgar PF, Kydd R, Faull RL, Cooper GJ. Proteomic analysis of the brain in Alzheimer's disease: molecular phenotype of a complex disease process. Proteomics. 2001;1:1519-28. doi: . PMID:11747211. [DOI] [PubMed] [Google Scholar]
- 72.Tsuji T, Shiozaki A, Kohno R, Yoshizato K, Shimohama S. Proteomic profiling and neurodegeneration in Alzheimer's disease. Neurochem Res. 2002;27:1245-53. doi: 10.1023/A:1020941929414. PMID:12462422. [DOI] [PubMed] [Google Scholar]
- 73.Liao L, Cheng D, Wang J, Duong DM, Losik TG, Gearing M, Rees HD, Lah JJ, Levey AI, Peng J. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J Biol Chem. 2004;279:37061-8. doi: 10.1074/jbc.M403672200. PMID:15220353. [DOI] [PubMed] [Google Scholar]
- 74.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss R a, Liotta L a. Laser capture microdissection. Science 1996;274:998-1001. [DOI] [PubMed] [Google Scholar]
- 75.Kryndushkin D, Pripuzova N, Burnett BG, Shewmaker F. Non-targeted identification of prions and amyloid-forming proteins from yeast and mammalian cells. J Biol Chem. 2013;288:27100-11. doi: 10.1074/jbc.M113.485359. PMID:23926098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nizhnikov AA, Alexandrov AI, Ryzhova TA, Mitkevich O V., Dergalev AA, Ter-Avanesyan MD, Galkin AP. Proteomic screening for amyloid proteins. PLoS One. 2014;9. doi: 10.1371/journal.pone.0116003. PMID:25549323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kushnirov V V., Alexandrov IM, Mitkevich O V., Shkundina IS, Ter-Avanesyan MD. Purification and analysis of prion and amyloid aggregates. Methods. 2006;39:50-5. doi: 10.1016/j.ymeth.2006.04.007. PMID:16774835. [DOI] [PubMed] [Google Scholar]
- 78.Antonets KS, Volkov K V, Maltseva AL, Arshakian LM, Galkin AP, Nizhnikov AA. Proteomic Analysis of Escherichia coli Protein Fractions Resistant to Solubilization by Ionic Detergents. Biochem. 2016;81:34-46. [DOI] [PubMed] [Google Scholar]
- 79.Nizhnikov AA, Ryzhova TA, Volkov K V, Zadorsky SP, Sopova J V, Inge-Vechtomov SG, Galkin AP. Interaction of Prions Causes Heritable Traits in Saccharomyces cerevisiae. PLOS Genet. 2016;12:e1006504. doi: 10.1371/journal.pgen.1006504. PMID:28027291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 1997;90:549-58. doi: 10.1016/S0092-8674(00)80514-0. PMID:9267034. [DOI] [PubMed] [Google Scholar]
- 81.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910-5. doi: 10.1073/pnas.97.22.11910. PMID:11050225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Colaco M, Park J, Blanch H. The kinetics of aggregation of poly-glutamic acid based polypeptides. Biophys Chem. 2008;136:74—86. doi: 10.1016/j.bpc.2008.04.008. PMID:18538463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Antonets KS, Nizhnikov AA. SARP: A Novel Algorithm to Assess Compositional Biases in Protein Sequences. Evol Bioinform Online. 2013;9:263-73. PMID:23919085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005;102:12825-30. doi: 10.1073/pnas.0506136102. PMID:16123127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hughes RE, Lo RS, Davis C, Strand AD, Neal CL, Olson JM, Fields S. Altered transcription in yeast expressing expanded polyglutamine. Proc Natl Acad Sci U S A. 2001;98:13201-6. doi: 10.1073/pnas.191498198. PMID:11687606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peters TW, Huang M. Protein aggregation and polyasparagine-mediated cellular toxicity in Saccharomyces cerevisiae. Prion. 2007;1:144-53. doi: 10.4161/pri.1.2.4630. PMID:19164913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Balakireva A V., Zamyatnin AA. Properties of gluten intolerance: Gluten structure, evolution, pathogenicity and detoxification capabilities. Nutrients. 2016;8:e644. doi: 10.3390/nu8100644. PMID:27763541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jackson P, Boulter D, Thurman DA. A comparison of some properties of vicilin and legumin isolated from seeds of Pisum sativum, Vicia faba and Cicer arietinum. New Phytol. 1969;68:25-33. doi: 10.1111/j.1469-8137.1969.tb06416.x. [DOI] [Google Scholar]
