Abstract
Purpose of Review
Autophagy is a conserved intracellular degradation system and plays a dual role in cell death, depending on context and phase. Ferroptosis is a new form of regulated cell death that mainly depends on iron accumulation and lipid peroxidation. In this review, we summarize the processes of autophagy and ferroptosis and discuss their crosstalk mechanisms at the molecular level.
Recent Findings
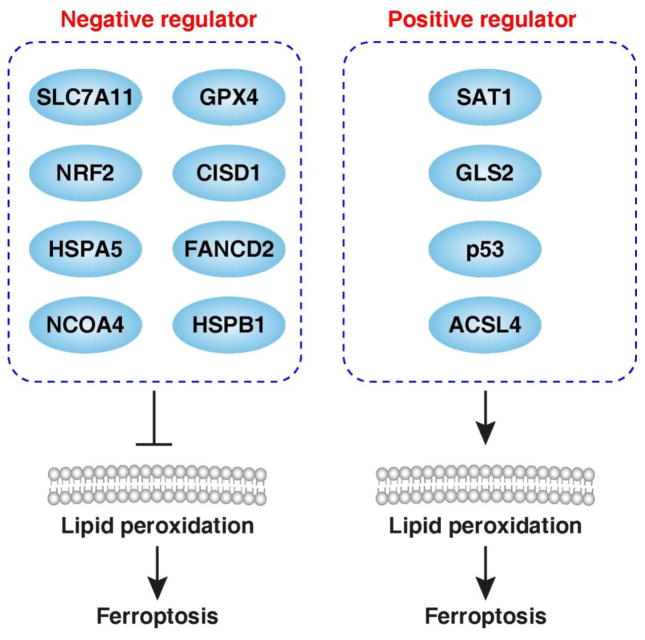
The original study shows that ferroptosis is morphologically, biochemically, and genetically distinct from autophagy and other types of cell death. However, recent studies demonstrate that activation of ferroptosis is indeed dependent on the induction of autophagy. Additionally, many ferroptosis regulators such as SLC7A11, GPX4, NRF2, p53, HSPB1, CISD1, FANCD2, and ACSL4 have been identified as potential regulators of autophagy.
Summary
This review not only highlights the importance of autophagy as an emerging mechanism of ferroptosis, but also raises new insights regarding regulated cell death.
Keywords: ferroptosis, autophagy, signal transduction, molecular interaction, lipid peroxidation, iron metabolism
Introduction
Different types of cell death are often defined by distinct morphological, biochemical, genetic, and functional mechanisms [1]. In recent years, the scientific world has discovered several new types of regulated cell death (RCD). These types of RCD are implicated in both physiological and pathological processes in human health and disease [2]. Research continues to focus on the molecular machinery and signaling pathways that control various types of RCD, as well as the crosstalk between different types of RCDs [3, 4]. The field of RCD research keeps advancing at such an alarming rate. At present, 11 different types of RCD have been identified: anoikis, autophagic cell death, apoptosis, cornification, entosis, ferroptosis, mitotic catastrophe, necroptosis, netosis, parthanatos, and pyroptosis [5]. Among them, ferroptosis, an iron-dependent form of RCD, is the latest to be identified in 2012 [6]. It has become increasingly evident that iron accumulation and subsequent lipid peroxidation play a critical role in mediating ferroptosis [7]. Thus, various molecules and signals involved in iron metabolism and lipid peroxidation contribute to ferroptosis regulation.
The original study shows that ferroptosis is distinct from apoptosis, necrosis, and autophagy in cancer cells [6]. However, recent studies demonstrate that activation of autophagy is required for the induction of ferroptosis not only in cancer cells, but also in normal cells [8–10]. Unraveling how autophagy regulates ferroptosis will not only reveal fundamental mechanistic insights into this type of RCD, but also provide new therapeutic targets for the treatment of ferroptosis-associated diseases. This review briefly introduces the processes of ferroptosis and autophagy and focuses on the emerging molecular interactions between these processes.
