Abstract
Structural Maintenance of Chromosomes (SMC) protein complexes are key determinants of chromosome conformation. Using Hi-C and polymer modeling, we study how cohesin and condensin, two deeply conserved SMC complexes, organize chromosomes in the budding yeast Saccharomyces cerevisiae. The canonical role of cohesin is to co-align sister chromatids whilst condensin generally compacts mitotic chromosomes. We find strikingly different roles for the two complexes in budding yeast mitosis. First, cohesin is responsible for compacting mitotic chromosome arms, independently of sister chromatid cohesion. Polymer simulations demonstrate this role can be fully accounted for through cis-looping of chromatin. Second, condensin is generally dispensable for compaction along chromosome arms. Instead it plays a targeted role compacting the rDNA proximal regions and promoting resolution of peri-centromeric regions. Our results argue that the conserved mechanism of SMC complexes is to form chromatin loops and that distinct SMC-dependent looping activities are selectively deployed to appropriately compact chromosomes.
Main Text
The extreme length of chromosomal DNA requires organizing mechanisms to both promote functional interactions between distal loci and ensure faithful chromosome segregation. Determining the unifying principles of functional organization requires understanding of how organizing mechanisms have converged and diverged across evolution. In metazoans, the polymer organization of both local interphase domains1,2 and entire mitotic chromosomes is well described by the presence of chromatin looping in cis2. The action of the SMC complexes cohesin and condensin is thought to be central to the formation of chromatin loops. Understanding how SMCs differentially orchestrate chromatin looping to develop functionally distinct chromatin structures in interphase and mitosis is a key question in cell biology.
In metazoans, SMCs play defined roles through the cell cycle. In interphase cohesin-dependent cis-looping is required for partitioning interphase chromosomes into domains3–5. During mitosis, metazoan chromosomes undergo chromosome-wide compaction leading to cytologically resolvable and longitudinally compacted structures. Both in vitro and in vivo studies suggest that condensin promotes the formation of cis-loops during this process6,7. Condensin is required for chromosome compaction in meiotic extracts and cells8,9. In early mitosis, cohesin is unloaded from chromosome arms10–12, with condensin I complexes only binding chromatin following nuclear envelope breakdown13. Therefore, condensin is considered central for chromatin looping during mitosis while cohesin’s looping activity is assumed to be confined to interphase.
Mitotic compaction is also detected in organisms such as the budding yeast Saccharomyces cerevisiae that have distinct mitotic contexts from those found in metazoans. Budding yeast have significantly shorter chromosomes than metazoans and segregate duplicated chromosomes without nuclear envelope breakdown. Condensin is present within nuclei throughout the cell cycle14 and cohesin maintained along chromosome arms until its cleavage in anaphase15. Budding yeast cells also rapidly progress from S to M without a definable G2 stage14,21, even undertaking mitosis-associated functions such as kinetochore bi-orientation, before DNA replication is completed16. The extent of compaction generally achieved in budding yeast in this compressed timeframe appears significantly less than in metazoans14. Resolvable and longitudinally compacted mitotic chromosomes are not readily apparent in metaphase arrested cells14,17. The only region to undergo readily visible compaction is the approximately 1Mb long array of rDNA repeats. This array condenses in early mitosis before becoming further longitudinally compacted post anaphase14. Pre-anaphase compaction at the rDNA array requires the concerted action of both cohesin and condensin18–21. A similar action of cohesin and condensin has been proposed to occur along chromosome arms18,22,23. However, interpreting locus-specific microscopic studies in cohesin and condensin mutants is impeded by the reduced mitotic compaction of the loci and the loss of sister chromatid arm cohesion that occurs in these backgrounds17,18,24. Therefore, fully assessing the role of cohesin and condensin in mitotic compaction along chromosomal arms requires an alternative approach to analyze mitotic structure.
Hi-C and computational modeling are ideal methodologies to study mitotic chromosome structure in budding yeast. Budding yeast have a relatively small and simple genome, a relative lack of repetitive regions, and are genetically tractable. Furthermore, their defined nuclear geometry makes them ideal for computational modeling of the chromosome structure underlying Hi-C contact maps25,26.
Here we use Hi-C and modeling to show that mitotic chromosome compaction in budding yeast is accounted for by cis-looping. Surprisingly, mitotic compaction of chromosome arms requires cohesin, but not sister chromatid cohesion or condensin. Therefore, our analysis indicates the deep conservation of chromosome compaction by SMC complexes whilst also demonstrating the divergent use of different SMC complexes in different contexts.
Results
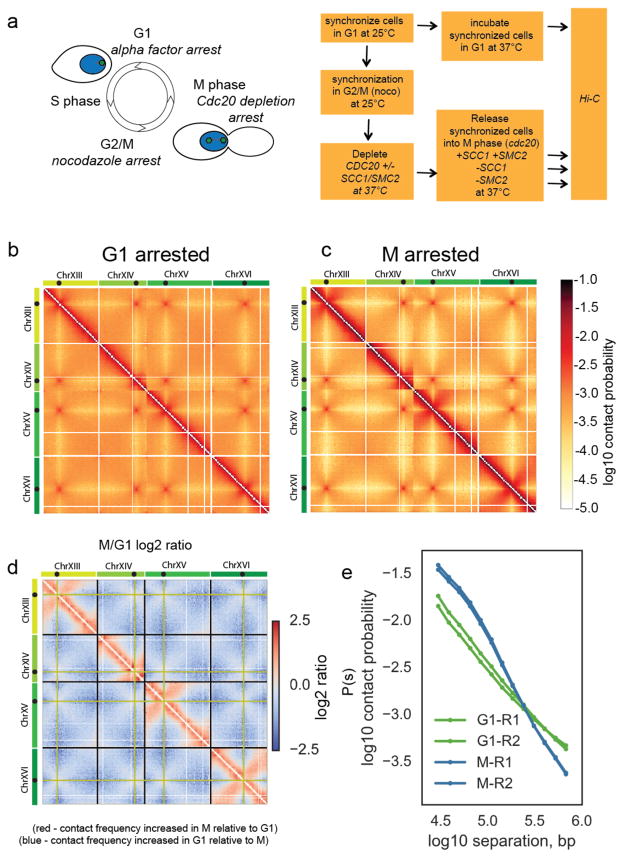
To study the mitotic organization of budding yeast chromosomes genome-wide, we used HiC on synchronized populations of budding yeast cells arrested in G1 or in metaphase (M) (Fig. 1a and Supplementary Fig. 1a). We fixed the synchronized populations with formaldehyde and prepared HiC libraries to assess chromatin conformation in each condition (Fig. 1a). We obtained on average 60 million unique valid pairs (pair-wise chromatin contacts) for each library (Supplementary Table 1). We binned contacts at 10kb resolution and removed intrinsic biases using iterative correction (ICE27).
