ABSTRACT
Komagataella phaffii (formerly Pichia pastoris) is a well-known fungal system for heterologous protein production in the context of modern biotechnology. To obtain higher protein titers in this system many researchers have sought to optimize gene expression by increasing the levels of transcription of the heterologous gene. This has been typically achieved by manipulating promoter sequences or by generating clones bearing multiple copies of the desired gene. The aim of this work is to describe how these different molecular strategies have been applied in K. phaffii presenting their advantages and drawbacks.
KEYWORDS: gene transcription, heterologous expression, Komagataella phaffii, multi-copy integration, promoter engineering
Introduction
The methylotrophic yeast Komagataella phaffii, still mostly known by its old name, Pichia pastoris, is considered one of the most important platforms in modern biotechnology for the production of heterologous proteins. This is due to several features including its high volumetric productivity and ability to perform some posttranslational modifications in a manner similar to mammalians cells (for review see 1,2). However, a clear limitation of this system is its low specific productivity which has prompted many researchers to pursue molecular strategies to improve protein production. The most popular approach to reach this goal is to offer to the translational machinery of the cell higher titers of a particular transcript. This is usually accomplished by driving the expression of the heterologous gene with a suitable promoter or by simply increasing the copy number of the target gene.3 One should consider that gene overexpression may pose a metabolic burden to the physiology of the cell thus resulting in disappointing outputs. Nonetheless, most researchers still consider these approaches (summarized in Fig. 1) as a starting point in the endeavor of obtaining higher protein titers in K. phaffii.
Figure 1.
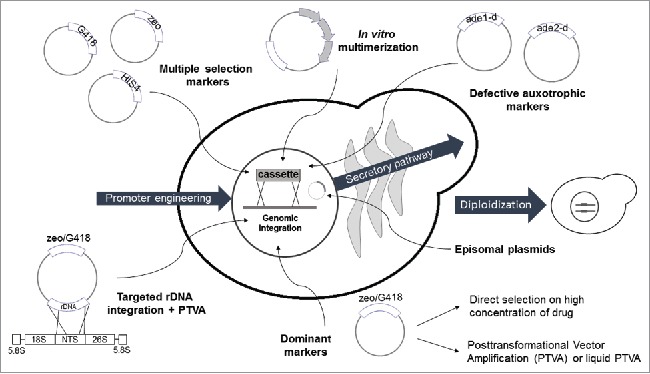
Molecular strategies used in K. phaffii to improve gene transcription. The titer of heterologous transcripts can be improved by using strong promoters (engineered or not) or by generating clones with extra doses of the desired gene integrated in the genome or present in episomal plasmids. The extra-copies may be introduced by different approaches: sequential transformation with vectors bearing different markers, in vitro multimerization, colony screening under drug selective pressure, gene amplification by PTVA (or liquid PTVA), use of defective auxotrophic markers in minimal medium and diploidization of selected clones. Abbreviations: zeo/G418 = dominant markers which confer resistance to zeocin or G418, respectively.
