ABSTRACT
Transforming growth factor-β1 (TGF-β1) signaling is involved in cell metabolism, growth, differentiation, carcinoma invasion and fibrosis development, which suggests TGF-β1 can be treated as a therapeutic target extensively. Because TGF-β1 receptor type α(TGFBR2) is the directed and essential mediator for TGF-β1 signals, the extracellular domain of TGFBR2 (eTGFBR2), binding partner for TGF-β1, has been produced in a series of expression systems to inhibit TGF-β1 signaling. However, eTGFBR2 is unstable with a short half-life predominantly because of enzymatic degradation and kidney clearance. In this study, a fusion protein consisting of human eTGFBR2 fused at the C-terminal of human serum albumin (HSA) was stably and highly expressed in Chinese Hamster Ovary (CHO) cells. The high and stable expression sub-clones with Ig kappa signal peptide were selected by Western blot analysis and used for suspension culture. After fed-batch culture over 8 d, the expression level of HSA-eTGFBR2 reached 180 mg/L. The fusion protein was then purified from culture medium using a 2-step chromatographic procedure that resulted in 39% recovery rate. The TGF-β1 binding assay revealed that HSA-eTGFBR2 could bind to TGF-β1 with the affinity constant (KD of 1.42 × 10−8 M) as determined by the ForteBio Octet System. In addition, our data suggested that HSA-eTGFBR2 exhibited a TGF-β1 neutralizing activity and maintained a long-term activity more than eTGFBR2. It concluded that the overexpressing CHO cell line supplied sufficient recombinant human HSA-eTGFBR2 for further research and other applications.
KEYWORDS: Antagonist, Chinese hamster ovary cells, eTGFBR2, human serum albumin, TGF-β1
Introduction
Transforming growth factor β (TGF-β) superfamily comprises TGF-βs, activins, bone morphogenetic proteins (BMPs) and inhibins,1-3 which regulate a variety of biologic functions including cell growth, development, differentiation, migration and carcinogenesis.4,5 TGF-β1, as the prominent member, has been extensively studied for its vital functions in pathophysiology. The deregulation of TGF-β1 signal is implicated in the pathogenesis of cancer, chondrodysplasia, pulmonary hypertension and fibrosis.6-8 TGF-β1 stimulates the proliferation of mesenchymal cells but inhibits the cells of epithelial and endothelial origin. For example, it exerts growth inhibition and apoptosis induction of hepatocytes by a paracrine manner.9,10 In addition, TGF-β1 involves in the actions of extracellular matrix accumulation and immunosuppression.11
The bioactivity of TGF-β1 is mediated by the activation of 2 cell surface receptors, type I and type II receptor, which are classified as the members of serine/threonine kinase receptor family.12-14 The TGF-β1 type II receptor consists of a signal sequence, a cysteine-rich extracellular TGF-β-binding domain, a transmembrane domain and cytoplasmic ser/thr kinase domain which is capable of autophosphorylation and constitutively active.15 TGF-β1 binds to the extracellular domain of the type II receptor, recruiting the type I receptor, which is incapable of ligand binding in the absence of type II receptor. Upon forming the large ligand-receptor complex, the active serine/threonine kinase of TβRII phosphorylates the type I receptor, followed by the activation of intracellular Smad signaling pathway which transmits signals from cell surface to the nucleus16-18 and non-Smad signaling pathway, such as PI3K/AKT, Rho GTPase and MAPK pathway.19
TGF-β signaling pathway plays crucial roles in carcinoma invasion and fibrosis development. Several therapeutic strategies have been used to disrupt TGF-β signal, including neutralizing antibodies, soluble receptors, anti-receptor antibodies, and inhibition of TGF-β receptor kinase or Smad signaling pathway.20,21 The soluble extracellular domain of TGF-β1 type II receptor (eTGFBR2) has been expressed in baculovirus-infected insect cells, murine myeloma cells,11 bacterial system22 and Pichia pastoris. The eTGFBR2 produced by Escherichia coli showed a more potent anti-TGF-β1 activity than eTGFBR2 produced by P. pastoris.23 However, these proteins expressed in bacterial systems were sensitive to proteolytic degradation and usually existed in the form of inclusion bodies.24,25 The baculovirus expression system has been the capable system for producing large amounts of eTGFBR2. But a heterogeneous population was usually obtained due to incomplete enzymatic deglycosylation and proteolytic digestion in baculovirus-infected insect cells.22
eTGFBR2 had been expressed in mammalian systems such as mouse myeloma NSO cells by transient transfection. However, due to the low yields, it can't be widely used for further studies. CHO cells are widely used for production of recombinant proteins because they have the capacity of efficient post-translational modification, which was close to the native proteins in biochemical properties, and easily adapt to grow in serum free suspension culture.26 Furthermore, CHO cells rarely secrete intrinsic proteins, facilitating the purification of foreign proteins.
TGFBR2 is unstable with a short metabolic half-life due to enzymatic degradation which limits its therapeutic application.27 A single-chain bivalent receptor trap consisted entirely of native receptor sequences, termed (TβRII)2, suggested that this trap had a half-life of less than 1 h due to kidney clearance and elimination in the urine.28 In previous reports, several methods have been attempted to prolong the half-life of small molecule protein, such as binding to or fusion with long-circulating serum proteins (albumin).29 With the long circulation persistence and little immunological function, human serum albumin (HSA) have been successfully used as a stabilizing agent to improve the potential of therapeutic proteins, such as Interleukin-2 (IL-2),30 Interferon(IFN),31,32 Vascular endothelial growth factor (VEGF),33 and Antibody Molecules.34,35 With a circulation half-life of approximately 19 d,36 HSA has been an ideal carrier protein to prolong the half-life of proteins on the basis of maintaining the bioactivity.37,38
In this study, we constructed the fusion protein composed of eTGFBR2 fused at the C-terminal of HSA and established a CHO cell line stable overexpressing HSA-eTGFBR2 fusion protein for large scale production. The yield of HSA-eTGFBR2 reached 180 mg/L during 8 d fed-batch culture. The TGF-β1-binding ability and biologic activities of recombinant human HSA-eTGFBR2 fusion protein were evaluated and compared with native eTGFBR2. Our results showed that the HSA-eTGFBR2 retained binding properties and antagonized growth inhibitory effects of TGF-β1. It speculated the prospect of HSA-eTGFBR2 fusion protein as a biologic TGF-β antagonist for therapy of fibrosis and cancer diseases.
Results
Construction of pMH3-HSA-eTGFBR2 expression vector
To effectively produce recombinant human HSA-eTGFBR2 in CHO-S cells, the hsa-etgfbr2 fusion gene with IgK signal sequence was inserted into the pMH3 plasmid, an UCOE containing expression vector. The structure of pMH3-HSA-eTGFBR2 expression vector was depicted in Fig. 1. It contains 3 highly GC-rich DNA structures that support the opening of chromatin and a neo gene used as a selection marker.
Figure 1.
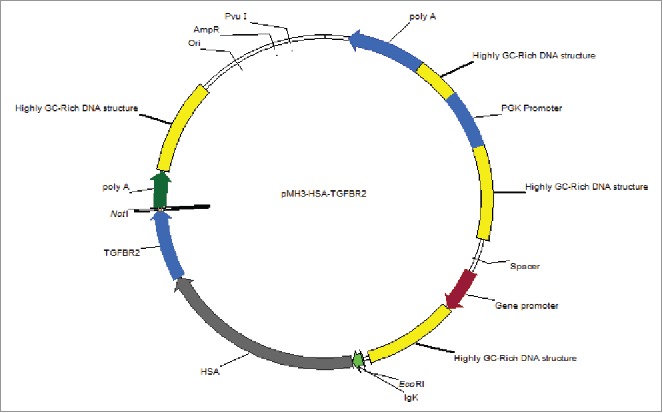
Schematic map of the recombinant pMH3-HSA-eTGFBR2 expression vector. The HSA-eTGFBR2 fusion gene was inserted in EcoRI and NotI sites of pHM3 plasmid.
Establishment of HSA-eTGFBR2 expression CHO-S cell line
The CHO-S cell line was transformed with recombinant pMH3-HSA-eTGFBR2 expression vector by electroporation. The survival cell clones under G418 were transferred into 96-well plate and the concentration of HSA-eTGFBR2 in supernatants were analyzed by Dot blot (Fig. 2A). The high expression sub-clones were selected and transferred into 24-well plate for further Western blot analysis (Fig. 2B). Two individual selected cell clones (number 7, 8) were performed a limited dilution method to obtain single cell derived sub-clones. The higher expression sub-clone was selected again as mentioned previously and used for further suspension culture.