Figure 1. Budding yeast chromosomes are compacted in mitosis.
a) Experimental procedure to synchronize cells in either G1 or M, with and without cohesin and condensin. Green spots in cartoon on the yeast nucleus (blue) represent spindle pole bodies.
b) Hi-C contact heatmap from G1 cells synchronously arrested at 37°C with alpha factor. The Hi-C contact map for chromosomes XIII to XVI is shown as representative of the whole genome. Contact maps for each condition were assembled from two independent experiments. Side bars show relative size of chromosomes XIII, XIV, XV and XVI with black dots on chromosomes showing locations of centromeres on each chromosome.
c) Hi-C contact heatmap from cells arrested in M phase by depletion of Cdc20 at 37°C. Both Hi-C maps have been normalized by iterative correction at 10kb resolution. Heatmap colour scale represents log10 number of normalized contacts. The Hi-C contact map for chromosomes XIII to XVI is shown as representative of the whole genome.
d) Log2 (M/G1) ratio of the data displayed in b and c. Regions where contact frequency was higher in M than G1 are shown in red, regions where contact frequency was lower in M than G1 in blue. Black guidelines show ends of chromosomes. Pale green lines indicate the bin containing the centromere.
e) Contact probability, P(s), as a function of genomic separation, s, for G1 and M phase cells averaged over all chromosomes. The P(s) from each of the two independent experiments for each condition are shown.
Hi-C contact maps in both G1 and M displayed the main features of budding yeast nuclear organization reported previously in asynchronous popualtions25,28: a Rabl-type organization with strong centromere clustering and arm length-dependent telomere clustering (Fig. 1b, c and Supplementary Fig. 1b). However, comparison of the G1 and M contact maps (Fig. 1b, c) and inspection of their log2 ratio (Fig. 1d) showed the global reduction of contacts between the arms of different chromosomes (inter-chromosomal contacts) as compared to contacts formed along chromosome arms (intra-chromosomal contacts). This change was not simply caused by normalization, since no loss of inter-chromosomal contacts was observed between centromeres (Fig. 1d). Rather, chromosome arms were resolved from one another in mitosis relative to their interphase state.
Concurrently, Hi-C contact maps changed locally along chromosome arms. In M cells, the frequency of intra-chromosomal contacts <100kb apart were markedly increased relative to G1 whilst longer-range intra-chromosomal contacts were reduced (Fig. 1d). Close analysis of chromosome arm regions did not reveal any distinct domain structure across chromosome arms in M cells relative to G1 (Supplementary Fig. 1c). Rather a general, chromosome-wide, increased frequency of contacts <100kb was apparent. We next analyzed the changes of intra-arm contact probability, P(s), with chromosomal distance s for all loci in the genome (Fig. 1e). P(s) analysis demonstrated that the G1 P(s) decayed at a similar rate at all distances, while M had a markedly slower decay in P(s) at short distances (<100kb), suggesting chromosome compaction at this scale29, followed by a more rapid loss of P(s) for larger genomic distances. Analysis of P(s) of each individual chromosome arm confirmed that these changes occurred uniformly across all chromosomes (Supplementary Fig. 1e). Interestingly, the two regimes of P(s) that we observed in budding yeast M, are reminiscent of Hi-C from mammalian mitotic cells which also displayed an initial slow decay in contact frequency followed by a more rapid decay at longer distances29. Therefore Hi-C analysis provides a distinct description of mitotic chromosome compaction in budding yeast chromosomes, demonstrating that all chromosomes undergo intra-chromosomal compaction in mitosis relative to their G1 state.
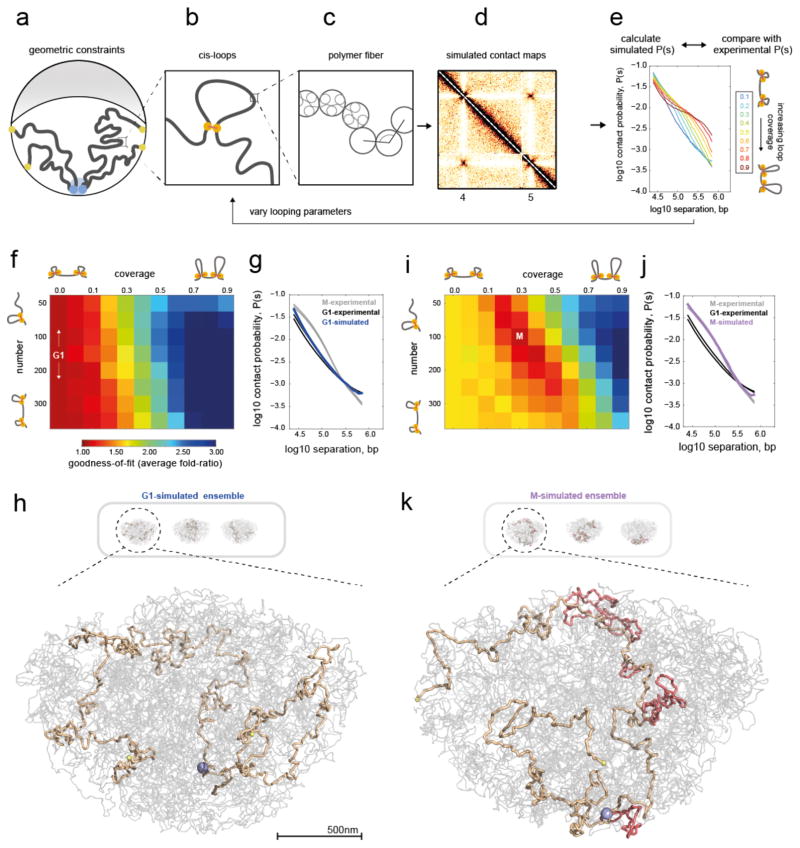
We next developed polymer models to test what changes to chromosomal structure can underlie the observed changes in the G1 and M Hi-C maps. Following previous simulations26,30 of yeast interphase Rabl organization, we modeled the genome as 16 long polymers confined to a spherical nucleus (Fig. 2a–e, Methods). Chromosomes are tethered by the centromeres to the spindle pole body, telomeres are held at the nuclear periphery, and the whole genome is excluded from the nucleolus, located opposite the spindle pole body (Fig. 2a). Following previous analysis of 3C and imaging data31 we modeled the chromatin fiber as a polymer of 20nm monomers (Fig. 2c), each representing 640bp (~4 nucleosomes), with excluded volume interactions and without topological constraints, subject to Langevin dynamics in OpenMM32,33. We additionally introduced intra-chromosomal (cis-) loops of varying number and coverage, i.e. the fraction of the genome spanned by all loops combined (Fig. 2b), motivated by previous models of mammalian mitotic29,34 and interphase4 chromosomes. Since changes occurred relatively uniformly along chromosome arms in M Hi-C maps, we introduced cis-loops stochastically from cell-to-cell at sequence-independent positions. For each combination of loop coverage and number, we collected conformations, simulated Hi-C maps and P(s) curves (Fig. 2d, e).