Inducible and constitutive promoters
K. phaffii vectors are derived from a few integrative plasmids which can carry expression cassettes under the control of inducible or constitutive promoters. A detailed review on the promoters available for protein production in K. phaffii is provided elsewhere.4 The first expression system for K. phaffii was based on the promoter of the alcohol oxidase gene (PAOX1) involved in the first step of methanol metabolism. This strong and methanol-induced promoter was partly responsible for the popularity of this yeast as a heterologous protein production platform since the 1980s.5,6 PAOX1 is repressed by glucose, glycerol and ethanol, therefore it represents a reliable tool when expressing toxic or growth-impairing proteins due to its tight regulation.7 Several other promoters involved in the methanol utilization pathway have been isolated and classified as strong, intermediate or weak according to its expression levels in comparison to PAOX1.8 Because induction by methanol represents a hazardous disadvantage for large-scale processes alternative inducible promoters have been considered. A novel regulated promoter has recently been identified from a high-affinity glucose transporter gene (GTH1) which is repressed by glycerol.9 Since PGTH1 is responsive to carbon source depletion it should represent an attractive and cheap alternative to other regulated systems that rely on the addition of an external source of the inducing agent. Recently, 2 inducible promoters from the K. phaffii rhamnose utilization pathway were proposed for the production of food-grade and therapeutically important recombinant proteins.10 Also, extensive transcriptome analysis has led to the identification of new promoters regulated by the carbon source which should represent a fine addition to the K. phaffii promoter toolbox.11
Another alternative to methanol-induction is the use of constitutive expression systems which may be more advantageous when considering the fermentation strategy used. Whereas inducible expression requires a growth phase before the addition of methanol (production phase) in a constitutive system protein production is concomitant to cell growth, thus favoring continuous fermentation processes.12 The promoter derived from the glycolytic glyceraldehyde 3-phosphate dehydrogenase gene (PGAP) is one of the most popular constitutive systems. Full transcription from PGAP is achieved when grown in glucose as a carbon source.13,14 In addition to PGAP other moderately strong constitutive systems may be used such as PTEF115 and PPGK1.16,17
Promoter engineering
A wide range of available promoters is an advantage for K. phaffii as a heterologous protein production platform since it allows the construction of fine-tuned systems and strains while also meeting requirements set by specific proteins.4 For protein production purposes, intermediate or weak promoters may be more advantageous when protein folding becomes an issue as a result of gene overexpression. Likewise, in metabolic engineering applications a diverse set of promoters with different strengths allows a more accurate tuning of the expression levels and metabolic flux.3 Transcription-factor binding sites (TFBS), upstream repression sites (URS) and upstream activation sequences (UAS) from different K. phaffii promoters may direct the design of new synthetic promoter sequences in the future.18 Promoter libraries have already been constructed using not only random mutagenesis methods but also with specific synthetic core promoter sequences derived from wild-type promoter alignments.9,19 The PGAP sequence has also been manipulated through error-prone PCR for the construction of a GAP-promoter library with strengths varying from ∼0,6% to 19,6-fold of that of the wild-type PGAP.20
Multi-copy number clones
One of the most common strategies to increase the levels of mRNA is by introducing several copies of the heterologous gene.3,21 In several cases this has led to a significant improvement in heterologous protein production, most notably when considering intracellular expression.22 However, in some cases involving secreted proteins, a higher gene copy number resulted in reduced heterologous protein production. For example, production of porcine insulin precursor (PIP) under inductive conditions was significantly affected in clones bearing >12 copies of the heterologous gene.23 This has been attributed to bottlenecks in the secretory pathway which may become overwhelmed by the effects of high titers of a particular heterologous protein.24 Strategies to overcome these barriers may include the use of new signal peptides based on the K. phaffii secretome25 and combinatorial engineering of secretion helper factors involved in protein folding and vesicle trafficking.26
In K. phaffii, expression vectors are typically integrated into the host chromosome by homologous recombination and non-homologous end joining events.27 Multiple integration events may occur at low frequency (5–6%) by the integration of expression cassettes in tandem in a head-to-tail configuration.28 These rare integration events can be favored by using vectors with dominant markers based on antibiotic resistance genes which allow the screening of desired clones under drug selective pressure. The use of dominant markers is the basis of the “posttransformational vector amplification” method (PTVA)29 or its recently described variant “liquid PTVA”.30 In PTVA, cells transformed with one or few copies of a vector carrying the marker that confers resistance to zeocin or G418 can be selected in higher concentrations of the antibiotic in a stepwise manner resulting in isolation of multi-copy clones several days after the initial transformation.