Figure 2.
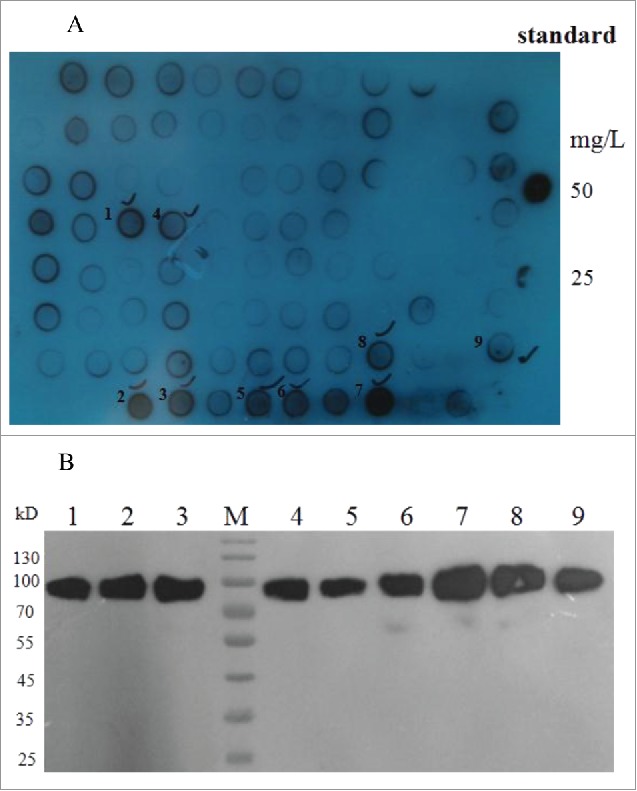
Selection of stable HSA-eTGFBR2 expressing cell clones by Dot blot (A) and Western blot (B) analysis. Standard: HSA concentration of 50, 25 mg/L; 1–9 were selected cell clones.
Production of recombinant human HSA-eTGFBR2 in fed-batch culture
The high expression cell clone was cultured by fed-batch suspension culture with serum-free medium B001 and the fusion protein was accumulated in a 3 L bioreactor with a working volume of 1 L. The feed medium F001 was added since the viable cell density reached 8 × 106 cells/mL. The viable cell density exhibited a peak of approximately 8.5 × 106 cells/mL and the cell viability exceeded 95% during whole culture process (Fig. 3A). After 8 d fed-batch culture, the yield of recombinant HSA-eTGFBR2 reached 180 mg/L (Fig. 3B).
Figure 3.
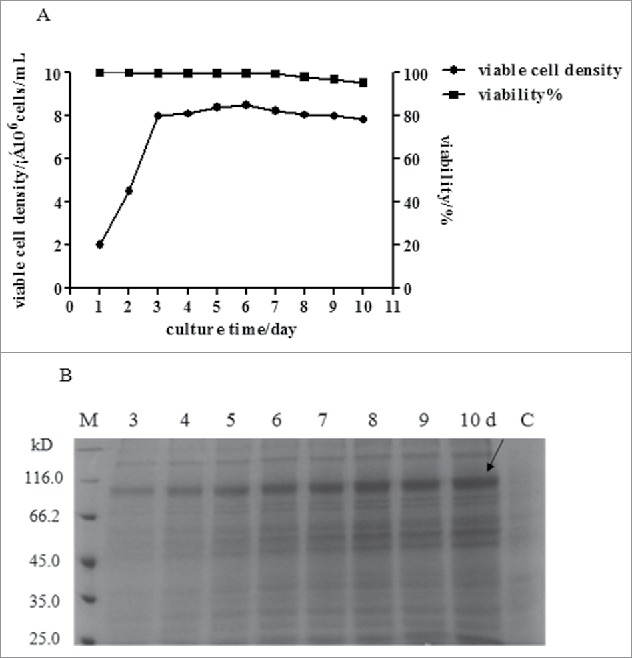
Cell growth and HSA-eTGFBR2 expression of in serum-free suspension fed-batch culture. (A) The growth curve of CHO cells in serum-free suspension fed batch culture; (B) The SDS-PAGE analysis of recombinant protein in fed batch culture, C: fed-batch culture of non-transferred CHO cells.
Purification of recombinant human HSA-eTGFBR2
The HSA-eTGFBR2 fusion protein in the culture supernatants was purified using Blue Sepharose Fast Flow affinity chromatography and Q Sepharose Fast Flow chromatography. Different fractions were obtained and analyzed by SDS-PAGE (Fig. 4A, B) and the purified HSA-eTGFBR2 was further confirmed by Western blot analysis (Fig. 4C). The result of Western blot showed a single band about 85kD that was close to theoretical value. As shown in Table 2, the recovery rate of HSA-eTGFBR2 reached 39% and 70 mg of HSA-eTGFBR2 fusion protein with purity more than 90% was obtained from 1L supernatants.
Figure 4.
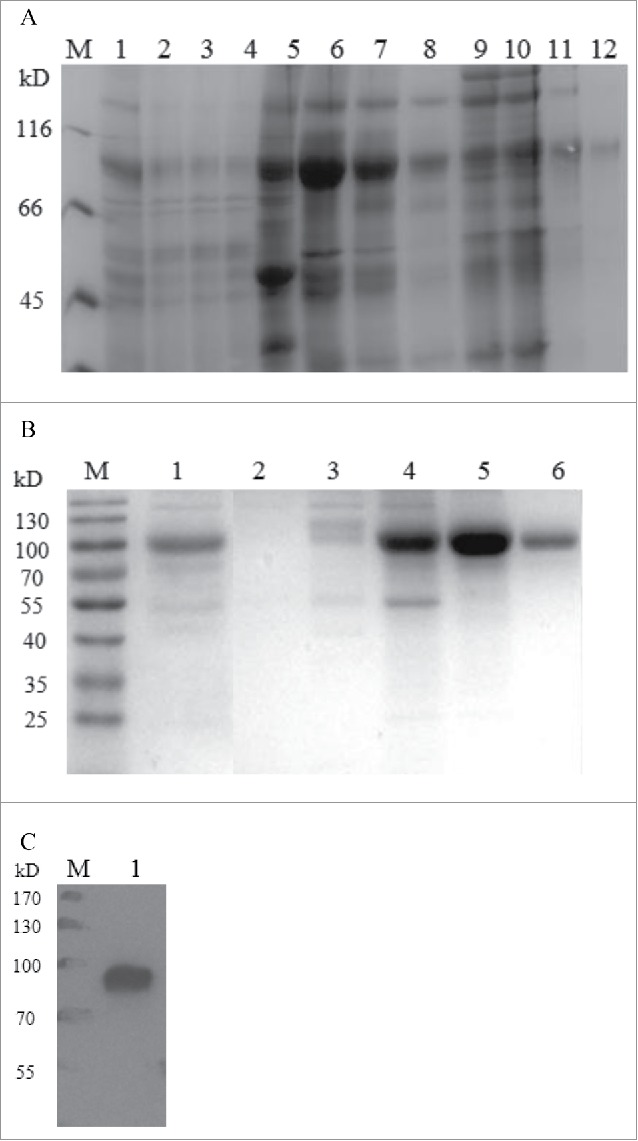
Purification of recombinant human HSA-eTGFBR2. (A) SDS-PAGE analysis of the fractions separated by Blue Sepharose column. M: marker; lane 1: the supernatants of fed-batch culture; lane 2–4: flow through; lane 5: fractions eluted by 30% buffer B; lane 6–8: fractions eluted by 100% buffer B; lane 9–12: fractions eluted by 1M Arg. (B) SDS-PAGE analysis of the fractions separated by Q Sepharose column. M: marker; lane 1: loading; lane 2: flow through; lane 3: washing fractions; lane 4–6: eluting fractions. (C) Western blot analysis of purified HSA-eTGFBR2 fusion protein.
Table 2.
Summary of purification step for recombinant human HSA- eTGFBR2.
| Step | Total protein(mg) | HSA-eTGFBR2(mg) | Recovery rate(%) | Purity(%) |
|---|---|---|---|---|
| Crude supernatants | 500 | 180 | 100 | 36 |
| Blue Sepharose Chromatography | 200 | 123 | 68 | 61.5 |
| Q Sepharose Chromatography | 77 | 70 | 38.9 | >90 |
Estimated by BCA protein assay, ELISA and coomassie blue-stained SDS–PAGE gel.
