ABSTRACT
Prions are misfolded, aggregated, infectious proteins found in a range of organisms from mammals to bacteria. In mammals, prion formation is difficult to study because misfolding and aggregation take place prior to symptom presentation. The study of the yeast prion [PSI+], which is the misfolded infectious form of Sup35p, provides a tractable system to monitor prion formation in real time. Recently, we showed that the de novo formation of prion aggregates begins with the appearance of highly mobile cytoplasmic foci, called early foci, which assemble into larger ring or dot structures. We also observed SDS-resistant oligomers during formation, and lysates containing newly formed oligomers can convert [psi−] cells to the [PSI+] state, suggesting that these oligomers have infectious potential. Here, we further characterize two aspects of prion formation: spatial sequestration of early foci and oligomerization of endogenous Sup35p. Our data provides important insights into the process of prion formation and explores the minimal oligomer requirement for infectivity.
KEYWORDS: [PSI+], amyloid, infectivity, inheritance, oligomer, prion, spatial sequestration, Sup35, yeast
INTRODUCTION
Prion formation involves the spontaneous conversion of native monomeric protein into misfolded infectious conformers. In mammalian systems, conversion events occur well before the presentation of symptoms, making study of these initial formation events extremely challenging. Several groups have used in vitro strategies and chemical manipulations to induce misfolding, which has provided some important insights into prion conversion (reviewed in ref.1). Yet, there is still much left unanswered about how conversion leads to infectivity.
Yeast prions have contributed immensely to our overall understanding of prion formation. Yeast prions share many characteristics with mammalian prions such as exhibiting detergent resistance, existing as strains or variants,2 spontaneously forming at very low frequencies within the population,3-5 and being infectious.6,7 The ability to increase rates of prion formation in yeast through “prion induction” provides a tractable means to study the formation process in vivo. The prion induction process involves the overexpression of a prion protein, which is thought to increase the chance of a misfolding event.8 De novo formation is further increased by the presence of a second prion, which likely allows for heterologous cross-seeding.9-11
The understanding of prion formation has been facilitated by the study of [PSI+], the prion form of the Sup35 protein. De novo [PSI+] formation can be monitored microscopically by fusing full-length Sup35p or it's prion domain (the N-terminus and middle domain of Sup35p; PrD) to a green fluorescent protein (Sup35PrD-GFP).12 The initial steps of prion induction have been shown to involve the assembly of Sup35PrD-GFP into large fluorescent ring and dot-like aggregates.12 These fluorescent aggregates are hallmarks of prion formation since cells containing these aggregates can result in a proportion of their progeny that are [PSI+], whereas cells lacking these aggregates remain [psi−].13 However, the process in which these aggregates form and the biochemical changes that occur over time to produce an infectious conformer is unclear.
Our recent work has provided insight into some of these unanswered questions. Using 3D time-lapse microscopy, we were able to capture the in vivo assembly of Sup35PrD-GFP ring and dot structures in real time. We observed that the formation process starts with one or several small fluorescent foci (which we term “early foci”). These early foci tend to coalesce into a larger aggregate that grows into a ring or dot shaped structure. We analyzed over 90 cells undergoing prion induction and found that there appeared to be no obvious preference for early foci to assemble into ring or dot structures, although cells containing either structure were able to give rise to [PSI+] colonies.14
We next asked how the Sup35PrD-GFP assembles into a prion oligomer during formation. Biochemical analysis of cell cultures taken at sequential time points in the aggregate formation process showed that the Sup35PrD-GFP protein formed SDS-resistant oligomers that change in size over time. For example, we observed a SDS-resistant high molecular weight band as early as 8 hours of induction, well before the first microscopic visualization of Sup35PrD-GFP early foci. By 24 hours, we observed distinct changes in the migration of the oligomer shifting from a single band to a large smear. Our data suggest that the Sup35PrD-GFP oligomer assembles into a diverse population of sizes during prion formation. In contrast, we noticed that endogenous Sup35p oligomers were only detectable at 24 hours, and had a slower migration compared to those that are oligomers associated with propagating [PSI+] cultures.14 It can be envisioned that the endogenous Sup35p oligomer must undergo remodeling over time to become [PSI+].
