Abstract
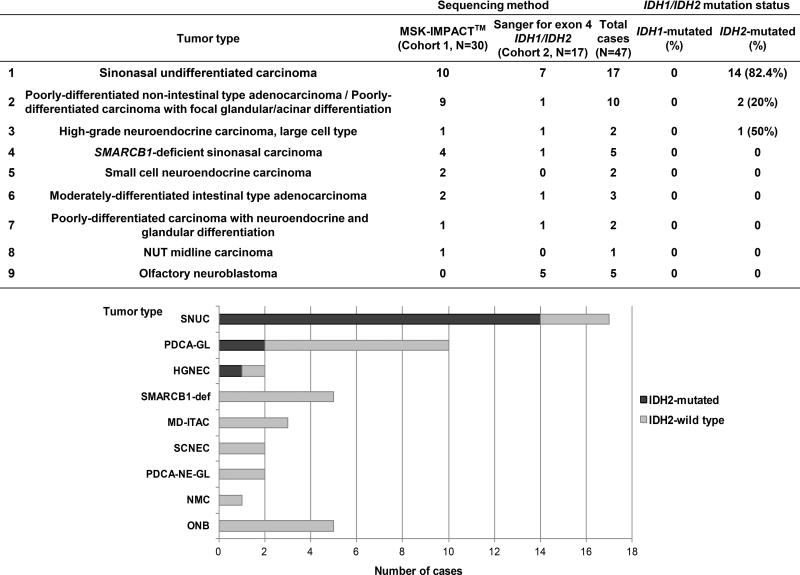
Sinonasal undifferentiated carcinoma (SNUC) is a high-grade malignancy with limited treatment options and poor outcome. A morphologic spectrum of 47 sinonasal tumors including 17 (36.2%) SNUCs were analyzed at genomic level. Thirty carcinomas (Cohort 1) were subjected to a hybridization exon-capture next-generation sequencing assay (MSK-IMPACT™) to interrogate somatic variants in 279 or 410 cancer-related genes. Seventeen sinonasal tumors (Cohort 2) were examined only for presence of IDH1/2 exon 4 mutations by Sanger sequencing. IDH2 R172 single nucleotide variants were overall detected in 14 (82.4%) SNUCs, in 2 (20%) poorly-differentiated carcinomas with glandular/acinar differentiation, and in one of 2 high-grade neuroendocrine carcinomas, large cell type (HGNEC). No IDH2 mutation was detected in any of 5 olfactory neuroblastomas or in any of 5 SMARCB1–deficient carcinomas.
Among 12 IDH2-mutated cases in Cohort 1, 5 (41.7%) harbored co-existing TP53 mutations, 4 (33.3%) CDKN2A/2B loss-of-function alterations, 4 (33.3%) MYC amplification, and 3 (25%) had concurrent SETD2 mutations. AKT1 E17K and KIT D816V hotspot variants were each detected in one IDH2-mutated SNUC. The vast majority of SNUCs and variable proportions of other poorly differentiated sinonasal carcinomas may be amenable to IDH2-targeted therapy.
Keywords: IDH2 R172, SNUC, sinonasal undifferentiated carcinoma
INTRODUCTION
Sinonasal undifferentiated carcinoma (SNUC) is a rare aggressive malignancy of the sinonasal tract first described by Frierson et al. in 1986. SNUC patients typically present with large, locally invasive lesions that often destruct the orbital and/or cranial bones [1]. A multimodal therapy including chemoradiation and surgery offer the best treatment results. However, the outcome remains poor with the overall 5-year survival ranging between 22% and 42% [2,3]. Here, we performed a comprehensive genomic profiling of a cohort of SNUCs and a morphologic spectrum of high-grade/poorly-differentiated sinonasal carcinomas aiming (1) to identify potential novel molecular therapeutic targets, and (2) to explore the genetic differences between SNUC and other high-grade/poorly-differentiated sinonasal carcinomas in order to elucidate the pathogenesis of these tumors.
MATERIALS AND METHODS
Cases
Upon obtaining the Institutional Review Board (IRB) approval the pathology files of Memorial Sloan Kettering Cancer Center (MSKCC) were searched for sinonasal carcinomas diagnosed from January 1996 to January 2014 using the following criteria: (1) sinonasal undifferentiated carcinoma and (2) non-salivary and non-squamous high-grade/poorly-differentiated carcinomas with or without neuroendocrine or glandular differentiation. Twenty-two cases were retrieved. Six additional cases prospectively sequenced as clinical samples between January 2014 and February 2017 were added to the study. An additional 19 cases were received from three collaborating institutions after obtaining their respective IRBs approvals. Details on pathology slides review and ancillary studies results are provided in Supplementary material, Supplementary materials and methods, and supplementary material, [DOGAN comment: “Supplementary material” is repeated 2×. Also, I am not sure if it should be capitalized or not – here it is, below in the text is not (?)] Table S1. Cohort 1 (N=30, including matched metastasis in 2 cases) was profiled by MSK-IMPACT™ (MSK-Integrated Mutation Profiling of Actionable Cancer Targets). Cohort 2 (N=17) was examined for presence of IDH1/2 exon 4 mutations by Sanger sequencing. The diagnostic spectrum of studied tumor categories is shown in Figure 1.