- 89.Zein Lawton JW.: A history of processing and use. Cereal Chem. 2002;79:1-18. doi: 10.1094/CCHEM.2002.79.1.1. [DOI] [Google Scholar]
- 90.Oliviero M, Di Maio E, Iannace S. Effect of Molecular Structure on Film Blowing Ability of Thermoplastic Zein. J Appl Polym Sci. 2010;115:277-87. doi: 10.1002/app.31116. [DOI] [Google Scholar]
- 91.Munialo CD, Martin AH, Van Der Linden E De Jongh HHJ. Fibril formation from pea protein and subsequent gel formation. J Agric Food Chem. 2014;62:2418-27. doi: 10.1021/jf4055215. PMID:24564788. [DOI] [PubMed] [Google Scholar]
- 92.Tang CH, Wang CS. Formation and characterization of amyloid-like fibrils from soy β-conglycinin and glycinin. J Agric Food Chem. 2010;58:11058-66. doi: 10.1021/jf1021658. PMID:20919718. [DOI] [PubMed] [Google Scholar]
- 93.Ridgley DM, Ebanks KC, Barone JR. Peptide mixtures can self-assemble into large amyloid fibers of varying size and morphology. Biomacromolecules. 2011;12:3770-9. doi: 10.1021/bm201005k. PMID:21879764. [DOI] [PubMed] [Google Scholar]
- 94.Maurer-Stroh S, Debulpaep M, Kuemmerer N, Lopez de la Paz M, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, et al.. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods. 2010;7:237-42. doi: 10.1038/nmeth.1432. PMID:20154676. [DOI] [PubMed] [Google Scholar]
- 95.van Zanten M Koini MA, Geyer R, Liu Y, Brambilla V, Bartels D, Koornneef M, Fransz P, Soppe WJJ. Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc Natl Acad Sci. 2011;108:20219-24. doi: 10.1073/pnas.1117726108. PMID:22123962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Manfre AJ, LaHatte GA, Climer CR, Marcotte WR. Seed dehydration and the establishment of desiccation tolerance during seed maturation is altered in the Arabidopsis thaliana mutant atem6-1. Plant Cell Physiol. 2009;50:243-53. doi: 10.1093/pcp/pcn185. PMID:19073649. [DOI] [PubMed] [Google Scholar]
- 97.Nizhnikov AA, Antonets KS, Inge-Vechtomov SG. Amyloids: from Pathogenesis to Function. Biochem. 2015;80:1127-44. [DOI] [PubMed] [Google Scholar]
- 98.Ahmed AB, Kajava A V. Breaking the amyloidogenicity code: methods to predict amyloids from amino acid sequence. FEBS Lett. 2013;587:1089-95. doi: 10.1016/j.febslet.2012.12.006. PMID:23262221. [DOI] [PubMed] [Google Scholar]
- 99.Maurer-Stroh S, Debulpaep M, Kuemmerer N, Lopez de la Paz M, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, et al.. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods. 2010;7:237-42. doi: 10.1038/nmeth.1432. PMID:20154676. [DOI] [PubMed] [Google Scholar]
- 100.Nawrot R, Barylski J, Nowicki G, Broniarczyk J, Buchwald W, Goździcka-Józefiak A. Plant antimicrobial peptides. Folia Microbiol (Praha). 2014;59:181-96. doi: 10.1007/s12223-013-0280-4. PMID:24092498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rai M, Pandit R, Gaikwad S, Kovics G. Antimicrobial peptides as natural bio-preservative to enhance the shelf-life of food. J. Food Sci. Technol. 2016;53:3381-94. doi: 10.1007/s13197-016-2318-5. PMID:27777445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hintz T, Matthews KK, Di R, Hintz T, Matthews KK, Di R. The Use of Plant Antimicrobial Compounds for Food Preservation. Biomed Res Int. 2015;2015:1-12. doi: 10.1155/2015/246264. [DOI] [PMC free article] [PubMed] [Google Scholar]