We also asked when do prions gain their infectious nature. To begin to answer this question, cell lysates obtained from cultures at different times of the induction process were transfected into [psi−] strains. We previously observed that lysates from cultures containing newly formed ring and dot structures were able to convert [psi−] recipient strains to [PSI+].14 Our data suggest that the infectious nature of the prion is not limited to the [PSI+] state, but appears to be acquired early during prion formation.
EARLY FOCI AND SPATIAL SEQUESTRATION
The age of the mother may influence how early foci form. We previously observed that early foci initially appear in G2/M phase cells. Interestingly, early foci rarely appeared in the daughter bud by herself, but rather in the mother alone or simultaneously in the mother cell and daughter bud.14 In young mothers, it has been shown that damaged proteins or unhealthy organelles are sequestered in the mother cell, leaving the daughter bud rejuvenated and free of potentially harmful material.15 In contrast, damaged or aggregation prone proteins are passed to daughter buds more frequently in mutants with a shortened life span.16,17 Therefore, it is possible that the age of the mother may correlate with the ability to form early foci in the daughter bud.
To explore how early foci are sequestered in the mother cell and daughter bud, we reanalyzed our 3D time-lapse microscopy videos. We observed that early foci behavior was different in the daughter bud compared to the corresponding mother cell. We analyzed 22 G2/M cells where early foci were present in both the mother cell and daughter bud. We found that early foci quickly coalesced into one single aggregate in the daughter bud, but remained as separate aggregates in the mother cell (Fig. 1A and B). The single aggregate in the daughter bud and the multiple distinct aggregates in the mother cell were maintained until the end of the time-lapse recordings, which went up to 6 hours. Quantification of early foci in mother cells and daughter buds at the end of the time-lapse recordings showed that mother cells had on average 2 aggregates versus the daughter that consistently had one (Fig. 1C). It is possible that age may play a role in the inability to fuse early foci into one central aggregate in the mother cell. Recently, Kumar et al.18 showed that actin/myosin mediated networks are used to move aggregates into a cellular inclusion called IPOD, Insoluble PrOtein Deposit. It is possible the rejuvenated daughter bud has robust actin/myosin networks to efficiently coalesce foci into IPOD but the mother cell does not. Since multiple aggregates were observed in the mother cell, it is possible that aggregates in aged cells are not held exclusively at IPOD. Recent evidence suggests that there are other protein accumulation sites in aged cells. For example, propagating [PSI+] aggregates in aged cells accumulate in a deposit called APOD,19 whereas protein accumulation on the surface of the ER increases with age.20 Further work with early foci will be required to understand the differences between how aggregates are sequestered in mother cells and daughter buds.
FIGURE 1.
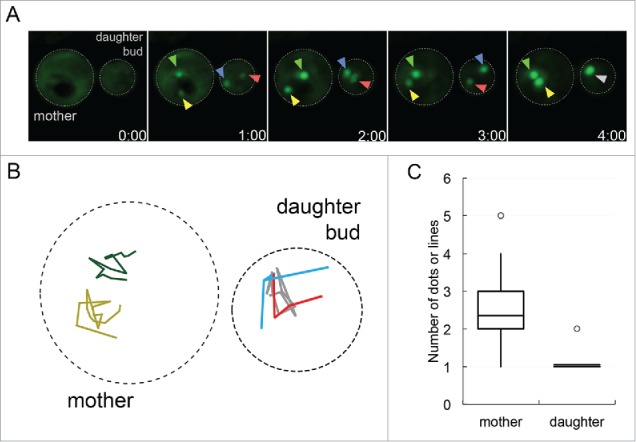
Protein aggregates coalesce in daughter buds but not in mother cells. A. Sup35PrD-GFP was overexpressed for approximately 18 hours, and then placed on 8-well glass bottom slides and imaged for an additional 12 hours according to Sharma et al. (2017).14 Of the 1382 cells that were imaged, 20.8% contained ring or line aggregates. Of these cells with aggregates, the initial formation of early foci was captured in 22 G2/M phase cells that contained early foci in both mother cell and daughter bud. These cells were followed until larger aggregates were assembled. Representative images of early foci initially appearing in both mother cell and daughter bud (01:00 minute) to their fusion in the daughter bud (04:00 minute) are shown. Yellow and green arrows point to the early foci in the mother cell, while the red and blue arrows are in the daughter bud, turning grey when they merge. B. Early foci trajectories (from part A) followed for 16 minutes are shown. The movement of each foci was tracked on a 2D plane, with colors corresponding to arrows in part A in the mother (left cell) and daughter bud (right cell). C. 22 time-lapse videos were analyzed for the number of aggregates in the mother cell and daughter bud present at the end of recordings, most of which ranged from one to six hours. The box-plot represents the mean (middle line), upper and lower quartiles and outliers (open circle). Statistically significant differences in aggregate formation in the mother cell and daughter bud were determined by a paired two-tailed t-test (p<0.0001).