Figure 1. Frequencies of IDH1 and IDH2 mutations in sinonasal carcinomas and olfactory neuroblastoma.
Abbreviations: SNUC=sinonasal undifferentiated carcinoma, PDCA-GL=poorly differentiated non-intestinal type adenocarcinoma/poorly-differentiated carcinoma with focal glandular differentiation, HGNEC=high-grade neuroendocrine carcinoma, large cell type, SCNEC=small cell neuroendocrine carcinoma, SMARCB1-def= SMARCB1-deficient sinonasal carcinoma, NMC=NUT midline carcinoma, MD-ITAC=moderately-differentiated intestinal type adenocarcinoma with signet-ring cells and mucinous features, PDCA-NE-GL=poorly differentiated carcinoma with neuroendocrine and glandular differentiation, ONB=olfactory neuroblastoma.
Immunohistochemistry and chromogenic in situ hybridization (CISH)
Immunohistochemistry and CISH were performed according to the manufacturer’s recommendations (Ventana medical systems, Tucson, AZ), (supplementary material, supplementary materials and methods) using antibodies and CISH probes listed in supplementary material, Table S2.
DNA extraction and hybridization exon-capture next-generation sequencing
Tumors were profiled for genomic alterations in 279 or 410 key cancer-associated genes in 13 and 17 cases respectively, using our clinically validated custom deep sequencing MSK-IMPACT™ assay [4]. Twenty samples with matched normal DNA were selected for allele-specific copy number and cancer cell fraction (CCF) analysis by FACETS [5] rendering successful results in 19 samples (supplementary material, supplementary materials and methods).
Sanger DNA sequencing of IDH1 and IDH2
Sanger DNA sequencing of IDH1/2 exon 4 was performed on ABI 3730 DNA sequencer (supplementary material, supplementary materials and methods) using primers listed in supplementary material, Table S3.
Statistical analysis
Statistical analyses were performed using SPSS software 22.0 (IBM Corporation, New York, NY). P values less than 0.05 were considered significant.
RESULTS AND DISCUSSION
Clinical features
Sinonasal carcinomas occurred in 29 men (61.7%) who were significantly younger than women presenting at the median age of 50 (range 33–95 years) and 72 (range 33–83 years), respectively (P=0.0006, two-tailed Student’s t-Test); (supplementary material, Table S1). Most (83.3%) cases were detected as multi-compartmental masses rendering the majority (86.7%) of patients to present at clinical stage IV. Multimodal treatment was administered in 83.3% cases and included surgery with radiation and/or chemotherapy (Table 1).
Table 1.
Clinical presentation and the outcome of patients with sinonasal carcinomas (Cohort 1).
| Case number |
Sex | Age | Location | Symptoms at presentation |
Diagnosis |
IDH2 status |
Size (cm) |
T stage |
N stage |
M stage |
Clinical Stage |
Primary treatment (post-recurrence treatment) |
Recurrence | Disease status at last follow- up (months) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 61 | Ethmoid sinus | Memory loss, personality changes | SNUC | Mutated | NA | cT4a | cN0 | cM0 | IVA | SX+RTCT | No | NED (102) |
