Abstract
Drug-drug interactions (DDIs) are known to be responsible for nearly a third of all adverse drug reactions. Hence several current efforts focus on extracting signal from EMRs to prioritize DDIs that need further exploration. To this end, being able to extract explicit mentions of DDIs in free text narratives is an important task. In this paper, we explore recurrent neural network (RNN) architectures to detect and classify DDIs from unstructured text using the DDIExtraction dataset from the SemEval 2013 (task 9) shared task. Our methods are in line with those used in other recent deep learning efforts for relation extraction including DDI extraction. However, to our knowledge, we are the first to investigate the potential of character-level RNNs (Char-RNNs) for DDI extraction (and relation extraction in general). Furthermore, we explore a simple but effective model bootstrapping method to (a). build model averaging ensembles, (b). derive confidence intervals around mean micro-F scores (MMF), and (c). assess the average behavior of our methods. Without any rule based filtering of negative examples, a popular heuristic used by most earlier efforts, we achieve an MMF of 69.13. By adding simple replicable heuristics to filter negative instances we are able to achieve an MMF of 70.38. Furthermore, our best ensembles produce micro F-scores of 70.81 (without filtering) and 72.13 (with filtering), which are superior to metrics reported in published results. Although Char-RNNs turnout to be inferior to regular word based RNN models in overall comparisons, we find that ensembling models from both architectures results in nontrivial gains over simply using either alone, indicating that they complement each other.
I. Introduction
Adverse drug reactions (ADRs) have been a major concern as polypharmacy became more common in modern medical practice [1]. ADRs may lead to hospitalization and/or extend the lengths of stay for already admitted in-patients [2]. Drug-drug interactions (DDIs) represent an important category of ADRs. Specifically, a drug interaction is said to occur “when the effects of one drug are changed by the presence of another drug, herbal medicine, food, drink or by some environmental chemical agent” [1]. The result of DDIs can be unexpected failure of therapy [3] due to reduction in efficacy or more direct harm due to increase in toxicity of a drug. Although manually curated databases that discuss DDIs exist [1], [4], most of the up-to-date information is still latent in unstructured text. Thus it is important to extract such interactions as they are presented as findings in research articles, warnings in drug labels, or observations in clinical notes [5]. The 2013 DDI extraction challenge [6], [7] introduced a new dataset and challenge to extract mentions of such interactions from free text narratives. A few recent efforts focused on employing deep neural networks (or deep nets) for extracting DDIs using the SemEval challenge dataset and demonstrated improvements over linear models. In this paper, we use character-level recurrent neural networks (Char-RNNs) along with conventional word-level RNNs to extract DDIs using the 2013 DDIExtraction dataset [6].
II. 2013 DDIExtraction Challenge Dataset
In this section, we briefly outline the characteristics of the dataset used in this task. Two different databases, Medline abstracts and DrugBank [4] narratives, were used to identify sentences on the subject of DDIs to create the DDI corpus used for the SemEval 2013 shared on DDI extraction [8]. Pharmacological substances and four different types of DDI manifestations in text were annotated for a total of 792 documents. The different types of interactions annotated in descending order of their frequencies are
mechanism, where the pharmacokinetic mechanism is explicitly discussed (e.g., “ Ethanol decreases the elimination of abacavir causing an increase in overall exposure”),
effect, where a consequence of an interaction (pharmacodynamic aspect) is specified (e.g., “the antihypertensive effect of losartan may be blunted by the non-steroidal anti-inflammatory drug indomethacin”)
advice, where suggestions regarding handling a drug interaction are made in text (e.g., “Patients should be warned of the potential danger of the self-administration of benzodiazepines while under treatment with Suboxone”)
int, where a DDI is discussed without any specific additional information.
All pairs of drugs mentioned in each sentence are separately annotated as either participating in any of these four types of interactions or simply declared as false indicating there is no interaction. For this study, following other efforts in DDI extraction, we assume the drug mention spans are already provided and thus the main task is to classify the type of interaction (any of the four types above or false if no interaction exists). The total number of candidate drug pairs in the training dataset is 27,792 out of which ≈ 15% are positive examples assigned to one of the four classes mentioned earlier. The test set has 5,716 candidate pairs and around 17% are positive examples.
III. Related Work
The DDI extraction task is a special case of binary relation extraction where (subject, predicate, object) triples are extracted from natural language. In this case both the subject and object are pharmacological substances and the predicate is the type of interaction discussed in Section II. The top two teams [9], [10] in the competition used a variety of techniques but the two commons themes were usage of interesting kernels for support vector machine (SVM) models and application of negative instance filtering using straightforward rules (more later). The top team [9] used a two stage approach where a binary SVM classifier first rules out sentences as “less informative” when they are not expected to contain valid DDIs. The second stage relation classifier uses a hybrid kernel that combines a feature-based, a shallow linguistic, and a tree kernel for the SVM model. The second ranking team [10] also followed the two stage setup (detection followed by classification) using a majority voting approach involving SVM models with interesting shallow linguistic and tree kernels. After the competition, Kim et al. [11] used a more involved set of features including n-grams based on shortest dependency and constituency paths connecting candidate pairs. They used these features with the one-against-one approach for muticlass modeling with SVMs using a linear kernel. Using a graph kernel based on the so called context vectors defined using the dependency graphs of sentences, Zheng et al. [12] used SVMs to extract DDIs.