THE FORMATION OF ENDOGENOUS SUP35P OLIGOMERS DURING DE NOVO PRION FORMATION AND THE POTENTIAL FOR INFECTIVITY
We recently observed that Sup35PrD-GFP forms SDS-resistant oligomers that change in size over time using Semi-Denaturing Detergent Agarose Gel Electrophoresis (SDD-AGE21) methods, yet the ability to resolve the endogenous Sup35p was more difficult.14 To better resolve endogenous Sup35p oligomerization during prion induction, we performed sedimentation analysis on induced cultures at incremental time points using sucrose gradient centrifugation. We found that as early as 8 hours of induction, Sup35p sediments to heavier fractions compared to uninduced and [psi−] controls (Fig. 2). Our data provides additional support that Sup35p forms complexes well before the formation of visible Sup35PrD-GFP aggregates. By 24 hours, the relative distribution of Sup35p shifts from lighter to heavier fractions. The change in sedimentation suggests that the endogenous Sup35p likely forms complexes that are remodeling to form a propagating prion in vivo.
FIGURE 2.
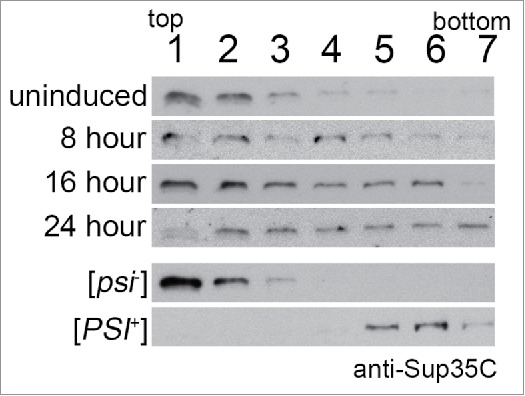
Endogenous Sup35p sediments in heavy fractions by 24 hours of induction. Sup35PrD-GFP was overexpressed for 8, 16, and 24 hours. Cultures were lysed using glass beads and protein was immediately separated by sucrose gradient centrifugation (20-60% sucrose). Samples were centrifuged at 4°C for 1 hour. Seven fractions were collected and run on SDS-PAGE for immunoblotting. Anti-Sup35C antibody was used in immunoblot analysis. An uninduced 24 hour culture, [PSI+] and [psi−] culture were also used as controls.
It is unclear when a prion gains its infectious quality. Previous work showed that recombinant derived Sup35p fibrils were able to convert [psi−] cells to [PSI+] cells, providing solid proof for the protein only hypothesis of prion infection.6,7 We recently asked at what point in the induction process would lysates contain infectious material. Surprisingly, we found that infectivity was observed from lysates generated from cultures induced for little as 16 hours, yet lysates from prior time points were not infectious. Since we observe a change in Sup35p sedimentation between 8 and 16 hours, it is possible that there is a transition state in which immature oligomers go from a non-infectious form to a mature infectious form. Conversely, it could be argued that the oligomer must reach a critical concentration for infectivity, implying that the initially formed oligomer has inherent infectious qualities. Regardless of the explanation, prion formation in yeast will continue to provide valuable insights into how and when a newly formed prion becomes infectious.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
ACKNOWLEDGMENTS
We thank Susan W. Liebman for the kind gift of strains and plasmids used in this study. The Sup35 (BE4) antibody was a kind gift from Viravan Prapapanich and Susan Liebman. We would also like to thank Jaya Sharma for her preliminary contributions to this project, and Jane Dorweiler and Brett Wisniewski for valuable discussions and helpful comments.
FUNDING
This work was supported by grant from the National Institutes of Health (GM109336) to A.L.M.