Autophagy: a lysosome-dependent degradation pathway
Autophagy (from the Greek for “self-eating”) is an evolutionarily-conserved homeostatic mechanism that includes three general subtypes: macroautophagy, microautophagy, and chaperone-mediated autophagy [11]. Macroautophagy (hereafter referred to as autophagy), as one of the lysosome-dependent degradation pathways, is involved in the removal of not only aging proteins, but also damaged organelles, as well as invading pathogens. To that end, autophagy is a critical regulator of cellular homeostasis and is involved in various pathologic conditions including infection, immunity, metabolism, and cancer [12]. The term “autophagy” was first introduced in the 1960s; however, the molecular mechanism of autophagy remained obscure until the 1990s. In 1993, Yoshinori Ohsumi, a Japanese cell biologist, identified the first autophagy-regulated (ATG) mutant that could not accumulate autophagic bodies in the vacuole and reported that 15 ATG genes are required for the activation of autophagy in eukaryotic cells [13]. He was therefore awarded the 2016 Nobel Prize in Physiology or Medicine for his pioneering studies on the molecular machinery of autophagy [14]. To date, 41 ATG genes have been identified as controlling autophagy in yeast; approximately half of these genes are clearly conserved in humans [15]. ATG proteins can form different complexes by posttranslational modifications to control the formation of three main subcellular structures of autophagy: the phagophore, the autophagosome, and the autolysosome [16, 17].
Autophagy plays a dual role either preventing or promoting death [18]. In many cases, autophagy-related stress tolerance can enable cell survival under various types of cellular death stimuli. In some cases, excessive or uncontrolled levels of autophagy can trigger autophagy-dependent cell death, termed “autophagic cell death” [19]. Distinguishing the various functions of autophagy in cell life and death remains challenging [20, 21].
Ferroptosis: an iron-dependent lipid peroxidation pathway
The molecular characterization of various cancers has shown that cancers with the same origins, pathologic stages, and clinical stages can be greatly heterogeneous in their patterns of genetic alterations, epigenetic changes, and gene expressions. One goal of precision medicine is to search for genotype-selective agents that become lethal to tumor cells only in the presence of specific genetic alterations [22]. Mutations in the small GTPase RAS of proto-oncogenes (e.g., K-RAS, H-RAS, and N-RAS) are very common in human cancer [23]. Ferroptosis was originally identified as a form of RAS mutation-dependent RCD by Brent Stockwell’s lab at Columbia University in 2012 [6, 24]. Ferroptosis can be induced by RAS-selective lethal (RSL) small molecular compounds such as erastin and RSL3 [25, 24]. Moreover, several anticancer drugs (e.g., sulfasalazine, sorafenib, and artesunate) also have the ability to induce ferroptosis [26–28]. In addition to cancer cells, ferroptosis can be observed in normal cells under some conditions [29–32]. Thus, ferroptosis is implicated in wide variety of physiological and pathological processes.
Stockwell’s group showed that ferroptosis is morphologically, biochemically, and genetically distinct from apoptosis, necroptosis, and autophagy. Transmission electron microscopy (TEM) analysis shows that ferroptotic cells undergo a different substructural change [6]. Onset of apoptosis (e.g., chromatin condensation), necrosis (e.g., rupture of the plasma membrane), and autophagy (e.g., double membrane structures) are not observed during ferroptosis by TEM assay [6]. In contrast, cells undergoing ferroptosis display characteristic changes in mitochondrial size (e.g., small size) and structure (e.g., reduced mitochondrial crista density and increased mitochondrial membrane potential) [6]. The activated caspases (the apoptosis effector) and receptor interacting serine/threonine kinase (“RIPK”, the necroptosis effector) are not observed in erastin-induced ferroptosis. ZVAD-FMK (an inhibitor of pan-caspase), necrostatin-1 (a selective inhibitor of RIPK1), and chloroquine (an inhibitor of autophagy by raising the lysosomal pH) cannot block ferroptosis [6]. In contrast, iron accumulation and lipid peroxidation are critical signaling events that drive ferroptosis [33]. Intracellular iron chelate (e.g., deferoxamine and desferrioxamine mesylate) and various antioxidants (e.g., vitamin E, liproxstatin-1, ferrostatin-1, and baicalein) prevent ferroptosis [6, 34, 32, 35]. Several genes and proteins have recently been demonstrated to be involved in ferroptosis by modulating the generation of reactive oxygen species (ROS)-mediated lipid peroxidation by iron and lipid (Figure 1). However, these critical players in ferroptosis factors also exhibit the ability to regulate autophagy and autophagic cell death (see below).
Figure 1.
Molecular regulators of ferroptosis.