Over the past couple of years, the resurgence of deep nets has renewed the interest in using such methods for DDI extraction, in particular using the DDIExtraction dataset. Here we outline some of these recent attempts. Liu et al. [13] represent the first team to work along these lines using a convolutional neural network (CNN) that uses dense word vectors to represent the document as a matrix that is subsequently convolved over using multiple convolution filters (CFs). They also used position vectors as additional information for each word based on its position relative to the two candidate drugs in the sentence. They use max-over-time pooling and finally predict the interaction type using a softmax output layer. The same team also extended this initial effort using convolutions over shortest dependency paths connecting the mentions of the drugs [14]. Zhao et al. [15] trained word embeddings using the shortest dependency paths and employed a CNN model with word, position, and additional part-of-speech (POS) tag embeddings. Recently, Suárez-Paniagua et al. [16] explored CNN architectures with different parameter settings including word and position vector dimensions and CF sizes. They, however, concluded that their results are not better than those achieved through more complex deep net models by other published efforts.
All prior efforts we discussed thus far move the state-of-the-art but we identify the following gaps:
Although CNNs are powerful, given the inherent sequential nature of sentences and differences in the relative importances of words closer to the drug mentions, recurrent neural networks (RNNs) also offer an important alternative. Moreover, RNNs are already popular for relation extraction in other domains [17], [18] but it appears there are no prior results on using them for DDI extraction.
Additionally, the role of Char-RNNs appears completely unexplored for relation extraction in general, not just for DDIs. Although character-level embeddings are intuitively more useful for morphologically richer languages unlike English, they might be more suitable for relation extraction given they can model tense/voice variations.
The training process of deep nets with many randomly initialized parameters is not deterministic in that for a fixed train and test set split of a dataset, different runs involving building a model with the train set and evaluating it on the test set do not result in the same performance. Depending on the initial parameters chosen, one may get (un)lucky in the eventual model's performance. Typically model averaging is employed to arrive at more stable models and may also be used to study the average behavior of deep nets' performance. These aspects are also missing in prior studies on DDI extraction.
In this paper, we address these gaps using the DDIExtraction dataset by exploring both regular RNN and Char-RNN based hierarchical architectures with model averaging and bootstrapping to study average behavior.
IV. Introduction to RNNs and LSTMs
Unlike feedforward networks like CNNs, RNNs have cyclical connections and are more suitable for natural language processing (NLP) tasks where the meaning of a text segment is naturally dependent on what occurred in the narrative before it. Typically, RNNs recurrently compose word vectors [19] of a sentence from left to right, effectively letting information persist from the history of previously seen words. There is usually an input layer, a hidden layer that is connected to itself, and an output layer. The hidden layer's output is fed back to itself at consecutive time steps (generally as many times as there are words in the narrative) and the output at any time step is generally the recurrent composition of information until that point. Parameter optimization is implemented through the so called back propagation through time because of the “unfolding” of the cyclical connections in the hidden layer through different time steps. For a thorough treatment of RNNs, we encourage the reader to refer to a popular resource by Graves [20, Chapter 3]. In the context of conventional RNNs for NLP, the input at each time step is the vector corresponding to the next word in the narrative. The output is the context vector that composes word vectors that include all previous words and itself using the RNN architecture.
To exploit signals that come from the future part of a sentence in interpreting the current word, running the RNN from right to left over the input text can yield additional contextual hints for eventual prediction tasks. This resulted in bi-directional RNNs (BiRNNs) which essentially have two separate RNNs, each with its own parameters, capturing the context at each position from both directions. The output at each time step is a combination of output vectors from both RNNs typically produced via concatenation. To handle the problem of vanishing gradients [21] in regular RNNs, a more involved hidden layer with the so called long short-term memory (LSTM) units [22], [23] has become popular, especially for NLP. The state representation in an LSTM unit includes an explicit memory cell access to and use of which is controlled through three gates – first to control how much of the next input to incorporate in the memory (input gate), second to determine to what extent the current memory is to be forgotten (forget gate), and third to limit the extent of information from the current memory cell to propagate to the output state (output gate). These three gates control the flow of information based on the previous output and cell state via sigmoid outputs ∈ [0, 1]. In this effort, we use BiRNNs with LSTM units (simply termed BiLSTMs) in the hidden layer as the main neural architecture with specific LSTM details outlined by Goldberg [24, Section 11].
V. Word and Character-Level RNNs
In this section, we present details of the two RNN architectures used in our study. The main architecture proposed is shown in Figure 1.
Fig. 1. Word and Character-Level RNN for DDI Extraction.