Figure 2. Polymer simulations of the yeast genome support compaction by intra-chromosomal loops in mitosis.
a–e) Overview of simulations.
a) Illustration of geometric constraints used in simulations: confinement to a spherical nucleus, clustering of centromeres (blue), localization of telomeres to the nuclear periphery (yellow), and exclusion of chromatin from the nucleolus (grey crescent).
b) Intra-chromosomal (cis-) loops, generated with a specified coverage and number per yeast genome.
c) Chromatin fiber, simulated as a flexible polymer.
d) Simulated contact maps are generated in simulations with the above constraints.
e) P(s) curves are then calculated from simulated contact maps. Simulations are run with systematically varied cis-loop parameters (coverage and number of loops), and the resulting P(s) curves are compared with experimental data. Shown here are P(s) curves for 150 loops, and a range of coverage.
f) Goodness-of-fit for simulated versus experimental intra-arm P(s) in G1. Goodness-of-fit represents the average fold deviation between simulated and experimental P(s) curves, best-fitting values indicated with white text. The coverage=0.0 column represents the fit for simulations without intra-chromosomal loops.
g) P(s) for best-fitting G1 simulations (coverage=0.0, i.e. no-loops) versus P(s) for each experimental replica of G1 and M.
h) Three sample conformations from ensemble generated in the no-loops simulations; one chromosome highlighted in light brown (from left to right: XI, V, III), with its centromere in blue, telomeres in yellow, and the rest of the genome in grey. Selected conformations show at higher zoom.
i) as F, but for experimental M Hi-C.
j) Best-fitting simulated P(s) for M has N=100–150, coverage=0.3–0.4.
k) Conformations for (N=100, coverage=0.3) with cis-loops additionally highlighted in light red.
Comparison of simulated and experimental P(s) curves allowed us to identify changes in chromosome organization upon G1→M transition (Fig. 2e). We found that in silico models with ~10 loops per Mb, ~35kb each, covering ~35% of the genome, closely reproduced the P(s) for experimental mitotic Hi-C data in M (Fig. 2i–k, and Supplementary Fig. 2a). In contrast, experimental G1 Hi-C data were best reproduced by models without cis-loops (coverage=0.0, Fig. 2f–h,). Interestingly, introducing sister chromatids to the best-fitting G1 models, either with or without sister chromatid cohesion between cognate loci, could not account for the differences we observe between G1 and M chromosomes (Supplementary Fig. 2b). Instead, the introduction of cis-loops into budding yeast chromosomes by a mitosis-specific activity best accounts for the compaction differences we observe between G1 and M chromosomes.
Next, we sought to identify the factors responsible for formation of these cis-loops in yeast mitosis. Mitotic compaction at the budding yeast rDNA array requires the concerted action of both cohesin and condensin19,20. In situ hybridization of individual loci has suggested that a similar process could occur on chromosome arms17,18,24. Cohesin only accumulates on chromatin in S and early mitosis, while condensin is activated by CDK in S and M phase15,19,35,36. Therefore, both SMC complexes are relatively inactive in G1 and active in M, consistent with both these complexes promoting chromosome compaction in mitosis. We genetically ablated cohesin or condensin activity in mitotically arrested cells using defined inducible mutations and assayed the changes in mitotic compaction as defined by Hi-C contacts.
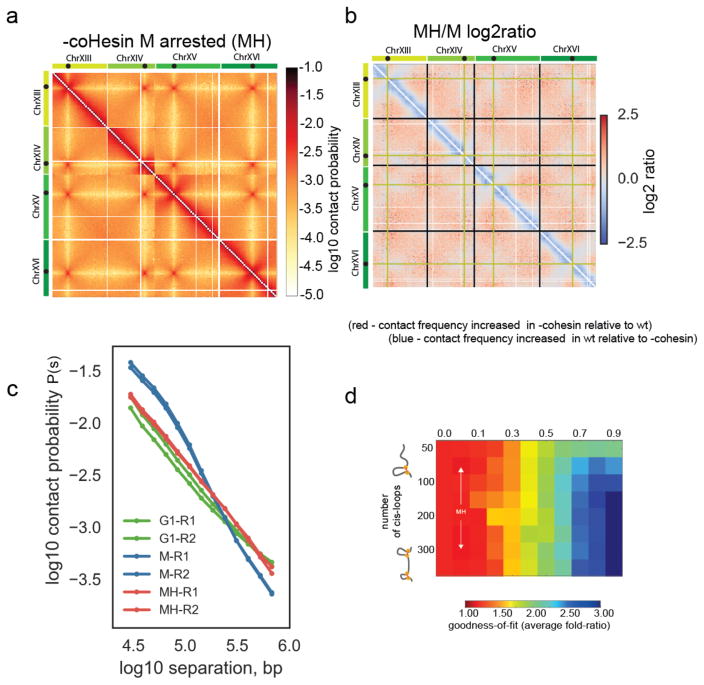
We first examined the role of cohesin in budding yeast mitotic chromosome condensation using the scc1-73 ts allele. Under the restrictive conditions of 37°C the scc1-73 encoded protein (S525N) loses its affinity for Smc1/3 resulting in loss of cohesin complex functions37,38. Disruption of cohesin function using scc1-73 led to a disappearance of the characteristic mitotic features, as determined by Hi-C (Fig. 3a–c), despite cells being maintained in metaphase by the depletion of Cdc20 (Supplementary Fig. 1a) and cultured in the same conditions as wildtype cells. Indeed, the two-regime M phase P(s) disappeared, becoming closer to that of G1 (Fig. 3c), with diminished short distance (<100kb) contacts and more frequent longer-range and inter-chromosomal contacts (Fig. 3a–c, Supplementary Fig. 3a). In contrast with the changes in the intra-arm organization, we found the Rabl conformation was maintained, as indicated by the persistence of centromere clustering contacts (Fig. 3a, b). Loss of cohesin activity also resulted in loss of short distance (<100kb) contacts and more frequent longer-range and inter-chromosomal contacts in the post-rDNA regions of chromosome XII (Supplementary Fig. 3a, b). This region is isolated from the centromere by the rDNA array and not subject to indirect effects resulting from cohesin action at the centromere. Consistently, modeling indicated that Hi-C maps for cohesin loss of function were well-fit by simulations with many fewer loops than wildtype mitotic Hi-C maps (Fig. 3d). Together our results indicate that cohesin is required for mitotic compaction in budding yeast, in a manner consistent with cohesin-dependent looping along chromosome arms.
Figure 3. Cohesin activity is required for mitotic compaction and cis-looping.
a) Hi-C data collected from M phase cells following disruption of coHesin using the scc1-73 allele (MH). Chromosomes XIII to XVI are shown as representative of the whole genome. Contact maps were assembled from two independent experiments.
b) Log2 ratio of –cohesin MH dataset over wt M dataset (MH/M), displayed in 3a and 1c, respectively.
c) Contact probability (P(s)) for wt metaphase M and cohesin depleted metaphase cells MH phase cells and G1 cells averaged over all chromosomes. The P(s) from each of the two independent experiments for each condition are shown.
d) Goodness-of-fit for models with variable cis-loop coverage (horizontal axis) and number (vertical axis). Best fitting parameter region indicated by arrows emanating from MH.