Autophagy contributes to ferroptosis by ferritinophagy
We and other groups recently provided confidence data that ferroptosis is an autophagic cell death process [9, 8, 10]. Mechanistically, selective autophagy (namely ferritinophagy) contributes to ferroptosis by mediating the degradation of ferritin. Ferritin is composed of ferritin heavy chain (FTH) and ferritin light chain (FTL). They regulate iron metabolism by sequestering and storing iron in non-toxic and bioavailable forms, which prevents harmful oxidative reactions. FTH catalyzes the first step in iron storage, the oxidation of Fe2+, whereas FTL promotes the nucleation of ferrihydrite, enabling storage of Fe3+. The expression of ferritin is downregulated in ferroptosis-sensitive cells compared to ferroptosis-resistant cells, suggesting that ferritin negatively regulates ferroptosis [25].
The clearance of aggregated proteins by autophagy requires various cargo receptor proteins linking cargo to growing autophagosomal membranes. Quantitative proteomics identifies nuclear receptor coactivator 4 (NCOA4) as the cargo receptor responsible for ferritin degradation by ferritinophagy [36]. Degradation of ferritin within autolysosomes ultimately results in the release of chelated iron, which is subsequently transported back into the cytosol to induce oxidative stress. Like knockout or knockdown of ATGs (e.g., ATG5, and ATG7), genetic depletion of NCOA4 by specific shRNA limits ferritin degradation and subsequent ferroptosis in normal fibroblasts and tumor cells [8], whereas forced expression of NCOA4 by cDNA transfection accelerates ferroptosis by induction of ferritin degradation [8]. These findings indicate that selective autophagy plays a specific role in mediating ferroptosis. In addition to iron buffering, FTH also enhances thymidine biosynthesis [37]. It is unclear whether thymidine biosynthesis is regulated by ferritinophagy, which affects the process of ferroptosis.
Molecular interactions between ferroptosis and autophagy
SlC7A11
System Xc−, an amino acid antiporter on the cellular surface, is involved in antioxidant defense through regulating glutamate, cysteine, and glutathione (GSH) metabolism. As a functional core component of system Xc−, solute carrier family 7 member 11 (SLC7A11) plays an important role in protection against ferroptosis by inhibition of lipid peroxidation [6]. In contrast, pharmacologic inhibition of SCL7A11 by erastin, sulfasalazine, and sorafenib significantly cause intracellular GSH depletion, which contribute to lipid peroxidation and subsequent ferroptosis [6]. Research studies have shown that GSH also can inhibit basal and induced autophagy by starvation or oxidative stress, suggesting a potential role of GSH in the modulation of crosstalk between ferroptosis and autophagy [38, 39].
GPX4
Glutathione peroxidase (GPX) is an antioxidant enzyme family that includes GPX1-8 in humans. They can scavenge hydrogen and lipid peroxides under oxidative stress. Compared with other GPX members, GPX4 plays a unique role in the inhibition of ferroptosis in vitro and in vivo. Loss of GPX4 expression or activity by genetic or drug approaches promotes ferroptosis through a lipid ROS-dependent manner [35]. In contrast, upregulation of GPX4 expression can diminish lipid ROS-induced ferroptosis [35]. In mice, GPX4 conditional knockout in kidney, T cells, or brain exhibits tissue injury and immune dysfunction associated with increased ferroptosis [32, 30, 40]. In contrast, ferroptosis inhibitors such as liproxstatin-1 can reverse these abnormalities including oxidative injuries in GPX4 conditional knockout mice [32, 30], supporting that excessive ferroptosis play a pathologic role in tissue injury, inflammation and immune response in vivo. However, GPX4 may have roles as modulators of non-ferroptotic RCD such as apoptosis, necroptosis, and autophagy. Consistent with their enhanced antioxidant ability, GPX4 overexpression has been shown to inhibit ROS-mediated autophagy as well as immunogical cell death [41]. In addition, it remains of great interest to determine whether autophagy contributes to GPX4 protein degradation in ferroptosis [42, 43].