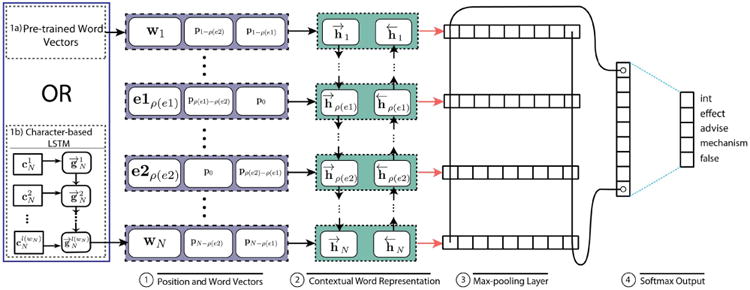
A. Instance Preprocessing
Before we proceed further, we discuss some basic preprocessing steps. Each instance for classification comes with a particular sentence and a pair of drugs mentioned in it. Given a sentence might contain more than two drugs, it might contain multiple candidate pairs (precisely n(n – 1)/2 pairs if n drugs are mentioned) for which classification is needed. We first convert all the sentences in the dataset into lowercase. Subsequently, we perform so called entity blinding by replacing the first drug (considered left to right) in the sentence with the upper case string “DRUGA” and the second drug span with the string “DRUGB”. All other drug mentions are replaced with “DRUGN”. For example, consider the drug pair benzodiazepines and subutex in the sentence “Patients should be warned of the potential danger of the intravenous self-administration of benzodiazepines while under treatment with suboxone or subutex”. The modified instance passed to the model is: “patients should be warned of the potential danger of the intravenous self-administration of DRUGA while under treatment with DRUGN or DRUGB”.
B. Word-Level RNNs with Position Vectors
In this model, the input to the neural network is an input sentence tokenized into N words including the special drug mention tokens as outlined in Section V-A. Such a sentence is represented as a sequence of word embeddings, [w1, …, e1, …, e2, …, wN], where wi is the word embedding for the i-th word and e1 and e2 are the special token vectors for DRUGA and DRUGB respectively. In addition to word vectors, we use position vectors first proposed by Zeng et al. [25] for relation classification and found useful by all deep net efforts discussed in Section III for DDI extraction. The idea is to use low dimensional vectors that represent the relative positions of a word with respect to the two drug mentions. These are denoted by pj where j is an integer that represents the positional difference with regards to a specific drug mention. For each word, we have two position vectors corresponding to its location with respect to the two candidate drugs. These vectors are concatenated to the word vector and input to the RNN at each time step. The position vectors are randomly initialized and are learned along with the word vectors during the training process. For the example sentence at the end Section V-A, the position vectors for the word “treatment” with respect to DRUGB and DRUGA are p−4 and p3 respectively. The position vectors concatenated with word vectors are as outlined in component ① of Figure 1, where ρ(e) ∈ ℤ+ is the position of the drug mention e. The final set of input vectors passed to the RNN is thus [x1, …, xN] where
| (1) |
with wi being the word vector for the word at position i and ‖ denoting the concatenation operation. We note that wi could be the vector for the tokens DRUGA, DRUGB, or DRUGN when the corresponding drug mention occurs at the i-th position.
With this setup, we first use a BiLSTM layer at the word level to capture the contextual information of the sentence with respect to each word. Concretely,
| (2) |
where h⃗i, h⃖i ∈ ℝd and hi ∈ ℝ2d such that d (a network hyperparameter) is the number of output units at each LSTM time step. Also, Word-LSTM is functionally identical to a traditional LSTM cell (without peepholes) and the prefix “Word” is used only to indicate that its input is based on word vectors. This forms the component ② of Figure 1.
Next, we perform a max-pooling operation over all hi vectors to produce the feature vector where represents the maximum value across all N BiLSTM word representations for the dimension i. This corresponds to the component ③ of Figure 1. Finally, we use a fully-connected output layer with m outputs, where m corresponds to the number of interaction types (here m = 5). The output is computed as
where q ∈ ℝm and 𝒲q ∈ ℝm×2d and bq ∈ ℝm are additional network parameters. In order to get a categorical distribution, we apply the softmax function to the vector q to obtain
where pj ∈ [0, 1] is the probability estimate of the label at index j forming the final component ④ of Figure 1.
C. Character-Level RNNs
Char-RNNs are popular for modeling morphologically richer languages [26] and solving NLP tasks such as POS tagging for such languages [27] often with substantially fewer network parameters. Although they were explored for text classification [28], we do not see prior studies applying them for relation extraction. To address this gap, in this effort, we assess their effectiveness for the DDI extraction use-case through a hierarchical character and word based RNN architecture.
