ABSTRACT
Alkaline pectinase has important applications in the pretreatment of waste water from food processing and in both the fabric and paper industries. In this study, a 2-level factorial design was used to screen significant factors that affect the activity of alkaline pectinase, and the response surface methodology (RSM) with a Box-Behnken design (BBD) was used to optimize their concentrations. Starch, peptone, KH2PO4 and K2HPO4·3H2O were found to significantly affect the activity of alkaline pectinase. Their optimal concentrations were as follows: 4.68% starch, 1.6% peptone, 0.26% KH2PO4 and 0.68% K2HPO4·3H2O. Under the above conditions, the activity of alkaline pectinase was significantly improved to 734.11 U/mL. Alkaline pectinase was purified to homogeneity with a recovery rate of 9.6% and a specific activity of 52372.52 U/mg. Its optimal temperature and pH were 50°C and 8.6, respectively. The purified enzyme showed strong thermo-stability and good alkali resistance. In addition, the activity of alkaline pectinase was improved in the presence of Mg2+. Cu2+, Mn2+, and Co2+ significantly inhibited its activity. This study provides an important basis for the future development and use of a heat-tolerant alkaline pectinase from B. subtilis ZGL14.
KEYWORDS: alkaline pectinase, Bacillus subtilis ZGL14, characterization, enzymatic properties, optimization, response surface methodology
Introduction
Pectic substances are a heterogeneous group of high molecular weight complex acidic structural polysaccharides that consist largely of D-galactopyranosyluronic acid, which is α-(1→4) glycosidically linked to polygalacturonic acid with small amounts of L-rhamnose (2–4%) and various side chains composed of L-arabinose, D-galactose and β-D-xylose.1-4 Recently, a significant interest in the degradation of pectic substances has been generated, which is evident from a vast range of industrial applications, such as the degumming of bast fibers,5-8 the treatment of alkaline pectic waste water,9 and the extraction, clarification and depectinization of fruit juices.10 The enzyme that hydrolyzes pectic substances is broadly known as pectinase, which includes polygalacturonase (exo-polygalacturonase and endo-polygalacturonase), pectin esterase, pectin lyase and pectate lyase on the basis of their modes of action. Pectinase is naturally produced by many organisms, including bacteria, fungi, yeasts, insects, nematodes, protozoan, and plants.11-15 Microbial pectinase is important in phyto-pathological processes, in plant-microorganism symbioses, and in the decomposition of dead plant materials, through which it contributes to the natural carbon cycle.8,16-21
Based on the pH requirement for optimal enzymatic activity, pectinase can be broadly classified into acidic and alkaline pectinase.22-25 Acidic pectinase has been extensively reported for the extraction and clarification of fruit juices and wines,10,26-28 the maceration of vegetables and fruits, the improvement of essential oil extraction and the enhancement of baby-food production,18,29 whereas alkaline pectinase is used for pretreating waste water from the food processing industry that contains pectinaceous waste9 and in the processing and degumming of bast fibers such as ramie (Boehmeria nivea),8 sunn hemp (Crotalaria juncea),8 buel (Grewia optiva)12 and jute (Chorchorus capsularis).30 It is also widely used in the fabric industry for the retting of plant fibers such as flax, hemp and jute, the biopreparation of cotton fabrics and the enzymatic polishing of jute/cotton blended fabrics; in the paper industry to solve the retention problem in mechanical pulp bleaching and in the treatment of pulp and paper mill effluents; and for improving the quality of black tea. However, the production of alkaline pectinase at a commercial scale remains quite low due to the paucity of cultures producing high enzyme yields.16,17,31-33
The conventional method for optimizing enzymatic production by the one factor at a time approach involves varying a single independent factor while maintaining the others at a constant level. This one-dimensional approach is laborious and time-consuming, especially for a large number of factors, and it does not consider interactions among factors. An alternative and more efficient approach is the use of statistical methods. Response surface methodology (RSM) involves a full factorial search by examining the simultaneous, systematic and efficient variation of all components, and it is useful for a large number of factors. It uses quantitative data from appropriate experiments to determine and simultaneously solve multivariate equations. It is a collection of statistical techniques that are used to design experiments, build models, evaluate the effects of factors, and analyze the optimum conditions of factors to obtain desirable responses.34-36 It has been successfully used to optimize compositions of fermentation medium, conditions of enzymatic hydrolysis, synthesis parameters for polymers, and parameters for food processes.37-39
In view of the industrial application of alkaline pectinase, the present study was performed to improve the production of a thermostable alkaline pectinase from Bacillus subtilis ZGL14 isolated from the soil. 2-level factorial design and RSM were used to identify critical factors and optimize them for the maximum production of alkaline pectinase. The purification and characterization of the alkaline pectinase from fermentation broth were also investigated in detail. This study provides new insight into the future development and use of a heat-tolerant alkaline pectinase from B. subtilis ZGL14. To the best of our knowledge, this is the first report regarding the optimization of conditions for the production of a heat-tolerant alkaline pectinase from B. subtilis ZGL14 and its purification and characterization.
Materials and methods
Chemicals
Pectin, barbituric acid, 3,5-dinitrosalicylic acid, N-cyclohexyl-3-aminopropanesulfonic acid and coomassie brilliant blue G-250 were purchased from Sangon Biotech Co., Ltd., Shanghai, China. DEAE-cellulose 52 and Sephadex G-100 were purchased from Boyun Biotech Co., Ltd., Shanghai, China. All other chemicals used in the experiments were of analytical grade and were used as the routine method.
Microorganism and culture conditions
B. subtilis ZGL14, isolated from the soil, was used as the source of alkaline pectinase in this study. This strain was first grown for 24 h at 37°C and 200 rpm in a 250 mL shake flask containing 50 mL of seed medium. The seed medium contained 1% glucose, 0.3% beef extract, 1% peptone and 0.5% NaCl, pH 8.0. Nine percent of each culture was inoculated into 50 mL of fermentation medium prepared under pre-designed conditions by 2-level factorial design and RSM with a Box-Behnken design (BBD) and was cultured for 72 h at 40°C and 200 r/min. The samples were taken out to analyze the activity of alkaline pectinase. The basic fermentation medium contained 4% glucose, 2% peptone, 0.5% yeast extract, 0.6% K2HPO4·3H2O and 0.3% KH2PO4, pH 9.0.