p53
p53, the most commonly mutated tumor-suppressor gene in human cancers, plays a critical role in the regulation of many aspects of cancer biology, including cell death. p53 regulates apoptosis and autophagy in both a transcription-dependent and -independent manner. For example, p53 in the cytosol blocks autophagy in a transcription-independent manner, whereas p53 in the nucleus activates autophagy in a transcription-dependent way [44]. These findings clearly demonstrate that the subcellular localization of p53 affects its activity in autophagy. Interestingly, recent studies indicate that nuclear p53 is required for ferroptosis in a transcription-dependent way. Nuclear p53 suppresses the expression of SLC7A11 and therefore increases lipid peroxidation [45]. Moreover, nuclear p53 can enhance spermidine/spermine N1-acetyltransferase 1 (SAT1) and glutaminase 2 (GLS2) expression in cancer cells, which results in ferroptosis by triggering lipid peroxidation [46]. This process of p53-mediated ferroptosis is highly regulated by p53 acetylation [47]. A better understanding of the transcription-dependent and -independent roles of p53 should give us key insight into the function of p53 in ferroptosis and tumor biology. The role of p53-mediated autophagy in p53-mediated ferroptosis remains unclear.
NRF2
The nuclear factor erythroid 2-related factor 2 (NRF2) is required for the activation of the cellular antioxidant response to oxidative or electrophilic stresses. This process is involved in increased NRF2 stabilization and subsequent NRF2-mediated gene transcription. Under normal conditions, NRF2 is mainly degraded by the kelch-like ECH-associated protein 1 (KEAP1)-mediated ubiquitin-proteasome pathway. Under oxidative or electrophilic stress, KEAP1-mediated degradation of NRF2 is inhibited; this process is also regulated by autophagy. More importantly, the cargo receptor p62 in autophagy activates NRF2 through inactivation of KEAP1 [48]. Our recent study also demonstrated that the binding of p62 to KEAP1 promotes NRF2 protein stability in hepatocellular carcinoma cells under sorafenib-induced ferroptosis [49]. As a result, activation of the p62-KEAP1-NRF2 pathway limits sorafenib-induced ferroptotic cell death. In particular, metallothionein-1G (a cysteine residues-rich protein) seems to be a key NRF2 target gene contributing to ferroptosis resistance in response to sorafenib in hepatocellular carcinoma cells [50]. These findings provide a potential molecular link between autophagy and ferroptosis in hepatocellular carcinoma cells.
HSPB1
Heat shock protein family B (small) member 1 (HSPB1, also termed HSP25 in mice or HSP27 in humans) is a member of the HSPs that is constitutively expressed in multiple cells or tissues. Under various types of environmental stress, the protein expression of HSPB1 is upregulated and confers cellular resistance to cell death types such as apoptosis. Moreover, phosphorylation of HSPB1 leads to a structural change of HSPB1 from a multimer to a dimer/monomer. This protein post-translational modification increases the ability of HSPB1-mediated actin polymerization and reorganization. Phosphorylated HSPB1 plays a different role in autophagy and apoptosis. Several studies have shown that phosphorylated HSPB1 is required for selective autophagy (e.g., mitophagy and lipophagy) [51–53]. However, phosphorylated HSPB1 induced by erastin blocks cytoskeleton-mediated iron uptake and subsequent lipid peroxidation under ferroptosis [54]. These functional differences may result from the different upstream kinase responses for the phosphorylation of HSPB1 at different sites.
CISD1
CDGSH iron sulfur domain 1 (CISD1, also called mitoNEET) is a mitochondrial protein located in the outer membrane. CISD1 plays an important role in mediating the crosstalk between mitochondrial iron uptake and oxidative stress in both normal and cancer cells. The human CISD1 is a novel target of pioglitazone, a type II diabetes drug. The upregulation of protein expression of CISD1 in cancer cells limits autophagic activity. In contrast, genetic or pharmacological inhibition of CISD1 remarkably increases autophagy in cancer cells [55, 56]. In addition to autophagy, upregulation of CISD1 also limits ferroptotic activity in cancer cells [57]. Knockdown of CISD1 by shRNA increased erastin-induced intramitochondrial lipid peroxidation and subsequent ferroptosis [57]. In contrast, the binding of pioglitazone to CISD1 stabilizes the Fe-S cluster, which blocks mitochondrial iron-mediated lipid peroxidation as well as ferroptosis [57]. These studies suggest that mitochondrial iron uptake through CISD1 is implicated in both autophagy and ferroptosis in cancer cells.