Multi-copy integration events can be enhanced by targeting vector insertion to repetitive sequences in the genome such as the rDNA cluster (rDNA) which comprises a 7450-bp repeated sequence organized in tandem. When a specific sequence is targeted to this cluster, a low copy number of integrated vectors is initially obtained, then, a strong selective pressure is applied for vector amplification.28 The use of the non-transcribed spacer region (NTS) of the rDNA as an integration target in combination with the PTVA method has led to the successful isolation of multi-copy clones in K. phaffii.22
In addition to the high costs of eukaryotic antibiotics, an important drawback in the use of dominant markers is that in several clones an increase in drug resistance does not necessarily reflect multi-copy integration or an improvement in protein production.3,22 Moreover, there are concerns that the accidental released of genetically modified organisms bearing drug-resistance markers may be horizontally transferred to environmental organisms.31 Marker-removal has been successfully accomplished in K. phaffii with the use of the Cre/loxP32 and Flp/FRT27 site-specific recombination systems. The use of the Escherichia coli counter-selectable toxin gene mazF may further improve the screening of marker-free clones.33 However, one should be aware that undesirable chromosomal rearrangement events may occur when using recombinase-based technology, especially when considering the simultaneous removal of multi-copy drug markers spread in the genome. The CRISPR-Cas9 system has recently been established in K. phaffii34 and may represent a powerful tool for marker-removal and generation of auxotrophic strains in this yeast.
The use of defective auxotrophic markers is an alternative to select multiple integration events. These markers generally represent biosynthetic pathway genes with a truncated promoter and, therefore, are transcribed at low levels. To compensate its low expression levels, transformed clones are selected to carry a high copy number of the defective marker to restore prototrophy. Defective markers have been extensively used in S. cerevisiae as a strategy to amplify expression plasmids.35,36 In K. phaffii, the use of defective ADE1 and ADE2 alleles as selection markers has favored multiple integration with the concomitant increase in heterologous protein production.37
Although the use of defective markers for multi-copy integration in K. phaffii requires the development of new auxotrophic mutant strains and the use of specific media it presents some clear advantages. Most importantly, clones with different copies of the desired gene are easily obtained on a single transformation event without the need of laborious replica plating steps as required in PTVA. Also, it is a cheap alternative to drug selection and recombinant clones that carry auxotrophic marker do not pose any significant threat to the environment.
Other less frequently used methods to obtain multi-copy clones have been described. One involves the in vitro construction of multimers of the expression cassette.38 In this case the size of the expression cassette may hamper DNA manipulation. Another method involves the sequential transformation of the host strain with different vectors bearing the expression cassettes, a laborious procedure which also requires the availability of strains with several genetic markers. However, the resulting strains may be further crossed to generate diploid cells thus increasing overall copy number of the desired sequence.39
Recently, an episomal plasmid carrying the panARS sequence was tested for recombinant protein production in K. phaffii.40 This autonomously replicating sequence derived from Kluyveromyces lactis conferred stable replicative maintenance to plasmids and allowed the selection of clones with 6 to 19 copies of the plasmid. Since the use of episomal vectors represent an approach that leads to increased gene copy number with a higher efficiency of transformation and clonal homogeneity it should draw more interest in the near future as a platform for heterologous protein production in K. phaffii.
Conclusion
It is clear that the improvement on protein production as a result of the use of any of the molecular strategies described in this work should be assessed on a case-by-case basis. In our experience, the use of a vector bearing a repetitive target sequence combined with an auxotrophic defective marker represents an interesting start point for the development of an expression platform in K. phaffii.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from Fundação de Amparo à Pesquisa do Distrito Federal (FAP/DF) and CNPq, Brazil.