The architecture of Char-RNNs we use is identical to that of the word based model (Figure 1) except the word vectors are derived using an LSTM on character embeddings instead of using pre-trained word embeddings. That is, the word embeddings are composed of embeddings for the constituent characters. Specifically, let the word
where is the j-th character of word wi and l(wi) is its length (number of characters). For wi, we feed its character embeddings into a forward LSTM with k (equal to the dimensionality of word vectors) output units such that
where is the output at time step t and Char-LSTM is a regular LSTM but indicates the particular instance that processes character embeddings in contrast with word level BiLSTM in equation (2). The output state at the last step encodes the left-to-right context accumulating at the last character and is used as the word embedding
| (3) |
for concatenation with position vectors in right hand side of equation (1). This is conveyed in step
 on the left bottom portion of Figure 1. Thus instead of pre-trained word vectors, component
on the left bottom portion of Figure 1. Thus instead of pre-trained word vectors, component
 of Figure 1, we use randomly initialized character embeddings, which are modified during the training process, to form word vectors. Except for these changes, the rest of the architecture of the end-to-end Char-RNN deep net for DDI extraction is identical to the components ② – ④ in Figure 1 with details as in Section V-B. This particular architecture is motivated by the hierarchical LSTM model (https://github.com/tensorflow/fold/blob/master/tensorflow_fold/g3doc/blocks.md) of the Tensor-Flow Fold framework [29]. As such, our setup is different from traditional approaches where Char-RNNs are used for language modeling or sequence tagging [26], [27].
of Figure 1, we use randomly initialized character embeddings, which are modified during the training process, to form word vectors. Except for these changes, the rest of the architecture of the end-to-end Char-RNN deep net for DDI extraction is identical to the components ② – ④ in Figure 1 with details as in Section V-B. This particular architecture is motivated by the hierarchical LSTM model (https://github.com/tensorflow/fold/blob/master/tensorflow_fold/g3doc/blocks.md) of the Tensor-Flow Fold framework [29]. As such, our setup is different from traditional approaches where Char-RNNs are used for language modeling or sequence tagging [26], [27].
VI. Experiments and Results
Next we describe our experimental setup and present results obtained using architectures described in Section V.
A. Negative Instance Filtering
All prior results from Section III on the DDI dataset (Section II) use some form of negative instance filtering. This is natural given a few straightforward types of drug pair mentions do not constitute DDIs. For example, when describing hypernymic relations using the construct “DRUGA such as DRUGB”, it is clear that this particular instance does not represent an interaction between the drugs. However, prior studies outline only general guidelines for filtering without unambiguously specifying all rules in an exhaustive manner. This hinders replicability for other follow-up efforts. In our current effort, we conduct experiments without any filtering and also apply some minimal filtering fully specified by the following patterns that capture certain coordination structures, hypernymic relations, and other constructs inspired from a prior effort by Kim et al. [11].
“^DRUGA :”, “^DRUGB :”,
“DRUGA, DRUGB”, “DRUGB, DRUGA”,
“DRUGA (DRUGB)”, “DRUGB (DRUGA)”,
“DRUGA, DRUGB, and DRUGN”,
“DRUGB, DRUGA, and DRUGN”,
“DRUGN, DRUGB, and DRUGA”,
“DRUGA, DRUGN, and DRUGB”,
“DRUGN, DRUGA, and DRUGB”,
“DRUGN, DRUGB, and DRUGA”,
“DRUGA such as DRUGB”,
“DRUGB such as DRUGA”,
“DRUGA such as DRUGN or DRUGB”, and
“DRUGB such as DRUGN or DRUGA”
The idea is to automatically classify any instance as false if it matches one of these patterns. However, this could lead to misclassification of some positive instances leading to false negatives (FNs). However, these specific rules weed out many more potential false positives (FPs) than the number of new FNs they incur. Table I shows the exact numbers of instances we have in the dataset before and after applying the filters. As we can see, these rules result in a major reduction in negative instances while incurring a very small dip in positive instances.
Table I. Counts before and after negative instance filtering.
| Class | Training | Test | ||
|---|---|---|---|---|
|
|
|
|||
| Before | After | Before | After | |
| Total Pairs | 27792 | 23338 | 5716 | 4872 |
| Negative DDIs | 23772 | 19342 | 4737 | 3896 |
| Positive DDIs | 4020 | 3996 | 979 | 976 |
|
| ||||
| Mechanism | 1319 | 1309 | 302 | 301 |
| Effect | 1687 | 1676 | 360 | 358 |
| Advise | 826 | 824 | 221 | 221 |
| Int | 188 | 187 | 96 | 96 |
B. Model Configuration Details
The following are the implementation choices made for the regular word based RNN model based on experimentation and best practices from other efforts. This architecture was implemented using the Theano [30] library.
We ran Google's word2vec [19] system on Medline citations (2014 PubMed baseline) to obtain 300-dimensional pre-trained word vectors, which are used as initial vectors to populate a sentence matrix. The tokenizer used is a simple splitter on non-word characters (those excluding the English alphabet, ten digits, and underscore symbol). For words in the training dataset that do not have pre-trained word vectors, we initialized each of their elements randomly by drawing from a uniform distribution with values ranging between the minimum and maximum values for that element among pre-trained vectors. The special tokens for DRUGA, DRUGB, and DRUGN were randomly initialized using the same approach.