ELISA assay for TGF-β1 binding activity
The TGF-β1 binding properties of recombinant human HSA-eTGFBR2 were detected by ELISA and compared with commercial eTGFBR2. Dose-response curves were measured by the absorbance of the horseradish peroxidase substrate TMB at 450 nm (Fig. 5). All non-coated wells and 0 nM TGF-β1 wells showed similar OD450 from 0.042 to 0.068 and these values have been subtracted in each point. Binding of TGF-β1 increased with the coating concentration of eTGFBR2 (Fig. 5A) or HSA-eTGFBR2 (Fig. 5B) in the range of 1.25 to 50 nM. These results showed that the recombinant human HSA-eTGFBR2 expressed in CHO cells demonstrated a good binding affinity to TGF-β1.
Figure 5.
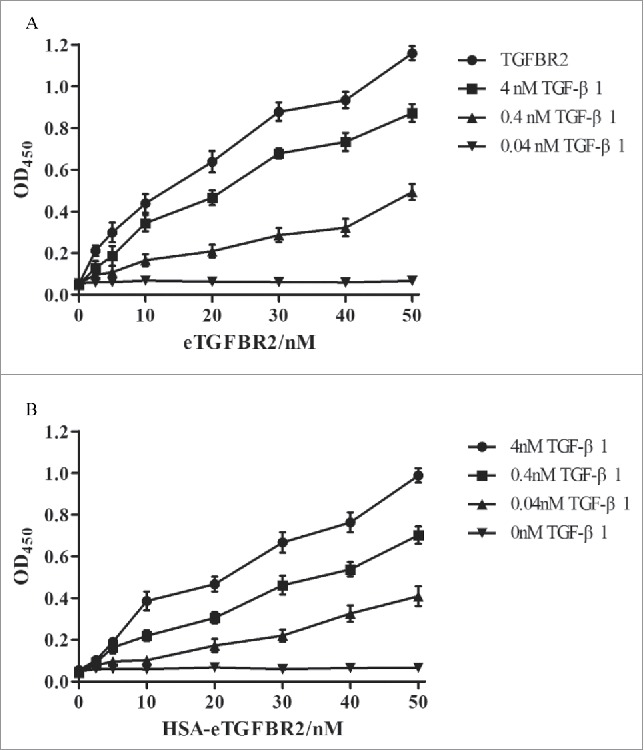
ELISA-type assay for affinity binding of TGF-β1 using eTGFBR2 (A) or HSA-eTGFBR2 (B) as a capture agent. (n = 3). Commercial eTGFBR2 or HSA-eTGFBR2 at concentrations ranging from 0 to 50 nM was precoated onto the 96-well plates. Results were detected by the microplate reader of OD450.
Determination of binding affinity of TGF-β1 to HSA-eTGFBR2
The binding affinities of TGF-β1 to eTGFBR2 or HSA-eTGFBR2 was determined by the ForteBio Octet System and calculated by instrument calculation software. As shown in Fig. 6, the adsorption capacity of HSA-eTGFBR2 to TGF-β1 rapidly increased at the beginning and then reached equilibrium at 120 s (Fig. 6B), whereas eTGFBR2 reached equilibrium at 40 s (Fig. 6A). The adsorption and dissociation equilibrium of eTGFBR2 was achieved more quickly than HSA-eTGFBR2. HSA was used as a negative control protein to confirm the competition reaction of specific binding. No binding affinity signal was detected to HSA because of its lack of affinity (KD < 1.0 × 10−12 M). After analysis of the raw data, the affinity (KD = 1.42 × 10−8 M) of HSA-eTGFBR2 for TGF-β1 was obtained, lower than that of eTGFBR2 for TGF-β1(KD = 9.49 × 10−9 M).
Figure 6.
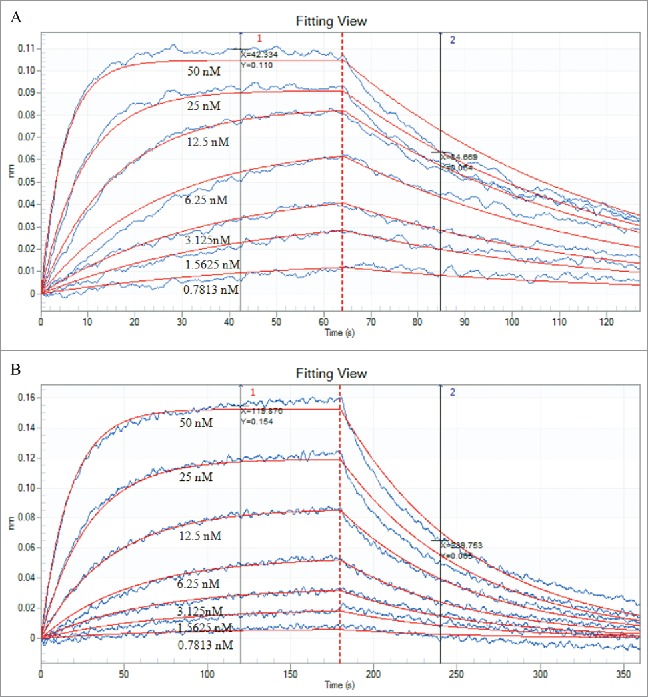
Data of affinity interactions between TGF-β1 and eTGFBR2 (A) or HSA-eTGFBR2 (B). The binding affinity of TGF-β1 was determined using 2-fold serial dilutions of eTGFBR2 and HSA-eTGFBR2 at concentrations of 0.7813–50 nM. Both of the TGF-β1 was in the same concentration of 1 μg/mL.
Neutralizing activity assays of HSA-eTGFBR2 for growth inhibitory effects of TGF-β1
The neutralizing activity of HSA-eTGFBR2 to TGF-β1 was tested using L-02 cells. As shown in Fig. 7, the proliferation of L-02 cells was inhibited by TGF-β1 at a concentration of 0.4 nM. The recombinant human HSA-eTGFBR2, as an antagonist, was able to partially reverse the inhibitory effects of TGF-β1 in a dose-dependent manner. A similar concentration of commercial eTGFBR2 was required for neutralization of TGF-β1. The results showed that recombinant human HSA-eTGFBR2 and commercial eTGFBR2 exhibit comparable antagonist activity in vitro cell proliferation inhibition experiment.
Figure 7.
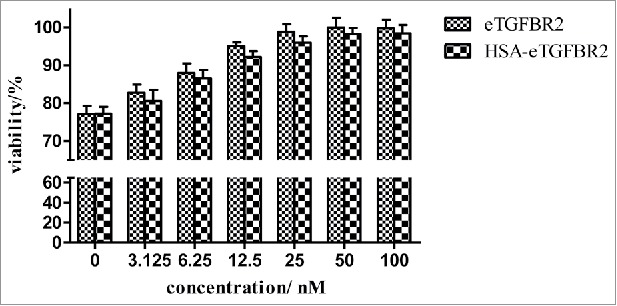
Neutralization of 0.4 nM TGF-β1 growth inhibitory activity by HSA-eTGFBR2. The values of viability% represent the relative value of fluorescence (560Ex/590Em) compared with the value of control group incubated without external protein.
To further investigate the effects of TGF-β1 and HSA-eTGFBR2 on L-02 cell cycle, 3 × 105 cells/well were inoculated in 6-well plates and divided into 4 groups: Control (RPMI-1640), 0.4 nM TGF-β1, TGF-β1+30 nM eTGFBR2, TGF-β1+30 nM HSA-eTGFBR2. Compared to control group (Fig. 8A), TGF-β1 group (Fig. 8B) brought increasing cell percentage in G0-G1 phase (from (65.35 ± 0.87)% to (78.4% ± 0.84)%) and the proliferation index (PI=(S+G2/M)/(G0/G1+S+G2/M)) reduced from (34.65 ± 0.87)% to (21.6 ± 0.84)% (Table 3). Whereas TGF-β1 in the presence of eTGFBR2 (Fig. 8C) or HSA-eTGFBR2 (Fig. 8D) resulted in a reverse of percentage of cells in G0-G1 phase (from (78.4% ± 0.84)% to (59.53 ± 1.53)% and (64.47 ± 1.39)% respectively), comparable to the negative control. These results further demonstrated that HSA-eTGFBR2 could neutralize the inhibitory effects of TGF-β1 on L-02 cells.