References
- [1].Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast 2005; 22(4):249-270; PMID:15704221; http://dx.doi.org/ 10.1002/yea.1208 [DOI] [PubMed] [Google Scholar]
- [2].Gasser B, Prielhofer R, Marx H, Maurer M, Nocon J, Steiger M, Puxbaum V, Sauer M, Mattanovich D. Pichia pastoris: protein production host and model organism for biomedical research. Future Microbiol 2013; 8(2):191-208; PMID:23374125; http://dx.doi.org/ 10.2217/fmb.12.133 [DOI] [PubMed] [Google Scholar]
- [3].Aw R, Polizzi KM. Can too many copies spoil the broth? Microb Cell Fact 2013; 12:128; PMID:24354594; http://dx.doi.org/ 10.1186/1475-2859-12-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vogl T, Glieder A. Regulation of Pichia pastoris promoters and its consequences for protein production. Nature Biotechnol 2013; 30(4):385-404. [DOI] [PubMed] [Google Scholar]
- [5].Ellis SB, Brust PF, Koutz PJ, Waters AF, Harpold MM, Gingeras TR. Isolation of alcohol oxidase and two other methanol regulatable genes from the yeast Pichia pastoris. Mol Cell Biol 1985; 5(5):1111-1121; PMID:3889590; http://dx.doi.org/ 10.1128/MCB.5.5.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cereghino GP, Cereghino JL, Ilgen C, Cregg JM. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol 2002; 13(4):329-332; PMID:12323354; http://dx.doi.org/ 10.1016/S0958-1669(02)00330-0 [DOI] [PubMed] [Google Scholar]
- [7].Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev 2000; 24(1):45-66; PMID:10640598; http://dx.doi.org/ 10.1111/j.1574-6976.2000.tb00532.x [DOI] [PubMed] [Google Scholar]
- [8].Vogl T, Sturmberger L, Kickenweiz T, Wasmayer R, Schmid C, Hatzl AM, Gerstmann MA, Pitzer J, Wagner M, Thallinger GG, et al. . A toolbox of diverse promoters related to methanol utilization: functionally verified parts for heterologous pathway expression in Pichia pastoris. ACS Synth Biol. 2015; 5(2):172-186; PMID:26592304; http://dx.doi.org/ 10.1021/acssynbio.5b00199 [DOI] [PubMed] [Google Scholar]
- [9].Prielhofer R, Maurer M, Klein J, Wenger J, Kiziak C, Gasser B, Mattanovich D. Induction without methanol: novel regulated promoters enable high-level expression in Pichia pastoris. Microb Cell Fact 2013; 12:5; PMID:23347568; http://dx.doi.org/ 10.1186/1475-2859-12-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu B, Zhang Y, Zhang X, Yan C, Zhang Y, Xu X, Zhang W. Discovery of a rhamnose utilization pathway and rhamnose-inducible promoters in Pichia pastoris. Sci Rep 2016; 6:27352; PMID:27256707; http://dx.doi.org/ 10.1038/srep27352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Love KR, Shah KA, Whittaker CA, Wu J, Bartlett MC, Ma D, Leeson RL, Priest M, Borowsky J, Young SK, Love JC. Comparative genomics and transcriptomics of Pichia pastoris. BMC Genom 2016; 17:550; http://dx.doi.org/ 10.1186/s12864-016-2876-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vassileva A, Chugh DA, Swaminathan S, Khanna N. Expression of hepatitis B surface antigen in the methylotrophic yeast Pichia pastoris using the GAP promoter. J Biotechnol 2001; 88(1):21-35; PMID:11377762; http://dx.doi.org/ 10.1016/S0168-1656(01)00254-1 [DOI] [PubMed] [Google Scholar]
- [13].Waterham HR, Digan ME, Koutz PJ, Lair SV, Cregg JM. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 1997; 186(1):37-44; PMID:9047342; http://dx.doi.org/ 10.1016/S0378-1119(96)00675-0 [DOI] [PubMed] [Google Scholar]
- [14].Zhang AL, Luo JX, Zhang TY, Pan YW, Tan YH, Fu CY, Tu FZ. Recent advances on the GAP promoter derived expression system of Pichia pastoris. Mol Biol Rep 2009; 36(6):1611-1619; PMID:18781398; http://dx.doi.org/ 10.1007/s11033-008-9359-4 [DOI] [PubMed] [Google Scholar]