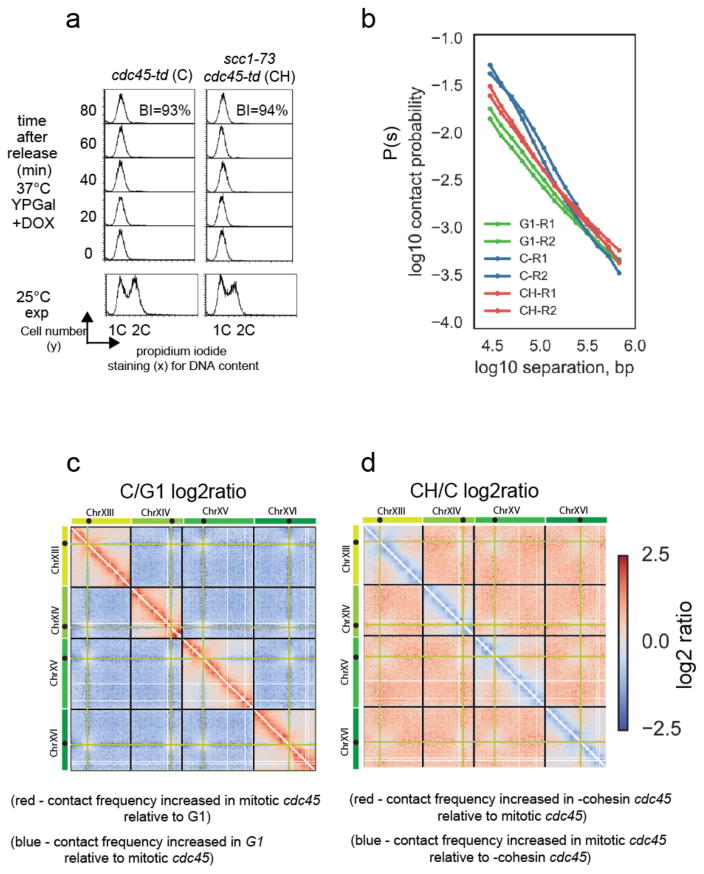
As a stringent test of the cis-looping function of cohesin, we next assessed whether cohesin could still compact mitotic chromosomes when no sister chromatid cohesion was present. We examined mitotic cells generated without a preceding round of DNA replication using a cdc45 degron allele. Cells depleted of Cdc45 fully activate CDK without replicating DNA (Fig. 4a and Supplementary Fig. 4a) and enter anaphase if not arrested in mitosis (Supplementary Fig. 4b), acting in an apparently identical manner to cdc6 mutants39. Therefore, this process generates mitotic chromosomes without sister chromatids40.
Figure 4. Mitotic cohesin-dependent conformational changes are independent of sister chromatid cohesion.
a) FACS analysis of DNA content and budding analysis of cdc45-td (C) and cdc45-td scc1-73 (CH) cells following release from G1 arrest into a nocodazole enforced mitotic block. Budding index (BI) confirmed that mitotic cells had activated CDK while FACS of DNA stained cells confirmed no DNA replication has taken place. Representative images shown from one of two independent experiments comparing C to CH.
b) Contact probability, P(s) versus genomic separation, s, for Hi-C of mitotic cdc45-td (C) mitotic cdc45-td scc1-73 (CH), and wt G1 cells (G1). The P(s) from each of the two independent experiments for each condition are shown.
c) Log2 ratio of C (cdc45 depleted cells arrested in mitosis with nocodazole) contact dataset over G1 dataset (C/G1). Contact maps for ratio plot were assembled from two independent experiments for each condition.
d) Log2 ratio of –cohesin C dataset over C dataset (CH/C).
Confirming our hypothesis regarding a role of cohesin in chromosome compaction, unreplicated mitotic chromosomes had contact frequencies distinct from G1, exhibiting P(s) with two regimes, similar to wildtype M phase Hi-C, and consistent with the presence of cis-loops (Fig. 4b, c). Moreover, this difference was cohesin-dependent (Fig. 4b, d, Supplementary Fig. 4c, d). Loss of cohesin function also resulted in the concurrent loss of <100kb intra-chromosomal contacts and increase in inter-chromosomal contacts between arms (Fig. 4d) as observed in cells with normal DNA replication. Therefore, cohesin activity was required for the mitotic resolution observed in mitotic cdc45 cells. Collectively, our data (Fig. 3 and Fig. 4) and simulations (Supplementary Fig. 2b) strongly indicate a function for cohesin in budding yeast chromosome compaction, independent of, and in addition to, its accepted role in sister chromatid cohesion.
We next considered the role of condensin in budding yeast mitotic chromosome structure. We first examined the consequence of degrading the condensin subunit Smc2 in mitosis using a degron allele of SMC2. We degraded Smc2 protein in G2/M before arresting the cells in M phase (Fig. 1a). In contrast to cohesin, loss of condensin activity had surprisingly mild effects on mitotic intra-arm chromosome organization (Supplementary Fig. 5a).
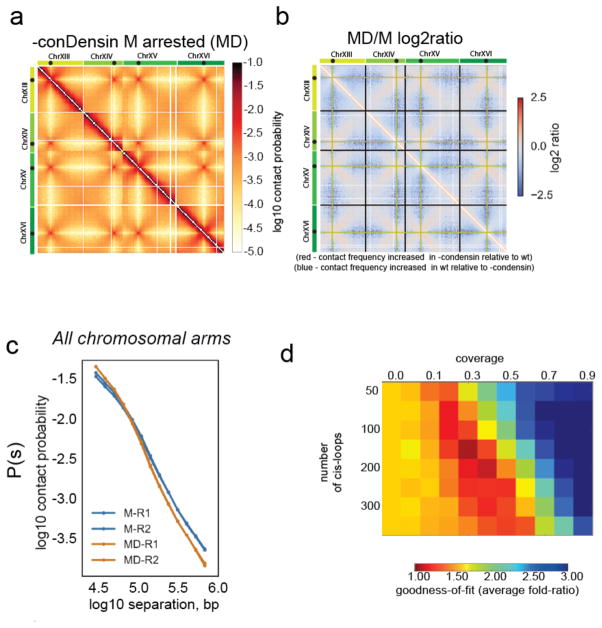
We considered the possibility that we were not completely ablating condensin function with the degron allele, and engineered a system predicted to cause a close-to-null condensin inactivation. We used a conditional depletion/expression system to express an enzymatically dead form of Smc2 (smc2K38I) in G2/M cells whilst also depleting active degron-tagged Smc2 before arresting the cells in M-phase (Supplementary Fig. 5b). Despite the increased penetrance of this allele, we did not observe any chromosome-wide loss of compaction or chromosome resolution (Fig. 5a, b) in metaphase arrested cells. Indeed, in contrast with cohesin depletion, the two regimes of mitotic P(s) persisted in condensin-depleted cells (Fig. 5c). Close examination of chromosome arms did not reveal loss of intra-chromosomal contacts (Supplementary Fig. 5c). Consistently, simulations did not support great differences in the amount of coverage by cis-loops (Fig. 5d). We conclude that condensin activity is not required for the chromosome compaction we observe along mitotic chromosome arms in cells arrested before anaphase.