Figure. 8.
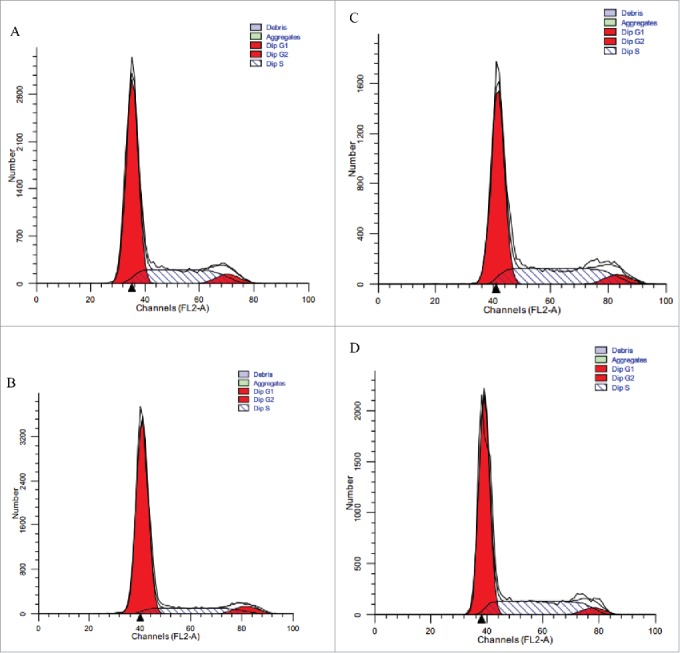
Flow cytometry analysis of L-02 cells in the presence of TGF-β1 with or without HSA-eTGFBR2 for 24 h. (A) RPMI-1640 medium for negative control; (B) 0.4 nM TGF-β1 for positive control; (C) TGF-β1 plus 30 nM eTGFBR2; (D) TGF-β1 plus 30 nM HSA-eTGFBR2.
Table 3.
The effects of TGF-β1 with eTGFBR2 or HSA-eTGFBR2 on cell cycle progression of L-02 cells. (n = 3).
| G0-G1% | S% | G2-M% | PtdIns% | |
|---|---|---|---|---|
| Control | 65.35 ± 0.87 | 27.66 ± 0.84 | 6.99 ± 0.66 | 34.65 ± 0.87 |
| TGF-β1 | 78.40 ± 0.84### | 16.26 ± 0.02### | 5.34 ± 0.84 | 21.6 ± 0.84### |
| eTGFBR2+TGF-β1 | 59.53 ± 1.53### | 34.80 ± 2.65### | 5.67 ± 1.17 | 40.47 ± 1.53### |
| HSA-eTGFBR2+TGF-β1 | 64.47 ± 1.39### | 30.28 ± 1.92### | 5.24 ± 0.66 | 35.53 ± 1.39### |
P < 0.001 vs control group;
P < 0.001 vs TGF-β1 group.
Phosphorylation of Smad3 analysis in HSC-T6 cells
The TGF-β1/Smad3 signaling pathway plays an important role in mediating TGF-β1 bioactivity.39 To investigate the inhibition activity of HSA-eTGFBR2, the levels of phosphorylated Smad3 were detected by different incubation time of eTGFBR2 (Fig. 9A) or HSA-eTGFBR2 (Fig. 9B). The results showed that HSA-eTGFBR2 fusion protein had more long-term bioactivity of inhibiting TGF-β1/Smad3 signaling pathway.
Figure 9.
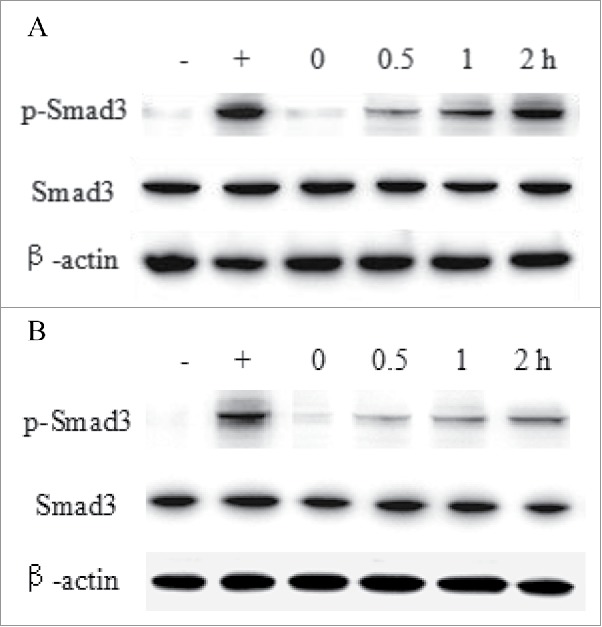
The Smad3 activation analysis of HSC-T6 cells. Cells were incubated with 30 nM eTGFBR2 (A) or HSA-eTGFBR2 (B) for 0, 0.5, 1, 2 h, and then exposed to 0.4 nM TGF-β1 for 30 min. -: DMEM medium for negative control; +: 0.4 nM TGF-β1 for positive control.
Half-life analysis of HSA-eTGFBR2 fusion protein in vivo
The circulating half-life of HSA-eTGFBR2 was compared with commercial eTGFBR2. Serum concentrations of eTGFBR2 and HSA-eTGFBR2 after a single intravenous dose of 3 nmol/kg in C57BL/6 mice were show in Fig. 10. After intravenous administration, HSA-eTGFBR2 fusion protein was cleared more slowly than eTGFBR2. The circulating half-life of eTGFBR2 was about 41.42 min (Fig. 10A), whereas intravenous administration of HSA-eTGFBR2 lead to a extended half-life about 11.84 h (Fig. 10B). This result suggested that HSA fusion remarkably improved the blood circulation profile of eTGFBR2.
Figure 10.
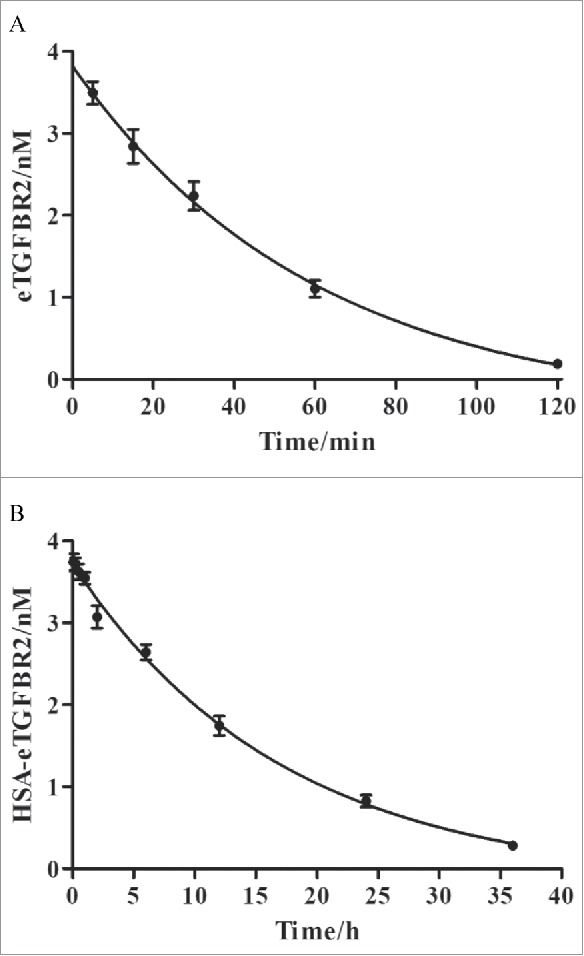
The pharmacokinetic properties of eTGFBR2 (A) and HSA-eTGFBR2 (B). Serum concentrations of eTGFBR2 and HSA-eTGFBR2 at different time points were determined after intravenous injection in C57BL/6 mice (n = 3).