| 2 | M | 57 | Ethmoid sinus and nasal cavity | Eye pain, proptosis | SNUC | Mutated | 5.1 | cT4b | cN0 | cM0 | IVB | SX+RTCX | Local recurrence and distant metastasis (bone, liver, abdominal lymph nodes) | DOD (6) |
| 3 | F | 49 | Ethmoid sinus and nasal cavity | Nasal congestion, headache | SNUC | Mutated | NA | cT4a | cN0 | cM0 | IVA | SX+RT | Regional metastasis (lymph nodes) and distant metastasis (liver) | AWD (13) |
| 4 | M | 48 | Nasal cavity | Double vision, swelling and proptosis of the eye, nasal obstruction | SNUC | Mutated | 6.2 | pT4b | cN2a | cM0 | IVB | SX+RTCX | Distant metastasis (bone) and regional metastasis (skin) | DOD (18) |
| 5 | M | 43 | Ethmoid sinus and nasal cavity | Nasal congestion, loss of smell and taste, supra-orbital headaches | SNUC | Mutated | 7 | cT4b | cN2c | cM0 | IVB | RTCX | No | NED (40) |
| 6 | M | 49 | Ethmoid sinus and nasal cavity | Nasal congestion, decrease in taste and smell | SNUC | Mutated | 5 | pT3 | cN0 | cM0 | III | SX+RTCX | No | NED (103) |
| 7 | F | 74 | Sphenoid sinus | Nasal congestion, epistaxis, headache | SNUC | wt | 3 | cT4a | cN0 | cM0 | IVA | NRTCX+SX | No | NED (96) |
| 8 | F | 72 | Nasal cavity | Visual changes, orbital pressure sensation | SNUC | Mutated | 4.5 | cT4b | cN0 | cM0 | IVB | RTCX | No | AWD (30) |
| 9 | M | 53 | Ethmoid sinus and nasal cavity | Nasal congestion | SNUC in situ | Mutated | N/A | pTis | cN0 | cM0 | 0 | SX (RTCX) | Local recurrence (soft palate) | NED (70) |
| 10 | M | 44 | Nasal cavity | Facial pain, swelling | SNUC | Mutated | 5.2 | cT4b | cN2 | cM0 | IVB | CX | Distant metastasis (adrenal gland) | DOD (15) |
| 11 | F | 83 | Ethmoid sinus and nasal cavity | Nasal obstruction, epistaxis | PD non-ITAC | Mutated | 2.5 | cT2 | cN0 | cM0 | II | SX+RT | No | DOUC (69) |
| 12 | M | 39 | Nasal cavity | Nasal obstruction | PDCA with focal glandular/acinar differentiation | Mutated | 5 | pT4a | cN0 | cM0 | IVA | NCX+SX+RTCX | Regional metastasis (lymph nodes) and local recurrence | DOD (33) |
| 13 | F | 65 | Nasal septum | Nasal obstruction | PDCA with focal glandular/acinar differentiation | wt | 2.8 | cT2 | cN0 | cM0 | II | SX+RT (RTCX for local recurrence) | Local recurrence, distant metastasis (bone) | DOD (34) |
| 14 | M | 49 | Maxillary sinus | Headaches, ear obstruction, maxillary and facial pain, diplopia, eye swelling | PDCA with focal glandular/acinar differentiation | wt | NA | cT4b | cN0 | cM0 | IVB | RTCX | No | AWD (14) |
| 15 | M | 47 | Ethmoid sinus | Headaches, blurred vision, facial pain, ear pain, eye swelling | PDCA with focal glandular/acinar differentiation | wt | 4.4 | cT4b | cN0 | cM0 | IVB | SX+RTCX | Local recurrence and distant metastasis (bone, liver, abdominal lymph nodes) | DOD (24) |
| 16 | M | 56 | Nasal cavity | Headache, weakness, poor appetite | PDCA with focal glandular/acinar differentiation | wt | 4.6 | cT4b | cN0 | cM1 | IVC | RTCX | Local recurrence and distant metastasis (liver) | DOD (16) |