2-level factorial design
Nine factors, starch, peptone, K2HPO4·3H2O, KH2PO4, temperature, pH, inoculation, liquid volume and fermentation time, were hypothesized to have an effect on the activity of alkaline pectinase. A 2-level factorial design to evaluate the main effect of the factors was performed as described by Bhunia and Dey.40
Box-Behnken design
Box-Behnken design was used to optimize the most significant factors to further improve the activity of alkaline pectinase. Each factor was studied at 3 different levels, and a set of 30 experiments was performed. For statistical calculations, the coding of factors was done according to the following equation:
| (1) |
where χi is the coded value of an independent factor, Xi is the actual value of an independent factor, X0 is the actual value of an independent factor at the center point, and ΔX is the step change. To predict the optimal point, a second-order polynomial equation was fitted to correlate the relationship between factor and response. The model equation used for the analysis is given below:
| (2) |
where Y is the predicted response, β0 is the intercept, βi is the linear coefficient, βii is the squared coefficient, and βij is the interaction coefficient. χi, χj represent independent factors in the form of coded values. The accuracy and general ability of the above polynomial model can be evaluated by the determination coefficient R2. Each design was performed in duplicate, and the mean value was given.
Determination of the activity of alkaline pectinase
The activity of alkaline pectinase was determined by the DNS method, using pectin as the substrate.41 Cultured cells were precipitated by centrifugation at 12000 rpm at 4°C for 20 min. The activity of alkaline pectinase in the supernatant was assayed by measuring the amount of D-galacturonic acid liberated from pectin. The reaction mixture containing 0.5 mL appropriately diluted alkaline pectinase and 0.5 mL of 0.5% pectin (pectin in N-cyclohexyl-3-aminopropanesulfonic acid buffer, pH 8.6) was incubated for 5 min at 50°C, and the end products were quantitated using the 3,5-dinitrosalicylic acid (DNS) reagent.42 One unit of the alkaline pectinase activity is defined as the amount of enzyme required to liberate 1 µmol of D-galacturonic acid mL−1 min−1 under the assay conditions.
Purification of alkaline pectinase
B. subtilis ZGL14 was first cultured in a 2000 mL shake flask with 1000 mL of fermentation medium prepared under the optimal conditions obtained by RSM. The purification of alkaline pectinase was performed as described by Bhunia et al.43 The purity of alkaline pectinase was determined by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) with 12% separation gel and 5% concentrated gel.44
Effect of temperature and pH on the activity of the purified alkaline pectinase
The optimal temperature for the activity of alkaline pectinase was investigated by incubating 0.5 mL of the reaction mixture for 30 min. The reaction mixture contained 0.2 mL of 20 mM Tris-HCl buffer (pH 8.6), 0.2 mL of 0.5% pectin and 0.1 mL of alkaline pectinase over the range of 25–80°C. The effect of pH on the activity of the purified alkaline pectinase was studied using 0.5 mL of the reaction mixture containing 0.1 mL of alkaline pectinase, 0.2 mL of buffer and 0.2 mL of 0.5% pectin as the substrate at 50°C for 30 min. The buffers used were a 20 mM phosphate buffer pH 6–7.5, a 20 mM Tris-HCl buffer pH 7.5–8.8, and a glycine-NaOH buffer pH 8.5–11. Water was used as a control, and the samples were analyzed in duplicate.
Effect of temperature and pH on the stability of the purified alkaline pectinase
The alkaline pectinase solutions were pre-incubated for 30 min at various temperatures ranging from 40 to 80°C and pH values ranging from 7 to 10 without the substrate and were immediately cooled to 4°C. The activity of alkaline pectinase was determined as described by Tang et al..41 The control sample was the purified alkaline pectinase solution at 4°C and pH 8.6, and its activity was defined as 100%. The relative activity was calculated as the ratio of the alkaline pectinase activity under a specific condition to the activity of the control sample.
Effect of metal ions on the activity of the purified alkaline pectinase
The alkaline pectinase was pre-incubated with various metal ions dissolved in a 20 mM Tris-HCl buffer (pH 8.6) at 50°C for 30 min. The activity of alkaline pectinase was determined as described by Tang et al.41 The control sample was the purified alkaline pectinase solution without metal ions, and its activity was defined as 100%. The relative activity was calculated as the ratio of the alkaline pectinase activity under a specific condition to the activity of the control sample.
Statistical analysis
Design expert, ver 8.0 (Statease Inc., Minneapolis, MN, USA) was used for experimental designs and the regression analysis of experimental data. Statistical analysis of the model was performed to evaluate the analysis of variance (ANOVA). The quality of the polynomial model equation was judged statistically by the determination coefficient R2, and its statistical significance was determined by F-test.
Results and discussion
2-level factorial design
The 2-level factorial design gives an effective screening method for finding the significant factors in one experiment. Table 1 shows 9 independent factors and their concentrations at different coded levels45 and gives the experimental results of 2-level factorial design for 20 runs using the activity of alkaline pectinase as the response. As shown in Table 1, the activity of alkaline pectinase showed considerable variation depending on the levels of the 9 factors. The regression results are shown in Table 2. Statistical analysis showed that starch, peptone, K2HPO4·3H2O and KH2PO4 had a very significant impact on the activity of alkaline pectinase with the confidence level above 95% (p < 0.05).
Table 1.
Experimental results of the 2-level factorial design.