Discussion
TGF-β1, as the important member of TGF-β family, is vital for cell growth, differentiation, apoptosis and motility. It plays important roles in normal liver and many hepatic diseases. In normal liver, TGF-β1 has the effects of growth inhibition40,41 associated with induction of apoptosis on hepatocytes.42-44 Furthermore, TGF-β1 is a master pro-fibrotic factor in chronic fibrotic diseases by triggering the hepatic stellate cells (HSCs) differentiation into myofibroblasts and the accumulation of extracellular matrices (ECMs).45 Numerous studies have indicated that overexpression of TGF-β1 has been implicated in the pathogenesis of many diseases,46,47 which suggests that TGF-β1 antagonists may be potential therapeutic agents in treatment of diseases such as keloids, glomerulonephritis, autoimmune, cancer and fibrosis.48
eTGFBR2 had been expressed in baculovirus-infected Sf9 insect cells and mouse myeloma cell lines via transient transfection with a low yield, following incomplete enzymatic deglycosylation and elimination of the N-terminal 22 amino acids by protease degradation. Moreover, as a small molecular protein, eTGFBR2 has a short half-life in vivo due to enzymatic degradation and kidney clearance, resulting in high doses and repeated injections on clinical utility. Albumin, mainly synthesized in the liver with a molecular weight of 66.5 kD, is the most abundant protein in blood plasma. It is highly stable with a long circulation half-life resulting from a recycling process mediated by the MHC-related Fc receptor for IgG (FcRn).49 HSA fusion technology extends the circulating half-life of recombinant proteins in vivo, resulting in enhanced pharmacodynamics of fusion proteins. Many studies have shown that HSA fused at the N-terminals of proteins resulted in a better activity than it fused at the C-terminal.50 This might be due to the steric hindrance effect or impact on the post-translational folding of HSA.
Our results demonstrate that the recombinant human HSA-eTGFBR2 expressed by CHO cells was capable of binding to TGF-β1, with an affinity value KD of 1.42 × 10−8 M. Whereas the affinity value KD of eTGFBR2 for TGF-β1 was 9.49 × 10−9 M. It suggested that the fusion protein HSA-eTGFBR2 retained binding properties and fusion of eTGFBR2 molecule to HSA didn't significantly impair their target binding capacity. The difference binding affinity of HSA-eTGFBR2 was speculated that the increase of molecular size after fusing with HSA might change the steric hindrance and even influence the binding rates of receptor dissociation and association, resulting in extended interaction time between the TGF-β1 and receptor.51 According to neutralizing activity assays, HSA-eTGFBR2 antagonized growth inhibitory effects of TGF-β1 in a dose-dependent manner. The ability of the HSA-eTGFBR2 acted as an antagonist for TGF-β1 suggested that the fusion protein bound to TGF-β1 with sufficient affinity to compete with the membrane-bound form of the receptor.
The blood circulation data indicated that HSA-eTGFBR2 had a prolonged half-life about 11.84 h. The short half-life protein eTGFBR2 fused with HSA could reduce the renal clearance and improve the stability characteristics.52 The favorable blood circulation profile of HSA-eTGFBR2 fusion protein will improve the clinical applicability, which would allow less injection frequency and maintain a more constant circulating level.
In conclusion, we inserted the 159 amino acid residues soluble extracellular domain of TGFBR2 fused to the C-terminal of HSA into pMH3 plasmid for expression by CHO-S cells, developed a fed-batch culture system with a yield of 180 mg/L and set up a 2-step purification system of Blue affinity chromatography and Q sepharose chromatography. The recombinant human HSA-eTGFBR2 exhibited affinity capacity to TGF-β1 (KD 1.42 × 10−8 M) and neutralized the growth inhibitory activity of TGF-β1 by blocking its signal transduction. Besides, HSA-eTGFBR2 had a prolonged circulating half-life in vivo, which has promising application prospects for therapy of diseases.
Materials and methods
Plasmid, cell line and culture medium
The plasmid pMH3 was purchased from Amprotein (Hangzhou, Zhejiang, China). The cell line CHO-S (Amprotein Co., Ltd, Hangzhou, China) were grown in DMEM/F12(1:1) medium (Gibco) supplemented with 10% (v/v) FBS (Gibco) in atmosphere of 5% CO2 at 37°C. The serum-free medium (SFM) B001 and feed medium F001 for suspension culture of CHO-S cells were provided by Amprotein (Hangzhou, Zhejiang, China). Viable cell density and cell viability were measured using trypan blue (Sigma-aldrich, St. louis, USA) stain method.
Construction of the pMH3-HSA-eTGFBR2 expression vectors
Eukaryotic expression vector pMH3 was used for high efficient and stable production of recombinant proteins. The pMH3 plasmid contains 3 non-coding GC-rich DNA regions that suitable for CHO expression system. The vector can improve the transcriptional activity of interesting genes no matter where they integrate into the chromosome.53 The encoding genes of human hsa and etgfbr2 were preserved in our laboratory and used as templates for fusion PCR. The hsa cDNA was amplified by PCR using primers P3 and P2. The etgfbr2 cDNA was amplified by PCR using primers P1 and P4. Then these 2 fragments were assembled and amplified by PCR with primers P3 and P4. Primers for this study were listed in Table 1. The hsa-etgfbr2 fusion fragments were inserted into the pMH3 plasmid using EcoRI and NotI sites to obtain the expression plasmid pMH3-HSA-eTGFBR2. The sequence in the digested plasmid pMH3-HSA-eTGFBR2 was confirmed by DNA sequencing (Shanghai Sangon, China).
Table 1.
Primer sequences for PCR.
| Primer | Sequences |
|---|---|
| Fusion forward (P1) | GTTGCTGCAAGTCAAGCTGCCTTAGGCTTAGACGACGACGACAAGacgatcccaccgcacgttcagaagtcggttaa |
| Fusion reverse (P2) | TTAACCGACTTCTGAACGTGCGGTGGGATCGTCTTGTCGTCGTCGTCTAAGCCTAAGGCAGCTTGACTTGCAGCAAC |
| HSA-eTGFBR2 forward (P3) | gcGAATTCcaccatggagacagacacactcctgctatgggtactgctgctctgggttccaggttccactggtGATGCACACAAGAGTGAGGTTGCTCATCGA TTTAAAGAT |
| HSA-eTGFBR2 reverse (P4) | AtGCGGCCGCCTAGTCAGGATTGCTGGTGTTATATTCTTCTGA |
Establishment of HSA-eTGFBR2 expression cell line
The expression vector pMH3-HSA-eTGFBR2 was transformed into CHO-S cells using electroporation with 400 V, 400 μS and repeated 3 times. The electroporation reaction mixture contained: 4 × 106 cells, 20 μg plasmid, 5 μg salmon sperm DNA (Invitrogen, Carlsbad, CA, USA). The cells were dispersed into a 100 mm plate and recovered for 24h. Then the medium was replaced with selective medium containing 2.8 mg/mL G418 (Sigma-aldrich, St. louis, USA). Subsequently, the surviving macroscopically visible single clones were picked and cultured in 96-well plate for 7 d. The high expression sub-clones were selected by Dot blot using anti-HSA antibody (Abcam, San Francisco, USA) and Western blot using anti-TGFBR2 antibody (R&D Systems, Minneapolis, MN, USA). Single cell derived clones were obtained by limited dilution method and selected again as mentioned previously. The final high expression sub-clones selected gradually from 96-well plate to 24-well plate and finally to 6-well plate were used for suspension culture in serum-free medium B001.
Fed-batch culture and purification
The high expression clone was seeded at a concentration of 2 × 106 cells/mL in a 3L bioreactor with a working volume of 1 L in a shaking incubator at 37°C, 100 rpm for 2 d. Then feed medium F001 were added daily to maintain the constant glucose concentration of 2.0–3.0 g/L. Besides, the culture temperature was shifted from 37°C to 34°C. The samples were taken every day to observe the cell growth, glucose concentration and protein accumulation. The concentration of HSA-eTGFBR2 was determined by Human TGF-β RII DuoSet ELISA kit (R&D).
To purify HSA-eTGFBR2 from the culture medium, we equilibrated a Blue Sepharose 6 Fast Flow column (GE) with buffer A (20 mM PB, 100 mM NaCl, pH7.2), then passed the supernatants through this Blue column. After a wash step, the target protein was eluted with buffer B (20 mM PB, 2 M NaCl, pH7.2). Then Q Sepharose Fast Flow chromatography (GE) was use for the next purification. The desalting fractions containing target protein was loaded to Q sepharose column pre-equilibrated with buffer C (20 mM PB, pH7.2) and eluted with 20 mM PB, 600 mM NaCl, pH7.2. The collected fractions were analyzed by SDS-PAGE and Western blot after purification.