| 17 | M | 51 | Sphenoid sinus | Headache, nasal congestion | PDCA with focal glandular/acinar differentiation | wt | 4.5 | pT4a | cN0 | cM0 | IVA | SX+RTCX | Local recurrence and distant metastasis (bone) | DOD (28) |
| 18 | M | 44 | Ethmoid sinus and nasal cavity | Nasal congestion, epistaxis | Non-ITAC, HG | wt | 6.5 | cT4a | cN0 | cM0 | IVA | SX+RTCX | No | NED (10) |
| 19 | F | 30 | Nasal cavity | Nasal congestion, visual changes, sensation deficits | PDCA with focal glandular/acinar differentiation | wt | 4.8 | pT4b | cN0 | cM0 | IVB | NCX+SX+RTCX (RT) | Local recurrence (orbit) and distant metastasis (spine, pleura, adrenal gland) | DOD (30) |
| 20 | M | 45 | Ethmoid sinus | Headache | HGNEC | Mutated | 3.1 | cT4a | pN2b | cM0 | IVA | SX (RTCX for local recurrence) | Local recurrence | NED (60) |
| 21 | F | 62 | Ethmoid sinus and nasal cavity | Sinus congestion, facial pain, occasional double vision | SCNEC | wt | 4.6 | cT4b | cN0 | cM1 | IVC | CX+RT | Local recurrence and distant metastasis (bone) | DOD (7) |
| 22 | F | 62 | Ethmoid sinus and nasal cavity | Nasal congestion, epistaxis | SCNEC | wt | 3.4 | pT4b | cN0 | cM0 | IVB | SX+RTCX | Local recurrence and distant metastasis (lung) | AWD (21) |
| 23 | M | 54 | Ethmoid sinus and nasal cavity | Nasal congestion | SMARCB1-deficient sinonasal carcinoma | wt | 2.6 | cT4a | cN0 | cM0 | IVA | SX (RTCX for local recurrence) | Distant metastasis (bone, brain) | DOD (39) |
| 24 | M | 47 | Ethmoid sinus | Nasal congestion, epistaxis, pain above eye | SMARCB1-deficient sinonasal carcinoma | wt | 4.0 | cT4b | cN0 | cM0 | IVB | NRTCX+SX | Local recurrence | AWD (7) |
| 25 | M | 54 | Ethmoid sinus and nasal cavity | Epistaxis | SMARCB1-deficient sinonasal carcinoma | wt | 4.5 | cT4b | cN0 | cM0 | IVB | NCX + SX | Local recurrence and distant metastasis (bone) | AWD (23) |
| 26 | M | 95 | Ethmoid sinus and nasal cavity | Progressive mental and gait deficit, behavioral changes | SMARCB1-deficient sinonasal carcinoma | wt | NA | cT4b | cN0 | cM0 | IVB | SX | N/A | N/A |
| 27 | F | 77 | Ethmoid sinus | Nasal congestion, obstruction, and purulent drainage | NUT midline carcinoma | wt | NA | cT4b | cN0 | cM0 | IVB | RT | Local recurrence and distant metastasis (bone, lung, mediastinum) | DOD (5) |
| 28 | F | 76 | Nasal cavity | Nasal congestion | MD-ITAC with signet-ring cells and mucinous features | wt | NA | cT4a | cN0 | cM0 | IVA | SX+RT | Local recurrence and regional metastasis (lymph nodes) | DOD (24) |
| 29 | M | 80 | Clivus/skull base | Visual changes, rhinorrhea | MD-ITAC | wt | 5.0 | cT4b | cN0 | cM0 | IVB | RTCX | No | AWD (7) |
| 30 | F | 81 | Nasal cavity | Epistaxis | PDCA with neuroendocrine and glandular differentiation | wt | 5.9 | cT4a | cN0 | cM0 | IVA | SX+RT | No | NED (37) |
Abbreviations: SNUC=sinonasal undifferentiated carcinoma, HGCA=high-grade carcinoma, PDCA=poorly-differentiated carcinoma, HGNEC=high-grade neuroendocrine carcinoma, large cell type, MD-ITAC=moderately-differentiated intestinal type adenocarcinoma, SCNEC=small cell neuroendocrine carcinoma, HG=high-grade,wt=wild type, SX=surgery, RT=radiation therapy, RTCX=chemoradiation, NCX=neoadjuvant chemotherapy, NRTCX=neoadjuvant chemoradiation, DOD=died of disease, AWD=alive with disease, NED=no evidence of disease, DOUC=died of unknown causes.