The position vector dimensionality was set to 32 with each element initialized from the uniform distribution U(0, 0.01). The number of position vectors was determined based on the maximum position variation observed in the training sentences. Positions with respect to DRUGA (DRUGB) that are beyond those observed during training were assigned the vector corresponding to the farthest position encountered during training for DRUGA (DRUGB).
The output dimensionality of each LSTM is 512, leading to 1024 features input to the softmax layer given there are two LSTMs. The BiLSTM weight matrices and the weight matrix 𝒲q were initialized to values drawn from a normal distribution with mean 0 and standard deviation where the input size is 364 (300 for word vectors and 32 each for two position vectors) for the LSTMs and 1024 for the softmax layer. The bias vector bq was initialized to the zero vector and the LSTM bias vectors were initialized to [1, …, 1].
AdaGrad [31] was used for optimization with an initial learning rate of 0.01. The mini-batch size was set to 50 instances and sentences were zero padded at the end based on the length of the longest sentence in the training dataset. The number of epochs used was 25 with an early stopping criterion where we stopped training if there were five consecutive epochs in the training procedure that did not increase the validation micro F-score. The dropout parameter was set to 0.5.
Next we discuss empirical implementation choices made for the Char-RNN based hierarchical LSTM model discussed in Section V-C and implemented using TensorFlow Fold [29].
Given word representations are based on character embeddings in this model, we don't have pre-trained word embeddings to incorporate. We used 128 ASCII characters and embedded them as 250 dimensional vectors. Although it is atypical to have the dimensionality to be higher the vocabulary size, our experiments indicated that higher dimensionality was essential to achieve reasonable performance. The dimensionality of Char-LSTM output was set to 250, which thus also becomes the word vector size given the setup in equation (3). The Word-LSTM output dimensionality was determined to be 250 and hence we have 500 input features for the softmax layer. All parameters were initialized according to TensorFlow's recommended guidelines based on the input size (https://www.tensorflow.org/api_docs/python/tf/uniform_unit_scaling_initializer).
The position vector dimensionality was set to 32 and positions farther than those observed during training were handled in the same way at test time as we outlined earlier for regular RNNs.
We used AdaGrad [31] for optimization with initial learning rate of 0.5 and initial gradient accumulator value of 0.1, the default value in TensorFlow. The mini-batch size was set to 50 instances and zero padding was not necessary given the dynamic computations allowed by TensorFlow Fold. We used 30 epochs and stored the best model that has the best micro F-score on the validation dataset. The dropout parameter was set to 0.2.
C. Bootstrapped Model Averaging
As discussed toward the end of Section III, due to large number of local minima owing to larger parameters spaces, a single end-to-end deep net tends to converge to different solutions under different parameter initializations. Although the error rates may be similar, the actual errors made might differ among the resulting models. Hence it is well known that ensembling several models built from the same architecture results in higher performance than using the constituent individual models [32], [33]. In our prior efforts in text classification with deep nets, we consistently observed performance gains with ensembling via voting [34] or through model averaging [35], [36]. In model averaging, we predict the final class based on the average of the class probability estimates of multiple (typically 10–20) deep net models trained using the same architecture but with different parameter initializations.
In this study, we build 20 regular RNN models and 20 Char-RNN based hierarchical models with different parameter initializations and subsets of the training data using 10% of it for validation and 90% for actual model building. To study the stability (and variance) of the model averaged ensembles, we randomly considered 10,000 such ensembles of ten models each from Char-RNN models, regular RNN models, and their union where five were selected from each group. Note that the number of combinations for ten models for the individual architectures is 20C10 and for the mixed setup is (20C5)2. The mean precision, mean recall, and mean micro-F score (MMF) with 95% confidence interval along with its standard deviation are shown in Table II. Among all ensembles, the table also shows the maximum and minimum F-scores achieved per ensemble type.
Table II. Mean scores of 10,000 different ten model ensembles on the DDIExtraction 2013 test set.
| Neg Filter | Ensemble Type | Mean P | Mean R | 95% CI of MMF | MMF σ | Min F | Max F |
|---|---|---|---|---|---|---|---|
| ✗ | Char-RNNs | 77.56 | 60.32 | 67.86 ± 0.00832 | 00.42 | 66.28 | 69.40 |
| Word-RNNs | 74.90 | 62.46 | 68.11 ± 0.00768 | 00.39 | 66.67 | 69.31 | |
| Both RNNs | 77.78 | 62.21 | 69.13 ± 0.00919 | 00.47 | 67.09 | 70.81 | |
|
| |||||||
| ✓ | Char-RNNs | 75.80 | 61.34 | 67.80 ± 0.00885 | 00.45 | 66.25 | 69.45 |
| Word-RNNs | 76.02 | 64.44 | 69.75 ± 0.01103 | 00.56 | 67.79 | 71.22 | |
| Both RNNs | 78.56 | 63.76 | 70.38 ± 0.00963 | 00.49 | 68.28 | 72.13 | |
We first make two interesting observations from Table II:
The standard deviation σ of micro F-scores is < 0.5 except for row five where it is just over 0.5. This indicates that ensemble models are highly consistent in their performance.