- [15].Ahn J, Hong J, Lee H, Park M, Lee E, Kim C, Choi E, Jung J. Translation elongation factor 1-alpha gene from Pichia pastoris: molecular cloning, sequence, and use of its promoter. Appl Microbiol Biotechnol 2007; 74(3):601-608; PMID:17124582; http://dx.doi.org/ 10.1007/s00253-006-0698-6 [DOI] [PubMed] [Google Scholar]
- [16].de Almeida JR, de Moraes LM, Torres FA. Molecular characterization of the 3-phosphoglycerate kinase gene (PGK1) from the methylotrophic yeast Pichia pastoris. Yeast 2005; 22(9):725-737; PMID:16034819; http://dx.doi.org/ 10.1002/yea.1243 [DOI] [PubMed] [Google Scholar]
- [17].Arruda A, Reis VC, Batista VD, Daher BS, Piva LC, De Marco JL, de Moraes LM, Torres FA. A constitutive expression system for Pichia pastoris based on the PGK1 promoter. Biotechnol Lett 2016; 38(3):509-517; PMID:26585331; http://dx.doi.org/ 10.1007/s10529-015-2002-2 [DOI] [PubMed] [Google Scholar]
- [18].Hartner FS, Ruth C, Langenegger D, Johnson SN, Hyka P, Lin-Cereghino GP, Lin-Cereghino J, Kovar K, Cregg JM, Glieder A. Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res 2008; 36(12):e76; PMID:18539608; http://dx.doi.org/ 10.1093/nar/gkn369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vogl T, Ruth C, Pitzer J, Kickenweiz T, Glieder A. Synthetic core promoters for Pichia pastoris. ACS Synth Biol 2014; 3(3):188-191; PMID:24187969; http://dx.doi.org/ 10.1021/sb400091p [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Qin X, Qian J, Yao G, Zhuang Y, Zhang S, Chu J. GAP promoter library for fine-tuning of gene expression in Pichia pastoris. Appl Environ Microbiol 2011; 77(11):3600-3608; PMID:21498769; http://dx.doi.org/ 10.1128/AEM.02843-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Romanos M, Scorer C, Sreekrishna K, Clare J. The generation of multicopy recombinant strains. Methods Mol Biol 1998; 103:55-72; PMID:9680633 [DOI] [PubMed] [Google Scholar]
- [22].Marx H, Mecklenbrauker A, Gasser B, Sauer M, Mattanovich D. Directed gene copy number amplification in Pichia pastoris by vector integration into the ribosomal DNA locus. FEMS Yeast Res 2009; 9(8):1260-1270; PMID:19799640; http://dx.doi.org/ 10.1111/j.1567-1364.2009.00561.x [DOI] [PubMed] [Google Scholar]
- [23].Zhu T, Guo M, Tang Z, Zhang M, Zhuang Y, Chu J, Zhang S. Efficient generation of multi-copy strains for optimizing secretory expression of porcine insulin precursor in yeast Pichia pastoris. J Appl Microbiol 2009; 107(3):954-963; PMID:19486418; http://dx.doi.org/ 10.1111/j.1365-2672.2009.04279.x [DOI] [PubMed] [Google Scholar]
- [24].Hohenblum H, Gasser B, Maurer M, Borth N, Mattanovich D. Effects of gene dosage, promoters, and substrates on unfolded protein stress of recombinant Pichia pastoris. Biotechnol Bioeng 2004; 85(4):367-375; PMID:14755554; http://dx.doi.org/ 10.1002/bit.10904 [DOI] [PubMed] [Google Scholar]
- [25].Massahi A, Calik P. In-silico determination of Pichia pastoris signal peptides for extracellular recombinant protein production. J Theor Biol 2015; 364:179-188; PMID:25218497; http://dx.doi.org/ 10.1016/j.jtbi.2014.08.048 [DOI] [PubMed] [Google Scholar]
- [26].Guan B, Chen F, Su S, Duan Z, Chen Y, Li H, Jin J. Effects of co-overexpression of secretion helper factors on the secretion of a HSA fusion protein (IL2-HSA) in Pichia pastoris. Yeast 2016; 33(11):587-600; http://dx.doi.org/ 10.1002/yea.3183 [DOI] [PubMed] [Google Scholar]
- [27].Naatsaari L, Mistlberger B, Ruth C, Hajek T, Hartner FS, Glieder A. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One 2012; 7(6):e39720; PMID:22768112; http://dx.doi.org/ 10.1371/journal.pone.0039720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lopes TS, Hakkaart GJ, Koerts BL, Raue HA, Planta RJ. Mechanism of high-copy-number integration of pMIRY-type vectors into the ribosomal DNA of Saccharomyces cerevisiae. Gene 1991; 105(1):83-90; PMID:1937009; http://dx.doi.org/ 10.1016/0378-1119(91)90516-E [DOI] [PubMed] [Google Scholar]