Genomic profile by MSK-IMPACT™
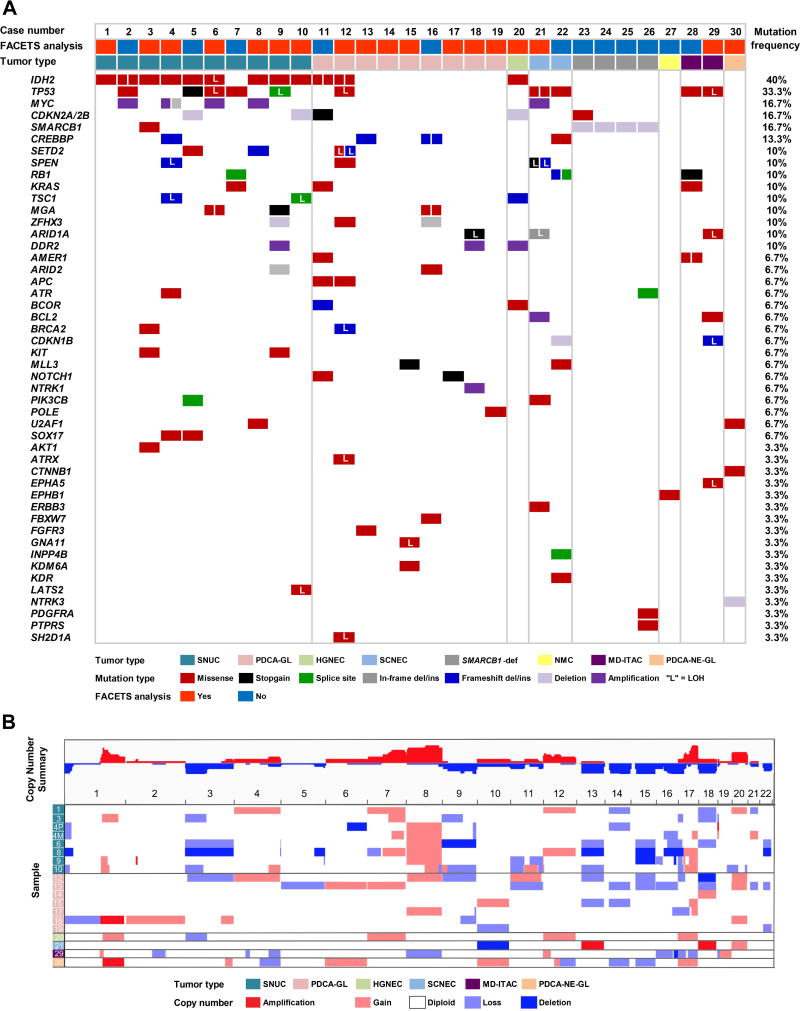
Most notably, IDH2 R172 mutations were detected in 12 (40%) carcinomas of various histologic types, and SMARCB1 deletions were detected in 4 (13.3%) cases. Frequent alterations involved tumor suppressor genes including TP53 (33.3%), CDKN2A/CDKN2B (16.7%), RB1 (10%) and TSC1 (10%). Loss of heterozygosity (LOH) was detected in 4 of 5 TP53-mutated cases, and in 2 of 3 TSC1-mutated carcinomas (Figure 2A). Aside from IDH2 and SMARCB1, recurrent alterations were detected in other epigenetically important genes including CREBBP (13.3%), and SETD2 (10%), and in the oncogenes MYC (16.7%) and KRAS (10%), including KRAS G12D variant in one SNUC; (Figure 2A). Copy number analysis revealed frequent gains involving chromosomes 8 and 20, and chromosome arms 1q and 17q; recurrent losses were detected in 3p, and in chromosomes 13, 14, 15, 16, and 18. Notably, the majority of SNUCs showed 8q gain (71.4%), and 3p loss (57.1%); (Figure 2B).
Figure 2.
Genetic alterations detected by MSK-IMPACT™ in sinonasal carcinomas (Cohort 1). A. Cases are represented in columns; genes are depicted in rows. Tumor types and mutation types are color-coded according to the legend. B. Spectrum of somatic copy number alterations identified in sinonasal carcinomas detected by FACETS [5]. Genome wide copy number alterations are color-coded according to the legend and summarized at the cohort level and by tumor type.
Abbreviations: SNUC=sinonasal undifferentiated carcinoma, PDCA-GL=poorly differentiated non-intestinal type adenocarcinoma/poorly-differentiated carcinoma with focal glandular/acinar differentiation, HGNEC=high-grade neuroendocrine carcinoma, large cell type, SCNEC=small cell neuroendocrine carcinoma, SMARCB1-def= SMARCB1-deficient sinonasal carcinoma, NMC=NUT midline carcinoma, MD-ITAC=moderately-differentiated intestinal type adenocarcinoma, PDCA-NE-GL=poorly differentiated carcinoma with neuroendocrine and glandular differentiation, LOH=loss of heterozygosity.
a. IDH2-mutated sinonasal carcinomas
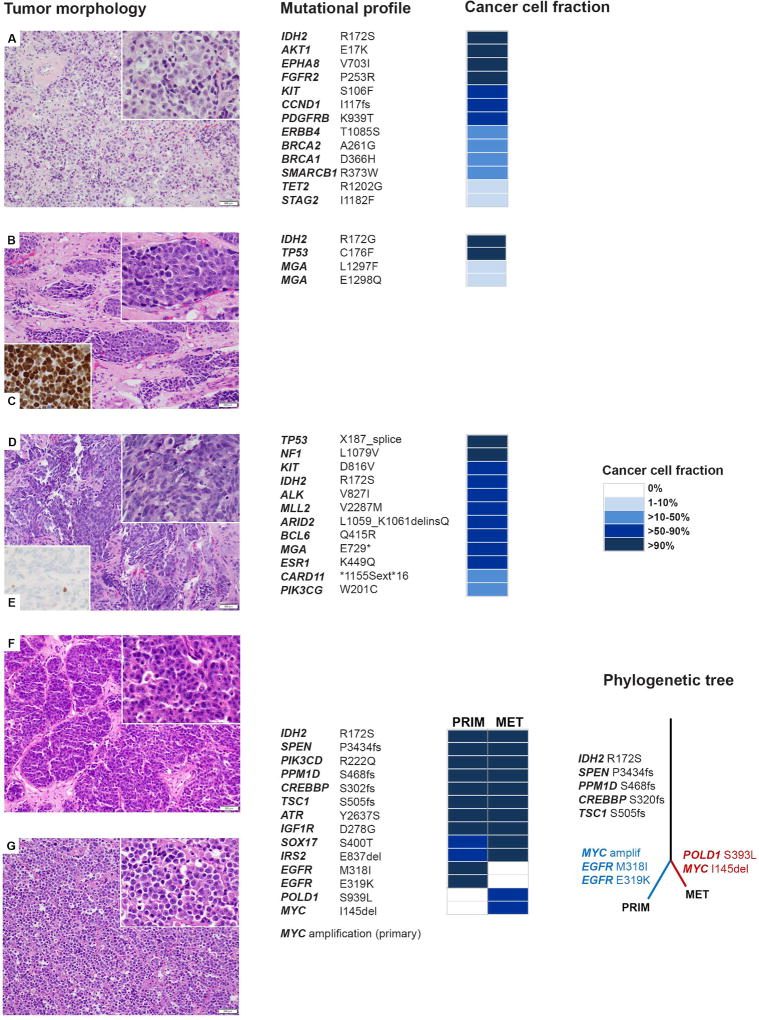
Activating hotspot IDH1/IDH2 mutations have been identified in various cancers. IDH1 mutations are predominant in solid tumors; gliomas [6], chondrosarcomas [7], and intrahepatic cholangiocarcinoma [8]. IDH2 mutations are more frequent in acute myeloid leukemia (AML) [9] and angioimmunoblastic T-cell lymphoma [10]. IDH1 (isocitrate dehydrogenase 1) and IDH2 (isocitrate dehydrogenase 2) are homodimeric enzymes that catalyze the conversion of isocitrate to α-ketoglutarate. The mutant protein, IDH1 or IDH2, loses its normal enzymatic activity and acquires a gain-of-function neomorphic enzymatic activity leading to abnormally increased levels of "oncometabolite" (R)-2-hydroxyglutarate [9]. In all except one low tumor purity sample, the CCF with IDH2 R172 mutation was high (average 0.87, range 0.4–1.0) (supplementary material, Table S4). In the case with matched metastasis, the CCF indicated the presence of IDH2 variant in (nearly) all cancer cells at both sites suggesting they are likely clonal (Figure 3). Among 12 IDH2-mutated cases, co-existing recurrent alterations were detected in TP53 (41.7%), CDKN2A/2B (33.3%), MYC (33.3%), SETD2 (25%), and SPEN (16.7%).