SDS-PAGE, Western blot and Dot blot
The samples were separated by 10% SDS-PAGE gel under reducing conditions and stained with Coomassie brilliant blue R-250. For Western blot analysis, the samples in gel were transferred to nitrocellulose membranes. For Dot blot, 5μL supernatants in well plates were dripped onto the nitrocellulose membranes. Then the membranes were blocked by TBST (10 mM Tris, 150 mM NaCl, pH7.5) containing 5% skimmed milk and incubated with the anti-HSA antibody or anti-eTGFBR2 antibody for 4 h at room temperature, followed by HRP-conjugated second antibody correspond to primary antibody. After washing 3 times with TBST, blots were detected with BeyoECL Plus reagent (Beyotime Biotechnology, Shanghai, China).
ELISA immunoassays for TGF-β1 binding assay
Binding properties of recombinant human HSA-eTGFBR2 to TGF-β1 were analyzed by sandwich enzyme-linked immunosorbent assay.54,55 96-well immunoassay plates were coated with 100 μL/well of recombinant human HSA-eTGFBR2 or commercial eTGFBR2 (R&D) at concentrations ranging from 0 to 50 nM overnight at 4°C. After washed 3 times with PBST (phosphate buffered saline, pH7.4 containing 0.05% Tween 20), the wells were blocked with 3% bovine serum albumin in PBST for 2 h at 37°C. Then 100 μL/well of TGF-β1(R&D) at different concentrations of 0, 0.004, 0.04, 0.4, 4 nM were added to the plate and incubated for 2 h at 37°C. After washing 3 times, 100 μL/well of mouse anti-TGF-β1 monoclonal antibodies (500 ng/mL, R&D) was added to the plates and incubated for 2 h at 37°C. Then the plate was washed with PBST and 100 μL of HRP-labeled goat anti-mouse IgG (1:2000, proteintech) was added to each well to react with anti-TGF-β1 antibodies for 2 h at 37°C. At the end of the incubation period, the plate was washed 3 times and 100 μL of TMB substrate was added to each well. After reaction for 20 min at room temperature, the color development was stopped with 50 μL of 2 M H2SO4. The optical density of each well was measured at 450 nm using a microplate reader.
ForteBio Octet system assay for affinity of TGF-β1 to HSA-eTGFBR2
The binding affinities of TGF-β1 to eTGFBR2 or HSA-eTGFBR2 were tested by the ForteBio Octet RED96 system (ForteBio Inc., CA, USA). The sensors (Anti-Mouse IgG Fc Capture Surface, AMC) were wetted in PBST for 10 min before detection. The anti-TGF-β1 antibody (20 μg/mL) was loaded onto the AMC sensors for 10 min. After equilibrated with PBST, the sensors were loaded with TGF-β1 at a concentration of 1 μg/mL. Then the processes of adsorption and desorption of the HSA-eTGFBR2 or eTGFBR2 molecule were monitored in parallel. After equilibrated with PBST for 5min, the sensors were transferred into HSA-eTGFBR2 or eTGFBR2 protein solution at the 2-fold serial dilutions concentration of 0.7813–50 nM. Then PBST was loaded to dissociate the nonspecifically bound components. The data were analyzed using the system software Data analysis 7.0 provided by Fortebio Instrument Co.
Antagonist ability of HSA-eTGFBR2 for growth inhibitory effects of TGF-β1
TGF-β1 inhibited the proliferation of hepatocytes and the bioactivity of HSA-eTGFBR2 as an antagonist of TGF-β1 was tested using L-02 cells. The cells were seeded in 96-well plate at a concentration of 4 × 103 cells/well and cultured in RPMI-1640 medium with 10% FBS for 24 h. Different concentrations of HSA-eTGFBR2 or eTGFBR2 (0, 1.56, 3.125, 6.25, 12.5, 25, 50 nM) with 0.4 nM TGF-β1 in RPMI-1640 were added to each well. After incubation at 37°C for 48 h, 10 μL of CellTiter-blue Reagent was added to each well. The 96-well plates were incubated for 2 h at 37°C and the viability of cells was measured with fluorescence (560Ex/590Em) using a microplate reader.
3 × 105 L-02 cells/well were inoculated in 6-well plate and incubated for 24 h, followed by 12 h of serum starvation. 0.4 nM TGF-β1 with or without HSA-eTGFBR2 in RPMI-1640 were added to the plate and incubated for 24 h, eTGFBR2 was used as the positive control. Then the cells were harvested and washed with ice cold PBS. After fixed with ice cold 70% ethanol overnight at −30°C, the cells were washed with ice cold PBS again and incubated with 50 μg/mL RNase solution for 30 min at 37°C. Finally, samples were stained with 50 μg/mL propidium iodide solution for 30 min at 4°C. Flow cytometry (BD Biosciences) was used to analyze the cell cycle. The data were analyzed with FACS Diva and ModFit LT software.
Phosphorylation of Smad3 analysis in HSC-T6 cells
HSC-T6 cells were seeded at the density of 3 × 105 cells/well in 6-well plate and incubated in DMEM with 10% FBS for 24 h. After serum starvation, the cells were incubated with 30 nM eTGFBR2 or HSA-eTGFBR2 for 0, 0.5, 1, 2 h, and then exposed to 0.4 nM TGF-β1 for 30 min. After incubation, cells in each plate were lysed by RIPA lysis buffer containing 1 mM PMSF (Beyotime Biotechnology). Equal amounts of proteins were separated on 12% SDS-PAGE gel and transferred onto nitrocellulose membrane. After blocking with 5% non-fat milk, the membranes were incubated with anti-p-Smad3 antibody or anti-Smad3 antibody (Cell Signaling technology) for 4 h at room temperature. Immunoblot analysis was performed using HRP-conjugated secondary antibody and the BeyoECL Plus Reagent.
Pharmacokinetics study
Animal experiments were performed in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals. C57BL/6 mice (male, weight between 18 and 20 g) were obtained from Comparative Medicine Center of Yangzhou University. Three mice were used for each time point. All mice were fasted for 12 h and received intraveneous injections of 3 nmol/kg of HSA-eTGFBR2 or eTGFBR2. Blood samples were collected through the eyes at 5 min, 15 min, 30 min, 60 min, 120 min (eTGFBR2) or 5 min, 15 min, 30 min, 1 h, 2 h, 6 h, 12 h, 24 h (HSA-eTGFBR2) after dosing and centrifuged at 1,500 g at 4°C for 10 min. The concentrations of HSA-eTGFBR2 or eTGFBR2 in serum at different time points were analyzed by Human TGF-β RII DuoSet ELISA kit. The half-life was calculated by nonlinear regression using Prism software.