| Run | X1 (%) | X2 (%) | X3 (%) | X4 (%) | X5 (°C) | X6 | X7 (mL) | X8 (mL) | X9 (mL) | alkaline pectinase activity (U/mL) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 1 | 0.9 | 0.2 | 37 | 9.5 | 8 | 75 | 84 | 579.14 |
| 2 | 5 | 3 | 0.3 | 0.2 | 37 | 9.5 | 10 | 25 | 84 | 674.1 |
| 3 | 3 | 3 | 0.9 | 0.2 | 37 | 8.5 | 10 | 75 | 84 | 447.76 |
| 4 | 5 | 3 | 0.3 | 0.2 | 43 | 8.5 | 10 | 75 | 60 | 579.36 |
| 5 | 3 | 3 | 0.3 | 0.4 | 37 | 9.5 | 10 | 75 | 60 | 633.86 |
| 6 | 5 | 1 | 0.9 | 0.2 | 43 | 8.5 | 8 | 25 | 60 | 669.43 |
| 7 | 3 | 1 | 0.3 | 0.2 | 37 | 8.5 | 8 | 25 | 84 | 652.34 |
| 8 | 5 | 3 | 0.3 | 0.4 | 37 | 8.5 | 8 | 75 | 60 | 612.95 |
| 9 | 5 | 1 | 0.9 | 0.4 | 37 | 8.5 | 10 | 25 | 60 | 259.45 |
| 10 | 3 | 1 | 0.9 | 0.2 | 43 | 9.5 | 10 | 25 | 60 | 666.44 |
| 11 | 4 | 2 | 0.6 | 0.3 | 40 | 9 | 9 | 50 | 72 | 684.43 |
| 12 | 5 | 3 | 0.9 | 0.4 | 43 | 9.5 | 10 | 75 | 84 | 576.34 |
| 13 | 3 | 3 | 0.3 | 0.2 | 43 | 9.5 | 8 | 75 | 60 | 520.79 |
| 14 | 4 | 2 | 0.6 | 0.3 | 40 | 9 | 9 | 50 | 72 | 692.69 |
| 15 | 5 | 1 | 0.3 | 0.4 | 43 | 9.5 | 8 | 25 | 84 | 269.47 |
| 16 | 4 | 2 | 0.6 | 0.4 | 40 | 9 | 9 | 50 | 72 | 676.32 |
| 17 | 3 | 3 | 0.9 | 0.4 | 37 | 9.5 | 8 | 25 | 60 | 622.97 |
| 18 | 3 | 1 | 0.9 | 0.4 | 43 | 8.5 | 8 | 75 | 84 | 610.38 |
| 19 | 3 | 3 | 0.3 | 0.4 | 43 | 8.5 | 10 | 25 | 84 | 671.99 |
| 20 | 4 | 2 | 0.6 | 0.3 | 40 | 9 | 9 | 50 | 60 | 699.06 |
X1. starch (%), X2. peptone (%), X3. K2HPO4·3H2O (%), X4. KH2PO4 (%), X5. temperature (°C),
X6. pH, X7. inoculation (mL), X8. liquid volume (mL) and X9. fermentation time (h)
Table 2.
ANOVA for the 2-level factorial design.
| Source | df | Sum of squares | Mean of square | F-value | p-value |
|---|---|---|---|---|---|
| X1. starch | 1 | 22936.35 | 22936.35 | 133.44 | 0.0003* |
| X2. peptone | 1 | 18658.88 | 18658.88 | 108.55 | 0.0005* |
| X3. K2HPO4·3H2O | 1 | 2103.37 | 2103.37 | 12.24 | 0.0249* |
| X4. KH2PO4 | 1 | 17718.94 | 17718.94 | 103.09 | 0.0005* |
| X5. temperature | 1 | 411.38 | 411.38 | 2.39 | 0.1968 |
| X6. pH | 1 | 99.75 | 99.75 | 0.58 | 0.4886 |
| X7. inoculation | 1 | 47.85 | 47.85 | 0.28 | 0.5346 |
| X8. liquid volume | 1 | 341.23 | 341.23 | 1.99 | 0.6257 |
| X9. fermentation time | 1 | 432.95 | 432.95 | 2.52 | 0.2316 |
| Lack of fit | 1 | 394.12 | 394.12 | 4.03 | 0.1383 |
| Pure error | 3 | 293.42 | 97.81 |
Statistically significant at 95% confidence level (p < 0.05).
Box-Behnken design and response surface methodology
Response surface methodology with a Box-Behnken design was used to determine the optimal concentrations of the 4 factors that were chosen by the 2-level factorial design. The coded and real values of the 4 factors and the results of the Box-Behnken design are shown in Table 3. The effects of the 4 factors on the activity of alkaline pectinase were predicted by the following second-order polynomial equation:
| (3) |
where Y is the predicted activity of alkaline pectinase, X1 is the concentration of starch, X2 is the concentration of peptone, X3 is the concentration of K2HPO4·3H2O, and X4 is the concentration of KH2PO4.
Table 3.
Experimental results of the Box-Behnken design.
| Coded levels |
Uncoded levels |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Run | X1 | X2 | X3 | X4 | X1 | X2 | X3 | X4 | alkaline pectinase activity (U/mL) |
| 1 | 0 | 0 | 0 | 0 | 2 | 1 | 0.3 | 0.15 | 694.69 |
| 2 | 0 | 0 | 0 | 0 | 2 | 1 | 0.3 | 0.15 | 712.25 |
| 3 | −1 | −1 | 1 | −1 | 1.5 | 0.5 | 0.45 | 0.1 | 476.65 |
| 4 | 1 | −1 | −1 | 1 | 2.5 | 0.5 | 0.15 | 0.2 | 521.01 |
| 5 | 0 | 0 | 0 | 0 | 2 | 1 | 0.3 | 0.15 | 707.65 |
| 6 | 0 | −2 | 0 | 0 | 2 | 0 | 0.3 | 0.15 | 587.88 |
| 7 | 1 | −1 | −1 | −1 | 2.5 | 0.5 | 0.15 | 0.1 | 552.12 |
| 8 | 0 | 0 | −2 | 0 | 2 | 1 | 0 | 0.15 | 228.56 |
| 9 | 0 | 0 | 0 | 2 | 2 | 1 | 0.3 | 0.25 | 521.6 |
| 10 | 0 | 0 | 0 | 0 | 2 | 1 | 0.3 | 0.15 | 689.58 |
| 11 | −1 | −1 | 1 | 1 | 1.5 | 0.5 | 0.45 | 0.2 | 317.34 |
| 12 | 1 | 1 | 1 | 1 | 2.5 | 1.5 | 0.45 | 0.2 | 590.61 |
| 13 | 0 | 0 | 0 | −2 | 2 | 1 | 0.3 | 0.05 | 628.28 |