Figure 3.
Phenotype and genetics of IDH2-mutated SNUC. Histology of the IDH2-mutated SNUCs including the matched metastasis in case 4 (G, 200× magnification) are shown with their respective genome plots and mutational profiles. Large undifferentiated epithelial tumor cells display high nuclear/cytoplasmic ratio, and in most cases they show open chromatin and prominent cherry-red nucleoli (insets A, B, D, F and G, 400× magnification). In case 6, somatic mutation in TP53 is consistent with an increased nuclear expression of p53 protein (C, 400× magnification), whereas in case 9, TP53 splice site variant with loss of heterozygosity is compatible with an absence of p53 protein (E, 400× magnification). In case 4, on the far right, the phylogenetic tree illustrates the clonal evolution of the primary tumor and the metastasis. The length of branches corresponds to the number of somatic mutations that distinguishes the primary or metastatic clone from their parental clone [22].
Key to cases: A=case 3, B, C=case 6, D, E=case 9, F=case 4, primary tumor, G=case 4, metastasis. Abbreviations: PRIM=primary tumor, MET= metastasis, amplif=amplification.
Three (25%) cases harbored a second, non-hotspot IDH2 mutation. AKT1 E17K and KIT D816V hotspot variants were each detected in one case. IDH1/2 pathogenic mutations were often found to co-exist with mutations affecting other oncogenic pathways. Studies on IDH1/2-mutated gliomas give a notion that the glial cells may become susceptible to mutant IDH1 through activation of MAPK, PI3K/Akt, and MYC, and loss of p53 signaling [11]. We hypothesize that the concurrent genetic alterations in IDH2-mutated sinonasal carcinomas may collaborate with mutant IDH2 to induce and promote carcinogenesis in these tumors.
High frequency of IDH2 mutations in Cohort 1 prompted a testing of an extended set of sinonasal tumors for presence of IDH1/2 hotspot mutations by Sanger sequencing (Cohort 2, N=17). The frequencies and distribution of IDH2 mutations in both Cohorts (N=47) are provided in Figure 1. IDH2 R172S (58.8%), R172T (23.5%), R172M (11.8%) and R172G (5.9%) were found in 12 (70.6%) men (median age 49, range 39–85 years), and 5 (29.4%) women (median age 62, range 49–83 years); (supplementary material, Table S1). In a recent study of 11 SNUCs, Jo et al. found IDH2 R172 mutations in 55% cases [12]. In addition to the higher IDH2 mutation detection rate in SNUC, the results of our larger, histologically diverse cohort show that (1) IDH2 mutations can also be found in poorly-differentiated sinonasal carcinomas other than SNUC, (2) they are likely clonal (in most cases), and (3) frequently co-exist with MYC amplifications.