- [29].Sunga AJ, Tolstorukov I, Cregg JM. Posttransformational vector amplification in the yeast Pichia pastoris. FEMS Yeast Res 2008; 8(6):870-876; PMID:18637138; http://dx.doi.org/ 10.1111/j.1567-1364.2008.00410.x [DOI] [PubMed] [Google Scholar]
- [30].Aw R, Polizzi KM. Liquid PTVA: a faster and cheaper alternative for generating multi-copy clones in Pichia pastoris. Microb Cell Fact 2016; 15:29; PMID:26849882; http://dx.doi.org/ 10.1186/s12934-016-0432-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Droge M, Puhler A, Selbitschka W. Horizontal gene transfer as a biosafety issue: a natural phenomenon of public concern. J Biotechnol 1998; 64(1):75-90; PMID:9823660; http://dx.doi.org/ 10.1016/S0168-1656(98)00105-9 [DOI] [PubMed] [Google Scholar]
- [32].Marx H, Mattanovich D, Sauer M. Overexpression of the riboflavin biosynthetic pathway in Pichia pastoris. Microb Cell Fact 2008; 7:23; PMID:18664246; http://dx.doi.org/ 10.1186/1475-2859-7-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yang J, Jiang W, Yang S. mazF as a counter-selectable marker for unmarked genetic modification of Pichia pastoris. FEMS Yeast Res 2009; 9(4):600-9; PMID:19416369; http://dx.doi.org/ 10.1111/j.1567-1364.2009.00503.x [DOI] [PubMed] [Google Scholar]
- [34].Weninger A, Hatzl AM, Schmid C, Vogl T, Glieder A. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J Biotechnol 2016. 235:139-49; PMID:27015975; http://dx.doi.org/ 10.1016/j.jbiotec.2016.03.027 [DOI] [PubMed] [Google Scholar]
- [35].Kazemi Seresht A, Norgaard P, Palmqvist EA, Andersen AS, Olsson L. Modulating heterologous protein production in yeast: the applicability of truncated auxotrophic markers. Appl Microbiol Biotechnol 2013; 97(9):3939-3948; PMID:22782252; http://dx.doi.org/ 10.1007/s00253-012-4263-1 [DOI] [PubMed] [Google Scholar]
- [36].Moon HY, Lee DW, Sim GH, Kim HJ, Hwang JY, Kwon MG, Kang BK, Kim JM, Kang HA. A new set of rDNA-NTS-based multiple integrative cassettes for the development of antibiotic-marker-free recombinant yeasts. J Biotechnol 2016; 233:190-199; PMID:27411901; http://dx.doi.org/ 10.1016/j.jbiotec.2016.07.006 [DOI] [PubMed] [Google Scholar]
- [37].Du M, Battles MB, Nett JH. A color-based stable multi-copy integrant selection system for Pichia pastoris using the attenuated ADE1 and ADE2 genes as auxotrophic markers. Bioeng Bugs 2012; 3(1):32-37; PMID:22126802 [DOI] [PubMed] [Google Scholar]
- [38].Li YT, Li MT, Fu CH, Zhou PP, Liu JM, Yu LJ. Improvement of arachidonic acid and eicosapentaenoic acid production by increasing the copy number of the genes encoding fatty acid desaturase and elongase into Pichia pastoris. Biotechnol Lett 2009; 31(7):1011-1017; PMID:19306085; http://dx.doi.org/ 10.1007/s10529-009-9970-z [DOI] [PubMed] [Google Scholar]
- [39].Chen MT, Lin S, Shandil I, Andrews D, Stadheim TA, Choi BK. Generation of diploid Pichia pastoris strains by mating and their application for recombinant protein production. Microb Cell Fact 2012; 11:91; PMID:22748191; http://dx.doi.org/ 10.1186/1475-2859-11-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Camattari A, Goh A, Yip LY, Tan AH, Ng SW, Tran A, Liu G, Liachko I, Dunham MJ, Rancati G. Characterization of a panARS-based episomal vector in the methylotrophic yeast Pichia pastoris for recombinant protein production and synthetic biology applications. Microb Cell Fact 2016; 15(1):139; PMID:27515025; http://dx.doi.org/ 10.1186/s12934-016-0540-5 [DOI] [PMC free article] [PubMed] [Google Scholar]