The widths of the 95% confidence intervals (CIs) around the MMFs are also very small (mostly < 0.01). This indicates that the true MMFs are expected to lie in extremely tight intervals around the corresponding sample MMFs. This implies the sample means are very reflective of the average behavior of ensemble models.
Rows 5–6 when compared with rows 2–3 show that negative instance filtering (from Section VI-A) consistently improves performance given the corresponding CIs do not overlap for Word-RNNs and the combined ensemble type. For the same reason, with or without filtering, we also observe consistent improvements when building ensembles with both character and word level models over those built just with either type. Furthermore, word model ensembles are better than their character counterparts, which is not surprising given direct representation of words is known to better capture their semantics at the expense of a large set of parameters to learn. Nevertheless it is interesting to see that by just using 128 character embeddings we are able to achieve scores close to those achieved by models where each word has its own representation. Also noteworthy is the ability of Char-RNNs to complement word based models in building better joint ensembles. From the last two columns we observe that the F-score values have a range of 3 to 4 points even if σ is small. Thus there could be better ensembles in the short tail portion. For instance, the best MMF value we have is 70.38 corresponding to the last row but the best micro F-score achieved for that ensemble type is 72.13 which is close to 2 points away from the mean.
In Table III, we compare our models with others described in Section III. The first two rows represent models that participated in the original competition and were blinded to the test set completely. The remaining rows represent efforts after the competition when the full dataset was released. We note that the winning team's entry [9] still has a high recall of 65.60 compared with rest of the subsequent attempts. Rows 1–4 are based on classical methods involving SVMs and different types of kernels while the rest deal with deep net models. Our models are shown in the last four rows of the table.
Table III. Comparison with prior efforts.
| Team | Precision | Recall | F-score |
|---|---|---|---|
| FBK-irst [9] | 64.60 | 65.60 | 65.10 |
| WBI [10] | 64.20 | 57.90 | 60.90 |
| Kim et al. [11] | – | – | 67.00 |
| Zheng et al. [12] | – | – | 68.40 |
|
| |||
| Liu et al. [13] | 75.70 | 64.60 | 69.75 |
| Liu et al. [14] | 78.24 | 64.66 | 70.81 |
| Zhao et al. [15] | 72.50 | 65.10 | 68.60 |
|
| |||
| Our mean scores (¬F) | 77.78 | 62.21 | 69.13 |
| Our best ensemble (¬F) | 79.34 | 63.94 | 70.81 |
|
| |||
| Our mean scores (+F) | 78.56 | 63.76 | 70.38 |
| Our best ensemble (+F) | 79.68 | 65.88 | 72.13 |
All teams used some form of negative instance filtering. But this process is subjective and the filtering rules differ from team to team and are often not fully specified. In some cases the counts after the filtering are not disclosed [13], [14]. Although Zhao et al. [15] present counts after filtering, it is not clear how the two rules they discuss lead to filtering 56% (over 2800) of the negative test instances while removing only 8 positive instances. Our rules (from Section VI-A) filter only 14% of negative instances while also removing 3 positive instances. If not implemented with few generic patterns, this process can be tedious and can result in rules that are potentially too specific to work for other datasets. Thus we wanted to assess the best we can do without any rule based filtering given the confusing variations noticed in earlier efforts. Rows 8 and 9 of Table III correspond our results in this scenario. Our mean F-score here 69.13 is already better or comparable with other models that filter negative instances. However, it trades off some recall for gains in precision to do that. Our best ensemble without filtering beats all models but has the same F-score as the model by Liu et al. [14].
Our ensemble performances with the mildly filtered dataset (see Section VI-A) are shown in the final two rows. Our mean performances in this case are better than reported by most other efforts that use elaborate yet underspecified negative instance filtering schemes. Our best ensemble with filtering achieves superior results when compared with prior results on the 2013 DDIExtraction dataset. However, we believe the mean measures are more reflective of the expected performance on new test sets given the best model was obtained by simply attempting various ensembles on the test set. This we believe is the common problem of all prior studies surveyed in this paper on the DDI extraction task. Without exception, all such efforts do not report their architecture's average behavior based on different instantiations of network parameters. In this context, it is not clear whether the results will be consistent when a different initialization is used. Hence, we refrain to follow suit and report both the best model's results and also the average behavior, which in our case turns out to be consistent with tight confidence intervals.
VII. Concluding Remarks
In this paper, we took a deliberate approach to evaluate the potential of word and character-level RNNs for DDI extraction. We used a small set of fully specified patterns that filter certain obvious types of negative instances. We conducted experiments to assess the utility and consistency of model averaging and the complementary aspects of regular and Char-RNNs. We demonstrated that our models are superior or on par with prior results even when considering average behavior with minimal filtering. As such, we believe, our effort throws new light on evaluating deep neural architectures. The following are some limitations of our current study discussed along with future research plans.