Clinical trials with established targeted therapies against IDH2 mutant proteins are in progress. Preliminary findings of an ongoing phase 1 clinical trial have shown that the mutant IDH2-inhibitor AG-221 produces clinical responses in about 40% of patients with AML and myelodysplastic syndrome [13]. These results may provide rationale for taking a similar approach in treatment of IDH2-mutated sinonasal carcinomas.
Morphologically, except for one poorly-differentiated non-intestinal type adenocarcinoma, all IDH2-mutated tumors were arranged in sheets or lobules. Irrespective of the final diagnosis, they shared very similar cytological features and consisted of large undifferentiated epithelial tumor cells in most cases (Figure 3, supplementary material, Figure S1). However, they were not notably morphologically distinct from their IDH2-wild type histological counterparts (supplementary material, Figure S2). Functional studies on different cell lines including murine bone marrow cells, adipocytes, and astrocytes showed that increased intracellular (R)-2-hydroxyglutarate lead to hypermethylation of target genes and the subsequent block in cellular differentiation [14–16]. IDH2 mutations in sinonasal carcinomas may have similar consequences leading to block in cellular differentiation and resulting in either undifferentiated or poorly-differentiated carcinomas in this location.
The outcome in respect to the IDH2 mutation status is provided in Table 1. Although IDH2-mutated sinonasal carcinomas were associated with a trend of improved disease free survival and overall survival, such trend did not reach significant level (log rank test, P=0.112 and P=0.145, respectively); (supplementary material, Figure S3).
b. SMARCB1-deficient sinonasal carcinomas
SMARCB1 deletions detected in Cohort 1 occurred in 4 men (age range 47–95 years); (Table 1). SMARCB1-deficient sinonasal carcinomas displayed a relative paucity of co-existing mutations emphasizing the oncogenic role of SMARCB1 loss in these tumors (Figure 2, supplementary material, Table S5).
c. Small cell neuroendocrine carcinoma (SCNEC) of the sinonasal tract
In one SCNEC, ERBB3 V104L co-existed with TP53 mutations, and BCL2 and MYC amplifications. In another, along with TP53 and CREBBP mutations, there were two pathogenic RB1 mutations and one likely loss-of-function INPP4B mutation (Figure 2, supplementary material, Figure S4). Mutations of TP53 and RB1, up-regulation of BCL2 signaling, and activation of MYC and PI3K pathways are frequent in pulmonary small cell carcinomas [17]. A notable similarity in the mutational profiles of sinonasal SCNEC and its lung counterpart supports their common genetic background. Novel findings, ERBB3 V104L oncogenic mutation [18] and INPP4B loss-of-function mutation may represent alternate mechanisms of PI3K/Akt pathway activation [19] in sinonasal SCNECs.
In conclusion, given the established knowledge on the biological significance of IDH1/2 mutations in other cancer types, our results suggest the IDH2 R172 mutations may play an important role in carcinogenesis of SNUC. IDH2-mutated sinonasal carcinomas may be amenable to IDH2-targeted therapy.
Supplementary Material
Figure S1. Phenotype and genetics of IDH2-mutated sinonasal carcinomas
Table S5. Genetic alterations with variant frequencies in sinonasal carcinomas detected by MSK-IMPACT™ in sinonasal carcinomas (Cohort 1).
Table S6. FACETS analysis: significant copy number variations at the gene level.
Table S7. FACETS analysis: significant copy number variations at the chromosome arm level.
Figure S2. Morphology and immunophenotype of IDH2 wild-type sinonasal carcinomas
Figure S3. Kaplan-Meier curves for disease specific survival (A) and overall survival (B) of sinonasal carcinoma patients (Cohort 1) in respect to the IDH2 mutation status
Figure S4. Morphology and genetics of small cell neuroendocrine carcinoma of the sinonasal tract
Table S1. Initially rendered diagnoses and revised (final) diagnoses with ancillary studies results (Cohort 1; case 1–30, and Cohort 2; case 31–47).
Table S2. Antibodies used for immunohistochemical studies and in situ hybridization probes.
Table S3. PCR and sequencing primers for exon 4 of IDH1 and IDH2.
Table S4. Mutational profile with clonal heterogeneity assessment and loss of heterozygosity status in sinonasal carcinomas (Cohort 1).
Acknowledgments
We would like to thank Dr. Diane L. Carlson from Cleveland Clinic, Weston, FL and Dr. William H. Westra from The Johns Hopkins Hospital, Baltimore, MD for their help in collecting the study cases. We also thank Allyne Manzo, our expert photographer at MSKCC for her assistance in arranging the microphotographs. Research reported in this publication was supported in part by the Farmer Family Foundation and the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure Statement:
No competing financial interests exist for all contributory authors.