| 14 | −2 | 0 | 0 | 0 | 1 | 1 | 0.3 | 0.15 | 387.33 |
| 15 | 1 | 1 | −1 | −1 | 2.5 | 1.5 | 0.15 | 0.1 | 363.5 |
| 16 | 1 | −1 | 1 | −1 | 2.5 | 0.5 | 0.45 | 0.1 | 653.44 |
| 17 | 2 | 0 | 0 | 0 | 3 | 1 | 0.3 | 0.15 | 637.81 |
| 18 | −1 | 1 | 1 | 1 | 1.5 | 1.5 | 0.45 | 0.2 | 371.83 |
| 19 | −1 | 1 | −1 | −1 | 1.5 | 1.5 | 0.15 | 0.1 | 398.46 |
| 20 | 1 | 1 | −1 | 1 | 2.5 | 1.5 | 0.15 | 0.2 | 405.35 |
| 21 | 0 | 2 | 0 | 0 | 2 | 2 | 0.3 | 0.15 | 473.69 |
| 22 | 0 | 0 | 0 | 0 | 2 | 1 | 0.3 | 0.15 | 729.48 |
| 23 | −1 | 1 | 1 | −1 | 1.5 | 1.5 | 0.45 | 0.1 | 468.59 |
| 24 | −1 | 1 | −1 | 1 | 1.5 | 1.5 | 0.15 | 0.2 | 307.9 |
| 25 | 0 | 0 | 2 | 0 | 2 | 1 | 0.6 | 0.15 | 374.62 |
| 26 | −1 | −1 | −1 | −1 | 1.5 | 0.5 | 0.15 | 0.1 | 525.63 |
| 27 | 1 | −1 | 1 | 1 | 2.5 | 0.5 | 0.4 | 0.2 | 576.12 |
| 28 | 0 | 0 | 0 | 0 | 2 | 1 | 0.3 | 0.15 | 697.58 |
| 29 | 1 | 1 | 1 | −1 | 2.5 | 1.5 | 0.45 | 0.1 | 597.01 |
| 30 | −1 | −1 | −1 | 1 | 1.5 | 0.5 | 0.15 | 0.2 | 399.98 |
The statistical significance of the above equation (3) was checked by F-test. The analysis of variance (ANOVA) for the second-order polynomial model is shown in Table 4. The analysis of factors showed that it was a highly significant model, as suggested by the model's F-value and low probability value (p < 0.0001). The analysis of factors also showed that the second-order polynomial equation was well adjusted to the experimental data. The low value, 2.41, of the coefficient of variation (CV) indicated a high degree of precision and a good deal of reliability of the experimental values because the coefficient of variation could indicate the degree of precision to which treatments are compared. Usually, a higher CV value indicates that the reliability of the experiment is lower.46 The precision of a model can be checked by the determination coefficient (R2) and correlation coefficient (R). Here, R2 was calculated to be 0.9921, indicating that 99.21% of the variability in the response could be explained by this model. Normally, a regression model with an R value higher than 0.9 is considered to exhibit very high correlation. It was considered that the closer the value of the correlation coefficient is to 1, the better the correlation is between the experimental values and predicted ones. R (0.9821) for the above equation (3) indicated close agreement between the experimental results and the predicted theoretical ones. p-values are used to check the significance of each coefficient, which may, in turn, indicate the pattern of interaction among the factors. The smaller the value of p is, the more significant the corresponding coefficient is.47,48 It can be seen that X1, X2, X3, X4, X1X2, X1X3, X1X4, X2X3, X2X4, X3X4, X12, X22, X32 and X42 were significant, with a small p-value (p < 0.05).
Table 4.
ANOVA of the Box-Behnken design.
| Source | df | Sum of squares | Mean of square | F-value | p-value |
|---|---|---|---|---|---|
| Model | 14 | 5.730E+005 | 40929.98 | 261.17 | <0.0001* |
| X1 | 1 | 92967.89 | 92967.89 | 593.23 | <0.0001* |
| X2 | 1 | 23277.15 | 23277.15 | 148.53 | <0.0001* |
| X3 | 1 | 31519.38 | 31519.38 | 201.12 | <0.0001* |
| X4 | 1 | 23978.71 | 23978.71 | 153.01 | <0.0001* |
| X1X2 | 1 | 1879.44 | 1879.44 | `11.99 | 0.0035* |
| X1X3 | 1 | 20502.66 | 20502.66 | 130.83 | <0.0001* |
| X1X4 | 1 | 9965.53 | 9965.53 | 63.59 | <0.0001* |
| X2X3 | 1 | 17424.66 | 17424.66 | 111.19 | <0.0001* |
| X2X4 | 1 | 3646.05 | 3646.05 | 23.27 | 0.0002* |
| X3X4 | 1 | 1127.45 | 1127.45 | 7.19 | 0.0171* |
| X12 | 1 | 67979.85 | 67979.85 | 433.78 | <0.0001* |
| X22 | 1 | 56112.34 | 56112.34 | 358.05 | <0.0001* |
| X32 | 1 | 2.883E+005 | 2.883E+005 | 1839.85 | <0.0001* |
| X42 | 1 | 32065.34 | 32065.34 | 204.61 | <0.0001* |
| Residual | 15 | 2350.74 | 156.72 | ||
| Lack of Fit | 10 | 1285.75 | 128.57 | 0.60 | 0.7678 |
| Pure Error | 5 | 1064.99 | 213.00 | ||
| Cor Total | 29 | 5.754E+005 | |||
| CV | 2.41 | ||||
| R-square | 0.9959 | ||||
| Adj R-square | 0.9921 |
Statistically significant at 95% confidence level (p < 0.05).