Figure 5. Condensin action is not required for mitotic cis-looping along chromosome arms.
a) Hi-C data collected from M phase cells following disruption of conDensin with smc2td GAL1-smc2K38I allele (MD). Chromosomes XIII to XVI are shown as representative of the whole genome. Contact maps were assembled from two independent experiments.
b) Log2 ratio of –condensin M dataset over wt M dataset (MD/M)
c) Contact probability (P(s)) for M and MD cells. The P(s) from each of the two independent experiments for each condition are shown.
d) Goodness-of-fit for simulated versus experimental intra-arm P(s), as in Figure 2, for conDensin depleted cells.
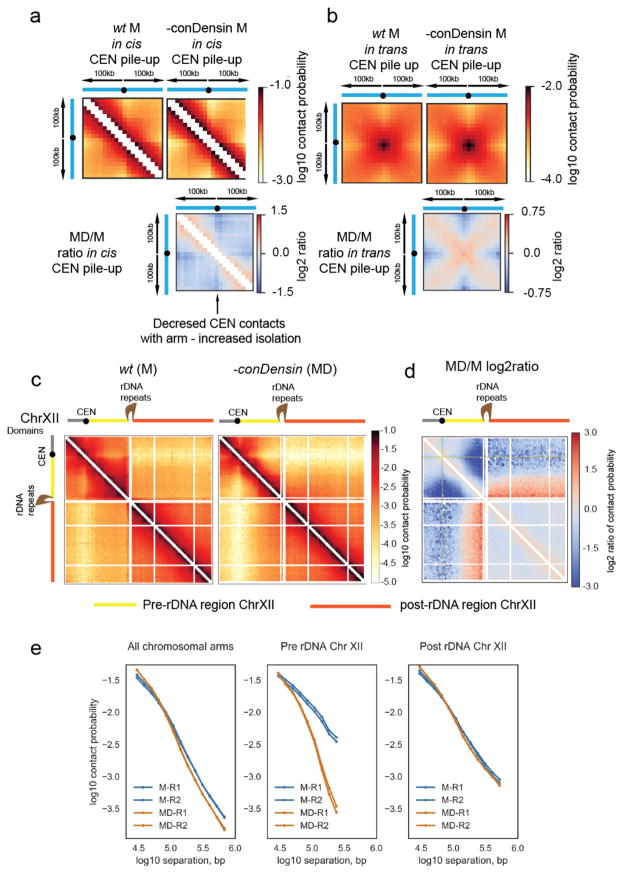
In contrast to the genome-wide role of cohesin, visual inspection of Hi-C maps revealed that condensin activity was relevant for higher order chromosome structure in specific genomic contexts. We observed condensin-dependent changes at centromeres and condensin-dependent compaction of the region between CENXII and the rDNA array on ChrXII. First, at centromeres, loss of condensin action led to an increased isolation of CEN-proximal regions from loci further down the chromosome arms in cis (Fig. 6a), concurrent with increased contacts between centromeres in trans (Fig. 5b, Fig. 6b). Others have shown that condensin has a focused role at centromeres in budding yeast41,42. The genome-wide visualization provided by Hi-C also suggests that condensin promotes resolution between the clustered centromeric regions. Condensin II has a similar role in neural stem cells43, suggesting that budding yeast condensin is functional for condensin II-like roles. Second, the pre-rDNA region, between centromere XII and the rDNA repeats, exhibited specific condensin-dependent compaction, with higher contact frequency at the same distance as compared with arm regions of other chromosomes in wildtype (Fig. 6c, d, e). While the repeated structure of the rDNA makes it refractory to direct analysis by Hi-C, we assume these changes are linked to the previously characterized loss of condensin-dependent compaction across the rDNA repeats20,23,42. Condensin acted in a distinct manner from cohesin across the pre-rDNA region. Loss of condensin led to loss of contacts >100kb (Fig. 6c, d, e) and left intact the <100kb contacts that were affected by cohesin loss (Supplementary Fig. 3b). In contrast to the pre-rDNA region, the post-rDNA region (from the rDNA repeats to the telomere), remained remarkably similar to wildtype mitotic cells following loss of condensin activity (Fig. 6c, d, e). This suggests that proximity to the centromere is a key facet of condensin-dependent changes in pre-anaphase cells.
Figure 6. Pre-anaphase condensin activity is focused on centromeres and proximal to the rDNA repeats.
a, b) Pile-ups of the contact heat-maps of the 100kb peri-centromeric regions a) in cis or b) in trans on either side of budding yeast CEN sequences. Bottom, log2 ratio of the different pile-ups in the mitotically arrested state.
c) Hi-C contact heat maps of ChrXII in M or MD. In the cartoon representation, ChrXII is separated into three regions, the pre-CEN region (grey), the region between CEN and the rDNA repeats (yellow) and post-rDNA (orange).
d) Log2 ratio of MD over M dataset (MD/M) for ChrXII.
e) Contact probability (P(s)) for M and MD cells for all chromosomes (taken from 5c) compared to contact probability (P(s)) of the pre-rDNA region and post-rDNA region of ChrXII for M and MD cells.
Finally we tested, and ruled out, the previously reported condensin-dependent tRNA gene clustering44,45. We did not observe any general preferential contact patterns associated with tRNA pairs, neither in wildtype nor in mutant cells (Supplementary Fig. 6a–c). We conclude that previously reported condensin-dependent tRNA clustering shown by FISH was likely an indirect consequence of of condensin action localized at the nuclear organizing hubs of yeast: the centromere cluster and the rDNA array. This potential for condensin to reorganize genomes globally by acting at a few specific locations could account for earlier reports of condensin-dependent changes in pre-anaphase chromosome structure17,22,46. These FISH and live cell studies focused on centromere-proximal loci that could be disproportionately affected by condensin-dependent changes within the centromere cluster. This issue highlights the usefulness of visualizing genome conformation with a genome-wide methodology such as Hi-C.
Discussion
In summary, our results support surprisingly different mitotic activities for both cohesin and condensin in budding yeast than those anticipated from their canonical functions in metazoans. For cohesin, our results indicate a genome-wide role in compacting mitotic chromosomes through the formation of intra-arm loops. For condensin, our results argue for a focused mitotic role in organizing centromeres and the vicinity of the rDNA locus. While cohesin has been previously reported to organize metazoan interphase chromosomes through looping4,5,47,48, our data additionally indicates that cohesin-dependent looping can be utilized to compact entire chromosomes in preparation for chromosome segregation. This functional coherence over long evolutionary timescales and contrasting cellular contexts argues for a fundamentally dual function of cohesin, both for the formation of DNA loops in cis, and holding sisters together in trans.
A compacting role for cohesin in addition to sister chromatid cohesion is consistent with numerous otherwise puzzling observations in budding yeast and prior in situ hybridization analysis of yeast mitotic chromosomes18. Cohesin only becomes maximally loaded onto chromosomes following bulk DNA replication36. Certain alleles of cohesin support rDNA condensation, but are defective in arm cohesion49–51. Two populations of chromatin bound cohesin are detected on mitotic chromosomes, one stable, one dynamic52,53. Such behavior would be consistent with the dynamic population of cohesin being engaged in chromatin looping and the stable population with sister chromatid cohesion (Supplementary Fig. 6d). However, other models would also be consistent with our data (Supplementary Fig. 6e). The lack of a definable G2 stage in budding yeast means we do not yet know if cohesin-dependent compaction is initiated as soon as cohesin is loaded following passage through START15 or is only initiated during the early stages of mitosis. The action of cohesin in interphase in other organisms suggests compaction initiates on loading4,5,47,48. However, the phosphorylation of cohesin prior to anaphase54 does provide a pathway for a mitosis-specific activity.