STATEMENT OF AUTHOR CONTRIBUTIONS
SD and MFB conceived the study, analyzed and interpreted the data. SD, DJC, NK, RAG, JAB, SIC, EBS provided samples. SD, NK, RAG and DJC performed histological review. RP performed FACETS analysis. BX performed statistical analysis. SD, DJC, BX, JAB, SIC, EBS, IG, DGP acquired the data. MFB, RC, KN and JC-M carried out experiments and analyzed the data. SD, MFB and RAG supervised the study. All authors were involved in writing the paper and had final approval of the manuscript.
References
* Reference cited in supplementary material only
- 1.Frierson HF, Mills SE, Fechner RE, et al. Sinonasal undifferentiated carcinoma. An aggressive neoplasm derived from the Schneiderian epithelium and distinct from olfactory neuroblastoma. Am J Surg Pathol. 1986;10:771–779. [PubMed] [Google Scholar]
- 2.Lin EM, Sparano A, Spalding A, et al. Sinonasal undifferentiated carcinoma: a 13-year experience at a single institution. Skull Base. 2010;20:61–67. doi: 10.1055/s-0029-1236165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuo P, Manes RP, Schwam ZG, et al. Survival outcomes for combined modality therapy for sinonasal undifferentiated carcinoma. Otolaryngol Head Neck Surg. 2017;156:132–136. doi: 10.1177/0194599816670146. [DOI] [PubMed] [Google Scholar]
- 4.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amary MF, Bacsi K, Maggiani F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 8.Borger DR, Tanabe KK, Fan KC, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns RA, Iqbal J, Lemonnier F, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119:1901–1903. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heuser M, Araujo Cruz MM, Goparaju R, et al. Enigmas of IDH mutations in hematology/oncology. Exp Hematol. 2015;43:685–697. doi: 10.1016/j.exphem.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Jo VY, Chau NG, Hornick JL, et al. Recurrent IDH2 R172X mutations in sinonasal undifferentiated carcinoma. Mod Pathol. 2017 Jan 13; doi: 10.1038/modpathol.2016.239. [DOI] [PubMed] [Google Scholar]
- 13.DiNardo C, Stein EM, Altman JK, et al. AG-221, an oral, selective, first-in-class, potent inhibitor of the IDH2 mutant enzyme, induced durable responses in a phase 1 study of IDH2 mutation-positive advanced hematologic malignancies [abstract]. Proceedings of the 20th Congress of the European Hematology Association; 2015 Jun 11–14; Vienna, Austria. The Hague (the Netherlands): EHA; 2015. Abstract nr P569. [Google Scholar]
- 14.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C, Venneti S, Akalin A, et al. Induction of sarcomas by mutant IDH2. Genes Dev. 2013;27:1986–1998. doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29:1447–1462. doi: 10.1101/gad.263145.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang MT, Asthana S, Gao SP, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ip LR, Poulogiannis G, Viciano FC, et al. Loss of INPP4B causes a DNA repair defect through loss of BRCA1, ATM and ATR and can be targeted with PARP inhibitor treatment. Oncotarget. 2015;6:10548–10562. doi: 10.18632/oncotarget.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *20.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *21.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Guerini-Rocco E, Piscuoglio S, Ng CK, et al. Microglandular adenosis associated with triple-negative breast cancer is a neoplastic lesion of triple-negative phenotype harbouring TP53 somatic mutations. J Pathol. 2016;238:677–688. doi: 10.1002/path.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phenotype and genetics of IDH2-mutated sinonasal carcinomas
Table S5. Genetic alterations with variant frequencies in sinonasal carcinomas detected by MSK-IMPACT™ in sinonasal carcinomas (Cohort 1).
Table S6. FACETS analysis: significant copy number variations at the gene level.
Table S7. FACETS analysis: significant copy number variations at the chromosome arm level.
Figure S2. Morphology and immunophenotype of IDH2 wild-type sinonasal carcinomas
Figure S3. Kaplan-Meier curves for disease specific survival (A) and overall survival (B) of sinonasal carcinoma patients (Cohort 1) in respect to the IDH2 mutation status
Figure S4. Morphology and genetics of small cell neuroendocrine carcinoma of the sinonasal tract
Table S1. Initially rendered diagnoses and revised (final) diagnoses with ancillary studies results (Cohort 1; case 1–30, and Cohort 2; case 31–47).
Table S2. Antibodies used for immunohistochemical studies and in situ hybridization probes.
Table S3. PCR and sequencing primers for exon 4 of IDH1 and IDH2.
Table S4. Mutational profile with clonal heterogeneity assessment and loss of heterozygosity status in sinonasal carcinomas (Cohort 1).