3.3. Response surface plot and contour plot
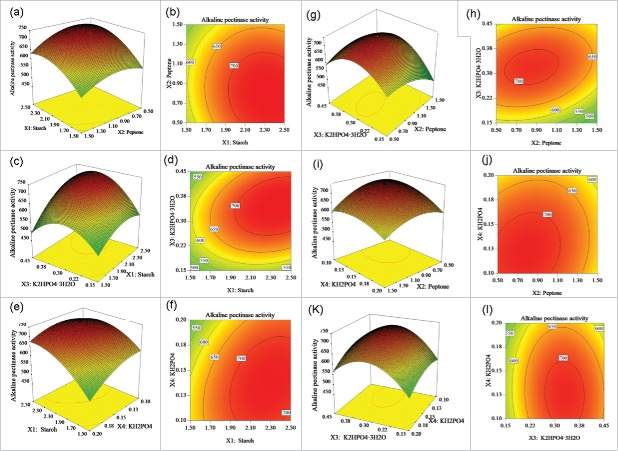
3D response surface plots and 2D contour plots were drawn to illustrate the interactive effect of each independent factor on the activity of alkaline pectinase, and the optimal concentration of each component required to maximize the activity of alkaline pectinase (Fig. 1). Each figure shows the effect of 2 factors, and the third factor is set at 0. As shown in Fig. 1a and b, when K2HPO4·3H2O (X3) and KH2PO4 (X4) were fixed at 0, starch (X1) and peptone (X2) had a significant impact on the activity of alkaline pectinase. The activity of alkaline pectinase was dramatically improved with an increase in starch content from 1.5% to 2.34%, and with a decrease in peptone content from 1.5% to 0.8%. Subsequently, it dropped slightly as the content of starch increased further and the content of peptone decreased further. The 2D contour plot showed that the interaction between starch and peptone was significant. It can thus be concluded from Fig. 1c and d that there was an obvious mutual interaction between starch (X1) and K2HPO4·3H2O (X3) when peptone (X2) and KH2PO4 (X4) were fixed at 0. The activity of alkaline pectinase increased with an increase in starch content and in the concentration of K2HPO4·3H2O. It reached a maximum when the concentrations of the 2 factors were approximately 2.34% and 0.34%, respectively. The 2D contour plot showed that the interaction between starch and K2HPO4·3H2O was significant. Fig. 1e and f show that starch (X1) and KH2PO4 (X4) had a significant mutual interaction. When the content of starch increased from 1.5% to 2.34% and the concentration of KH2PO4 decreased from 0.2% to 0.13%, the activity of alkaline pectinase increased. Fig. 1g and h show an obvious interaction between peptone (X2) and K2HPO4·3H2O (X3). When starch (X1) and KH2PO4 (X4) were fixed at 0, the activity of alkaline pectinase increased gradually with an increase in peptone content from 0.5% to 0.8%, and with an increase in K2HPO4·3H2O concentration from 0.15% to 0.34%. However, higher concentrations led to a decrease in the alkaline pectinase activity. The 2D contour plot showed that the interaction between peptone and K2HPO4·3H2O was significant. When starch (X1) and K2HPO4·3H2O (X3) were fixed at 0, as shown in Fig. 1i and j, the interaction of peptone (X2) and KH2PO4 (X4) was significant. When the concentrations of peptone and KH2PO4 were reduced from 1.5% to 0.8% and from 0.2% to 0.13%, respectively, the activity of alkaline pectinase increased gradually. Thus, lower concentrations were not conducive to improving the activity of alkaline pectinase. From Fig. 1k and l, it can be seen that K2HPO4·3H2O (X3) and KH2PO4 (X4) were significantly positive to improve the activity of alkaline pectinase. When starch (X1) and peptone (X2) were fixed at 0, an increase in the content of K2HPO4·3H2O and a decrease in the content of KH2PO4 led to a rise in the alkaline pectinase activity. The activity of alkaline pectinase reached a maximum when the concentrations of K2HPO4·3H2O and KH2PO4 were 0.34% and 0.13%, respectively.
Figure 1.
3D response surface plots and 2D contour plots showing the effect of starch (X1), peptone (X2), K2HPO4·3H2O (X3) and KH2PO4 (X4) on the activity of alkaline pectinase. (A) Y = f (X1, X2), (B) Y = f(X1, X3), (E) Y = f(X1, X4), (G) Y = f(X2, X3), (I) Y = f(X2, X4) and (K) Y = f(X3, X4).
By using Design Expert 8.0, the optimal concentrations of the 4 key factors predicted by the model were as follows: 4.68% starch, 1.6% peptone, 0.26% KH2PO4, 0.68% K2HPO4·3H2O. Under the above conditions, the predicted maximal activity of alkaline pectinase was 742 U/mL. To verify the predicted value, 3 tests in shake flasks were performed under the optimal medium composition. The mean value of the activity of alkaline pectinase in these experiments was 734.11 U/mL, which is in good agreement with the predicted value.
Purification of alkaline pectinase
The purification of alkaline pectinase was performed sequentially using ammonium sulfate precipitation, PEG20000 concentration, DE-52 anion-exchange column chromatography and Sephadex G-100 size-exclusion chromatography. The purification results are presented in Fig. 2 and Table 5. After (NH4)2SO4 precipitation, 56% of the total protein was removed. The residual protein was concentrated by PEG20000, giving 14.4% of the total protein. The concentrated protein was subject to DE-52 anion-exchange column chromatography (Fig. 2a), and the protein from the main peak was collected and pooled, giving 1.1% of the total protein. The protein was subjected to further purification using Sephadex G-100 column chromatography (Fig. 2b). A big peak (fractions 8–16) with higher activity of alkaline pectinase was presented (Fig. 2b). The protein from this peak was collected and pooled, giving 0.3% of the total protein. The fold purification of alkaline pectinase from the above 4 steps was 1.46, 3.08, 13.23 and 28.67, respectively. SDS-PAGE analysis of the purified protein is shown in Fig. 2c. Only a single protein band with an estimated molecular weight of approximately 65 kDa was presented after Sephadex G-100 column chromatography. This indicates that the alkaline pectinase obtained is electrophoretically pure and may be used to analyze its enzymatic properties.
Figure 2.

Elution curves of alkaline pectinase and the SDS-PAGE analysis. (A) Elution curve of the DE-52 ion-exchange chromatography purification step of the sample. (B) Elution curve of the Sephadex G-100 size-exclusion chromatography purification step of the sample. (C) SDS-PAGE analysis of alkaline pectinase. Lane M: protein molecular weight marker; lane 1: the sample after PEG20000 concentration; lane 2: the sample after DE-52 ion-exchange chromatography; lane 3: the sample after Sephadex G-100 size-exclusion chromatography.
Table 5.
Purification results for alkaline pectinase.
| Steps | Total volume (mL) | Total protein (mg) | Enzyme activity (U) | Specific activity (U/mg) | Purification fold | Recovery (%) |
|---|---|---|---|---|---|---|
| Crude enzyme | 940 | 364.41 | 665632.8 | 1826.60 | 1 | 100 |
| (NH4)2SO4 precipitation | 100 | 159.76 | 425722.64 | 2664.76 | 1.46 | 64 |
| PEG20000 concentration | 22 | 52.31 | 293967.46 | 5619.72 | 3.08 | 44.2 |
| Cellulose DE-52 chromatography | 4 | 4.05 | 97873.53 | 24166.30 | 13.23 | 14.7 |
| Sephadex G-100 chromatography | 4 | 1.22 | 63894.47 | 52372.52 | 28.67 | 9.6 |
Enzymatic properties of the purified alkaline pectinase
The enzymatic properties of the purified alkaline pectinase are shown in Fig. 3. The activity of alkaline pectinase increased when the temperature was increased from 25 to 50°C, and reached a maximum at 50°C. Then, it decreased rapidly as the temperature increased beyond 50°C. This shows that the optimal temperature of the purified alkaline pectinase is 50°C (Fig. 3a). The activity of alkaline pectinase varied as a function of pH. The highest activity of alkaline pectinase was observed at pH 8.6 (Fig. 3b). For alkaline pectinase from B. subtilis ZGL14, strong thermal stability was observed at 40 and 50°C. The activity of alkaline pectinase showed a negligible change when incubated at either of these temperatures for 40 min. The residual activity of alkaline pectinase was 51, 40 and 22% when it was incubated at 80°C for 60, 80 and 100 min, respectively (Fig. 3c), which suggested that alkaline pectinase had good thermal stability. High pH stability of alkaline pectinase was observed when it was incubated at pH 8.6 and 10.0. The enzymatic activity declined rapidly to 12.2% of the maximal activity at pH 7.0 for 120 min. When pH was 8.0, the enzymatic activity was 93.9% of the maximum after 60 min. The activity of alkaline pectinase was retained at approximately 49.4% after 120 min at pH 10.0, which indicated that the purified pectinase had good alkali resistance (Fig. 3d).