Disclosure of potential Conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Chinese New Medicine Research Fund (No. 2013ZX09102033), Chinese National High-Tech Programs of China (No.2014AA021003 and 2015AA020802), Chinese Natural Science Fund (No. 81273437) and Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
- [1].Philip GK, Claire G. Local roles of TGF-β superfamily members in the control of ovarian follicle development. Animal Reproduction Science 2003; 78:165-183; PMID:12818643; http://dx.doi.org/ 10.1016/S0378-4320(03)00089-7 [DOI] [PubMed] [Google Scholar]
- [2].Sporn MB, Roberts AB. Transforming growth factor beta: recent progress and new challenges. J Cell Biol 1992; 119(5):1017-1121; PMID:1332976; http://dx.doi.org/ 10.1083/jcb.119.5.1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Polata IM, Alçiğirb E, Pekcanc M, Vural SA, Özenç E, Canatan HE, Küplülü Ş, Dal GE, Yazlik MO, Baklaci C, et al.. Characterization of transforming growth factor beta superfamily, growth factors, transcriptional factors, and lipopolysaccharide in bovine cystic ovarian follicles. Theriogenology 2015; 84(6):1043-1052; PMID:26166168; http://dx.doi.org/ 10.1016/j.theriogenology.2015.06.003 [DOI] [PubMed] [Google Scholar]
- [4].Shi Y, Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 2003; 113(6):685-700; PMID:12809600; http://dx.doi.org/ 10.1016/S0092-8674(03)00432-X [DOI] [PubMed] [Google Scholar]
- [5].Massague J. TGF-β signal transduction. Annu Rev Biochem 1998; 67:753-791; PMID:9759503; http://dx.doi.org/ 10.1146/annurev.biochem.67.1.753 [DOI] [PubMed] [Google Scholar]
- [6].Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci 2003; 100(15):8621-8623; http://dx.doi.org/ 10.1073/pnas.1633291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev 2002; 12(1):22-29; PMID:11790550; http://dx.doi.org/ 10.1016/S0959-437X(01)00259-3 [DOI] [PubMed] [Google Scholar]
- [8].Danielpour D. Functions and regulation of transforming growth factorbeta (TGF-beta) in the prostate. Eur J Cancer 2005; 41(6):846-57; PMID:15808954; http://dx.doi.org/ 10.1016/j.ejca.2004.12.027 [DOI] [PubMed] [Google Scholar]
- [9].Russell WE, Coffey RJ, Ouellette AJ, Moses HL. Type beta transforming growth factor reversibly inhibits the early proliferative response to partial hepatectomy in the rat. Proc Nat Acad Sci USA 1988; 85:5126-5130; PMID:3164865; http://dx.doi.org/ 10.1073/pnas.85.14.5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Oberhammer FA, Pavelka M, Sharma S, Tiefenbacher R, Purchio AF, Bursch W, Schulte-Hermann R. Induction of apoptosis in cultured hepatocytes and in regressing liver by transforming growth factor beta 1. Proc Nat Acad Sci USA 1992; 89:5408-5412; PMID:1608949; http://dx.doi.org/ 10.1073/pnas.89.12.5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tsang ML, Zhou L, Zheng BL, Wenker J, Fransen G, Humphrey J, Smith JM, O'Connor-McCourt M, Lucas R, Weatherbee JA. Characterization of recombinant soluble human transforming growth factor-beta receptor type II (rhTGF-beta sRII). Cytokine 1995; 7(5):389-397; PMID:7578976; http://dx.doi.org/ 10.1006/cyto.1995.0054 [DOI] [PubMed] [Google Scholar]
- [12].Wrana JL. TGF-beta receptors and signalling mechanisms. Miner Electrolyte Metab 1998; 24(3):120-130; PMID:9525694; http://dx.doi.org/ 10.1159/000057359 [DOI] [PubMed] [Google Scholar]
- [13].Wrana JL. Regulation of Smad activity. Cell 2000; 100(2):189-192; PMID:10660041; http://dx.doi.org/ 10.1016/S0092-8674(00)81556-1 [DOI] [PubMed] [Google Scholar]
- [14].Wrana JL, Attisano L. MAD-related proteins in TGF-beta signalling. Trends Genet 1996; 12(12):493-496; PMID:9257525; http://dx.doi.org/ 10.1016/S0168-9525(96)30109-1 [DOI] [PubMed] [Google Scholar]
- [15].Mehra A, Wrana JL. TGF-β and the Smad signal transduction pathway. Biochem Cell Biol 2002; 80(5):605-622; http://dx.doi.org/ 10.1139/o02-161 [DOI] [PubMed] [Google Scholar]
- [16].Heldin CH, Miyazono K. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 1997; 390(6659):465-471; http://dx.doi.org/ 10.1038/37284 [DOI] [PubMed] [Google Scholar]
- [17].Kang JS, Liu C, Derynck R. New regulatory mechanisms of TGF-β receptor function. Trends Cell Biol 2009; 198:385-394; http://dx.doi.org/ 10.1016/j.tcb.2009.05.008 [DOI] [PubMed] [Google Scholar]
- [18].Inagaki Y, Okazaki I. Emerging insights into Transforming growth factor β Smad signal in hepatic fibrogenesis. Gut 2007; 56(2):284-292; PMID:17303605; http://dx.doi.org/ 10.1136/gut.2005.088690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Biernacka A, Dobaczewski M, Frangogiannis NG. TGF-β signaling in fibrosis. Growth Factors 2011; 29(5):196-202; http://dx.doi.org/ 10.3109/08977194.2011.595714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Padua D, Massagué J. Roles of TGFβ in metastasis. Cell Res 2009; 19(1):89-102; http://dx.doi.org/ 10.1038/cr.2008.316 [DOI] [PubMed] [Google Scholar]
- [21].Yingling JM, Blanchard KL, Sawyer JS. Development of TGF-β signaling inhibitors for cancer therapy. Nat Rev Drug Discovery 2004; 3(12):1011-1022; PMID:15573100; http://dx.doi.org/ 10.1038/nrd1580 [DOI] [PubMed] [Google Scholar]
- [22].Boesen CC, Motyka SA, Patamawenu A, Sun PD. Development of a recombinant bacterial expression system for the active form of a human transforming growth factor β Type II receptor ligand binding domain. Protein Expression Purification 2000; 20(1):98-104; PMID:11035957; http://dx.doi.org/ 10.1006/prep.2000.1306 [DOI] [PubMed] [Google Scholar]
- [23].Glansbeek HL, Beuningen HMV, Vitters EL, van der Kraan PM, van den Berg WB. Expression of recombinant human soluble Type II Transforming Growth Factor-b receptor in Pichia pastoris and Escherichia coli: Two powerful systems to express a potent inhibitor of transforming growth Factor-β. Protein Expression Purification 1998; 12(2):201-207; PMID:9518461; http://dx.doi.org/ 10.1006/prep.1997.0819 [DOI] [PubMed] [Google Scholar]
- [24].Chaudhuri JB. Refolding recombinant proteins: process strategies and novel approaches. Annals N York Acad Sci 1994; 721:374-385; PMID:8010686; http://dx.doi.org/ 10.1111/j.1749-6632.1994.tb47409.x [DOI] [PubMed] [Google Scholar]
- [25].Gasparian ME, Elistratov PA, Yakimov SA, Dolgikh DA, Kirpichnikov MP. An efficient method for expression in Escherichia coli and purification of the extracellular ligand binding domain of the human TGF beta type II receptor. J Biotechnol 2010; 148(2–3):113-118; PMID:20451568; http://dx.doi.org/ 10.1016/j.jbiotec.2010.04.013 [DOI] [PubMed] [Google Scholar]
- [26].Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol 2004; 22(11):1393-1398; PMID:15529164; http://dx.doi.org/ 10.1038/nbt1026 [DOI] [PubMed] [Google Scholar]
- [27].Wells RG, Yankelev H, Lin HY, Lodish HF. Biosynthesis of the type I and type II TGF-beta receptors. Implications for complex formation[J]. J Biol Chem 1997; 272(17):11444-11451; PMID:9111056; http://dx.doi.org/ 10.1074/jbc.272.17.11444 [DOI] [PubMed] [Google Scholar]
- [28].Zwaagstra JC, Sulea T, Baardsnes J, Lenferink AE, Collins C, Cantin C, Paul-Roc B, Grothe S, Hossain S, Richer LP, et al.. Engineering and therapeutic application of single-chain Bivalent TGF-β family traps. Mol Cancer Therapeutics 2012; 11(7):1477-1487; PMID:22562986; http://dx.doi.org/ 10.1158/1535-7163.MCT-12-0060 [DOI] [PubMed] [Google Scholar]
- [29].Lei JY, Guan B, Li B, Duan ZY, Chen Y, Li H, Jin J. Expression, purification and characterization of recombinant human interleukin-2-serum albumin (rhIL-2-HSA) fusion protein in Pichia pastoris. Protein Expr Purif 2012; 84(1):154-160; PMID:22609631; http://dx.doi.org/ 10.1016/j.pep.2012.05.003 [DOI] [PubMed] [Google Scholar]