FANCD2
Fanconi anemia (FA) is a genetically heterogeneous recessive disorder due to defective DNA repair. Disruption of iron metabolism plays a key role in the etiology of Fanconi anemia. FANCD2, the central protein of the FA pathway, is activated by mono-ubiquitination in the DNA damage response. Loss of FANCD2 increases DNA damage and cancer incidence. Previous studies show that FANCD2-deficient cells are hypersensitive to oxidative stress and DNA crosses links due to impaired autophagy [58]. Despite that FANCD2-deficient bone marrow stromal cells (BMSCs) are hypersensitive to erastin-induced ferroptosis, the autophagic activity is not significantly changed by FANCD2 deficiency. These findings suggest an autophagy-independent role of FANCD2 in the regulation of ferroptosis in BMSCs. FANCD2-deficient BMSCs exhibit iron overload and lipid peroxidation in response to erastin [59]. In particular, loss of FANCD2 is associated with increased gene expression for iron uptake (e.g., transferrin, transferrin receptor, and HSPB1) and decreased gene expression for iron storage (e.g., FTH) and iron export (e.g., hepcidin antimicrobial peptide) in ferroptosis [59]. These data indicate a critical role of FANCD2 in the protection against ferroptosis in BMSCs.
ACSL4
Lipid peroxidation can be described generally as a core event of ferroptosis. However, the source and identity of lipid death signals that cause ferroptosis are poorly defined. We and other groups recently identified that the acyl-CoA synthetase long-chain family member 4 (ACSL4) plays a key role in the execution of ferroptosis [60–62]. ACSL4 (but not other ACSLs including ACSL1, ACSL3, ACSL5, and ACSL6) expression correlates with cellular sensitivity to erastin-induced ferroptosis [60]. Mechanically, ACSL4-mediated production of 5-hydroxyeicosatetraenoic acid and subsequent production of oxidized species of phosphatidylethanolamines is essential for ferroptosis [60–62]. These findings indicate that ACSL4 is a not only a biomarker, but also a contributor of ferroptosis [60]. Interestingly, ACSL4 is also involved in the regulation of the activity of mechanistic target of rapamycin (mTOR) complex I (mTORC1) and mTOR complex II (mTORC2) [63]. These two mTOR complexes have different functions in cell growth, metabolism, and autophagy. mTORC2 can inhibit autophagy by activation of mTORC1 [64]. The direct contribution of mTOR in ACSL4-mediated ferroptosis needs further investigation.
HSPA5
The heat shock 70kDa protein 5 (HSPA5, also termed GRP78 or BIP) is a member of the molecular chaperones expressed primarily in the endoplasmic reticulum. As a key component of the unfolded protein response, HSPA5 promotes cell survival under conditions of endoplasmic reticulum stress-induced autophagy. A recent study demonstrated that upregulation of HSPA5 is a negative regulator of ferroptosis in pancreatic cancer cells [65]. Increased HSPA5 expression limits lipid peroxidation in ferroptosis by directly protecting against GPX4 degradation [65].
Conclusion
In the context of biological diversity, multiple types of cell death exist and have been reported. As a novel RCD, ferroptosis occurs in cells when iron accumulation and lipid peroxidation is activated. A high level of ferroptosis not only selectively kills cancer cells, but also causes tissue injury. Autophagy as a degradation pathway plays a dual role - either pro-survival or pro-death - depending on many conditions. Activation of autophagy seems to contribute to ferroptosis. However, the mechanism of autophagy-mediated ferroptosis remains largely unknown. Another important area is delineating whether and how lipid peroxidation impacts autophagosome formation as a feedback loop.
Acknowledgments
We thank Christine Heiner (Department of Surgery, University of Pittsburgh) for her critical reading of the manuscript. This work was supported by the National Institutes of Health of the USA (R01GM115366 and R01CA160417), the National Natural Science Foundation of China (31671435), the National Natural Science Foundation of Guangdong (2016A030308.), and a Research Scholar Grant from the American Cancer Society (RSG-16-014-01-CDD).