In contrast to cohesin, condensin is not required for the chromosome-wide compaction prior to anaphase. Instead, condensin has a more focused role to prevent excessive clustering of centromeres and compacting the regions between the rDNA array and its proximal centromere, in addition to its well established role across the rDNA repeats. While this manuscript was under review and available as a preprint, others have also reported that cohesin is required for the normal conformation of mitotic chromosomes and that condensin is required for restructuring the pre-rDNA region55.
A general role of cohesin in mitotic chromosome compaction acting alongside a focused activity of condensin would appear to be at odds with their roles during mitosis in higher eukaryotes. In metazoans, cohesin is removed from mitotic chromosomes during prophase11, while condensin appears to act across whole chromosomes during mitosis6, leading to the formation of densely looped, compacted chromosomes29. A key difference between the two SMC-mediated mitotic chromatin states is the density of chromatin looping. Our modeling predicts that loops cover 30–40% of mitotic budding yeast chromosomes, significantly lower than the 100% coverage predicted for mammalian mitotic chromosomes29. We speculate that cohesin-dependent looping is generally sufficient for the lower level of metaphase compaction required for chromosome segregation in budding yeast. In this model, budding yeast condensin activity provides an auxiliary compaction mechanism, deployed when compaction provided by cohesin-dependent looping is either insufficient or inappropriate for segregation. Indeed, the focused action of condensin at and adjacent to the rDNA is consistent with the exceptional segregation requirements of this region in yeast; the rDNA accumulates excessive levels of sister chromatid intertwines56 and requires extra longitudinal compaction to segregate its exceptional length57. This is additionally consistent with ‘adaptive hypercondensation’, where condensin is deployed along other chromosomal arms specifically in anaphase as an emergency measure to resolve persistent entanglements58,59. In this framework, the longer and more repetitive chromosomes of higher eukaryotes, not only require functional compaction during interphase, imposed via cohesin, but will also require the additional compaction offered by condensin through mitosis.
Understanding how different SMC complexes promote distinct chromatin looping states is a crucial question for the future. There are clear differences in the form and function of mitotic chromosomes in metazoans and budding yeast14,60,61. Potentially these differences reflect the functional consequences of compaction via either cohesin or condensin promoted looping. We speculate that the conserved mechanism of SMC action has been adapted within the different complexes to cope with the varying requirements for chromatin looping in different organisms and contexts. Unraveling how the baton of SMC function has been passed through evolution presents a fascinating topic for future research, and promises to shed light on the pleiotropic consequences of mutations to these key chromosome organizers in human disease62.
Methods
Statistics and reproducibility
No statistical methods were used to predetermine sample size. The replicates of the conditions are shown individually for all P(s) histograms. For contact heatmaps data from replicates was pooled. Details of individual libraries available in Supplementary Table 1. Number of times each condition tested is indicated in each figure legend.
Yeast Strains
Yeast strains were derived from W303-1a (MATa ade2-1 ura3-1 his3-11, trp1-1 leu2-3, can1-100) adapted for use with the degron system. Full details in Supplemental Table 2.
Media and cell cycle synchronization for Hi-C experiments
Yeast cells were grown in YP + 2% Glucose at 25 °C, transferred to YP + 2% Raffinose (YPR) and grown over night to log phase. Cells were then arrested in G1 with 10 μg/ml alpha factor until 90% of cells were unbudded (120 min). The cells were washed three times with YPR and released in YPR. After 30 min 10 μg/ml of nocodazole was added and budding was checked after 1h. After cells entered G2 (60–70 min after release), Galactose at 2% final concentration and 15 min later Doxycycline at 50μg/ml final concentration were added. 30 min after addition of Galactose, the temperature was shifted to 37 °C. Cells were grown at 37 °C for 1h and then washed three times in YPR + Galactose + Doxycycline and released in the same media to allow spindles to reform in cells depleted of Cdc20. Cells were then collected after 30 min for cdc20 metaphase arrest.
Cell fixation and Hi-C library preparation
Carried out as described63 with the variation that cells were fixed at 37 °C. Conditions and number of replicates used for each state shown in Table S1.
FACS, Nuclear morphology, Western blotting and Antibodies used
Carried out as described64. Smc4 phospoS4 antibody was a kind gift from Damien D’Amours35. Anti-HA antibody (12Ca5 mouse monoclonal IgG2βK . Roche, Fisher scientific 10026563). Anti-V5 antibody (MCA1360, abD Serotec) used for ChIP.
Chromatin Immunoprecipitation (ChIP)
Fixed cells were defrosted and re-suspended in 100 μl ChIP buffer (150 mM NaCl, 50 mM Tris HCl, 5mM EDTA, 0.5 % NP-40 (IGEPAL), 7 % Triton X-100, cOmplete Tablets, Mini EDTA-free EASYpack (Roche). Cells were lysed in a FASTPREP machine, 6 rounds of 30 seconds at 6.5 power, with1 ml of 0.5 mm silica beads The bottom. Lysate spun out and resuspended in 1ml ChIP buffer. Sonicated for 15x 30 seconds (Biorupter Pico, Diagenode). 100 μl of sonicate was processed as an input control, 200 μl was incubated with 12.5μg/ml anti-V5 antibody (MCA1360, abD Serotec). For immunoprecipitation (IP) tubes were agitated for an hour and a half at 4°C. 45μl magnetic beads, (Dynabeads protein G – Life Technologies), washed 3 times in 1ml ChIP buffer were added and tubesincubated at 4°C for 2 hours.
Magnetic beads were isolated and washed four times in ChIP buffer, and a fifth time in ChIP buffer minus protease-inhibiting supplement. To reverse cross-linking, magnetic beads were incubated with 10% Chelex100 resin beads (BioRad 142-1253), in purified water at 95°C for 30 min. Samples were spun down and the supernatant kept at −20°C prior to analysis by qPCR. Input controls were precipitated using 0.1x vol 3M NaAC pH5.2 and 2.5 x vol 100% EtOH and then cross-linking reversed as above, before purification with Nucleospin PCR clean up kit and eluted in nuclease-free purified water.
The immune-precipitated DNA was analysed using 2X AB-1323/B ABsolute™ QPCR SYBR. Green Low ROX Mix and processed in an MX3005p qPCR machine. Primers used for RT-PCR of DNA at CENIV are (forward) TGCTTGCAAAAGGTCACATGCTTAT and (reverse) CATTTTGGCCGCTCCTAGGTAGTG.