In this paper, we do not report any qualitative error analyses of our results in terms of patterns found in FPs and potential reasons for FNs. Furthermore, it is not clear why and how Char-RNNs complement regular RNNs. While we notice the effect, the underlying linguistic insights are not apparent. We wish to pursue attention incorporated architectures to analyze these phenomena based on our prior experiences [35].
Although we considered 10,000 model averaging ensembles, we trained only 20 models for word based and Char-RNN based hierarchical architectures. So the variety of models within the ensembles is limited and we plan to increase it by training more models per architecture to generate more diverse ensembles.
The main focus of this paper was on classifying the type of interaction between a pair of drugs including the case when there is no interaction captured by the false class (in addition to the four specific types). However, it might be more useful just to detect the interaction without having to identify the specific type. Although this is a simpler binary classification problem, it nevertheless warrants a separate architecture tuned to maximize the detection F-score, which is going to be part of our future work.
We are aware of more complex neural architectures that combine RNNs and CNNs [17], employ hierarchical attention over the three sentence segments separated by the entity pair [18], and use Tree-RNNs (also known as recursive neural networks) [37] for relation classification. Our initial attempts in using these did not improve over results in this paper. We will take a more thorough approach in evaluating the suitability and assessing the modifications needed to adapt them to the DDI extraction task.
Acknowledgments
Research reported in this publication was primarily supported by the National Library Of Medicine of the National Institutes of Health under Award Number R21LM012274. Additional support is provided by the National Center for Advancing Translational Sciences through grant UL1TR001998 and the Kentucky Lung Cancer Research Program through grant PO2 41514000040001. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Ramakanth Kavuluru, Email: ramakanth.kavuluru@uky.edu.
Anthony Rios, Email: anthony.rios1@uky.edu.
Tung Tran, Email: tung.tran@uky.edu.
References
- 1.Preston CL. Stockley's drug interactions: A source book of interactions, their mechanisms, clinical importance and management. Pharmaceutical Press; 2016. [Google Scholar]
- 2.Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, Farrar K, Park BK, Breckenridge AM. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ. 2004;329(7456):15–19. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies EC, Green CF, Taylor S, Williamson PR, Mottram DR, Pirmohamed M. Adverse drug reactions in hospital in-patients: a prospective analysis of 3695 patient-episodes. PLoS one. 2009;4(2):e4439. doi: 10.1371/journal.pone.0004439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al. Drugbank 4.0: shedding new light on drug metabolism. Nucleic acids research. 2014;42(D1):D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Percha B, Altman RB. Informatics confronts drug–drug interactions. Trends in pharmacological sciences. 2013;34(3):178–184. doi: 10.1016/j.tips.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrero-Zazo M, Segura-Bedmar I, Martínez P, Declerck T. The ddi corpus: An annotated corpus with pharmacological substances and drug–drug interactions. Journal of biomedical informatics. 2013;46(5):914–920. doi: 10.1016/j.jbi.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Segura-Bedmar I, Martínez P, Herrero-Zazo M. SemEval-2013 task 9 : Extraction of drug-drug interactions from biomedical texts (DDIExtraction 2013) Second Joint Conference on Lexical and Computational Semantics (*SEM), Volume 2: Seventh International Workshop on Semantic Evaluation. 2013:341–350. [Google Scholar]
- 8.Segura-Bedmar I, Martínez P, Herrero-Zazo M. Lessons learnt from the ddiextraction-2013 shared task. Journal of biomedical informatics. 2014;51:152–164. doi: 10.1016/j.jbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury MFM, Lavelli S. FBK-irst: a multi-phase kernel based approach for drug-drug interaction detection and classification that exploits linguistic information. Second Joint Conference on Lexical and Computational Semantics (*SEM), Volume 2: Seventh International Workshop on Semantic Evaluation. 2013:351–355. [Google Scholar]
- 10.Thomas P, Neves M, Rocktäschel T, Leser U. Wbi-ddi: drug-drug interaction extraction using majority voting. Second Joint Conference on Lexical and Computational Semantics (*SEM), Volume 2: Seventh International Workshop on Semantic Evaluation. 2013:628–635. [Google Scholar]