Figure 3.
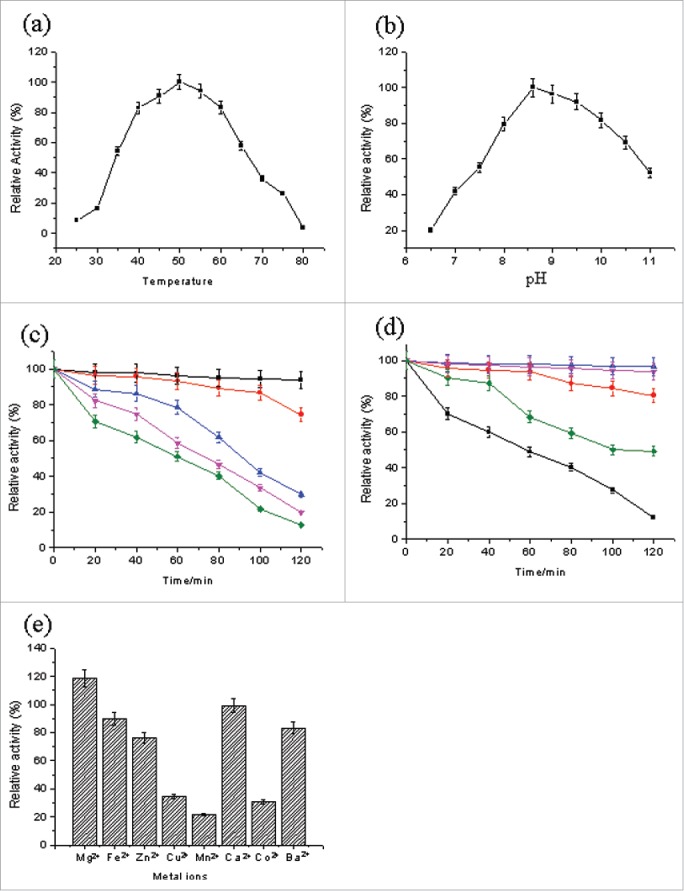
Enzymatic properties of the purified alkaline pectinase. (A) Effect of temperature on the activity of the purified alkaline pectinase. (B) Effect of pH on the activity of the purified alkaline pectinase. (C) Thermal stability of the purified alkaline pectinase. (D) pH stability of the purified alkaline pectinase. (E) Effect of different metal ions on the activity of the purified alkaline pectinase.
The effect of different metal ions on the activity of the purified alkaline pectinase is shown in Fig. 3e. It was obvious that metal ions had a certain effect on the activity of alkaline pectinase. The addition of Mg2+ could obviously promote the activity of alkaline pectinase, which was increased by 18.9% in the presence of 1 mM Mg2+. Ca2+ had no significant effect on the activity of alkaline pectinase, while Cu2+, Mn2+, and Co2+ significantly inhibited the activity of alkaline pectinase. Zn2+, Fe2+ and Ba2+ had a moderate inhibitory effect on the activity of alkaline pectinase.
Conclusions
The conditions for the production of alkaline pectinase from B. subtilis ZGL14 were optimized by RSM. The optimal concentrations of the 4 key factors screened were as follows: 4.68% starch, 1.6% peptone, 0.26% KH2PO4, and 0.68% K2HPO4·3H2O. Under the above conditions, the activity of alkaline pectinase reached 734.11 U/mL. The molecular weight of the purified alkaline pectinase was approximately 65 kDa. The final specific activity of the purified alkaline pectinase was 52372.52 U/mg. The optimal temperature and pH was 50°C and 8.6, respectively. The purified alkaline pectinase had strong thermo-stability and good alkali resistance. This enzyme had an absolute requirement for Mg2+ to stimulate its enzymatic activity, but it did not need other metal ions such as Cu2+, Mn2+, Co2+, Fe2+, Zn2+ and Ba2+. The study provides an important basis for the future industrial production of heat-tolerant alkaline pectinase.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study is supported by the National Natural Science Foundation of China (No.31171658) and the Zhejiang Province Education Committee (No.Z201010029).