Data was analysed using the ‘Percentage Input Method’ where the CT values obtained from the ChIP are divided by the CT values obtained from the Input control samples. Since 1% of starting chromatin was used for each input sample, in order to adjust the CT value of input samples to 100% 6.644 (log2 of 100) was subtracted from it. Then the following formula was used to calculate the percentage input for each IP sample:
2^(adjusted ChIP input CT value – IP CT value) x 100.
Computational Analysis of Hi-C maps
Mapping and filtering contacts
We mapped sequenced read pairs to W303 yeast genome using Bowtie 2.1.0 and the previously described method of iterative mapping27. To generate contact lists, we assigned each mapped side to a HindIII fragment and removed the contacts with both sides assigned to the same HindIII fragment, reads with one unmapped side as well as PCR duplicates, i.e. identical contact pairs.
Aggregating contact maps
To generate Hi-C contact maps, we aggregated the filtered contact lists into 10kb genomic bins using the cooler Python package for Hi-C data analysis, publicly available at https://github.com/mirnylab/cooler/. We filtered out low coverage genomic bins using the MAD-max (maximum allowed median absolute deviation from the median coverage) filter on the total number of interactions per bin, set to 7.4 median absolute deviations (corresponding to 5 standard deviations in the case of a normal distribution). We also removed the contacts within the first two diagonals of the contact maps as they are contaminated by uninformative Hi-C artifacts, unligated and self-ligated DNA fragments. Finally, we iteratively corrected the resulting maps to equalize genomic coverage.
Contact map analyses
We calculated the curves of intra-arm contact probability P(s) vs genomic separation, s, from the 10kb contact maps using 15 logarithmically spaced bins27 spanning distances between 20kb to 1Mb. We excluded Chromosomes IV and XII from these analyses as well as the bins within 40kb distance from the nearest centromere and telomere. The post-rDNA scalings were generated using the same approach on the region between the rDNA locus on chromosome XII and the telomere of the corresponding arm. We generated the centromere and tRNA pile-ups by averaging the contact maps of regions surrounding the respective genomic features. To exclude a possible contribution of telomeric conformations, we only used genomic features that were separated from the telomeres by more than 200kb.
Polymer models
We modeled the yeast genome as 16 polymers, subject to additional constraints imposed by yeast chromosome organization, and intra-chromosomal loops generated specified number and coverage (Fig. 2a–e). We then obtained conformations from simulations and calculated simulated contact maps. For a range of looping parameters, intra-arm P(s) was calculated from these contact maps and compared with experimental P(s) to determine the best-fitting parameter sets for each experimental condition.
Polymer Fiber
Following31 we simulate the yeast genome as 16 chromatin fibers of 20nm monomers, each monomer representing 640bp (~4 nucleosomes). Polymer connectivity was implemented by connecting adjacent monomers with harmonic bonds using a potential U = 100*(r – 1)2 (energy in units of kT, distances in monomers). The stiffness of the fiber was implemented using a three point interaction term, with the potential U = 2.5*(1−cos(α)). Excluded volume interactions between monomers were implemented using a soft-core repulsive potential with a maximum repulsion U(0)=1.5 that goes to zero at a distance of 1.05 (i.e. U(1.05) = 0), as in 4.
Geometric constraints
Geometric constraints were implemented as in30 with slight modifications:
Confinement to the nucleus: harmonically increasing potential (1kT /monomer) when monomer radial position exceeds the confinement radius (1000nm).
Centromere tethering to the SPB: centromeres were confined to a spherical volume (radius of 250nm) with a harmonically increasing potential (1kT /monomer) when monomer radial position exceeds the confinement radius.
Tethering of telomeres to the nuclear periphery: telomeres were subject to a harmonic potential pushing them to the periphery (1kT/monomer distance) when their radial position was less than .95 of the nucleus radius.
Repulsion from the nucleolus: monomers entering the nucleolus were subject to a harmonically increasing potential (1kT/monomer distance). Both sides of the rDNA locus on chromosome XII were attracted to the periphery of the nucleolus, imposed with harmonic potentials (1kT/monomer distance).
Intra-chromosomal loops
To generate sets of intra-chromosomal (cis-) loops with a desired coverage and number, we used 1D loop extrusion simulations (see4,34) using a genome-wide lattice at monomer resolution with boundaries at chromosome telomeres, centromeres, and the rDNA locus on chrXII. This ensures that loops are generated within chromosomes, are non-overlapping, do not cross centromeres, or link two regions together on opposite sides of the rDNA locus. Note that other than these requirements, we allow positions of loops to be variable along the chromosome arm. In polymer simulations, intra-chromosomal loops are then imposed with harmonic bonds (as in 4,65).
Code Availability
All libraries used for analysis and simulations are publically available at https://bitbucket.org/mirnylab/hiclib and https://bitbucket.org/mirnylab/openmm-polymer
Data availability statement
Full details of libraries and raw sequencing available at GEO number: GSE87311. Genomic sequencing of each test strain available from the authors on request. All data that support the conclusions are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We thank Bryan Lajoie and Johan Gibcus for aid in processing the Hi-C datasets. We thank Luis Aragon, Kim Nasmyth and John Diffley for yeast strains. This work was funded by the Biotechnology and Biological Sciences Research Council United Kingdom, (BBSRC UK) Grant ref BB/J018554/1 (S.A.S., M. Y.), the Royal Society U.K. (J. B.), NIH U54 4D Nucleome grant (DK107980) and NIH R01 (HG003143) (A. G., G. F, J. M. B., L.M., and J. D.). J.D. in an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
S. A. S. performed all cell culture and generated Hi-C libraries. A. G. analysed sequenced libraries and Hi-C data sets. G. F. modelled chromosome conformation of the budding yeast nucleus with help from A.G. J. M. B. guided Hi-C library instruction and analysed sequenced libraries. M. Y. and C. M. constructed and characterized the inducible condensin alleles. J. D. advised on study construction and guided processing and analysis of Hi-C datasets. L. M. guided the modeling of the Hi-C data. J.B. conceived and co-ordinated the study. J.B., G. F and A. G. wrote the manuscript with input from S. A. S., L.M. and J. D.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. CellReports. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proceedings of the National Academy of Sciences. 2015;112:E6456–65. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes & Development. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 8.Hirano T. Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell. 2016;164:847–857. doi: 10.1016/j.cell.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 9.Hudson DF, Marshall KM, Earnshaw WC. Condensin: Architect of mitotic chromosomes. Genome Research. 2009;17:131–144. doi: 10.1007/s10577-008-9009-7. [DOI] [PubMed] [Google Scholar]