REFERENCES
- 1.Diaz-Espinoza R, Soto C. Generation of prions in vitro and the protein-only hypothesis. Prion. 2010;4(2):53–9. doi: 10.4161/pri.4.2.11960. PMID:20448454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191(4):1041–72. doi: 10.1534/genetics.111.137760. PMID:22879407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aigle M, Lacroute F. Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol Gen Genet. 1975;136(4):327–35. doi: 10.1007/BF00341717. PMID:16095000. [DOI] [PubMed] [Google Scholar]
- 4.Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184(2):393–400. doi: 10.1534/genetics.109.110213. PMID:19917766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lund PM, Cox BS. Reversion analysis of [psi-] mutations in Saccharomyces cerevisiae. Genet Res. 1981;37(2):173–82. doi: 10.1017/S0016672300020140. PMID:7021322. [DOI] [PubMed] [Google Scholar]
- 6.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428(6980):319–23. doi: 10.1038/nature02391. PMID:15029195. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428(6980):323–8. doi: 10.1038/nature02392. PMID:15029196. [DOI] [PubMed] [Google Scholar]
- 8.Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24(3):268–70. doi: 10.1007/BF00351802. PMID:8221937. [DOI] [PubMed] [Google Scholar]
- 9.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: The story of [PIN(+)]. Cell. 2001;106(2):171–82.S0092-8674(01)00427-5 [pii] doi: 10.1016/S0092-8674(01)00427-5. PMID:11511345. [DOI] [PubMed] [Google Scholar]
- 10.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106(2):183–94. doi: 10.1016/S0092-8674(01)00440-8. PMID:11511346. [DOI] [PubMed] [Google Scholar]
- 11.Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell. 2000;5(1):163–72. doi: 10.1016/S1097-2765(00)80412-8. PMID:10678178. [DOI] [PubMed] [Google Scholar]
- 12.Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)]. Mol Microbiol. 2001;39(1):37–46.mmi2224 [pii] doi: 10.1046/j.1365-2958.2001.02224.x. PMID:11123686. [DOI] [PubMed] [Google Scholar]
- 13.Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, Chernoff YO. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol. 2006;26(2):617–29. doi: 10.1128/MCB.26.2.617-629.2006. PMID:16382152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma J, Wisniewski BT, Paulson E, Obaoye JO, Merrill SJ, Manogaran AL. De novo [PSI +] prion formation involves multiple pathways to form infectious oligomers. Sci Rep. 2017;7(1):76. doi: 10.1038/s41598-017-00135-6. PMID:28250435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higuchi-Sanabria R, Pernice WM, Vevea JD, Alessi Wolken DM, Boldogh IR, Pon LA. Role of asymmetric cell division in lifespan control in Saccharomyces cerevisiae. FEMS Yeast Res. 2014;14(8):1133–46. doi: 10.1111/1567-1364.12216. PMID:25263578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erjavec N, Larsson L, Grantham J, Nyström T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21(19):2410–21. doi: 10.1101/gad.439307. PMID:17908928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song J, Yang Q, Yang J, Larsson L, Hao X, Zhu X, Malmgren-Hill S, Cvijovic M, Fernandez-Rodriguez J, Grantham J, et al.. Essential genetic interactors of SIR2 required for spatial sequestration and asymmetrical inheritance of protein aggregates. PLoS Genet. 2014;10(7):e1004539. doi: 10.1371/journal.pgen.1004539. PMID:25079602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar R, Nawroth PP, Tyedmers J. Prion aggregates are recruited to the insoluble protein deposit (IPOD) via myosin 2-based vesicular transport. PLoS Genet. 2016;12(9):e1006324. doi: 10.1371/journal.pgen.1006324. PMID:27689885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saarikangas J, Barral Y. Protein aggregates are associated with replicative aging without compromising protein quality control. Elife. 2015;4:e06197. doi: 10.7554/eLife.06197. PMID:26544680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saarikangas J, Caudron F, Prasad R, Moreno DF, Bolognesi A, Aldea M, Barral Y. Compartmentalization of ER-bound chaperone confines protein deposit formation to the aging yeast cell. Curr Biol. 2017;27(6):773–783. doi: 10.1016/j.cub.2017.01.069. PMID:28262489. [DOI] [PubMed] [Google Scholar]
- 21.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278(49):49636–43. doi: 10.1074/jbc.M307996200. PMID:14507919. [DOI] [PubMed] [Google Scholar]