List of Abbreviations
- RCD
regulated cell death
- ATG
autophagy-regulated
- TEM
transmission electron microscopy
- RSL
RAS-selective lethal
- RIPK
receptor interacting serine/threonine kinase
- ROS
reactive oxygen species
- NCOA4
nuclear receptor coactivator 4
- GSH
glutathione
- SLC7A11
solute carrier family 7 member 11
- GPX
glutathione peroxidase
- SAT1
spermidine/spermine N1-acetyltransferase 1
- GLS2
glutaminase 2
- NRF2
nuclear factor erythroid 2-related factor 2
- KEAP1
kelch-like ECH-associated protein 1
- HSPB1
heat shock protein family B (small) member 1
- CISD1
CDGSH iron sulfur domain 1
- FA
fanconi anemia
- ACSL4
acyl-CoA synthetase long-chain family member 4
- mTOR
mechanistic target of rapamycin
- FTH
ferritin heavy chain
- FTL
ferritin light chain
- BMSCs
bone marrow stromal cells
- HSPA5
the heat shock 70kDa protein 5
Footnotes
Compliance with Ethical Guidelines
Conflict of Interest: Rui Kang and Daolin Tang declare that they have no conflicts of interest.
Human and Animal Rights and Informed Consent: This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, are highlighted as follows:
• of importance
• of major importance
- 1.Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell death and differentiation. 2012;19(1):107–20. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conrad M, Angeli JP, Vandenabeele P, Stockwell BR. Regulated necrosis: disease relevance and therapeutic opportunities. Nature reviews Drug discovery. 2016;15(5):348–66. doi: 10.1038/nrd.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galluzzi L, Bravo-San Pedro JM, Kepp O, Kroemer G. Regulated cell death and adaptive stress responses. Cellular and molecular life sciences : CMLS. 2016;73(11–12):2405–10. doi: 10.1007/s00018-016-2209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nature reviews Molecular cell biology. 2014;15(2):135–47. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 5.Galluzzi L, Bravo-San Pedro JM, Vitale I, Aaronson SA, Abrams JM, Adam D, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell death and differentiation. 2015;22(1):58–73. doi: 10.1038/cdd.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Y, Hou W, Song X, Yu Y, Huang J, Sun X, et al. Ferroptosis: process and function. Cell death and differentiation. 2016;23(3):369–79. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Hou W, Xie Y, Song X, Sun X, Lotze MT, Zeh HJ, 3rd, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12(8):1425–8. doi: 10.1080/15548627.2016.1187366. This study demonstrated the importance of autopahgy in the induciton of ferroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Gao M, Monian P, Pan Q, Zhang W, Xiang J, Jiang X. Ferroptosis is an autophagic cell death process. Cell research. 2016;26(9):1021–32. doi: 10.1038/cr.2016.95. This study demonstrated the importance of autopahgy in the induciton of ferroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Torii S, Shintoku R, Kubota C, Yaegashi M, Torii R, Sasaki M, et al. An essential role for functional lysosomes in ferroptosis of cancer cells. The Biochemical journal. 2016;473(6):769–77. doi: 10.1042/BJ20150658. This study demonstrated the importance of lysosomes in the induciton of ferroptosis. [DOI] [PubMed] [Google Scholar]
- 11.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS letters. 1993;333(1–2):169–74. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 14.Mohammadi D. 2016 Nobel prize in medicine goes to Japanese scientist. Lancet. 2016;388(10054):1870. doi: 10.1016/S0140-6736(16)31797-4. [DOI] [PubMed] [Google Scholar]
- 15.Yao Z, Delorme-Axford E, Backues SK, Klionsky DJ. Atg41/Icy2 regulates autophagosome formation. Autophagy. 2015;11(12):2288–99. doi: 10.1080/15548627.2015.1107692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, et al. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy. 2015;11(1):28–45. doi: 10.4161/15548627.2014.984267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell death and differentiation. 2011;18(4):571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzwalter BE, Thorburn A. Recent insights into cell death and autophagy. The FEBS journal. 2015;282(22):4279–88. doi: 10.1111/febs.13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell death and differentiation. 2015;22(3):367–76. doi: 10.1038/cdd.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nature reviews Molecular cell biology. 2014;15(2):81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157(1):65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nature reviews Cancer. 2015;15(12):747–56. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nature reviews Drug discovery. 2014;13(11):828–51. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer cell. 2003;3(3):285–96. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 25.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chemistry & biology. 2008;15(3):234–45. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dixon SJ, Patel DN, Welsch M, Skouta R, Lee ED, Hayano M, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife. 2014;3:e02523. doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eling N, Reuter L, Hazin J, Hamacher-Brady A, Brady NR. Identification of artesunate as a specific activator of ferroptosis in pancreatic cancer cells. Oncoscience. 2015;2(5):517–32. doi: 10.18632/oncoscience.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louandre C, Ezzoukhry Z, Godin C, Barbare JC, Maziere JC, Chauffert B, et al. Iron-dependent cell death of hepatocellular carcinoma cells exposed to sorafenib. International journal of cancer Journal international du cancer. 2013;133(7):1732–42. doi: 10.1002/ijc.28159. [DOI] [PubMed] [Google Scholar]
- 29.Linkermann A, Skouta R, Himmerkus N, Mulay SR, Dewitz C, De Zen F, et al. Synchronized renal tubular cell death involves ferroptosis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(47):16836–41. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsushita M, Freigang S, Schneider C, Conrad M, Bornkamm GW, Kopf M. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. The Journal of experimental medicine. 2015;212(4):555–68. doi: 10.1084/jem.20140857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and Transferrin Regulate Ferroptosis. Molecular cell. 2015;59(2):298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nature cell biology. 2014;16(12):1180–91. doi: 10.1038/ncb3064. This study demonstrated the importance of ferroptosis in tissue injury. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends in cell biology. 2016;26(3):165–76. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Y, Song X, Sun X, Huang J, Zhong M, Lotze MT, et al. Identification of baicalein as a ferroptosis inhibitor by natural product library screening. Biochemical and biophysical research communications. 2016;473(4):775–80. doi: 10.1016/j.bbrc.2016.03.052. [DOI] [PubMed] [Google Scholar]
- 35••.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–31. doi: 10.1016/j.cell.2013.12.010. This study demonstrated the importance of GPX4 in the regualtion of ferroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509(7498):105–9. doi: 10.1038/nature13148. This study demonstrated the importance of NCOA4 in the regualtion of ferritinophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oppenheim EW, Adelman C, Liu X, Stover PJ. Heavy chain ferritin enhances serine hydroxymethyltransferase expression and de novo thymidine biosynthesis. The Journal of biological chemistry. 2001;276(23):19855–61. doi: 10.1074/jbc.M100039200. [DOI] [PubMed] [Google Scholar]
- 38.Desideri E, Filomeni G, Ciriolo MR. Glutathione participates in the modulation of starvation-induced autophagy in carcinoma cells. Autophagy. 2012;8(12):1769–81. doi: 10.4161/auto.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mancilla H, Maldonado R, Cereceda K, Villarroel-Espindola F, Montes de Oca M, Angulo C, et al. Glutathione Depletion Induces Spermatogonial Cell Autophagy. Journal of cellular biochemistry. 2015;116(10):2283–92. doi: 10.1002/jcb.25178. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Hambright WS, Na R, Ran Q. Ablation of the Ferroptosis Inhibitor Glutathione Peroxidase 4 in Neurons Results in Rapid Motor Neuron Degeneration and Paralysis. The Journal of biological chemistry. 2015;290(47):28097–106. doi: 10.1074/jbc.M115.680090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garg AD, Dudek AM, Ferreira GB, Verfaillie T, Vandenabeele P, Krysko DV, et al. ROS-induced autophagy in cancer cells assists in evasion from determinants of immunogenic cell death. Autophagy. 2013;9(9):1292–307. doi: 10.4161/auto.25399. [DOI] [PubMed] [Google Scholar]
- 42.Yu Y, Xie Y, Cao L, Yang L, Yang M, Lotze MT, et al. The ferroptosis inducer erastin enhances sensitivity of acute myeloid leukemia cells to chemotherapeutic agents. Molecular & cellular oncology. 2015;2(4):e1054549. doi: 10.1080/23723556.2015.1054549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimada K, Skouta R, Kaplan A, Yang WS, Hayano M, Dixon SJ, et al. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nature chemical biology. 2016;12(7):497–503. doi: 10.1038/nchembio.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tasdemir E, Chiara Maiuri M, Morselli E, Criollo A, D'Amelio M, Djavaheri-Mergny M, et al. A dual role of p53 in the control of autophagy. Autophagy. 2008;4(6):810–4. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- 45••.Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57–62. doi: 10.1038/nature14344. This study demonstrated the importance of p53 in the regualtion of ferroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ou Y, Wang SJ, Li D, Chu B, Gu W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(44):E6806–E12. doi: 10.1073/pnas.1607152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang SJ, Li D, Ou Y, Jiang L, Chen Y, Zhao Y, et al. Acetylation Is Crucial for p53-Mediated Ferroptosis and Tumor Suppression. Cell reports. 