- 10.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Development. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 12.Liang Z, et al. Chromosomes Progress to Metaphase in Multiple Discrete Steps via Global Compaction/Expansion Cycles. Cell. 2015;161:1124–1137. doi: 10.1016/j.cell.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ono T, Fang Y, Spector DL, Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Molecular Biology of the Cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guacci V, Hogan E, Koshland DE. Chromosome condensation and sister chromatid pairing in budding yeast. The Journal of Cell Biology. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 16.Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes & Development. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vas ACJ, Andrews CA, Kirkland Matesky K, Clarke DJ. In vivo analysis of chromosome condensation in Saccharomyces cerevisiae. Molecular Biology of the Cell. 2007;18:557–568. doi: 10.1091/mbc.E06-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guacci V, Koshland DE, Strunnikov AV. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavoie BD, Hogan E, Koshland DE. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes & Development. 2004;18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavoie BD, Hogan E, Koshland DE. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. The Journal of Cell Biology. 2002;156:805–815. doi: 10.1083/jcb.200109056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Serra L, Lengronne A, Borges V, Kelly G, Uhlmann F. Budding yeast Wapl controls sister chromatid cohesion maintenance and chromosome condensation. Curr Biol. 2013;23:64–69. doi: 10.1016/j.cub.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 22.Strunnikov AV, Hogan E, Koshland DE. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes & Development. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- 23.Freeman L, Aragon-Alcaide L, Strunnikov AV. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. The Journal of Cell Biology. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam WW, Peterson EA, Yeung M, Lavoie BD. Condensin is required for chromosome arm cohesion during mitosis. Genes & Development. 2006;20:2973–2984. doi: 10.1101/gad.1468806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong H, et al. A predictive computational model of the dynamic 3D interphase yeast nucleus. Curr Biol. 2012;22:1881–1890. doi: 10.1016/j.cub.2012.07.069. [DOI] [PubMed] [Google Scholar]
- 27.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Meth. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin QW, Fuchs J, Loidl J. Centromere clustering is a major determinant of yeast interphase nuclear organization. Journal of Cell Science. 2000;113( Pt 11):1903–1912. doi: 10.1242/jcs.113.11.1903. [DOI] [PubMed] [Google Scholar]
- 29.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tjong H, Gong K, Chen L, Alber F. Physical tethering and volume exclusion determine higher-order genome organization in budding yeast. Genome Research. 2012;22:1295–1305. doi: 10.1101/gr.129437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dekker J. Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction. J Biol Chem. 2008;283:34532–34540. doi: 10.1074/jbc.M806479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eastman P, Pande VS. Efficient nonbonded interactions for molecular dynamics on a graphics processing unit. J Comput Chem. 2010;31:1268–1272. doi: 10.1002/jcc.21413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eastman P, et al. OpenMM 4: A Reusable, Extensible, Hardware Independent Library for High Performance Molecular Simulation. J Chem Theory Comput. 2013;9:461–469. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goloborodko A, Marko JF, Mirny LA. Chromosome Compaction by Active Loop Extrusion. Biophysical Journal. 2016;110:2162–2168. doi: 10.1016/j.bpj.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robellet X, et al. A high-sensitivity phospho-switch triggered by Cdk1 governs chromosome morphogenesis during cell division. Genes & Development. 2015;29:426–439. doi: 10.1101/gad.253294.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu B, et al. Biological chromodynamics: a general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic Acids Research. 2015;43:e132. doi: 10.1093/nar/gkv670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 38.Haering CH, et al. Structure and stability of cohesin's Smc1-kleisin interaction. Molecular Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 39.Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. The EMBO Journal. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tercero JA, Labib K, Diffley JF. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. The EMBO Journal. 2000;19:2082–2093. doi: 10.1093/emboj/19.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verzijlbergen KF, et al. Shugoshin biases chromosomes for biorientation through condensin recruitment to the pericentromere. Elife. 2014;3:e01374. doi: 10.7554/eLife.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephens AD, Haase J, Vicci L, Taylor RM, Bloom KS. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. The Journal of Cell Biology. 2011;193:1167–1180. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishide K, Hirano T. Overlapping and non-overlapping functions of condensins I and II in neural stem cell divisions. PLoS Genet. 2014;10:e1004847. doi: 10.1371/journal.pgen.1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes & Development. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thadani R, Uhlmann F, Heeger S. Condensin, Chromatin Crossbarring Review and Chromosome Condensation. Current Biology. 2012;22:R1012–R1021. doi: 10.1016/j.cub.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 46.D'Ambrosio C, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes & Development. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizuguchi T, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature. 2014;516:432–435. doi: 10.1038/nature13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. The EMBO Journal. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guacci V, Koshland DE. Cohesin-independent segregation of sister chromatids in budding yeast. Molecular Biology of the Cell. 2012;23:729–739. doi: 10.1091/mbc.E11-08-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orgil O, et al. A conserved domain in the scc3 subunit of cohesin mediates the interaction with both mcd1 and the cohesin loader complex. PLoS Genet. 2015;11:e1005036. doi: 10.1371/journal.pgen.1005036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eng T, Guacci V, Koshland D. ROCC, a conserved region in cohesin's Mcd1 subunit, is essential for the proper regulation of the maintenance of cohesion and establishment of condensation. Molecular Biology of the Cell. 2014;25:2351–2364. doi: 10.1091/mbc.E14-04-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McNairn AJ, Gerton JL. Intersection of ChIP and FLIP, genomic methods to study the dynamics of the cohesin proteins. Chromosome Res. 2009;17:155–163. doi: 10.1007/s10577-008-9007-9. [DOI] [PubMed] [Google Scholar]
- 53.Chan KL, et al. Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell. 2012;150:961–974. doi: 10.1016/j.cell.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 55.Lazar-Stefanita L, et al. Choreography of budding yeast chromosomes during the cell cycle. bioRxiv. 2016:096826. doi: 10.1101/096826. [DOI] [Google Scholar]
- 56.D'Ambrosio C, Kelly G, Shirahige K, Uhlmann F. Condensin-dependent rDNA decatenation introduces a temporal pattern to chromosome segregation. Curr Biol. 2008;18:1084–1089. doi: 10.1016/j.cub.2008.06.058. [DOI] [PubMed] [Google Scholar]
- 57.Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- 58.Renshaw MJ, et al. Condensins Promote Chromosome Recoiling during Early Anaphase to Complete Sister Chromatid Separation. Developmental Cell. 2010;19:232–244. doi: 10.1016/j.devcel.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neurohr G, et al. A Midzone-Based Ruler Adjusts Chromosome Compaction to Anaphase Spindle Length. Science. 2011;332:465–468. doi: 10.1126/science.1201578. [DOI] [PubMed] [Google Scholar]
- 60.Elliott SG, McLaughlin CS. Regulation of RNA synthesis in yeast. III. Synthesis during the cell cycle. Mol Gen Genet. 1979;169:237–243. doi: 10.1007/BF00382269. [DOI] [PubMed] [Google Scholar]
- 61.Clemente-Blanco A, et al. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature. 2009;457:219–222. doi: 10.1038/nature07652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watrin E, Kaiser FJ, Wendt KS. Gene regulation and chromatin organization: relevance of cohesin mutations to human disease. Current Opinion in Genetics & Development. 2016;37:59–66. doi: 10.1016/j.gde.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 63.Belton JM, Dekker J. Hi-C in Budding Yeast. Cold Spring Harb Protoc. 2015;2015:649–661. doi: 10.1101/pdb.prot085209. [DOI] [PubMed] [Google Scholar]
- 64.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Molecular Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 65.Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. Elife. 2016;5:e14864. doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full details of libraries and raw sequencing available at GEO number: GSE87311. Genomic sequencing of each test strain available from the authors on request. All data that support the conclusions are available from the corresponding author on reasonable request.