- [30].Melder RJ, Osborn BL, Riccobene T, Kanakaraj P, Wei P, Chen G, Stolow D, Halpern WG, Migone TS, Wang Q. Pharmacokinetics and in vitro and in vivo anti-tumor response of an interleukin-2-human serum albumin fusion protein in mice. Cancer Immunol Immunotherapy 2005; 54(6):535-547; PMID:15592670; http://dx.doi.org/ 10.1007/s00262-004-0624-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tian S, Li Q, Yao W, Xu C. Construction and characterization of a potent, long-lasting recombinant human serum albumin-interferon α1 fusion protein expressed in Pichia pastoris. Protein Expression Purification 2013; 90(2):124-128; PMID:23748141; http://dx.doi.org/ 10.1016/j.pep.2013.05.002 [DOI] [PubMed] [Google Scholar]
- [32].Zhao HL, Xue C, Wang Y, Sun B, Yao XQ, Liu ZM. Elimination of the free sulfhydryl group in the human serum albumin (HSA) moiety of human interferon-α2b and HSA fusion protein increases its stability against mechanical and thermal stresses. Eur J Pharmaceutics Biopharmaceutics 2009; 72(2):405-411; PMID:19462475; http://dx.doi.org/ 10.1016/j.ejpb.2009.01.008 [DOI] [PubMed] [Google Scholar]
- [33].Zhu RY, Xin X, Dai HY, Li Q, Lei JY, Chen Y, Jin J. Expression and purification of recombinant human serum albumin fusion protein with VEGF165b in Pichia pastoris. Protein Expression Purification 2012; 85(1):32-37; PMID:22750397; http://dx.doi.org/ 10.1016/j.pep.2012.06.009 [DOI] [PubMed] [Google Scholar]
- [34].Müller D, Kane A, Meißburger B, Höfig I, Stork R, Kontermann RE. Improved pharmacokinetics of recombinant bispecific antibody molecules by fusion to human serum albumin. J Biol Chem 2007; 282(17):12650-12660; PMID:17347147; http://dx.doi.org/ 10.1074/jbc.M700820200 [DOI] [PubMed] [Google Scholar]
- [35].Berger V, Richter F, Zettlitz K, Unverdorben F, Scheurich P, Herrmann A, Pfizenmaier K, Kontermann RE. An anti-TNFR1 scFv-HSA fusion protein as selective antagonist of TNF action. Protein Engineering Design Selection 2013; 26 (10):581-587; http://dx.doi.org/ 10.1093/protein/gzt044 [DOI] [PubMed] [Google Scholar]
- [36].Peters T. Serum Albumin. Adv Protein Chem 1985; 37(37): 161-245;PMID:3904348; http://dx.doi.org/27115755 10.1016/S0065-3233(08)60065-0 [DOI] [PubMed] [Google Scholar]
- [37].Ru Y, Zhi D, Guo D, Wang Y, Li Y, Wang M, Wei S, Wang H, Wang N, Che J, et al.. Expression and bioactivity of recombinant human serum albumin and dTMP fusion proteins in CHO cells. Appl Microbiol Biotechnol 2016; 100(17):7565-7575; PMID:27115755; http://dx.doi.org/ 10.1007/s00253-016-7447-2 [DOI] [PubMed] [Google Scholar]
- [38].Kim YM, Lee SM, Chung HS. Novel AGLP-1 albumin fusion protein as a long-lasting agent for type 2 diabetes. BMB Rep 2013; 46(12):606-610; PMID:24195794; http://dx.doi.org/ 10.5483/BMBRep.2013.46.12.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kitamura Y, Ninomiya H. Smad expression of hepatic stellate cells in liver cirrhosis in vivo and hepatic stellate cell line in vitro. Pathol Int 2003; 53:18-26; PMID:12558865; http://dx.doi.org/ 10.1046/j.1440-1827.2003.01431.x [DOI] [PubMed] [Google Scholar]
- [40].Nguyen LN, Furuya MH, Wolfraim LA, Nguyen AP, Holdren MS, Campbell JS, Knight B, Yeoh GC, Fausto N, Parks WT. Transforming growth Factor-Beta differentially regulates oval cell and hepatocyte proliferation. Hepatology 2007; 45(1):31-41; PMID:17187411; http://dx.doi.org/ 10.1002/hep.21466 [DOI] [PubMed] [Google Scholar]
- [41].Carr BI, Hayashi I, Branum EL, Moses HL. Inhibition of DNA synthesis in rat hepatocytes by Platelet-derived Type β transforming growth factor. Cancer Res 1986; 46(5):2330-2334; PMID:3008986 [PubMed] [Google Scholar]
- [42].Oberhammer F, Bursch W, Parzefall W, Breit P, Erber E, Stadler M, Schulte-Hermann R. Effect of transforming growth factor beta on cell death of cultured rat hepatocytes. Cancer Res 1991; 51(9):2478-2485; PMID:2015607 [PubMed] [Google Scholar]
- [43].Bursch W, Oberhammer F, Jirtle RL, Askari M, Sedivy R, Grasl-Kraupp B, Purchio AF, Schulte-Hermann R. Transforming growth factor-β1 as a signal for induction of cell death by apoptosis. Br J Cancer 1993; 67(3):531-536; PMID:8439503; http://dx.doi.org/ 10.1038/bjc.1993.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Oberhammer FA, Pavelka M, Sharma S, Tiefenbacher R, Purchio AF, Bursch W, Schulte-Hermann R. Induction of apoptosis in cultured hepatocytes and in regressing liver by transforming growth factor beta 1. Proc Natl Acad Sci U S A 1992; 89(12):5408-5412; http://dx.doi.org/ 10.1073/pnas.89.12.5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sakai K, Jawaid S, Sasaki T, Bou-Gharios G, Sakai T. Transforming Growth Factor-beIndependent role of connective tissue growth factor in the development of Liver Fibrosis. Am J Pathol 2014; 184(10):2611-2617; PMID:25108224; http://dx.doi.org/ 10.1016/j.ajpath.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yingling JM, Blanchard KL, Sawyer JS. Development of TGF-beta signalling inhibitors for cancer therapy. Nat Rev Drug Discov 2004; 3(12):1011-1022; PMID:15573100; http://dx.doi.org/ 10.1038/nrd1580 [DOI] [PubMed] [Google Scholar]
- [47].Pinkas J, Teicher BA. TGF-β in cancer and as a therapeutic target. Biochem Pharmacol 2006; 72(5):523-529; PMID:16620790; http://dx.doi.org/ 10.1016/j.bcp.2006.03.004 [DOI] [PubMed] [Google Scholar]
- [48].Kathleen C, James K. Medical applications of transforming growth factor-beta. Clin Med Res 2003; 1(1):13-20; http://dx.doi.org/ 10.3121/cmr.1.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chaudhury C, Brooks CL, Carter DC, Robinson JM, Anderson CL. Albumin binding to FcRn: Distinct from the FcRn-IgG interaction. Biochemistry 2006; 45(15):4983-4990; PMID:16605266; http://dx.doi.org/ 10.1021/bi052628y [DOI] [PubMed] [Google Scholar]
- [50].Ding Y, Peng Y, Deng L, Wu Y, Fu Q, Jin J. The effects of fusion structure on the expression and bioactivity of human brain natriuretic peptide (BNP) albumin fusion proteins. Curr Pharmaceutical Biotechnol 2014; 15(9):856-863; PMID:25307015; http://dx.doi.org/ 10.2174/1389201015666141012182106 [DOI] [PubMed] [Google Scholar]
- [51].Peng Y, Deng LL, Ding YD, Chen Q, Wu Y, Yang M, Wang Y, Fu Q. Comparative study of Somatostatin-Human Serum Albumin fusion proteins and natural somatostatin on receptor binding, internalization and activation. Plos One 2014; 9(2):e89932; PMID:24587133; http://dx.doi.org/ 10.1371/journal.pone.0089932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Syed S, Schuyler PD, Kulczycky M, Sheffield WP. Potent antithrombin activity and delayed clearance from the circulation characterize recombinant hirudin genetically fused to albumin. Blood 1997; 89(9):3243-3252; PMID:9129029 [PubMed] [Google Scholar]
- [53].Jia Q, Wu HT, Zhou XJ, Gao J, Zhao W, Aziz J, Wei J, Hou L, Wu S, Zhang Y, et al.. A “GC-rich” method for mammalian gene expression: A dominant role of non-coding DNA GC content in regulation of mammalian gene expression. Sci China. Series C: Life Sci 2010; 53(1):94-100; http://dx.doi.org/ 10.1007/s11427-010-0003-x [DOI] [PubMed] [Google Scholar]
- [54].Danielpou D, Kim KU, Dart L. Sandwich Enzyme-Linked Imrnunosorbent Assays (SELISAs) Quantitate and distinguish two forms of transforming growth Factor-Beta (TGF-bl and TGF-P2) in complex bioiogical fluids. Growth Factors 1989; 2:61-71; PMID:2483947; http://dx.doi.org/ 10.3109/08977198909069082 [DOI] [PubMed] [Google Scholar]
- [55].Tsang ML, Zhou L, Zheng BL, Wenker J, Fransen G, Humphrey J, Smith JM, O'Connor-McCourt M, Lucas R, Weatherbee JA. Characterization of recombinant soluble human transforming growth factor-beta receptor type II (rhTGF-beta sRII). Cytokine 1995; 7(5):389-397; PMID:7578976; http://dx.doi.org/ 10.1006/cyto.1995.0054 [DOI] [PubMed] [Google Scholar]