References
- [1].Esquivel JCC, Hours RA, Voget CE, Mignone CF. Aspergillus kawachii produces an acidic pectin releasing enzyme activity. J Biosci Bioeng 1999; 88:48-52; PMID:16232572; http://dx.doi.org/ 10.1016/S1389-1723(99)80174-1 [DOI] [PubMed] [Google Scholar]
- [2].Singh SA, Plattner H, Diekmann H. Exopolygalacturonate lyase from a thermophilic Bacillus sp. Enzyme Microb Ttechnol 1999; 25:420-425; http://dx.doi.org/ 10.1016/S0141-0229(99)00066-6 [DOI] [Google Scholar]
- [3].Rehman HU, Aman A, Zohra RR, Qader SAU. Immobilization of pectin degrading enzyme from Bacillus licheniformis KIBGE IB-21 using agar-agar as a support. Carbohydr Polym 2014; 102:622-626; PMID:24507327; http://dx.doi.org/ 10.1016/j.carbpol.2013.11.073 [DOI] [PubMed] [Google Scholar]
- [4].Gan CY, Latiff AA. Extraction of antioxidant pectic-polysaccharide from mangosteen (Garcinia mangostana) rind: Optimization using response surface methodology. Carbohydr Polym 2011; 83:600-607; http://dx.doi.org/ 10.1016/j.carbpol.2010.08.025 [DOI] [Google Scholar]
- [5].Sharma H. Enzymatic degradation of residual non-cellulosic polysaccharides present on dew-retted flax fibres. Appl Microbiol Biotechnol 1987; 26:358-362; http://dx.doi.org/ 10.1007/BF00256669 [DOI] [Google Scholar]
- [6].Baracat M, Valentim C, Muchovej J, Silva D. Selection of pectinolytic fungi for degumming of natural fibers. Biotechnol Lett 1989; 11:899-902; http://dx.doi.org/ 10.1007/BF01026849 [DOI] [Google Scholar]
- [7].Brühlmann F, Kim KS, Zimmerman W, Fiechter A. Pectinolytic enzymes from actinomycetes for the degumming of ramie bast fibers. Appl Environ Microbiol 1994; 60:2107-2112; PMID:16349296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kapoor M, Beg QK, Bhushan B, Singh K, Dadhich K, Hoondal G. Application of an alkaline and thermostable polygalacturonase from Bacillus sp. MG-cp-2 in degumming of ramie (Boehmeria nivea) and sunn hemp (Crotalaria juncea) bast fibres. Process Biochem 2001; 36:803-807; http://dx.doi.org/ 10.1016/S0032-9592(00)00282-X [DOI] [Google Scholar]
- [9].Tanabe H, Kobayashi Y, Akamatsu I. Pretreatment of pectic wastewater with pectate lyase from an alkalophilic Bacillus sp. Agric Biol Chem 1988; 52:1855-1856; http://dx.doi.org/ 10.1080/00021369.1988.10868946 [DOI] [Google Scholar]
- [10].Alkorta I, Garbisu C, Llama MJ, Serra JL. Industrial applications of pectic enzymes: a review. Process Biochem 1998; 33:21-28; http://dx.doi.org/ 10.1016/S0032-9592(97)00046-0 [DOI] [Google Scholar]
- [11].Kapoor M, Kuhad RC. Improved polygalacturonase production from Bacillus sp. MG-cp-2 under submerged (SmF) and solid state (SSF) fermentation. Lett Appl Microbiol 2002; 34:317-322; PMID:11967052; http://dx.doi.org/ 10.1046/j.1472-765X.2002.01107.x [DOI] [PubMed] [Google Scholar]
- [12].Kashyap DR, Vohra PK, Chopra S, Tewari R. Applications of pectinases in the commercial sector: a review. Bioresourc Technology 2001; 77:215-227; http://dx.doi.org/ 10.1016/S0960-8524(00)00118-8 [DOI] [PubMed] [Google Scholar]
- [13].Niture SK, Pant A. Purification and biochemical characterization of polygalacturonase II produced in semi-solid medium by a strain of Fusarium moniliforme. Microbiol Res 2004; 159:305-314; PMID:15462530; http://dx.doi.org/ 10.1016/j.micres.2004.06.002 [DOI] [PubMed] [Google Scholar]
- [14].Niture SK, Pant A, Kumar AR. Active site characterization of the single endo-polygalacturonase produced by Fusarium moniliforme NCIM 1276. Eur J Biochem/FEBS 2001; 268:832-840; http://dx.doi.org/ 10.1046/j.1432-1327.2001.01959.x [DOI] [PubMed] [Google Scholar]
- [15].Pedrolli DB, Carmona EC. Purification and characterization of the exopolygalacturonase produced by Aspergillus giganteus in submerged cultures. J Ind Microbiol Biotechnol 2010; 37:567-573; PMID:20204453; http://dx.doi.org/ 10.1007/s10295-010-0702-0 [DOI] [PubMed] [Google Scholar]
- [16].Hoondal G, Tiwari R, Tewari R, Dahiya N, Beg Q. Microbial alkaline pectinases and their industrial applications: a review. Appl Microbiol Biotechnol 2002; 59:409-418; PMID:12172603; http://dx.doi.org/ 10.1007/s00253-002-1061-1 [DOI] [PubMed] [Google Scholar]
- [17].Kuhad RC, Kapoor M, Rustagi R. Enhanced production of an alkaline pectinase from Streptomyces sp. RCK-SC by whole-cell immobilization and solid-state cultivation. World J Microbiol Biotechnol 2004; 20:257-263; http://dx.doi.org/ 10.1023/B:WIBI.0000023833.15866.45 [DOI] [Google Scholar]
- [18].Jayani RS, Saxena S, Gupta R. Microbial pectinolytic enzymes: a review. Process Biochem 2005; 40:2931-2944; http://dx.doi.org/ 10.1016/j.procbio.2005.03.026 [DOI] [Google Scholar]
- [19].Favela-Torres E, Volke-Sepúlveda T, Viniegra-González G. Production of hydrolytic depolymerising pectinases. Food Technol Biotechnol 2006; 44:221 [Google Scholar]
- [20].Ruiz HA, Rodríguez-Jasso RM, Rodríguez R, Contreras-Esquivel JC, Aguilar CN. Pectinase production from lemon peel pomace as support and carbon source in solid-state fermentation column-tray bioreactor. Biochem Eng J 2012; 65:90-95; http://dx.doi.org/ 10.1016/j.bej.2012.03.007 [DOI] [Google Scholar]
- [21].Chen J, Yang R, Chen M, Wang S, Li P, Xia Y, Zhou L, Xie J, Wei D. Production optimization and expression of pectin releasing enzyme from Aspergillus oryzae PO. Carbohydr Polym 2014; 101:89-95; PMID:24299753; http://dx.doi.org/ 10.1016/j.carbpol.2013.09.011 [DOI] [PubMed] [Google Scholar]
- [22].Esquivel JC, Voget C. Purification and partial characterization of an acidic polygalacturonase from Aspergillus kawachii. J Biotechnol 2004; 110:21-28; PMID:15099902; http://dx.doi.org/ 10.1016/j.jbiotec.2004.01.010 [DOI] [PubMed] [Google Scholar]
- [23].Schnitzhofer W, Weber HJ, Vršanská M, Biely P, Cavaco-Paulo A, Guebitz G. Purification and mechanistic characterisation of two polygalacturonases from Sclerotium rolfsii. Enzyme Microb Technol 2007; 40:1739-1747; http://dx.doi.org/ 10.1016/j.enzmictec.2006.11.005 [DOI] [Google Scholar]