- 11.Kim S, Liu H, Yeganova L, Wilbur WJ. Extracting drug–drug interactions from literature using a rich feature-based linear kernel approach. Journal of biomedical informatics. 2015;55:23–30. doi: 10.1016/j.jbi.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng W, Lin H, Zhao Z, Xu B, Zhang Y, Yang Z, Wang J. A graph kernel based on context vectors for extracting drug–drug interactions. Journal of biomedical informatics. 2016;61:34–43. doi: 10.1016/j.jbi.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 13.Liu S, Tang B, Chen Q, Wang X. Drug-drug interaction extraction via convolutional neural networks. Computational and mathematical methods in medicine. 2016;2016 doi: 10.1155/2016/6918381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, Chen K, Chen Q, Tang B. Bioinformatics and Biomedicine (BIBM), 2016 IEEE International Conference on. IEEE; 2016. Dependency-based convolutional neural network for drug-drug interaction extraction; pp. 1074–1080. [Google Scholar]
- 15.Zhao Z, Yang Z, Luo L, Lin H, Wang J. Drug drug interaction extraction from biomedical literature using syntax convolutional neural network. Bioinformatics. 2016;32(22):3444–3453. doi: 10.1093/bioinformatics/btw486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suárez-Paniagua V, Segura-Bedmar I, Martínez P. Exploring convolutional neural networks for drug–drug interaction extraction. Database. 2017;2017 doi: 10.1093/database/bax019. bax019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vu NT, Adel H, Gupta P, Schütze H. Combining recurrent and convolutional neural networks for relation classification. Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. 2016 June;:534–539. [Google Scholar]
- 18.Xiao M, Liu C. Semantic relation classification via hierarchical recurrent neural network with attention. Proceedings of COLING 2016, the 26th International Conference on Computational Linguistics: Technical Papers. 2016:1254–1263. [Google Scholar]
- 19.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Advances in Neural Information Processing Systems. 2013:3111–3119. [Google Scholar]
- 20.Graves A. ser Studies in Computational Intelligence. Vol. 385 Springer; 2012. Supervised Sequence Labelling with Recurrent Neural Networks. [Google Scholar]
- 21.Pascanu R, Mikolov T, Bengio Y. On the difficulty of training recurrent neural networks. Proceedings of the 30th International Conference on Machine Learning. 2013;28:1310–1318. [Google Scholar]
- 22.Gers FA, Schmidhuber J, Cummins F. Learning to forget: Continual prediction with LSTM. Neural computation. 2000;12(10):2451–2471. doi: 10.1162/089976600300015015. [DOI] [PubMed] [Google Scholar]
- 23.Hochreiter S, Schmidhuber J. Long short-term memory. Neural computation. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg Y. A primer on neural network models for natural language processing. Journal of Artificial Intelligence Research. 2016;57:345–420. [Google Scholar]
- 25.Zeng D, Liu K, Lai S, Zhou G, Zhao J. Relation classification via convolutional deep neural network. Proceedings of COLING 2014, the 25th International Conference on Computational Linguistics: Technical Papers. 2014:2335–2344. [Google Scholar]
- 26.Kim Y, Jernite Y, Sontag D, Rush AM. Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence. AAAI Press; 2016. Character-aware neural language models; pp. 2741–2749. [Google Scholar]
- 27.Santos CD, Zadrozny B. Learning character-level representations for part-of-speech tagging. Proceedings of The 31st International Conference on Machine Learning. 2014:1818–1826. [Google Scholar]
- 28.Zhang X, Zhao J, LeCun Y. Character-level convolutional networks for text classification. Advances in neural information processing systems. 2015:649–657. [PMC free article] [PubMed] [Google Scholar]
- 29.Looks M, Herreshoff M, Hutchins D, Norvig P. Deep learning with dynamic computation graphs. Proceedings of International Conference on Learning Representations. 2017 [Google Scholar]
- 30.Bergstra J, Breuleux O, Bastien F, Lamblin P, Pascanu R, Desjardins G, Turian J, Warde-Farley D, Bengio Y. Theano: a CPU and GPU math expression compiler. Proceedings of the Python for Scientific Computing Conference (SciPy) 2010 Jun; oral Presentation. [Google Scholar]
- 31.Duchi J, Hazan E, Singer Y. Adaptive subgradient methods for online learning and stochastic optimization. Journal of Machine Learning Research. 2011 Jul;12:2121–2159. [Google Scholar]
- 32.Rokach L. Ensemble-based classifiers. Artificial Intelligence Review. 2010;33(1):1–39. [Google Scholar]
- 33.Huang G, Li Y, Pleiss G, Liu Z, Hopcroft JE, Weinberger KQ. Snapshot ensembles: Train 1, get M for free. Proceedings of International Conference on Learning Representations. 2017 [Google Scholar]
- 34.Rios A, Kavuluru R. Proceedings of the 6th ACM Conference on Bioinformatics, Computational Biology and Health Informatics. ACM; 2015. Convolutional neural networks for biomedical text classification: application in indexing biomedical articles; pp. 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran T, Kavuluru R. Predicting mental conditions based on “history of present illness“ in psychiatric notes with deep neural networks. Journal of Biomedical Informatics. 2017 doi: 10.1016/j.jbi.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rios A, Kavuluru R. Ordinal convolutional neural networks for predicting RDoC positive valence psychiatric symptom severity scores. Journal of Biomedical Informatics. 2017 doi: 10.1016/j.jbi.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ebrahimi J, Dou D. Chain based rnn for relation classification. Proceedings of the 2015 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. 2015:1244–1249. [Google Scholar]