2016;17(2):366–73. doi: 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature cell biology. 2010;12(3):213–23. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 49••.Sun X, Ou Z, Chen R, Niu X, Chen, Kang R, et al. Activation of the p62-Keap1-NRF2 Pathway Protects against Ferroptosis in Hepatocellular Carcinoma Cells. Hepatology. 2015 doi: 10.1002/hep.28251. This study demonstrated the importance of NRF2 in the regualtion of ferroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Sun X, Niu X, Chen R, He W, Chen D, Kang R, et al. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology. 2016;64(2):488–500. doi: 10.1002/hep.28574. This study demonstrated the importance of MT-1G in the regualtion of ferroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B, et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell metabolism. 2011;13(6):701–11. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen L, Qi Z, Zhu Y, Song X, Xuan C, Ben P, et al. Phosphorylated heat shock protein 27 promotes lipid clearance in hepatic cells through interacting with STAT3 and activating autophagy. Cellular signalling. 2016;28(8):1086–98. doi: 10.1016/j.cellsig.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 53.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, et al. Endogenous HMGB1 regulates autophagy. The Journal of cell biology. 2010;190(5):881–92. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Sun X, Ou Z, Xie M, Kang R, Fan Y, Niu X, et al. HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene. 2015;34(45):5617–25. doi: 10.1038/onc.2015.32. This study demonstrated the importance of HSPB1in the regualtion of ferroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamir S, Rotem-Bamberger S, Katz C, Morcos F, Hailey KL, Zuris JA, et al. Integrated strategy reveals the protein interface between cancer targets Bcl-2 and NAF-1. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(14):5177–82. doi: 10.1073/pnas.1403770111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sohn YS, Tamir S, Song L, Michaeli D, Matouk I, Conlan AR, et al. NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(36):14676–81. doi: 10.1073/pnas.1313198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Yuan H, Li X, Zhang X, Kang R, Tang D. CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochemical and biophysical research communications. 2016;478(2):838–44. doi: 10.1016/j.bbrc.2016.08.034. This study demonstrated the importance of CISD1in the regualtion of ferroptosis. [DOI] [PubMed] [Google Scholar]
- 58••.Sumpter R, Jr, Sirasanagandla S, Fernandez AF, Wei Y, Dong X, Franco L, et al. Fanconi Anemia Proteins Function in Mitophagy and Immunity. Cell. 2016;165(4):867–81. doi: 10.1016/j.cell.2016.04.006. This study demonstrated the importance of FA proteins in the regualtion of autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59•.Song X, Xie Y, Kang R, Hou W, Sun X, Epperly MW, et al. FANCD2 protects against bone marrow injury from ferroptosis. Biochemical and biophysical research communications. 2016;480(3):443–9. doi: 10.1016/j.bbrc.2016.10.068. This study demonstrated the importance of FANCD2in the regualtion of ferroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60••.Yuan H, Li X, Zhang X, Kang R, Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochemical and biophysical research communications. 2016;478(3):1338–43. doi: 10.1016/j.bbrc.2016.08.124. This study demonstrated the importance of ACSL4 in the regualtion of ferroptosis. [DOI] [PubMed] [Google Scholar]
- 61••.Kagan VE, Mao G, Qu F, Angeli JP, Doll S, Croix CS, et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nature chemical biology. 2016 doi: 10.1038/nchembio.2238. This study demonstrated the importance of ACSL4 in the regualtion of ferroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62••.Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nature chemical biology. 2016 doi: 10.1038/nchembio.2239. This study demonstrated the importance of ACSL4 in the regualtion of ferroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orlando UD, Castillo AF, Dattilo MA, Solano AR, Maloberti PM, Podesta EJ. Acyl-CoA synthetase-4, a new regulator of mTOR and a potential therapeutic target for enhanced estrogen receptor function in receptor-positive and -negative breast cancer. Oncotarget. 2015;6(40):42632–50. doi: 10.18632/oncotarget.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS letters. 2010;584(7):1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu S, Zhang Q, Sun X, Zeh HJ, Lotze MT, Kang R, et al. HSPA5 Regulates Ferroptotic Cell Death in Cancer Cells. Cancer research. 2017 doi: 10.1158/0008-5472.CAN-16-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]