- [24].Raj Kashyap D, Kumar Soni S, Tewari R. Enhanced production of pectinase by Bacillus sp. DT7 using solid state fermentation. Bioresourc Technol 2003; 88:251-254; http://dx.doi.org/ 10.1016/S0960-8524(02)00206-7 [DOI] [PubMed] [Google Scholar]
- [25].Romdhane ZB, Tounsi H, Hadj-Sassi A, Hadj-Taieb N, Gargouri A. The constitutive production of pectinase by the CT1 mutant of Penicillium occitainis is modulated by pH. Appl Biochem Biotechnol 2013; 169:215-227; PMID:23179280; http://dx.doi.org/ 10.1007/s12010-012-9971-6 [DOI] [PubMed] [Google Scholar]
- [26].Ortega N, De Diego S, Perez-Mateos M, Busto M. Kinetic properties and thermal behaviour of polygalacturonase used in fruit juice clarification. Food Chem 2004; 88:209-217; http://dx.doi.org/ 10.1016/j.foodchem.2004.01.035 [DOI] [Google Scholar]
- [27].Vaillant F, Cisse M, Chaverri M, Perez A, Dornier M, Viquez F. Clarification and concentration of melon juice using membrane processes. Innov Food Sci Emerg Technol 2005; 6:213-220; http://dx.doi.org/ 10.1016/j.ifset.2004.11.004 [DOI] [Google Scholar]
- [28].Ingallinera B, Barbagallo RN, Spagna G, Palmeri R, Todaro A. Effects of thermal treatments on pectinesterase activity determined in blood oranges juices. Enzyme Microb Technol 2005; 36:258-263; http://dx.doi.org/ 10.1016/j.enzmictec.2004.08.041 [DOI] [Google Scholar]
- [29].Benen JAE, Visser J. Pectate and pectinase lyases. In: Whitaker JR, Voragen AGJ, Wong DJS, editors. Handbook of food enzymology. New York, NY: Dekker, 2003. [Google Scholar]
- [30].Sreenath HK, Shah AB, Yang VW, Gharia MM, Jeffries TW. Enzymatic polishing of jute/cotton blended fabrics. J Ferment Bioeng 1996; 81:18-20; http://dx.doi.org/ 10.1016/0922-338X(96)83113-8 [DOI] [Google Scholar]
- [31].Kashyap DR, Soni SK, Tewari R. Enhanced production of pectinase by Bacillus sp. DT7 using solid state fermentation. Bioresourc Technol 2003; 88:251-254; http://dx.doi.org/ 10.1016/S0960-8524(02)00206-7 [DOI] [PubMed] [Google Scholar]
- [32].Li Z, Bai Z, Zhang B, Xie H, Hu Q, Hao C. Newly isolated Bacillus gibsonii S-2 capable of using sugar beet pulp for alkaline pectinase production. World J Microbiol Biotechnol 2005; 21:1483-1486; http://dx.doi.org/ 10.1007/s11274-005-7025-8 [DOI] [Google Scholar]
- [33].Sharma D, Satyanarayana T. A marked enhancement in the production of a highly alkaline and thermostable pectinase by Bacillus pumilus dcsr1 in submerged fermentation by using statistical methods. Bioresourc Technol 2006; 97:727-733; http://dx.doi.org/ 10.1016/j.biortech.2005.04.012 [DOI] [PubMed] [Google Scholar]
- [34].Uday US, Bandyopadhyay TK, Bhunia B. Rapid development of xylanase assay conditions using Taguchi methodology. Bioengineered 2016; 7:424-431; PMID:27435915; http://dx.doi.org/ 10.1080/21655979.2016.1180486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bhunia B, Dutta D, Chaudhuri S. Optimization of enzyme activity determination and partial characterization of extracellular alkaline protease from Bacillus licheniformis NCIM-2042. Eng Life Sci 2011; 11:207-215; http://dx.doi.org/ 10.1002/elsc.201000020 [DOI] [Google Scholar]
- [36].Anjum MF, Tasadduq I, Al-Sultan K. Response surface methodology: A neural network approach. Eur J Operat Res 1997; 101:65-73; http://dx.doi.org/ 10.1016/S0377-2217(96)00232-9 [DOI] [Google Scholar]
- [37].Baş D, Boyacı İH. Modeling and optimization I: Usability of response surface methodology. J Food Eng 2007; 78:836-845; http://dx.doi.org/ 10.1016/j.jfoodeng.2005.11.024 [DOI] [Google Scholar]
- [38].Baş D, Boyacı İH. Modeling and optimization II: Comparison of estimation capabilities of response surface methodology with artificial neural networks in a biochemical reaction. J Food Eng 2007; 78:846-854; http://dx.doi.org/ 10.1016/j.jfoodeng.2005.11.025 [DOI] [Google Scholar]
- [39].Paseephol T, Small D, Sherkat F. Process optimisation for fractionating Jerusalem artichoke fructans with ethanol using response surface methodology. Food Chem 2007; 104:73-80; http://dx.doi.org/ 10.1016/j.foodchem.2006.10.078 [DOI] [Google Scholar]
- [40].Bhunia B, Dey A. Statistical approach for optimization of physiochemical requirements on alkaline protease production from Bacillus licheniformis NCIM 2042. Enzyme Res 2012; 2012:905804; PMID:22347624; http://dx.doi.org/ 10.1155/2012/905804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tang M, Bian M, Li Q. Study on solid fermentation production conditions of pectinase from Aspergillus oryzae. J Fuqing Branch Fujian Norm Univ 2006; 2:6-9 [Google Scholar]
- [42].Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 1959; 31:426-428; http://dx.doi.org/ 10.1021/ac60147a030 [DOI] [Google Scholar]
- [43].Bhunia B, Basak B, Mandal T, Bhattacharya P, Dey A. Effect of pH and temperature on stability and kinetics of novel extracellular serine alkaline protease (70kDa). Int J Biol Macromol 2013; 54:1-8; PMID:23219732; http://dx.doi.org/ 10.1016/j.ijbiomac.2012.11.024 [DOI] [PubMed] [Google Scholar]
- [44].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680-685; PMID:5432063; http://dx.doi.org/ 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- [45].Li Q. Breeding of high pectinase-producing strain and optimization of its fermentation conditions [Dissertation]. Hangzhou (China): Zhejiang Gongshang University 2014. [Google Scholar]
- [46].Box GEP, Hunter JS, Hunter WG. Statistics for experimenters: design, innovation and discovery. Hoboken, NJ: Wiley-Interscience Press, 2005. [Google Scholar]
- [47].Li Y, Lu J, Gu G, Mao Z. Characterization of the enzymatic degradation of arabinoxylans in grist containing wheat malt using response surface methodology. J Am Soc Brew Chem 2005; 63:171-176 [Google Scholar]
- [48].Li Y, Cui F, Liu Z, Xu Y, Zhao H. Improvement of xylanase production by Penicillium oxalicum ZH-30 using response surface methodology. Enzyme Microb Technol 2007; 40:1381-1388; http://dx.doi.org/ 10.1016/j.enzmictec.2006.10.015 [DOI] [Google Scholar]