ABSTRACT
Discovery of yeast autophagy-related (ATG) genes and subsequent identification of their homologs in other organisms have enabled researchers to investigate physiological functions of macroautophagy/autophagy using genetic techniques. Specific identification of autophagy-related structures is important to evaluate autophagic activity, and specific ablation of autophagy-related genes is a critical means to determine the requirements of autophagy. Here, we review currently available mouse models, particularly focusing on autophagy (and mitophagy) indicator models and systemic autophagy-related gene-knockout mouse models.
KEYWORDS: autophagy, knockout mouse, mitophagy, reporter mouse, selective autophagy
Introduction
In the past decade, knowledge of the physiological roles of autophagy in mammals has increased. Autophagy plays important roles in many physiological and disease processes, including starvation adaptation, quality control of intracellular proteins and organelles, development, tumor suppression, prevention of neurodegeneration, and immunity.1 For analysis of autophagy in vivo, 2 types of gene-modified mouse models have played critical roles: “autophagy-monitoring mice” and “autophagy-deficient mice." Monitoring the autophagic process and measuring autophagic activity are critical to investigate the function of autophagy. In the past, electron microscopy was mainly used to monitor autophagy because specific autophagosome markers were lacking. Soon after the identification of MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3), a mammalian homolog of yeast Atg8, as an autophagosome marker,2 transgenic mice systemically expressing EGFP-LC3 (GFP-LC3 mice) were generated.3 By using GFP-LC3 mice, observation and quantification of autophagy in vivo became drastically simplified, and this transgenic mouse line has been widely used for monitoring autophagy.
Another tool that has greatly contributed to understanding the role of autophagy in vivo is mice deficient in specific Atg genes. It is notable that analysis of autophagy-deficient mice has led to conceptual findings in this field. For example, the concept of “intracellular quality control” has been established based on the observation of mice with neuron-specific and liver-specific deletion of Atg5 or Atg7. The importance of autophagy in quality control of proteins and organelles is difficult to observe in yeast, probably due to dilution of damaged cellular components because of their rapid cell division. In mouse models, autophagy deficiency in the brain and liver causes accumulation of ubiquitinated proteins, damaged organelles, and autophagy-specific substrates such as SQSTM1/p62. These findings suggest that autophagy is critical for preventing the toxic accumulation of damaged proteins and organelles in mammals. In addition, autophagic dysfunction has been implicated in many pathological processes including human diseases. Here, we discuss the advanced utilization of autophagy-monitoring and autophagy-deficient mice for analysis of the physiological roles of autophagy in vivo.
Monitoring and measuring autophagy in vivo
The ‘core’ ATG genes
From yeast genetic studies, 41 ATG genes have been identified. Among them, ATG1 through ATG10, ATG12 through ATG14, ATG16 through ATG18, ATG29, and ATG31 are required for efficient autophagosome formation. These 18 ‘core’ ATG genes consist of several functional units; (i) Atg1 kinase and its regulators (Atg1, Atg13, Atg17, Atg29, Atg31), (ii) the phosphatidylinositol 3-kinase (PtdIns3K) complex (Vps30/Atg6, Atg14), (iii) the Atg12 conjugation system (Atg5, Atg7, Atg10, Atg12, Atg16), (iv) the Atg8 conjugation system (Atg3, Atg4, Atg7, Atg8), (v) Atg9, and (vi) the Atg2-Atg18 complex. Many of these proteins are conserved in mammals, and identification of the corresponding genes provides the means to assess autophagic activity and study the role of autophagy. There are several steps for the complete pathway of autophagy: initiation, phagophore expansion, autophagosome maturation, fusion with the vacuole/lysosome, cargo degradation in the lysosome and efflux. Therefore, it is important to assess autophagic activity at each step or to monitor flux (from initiation to degradation). Among the ATG proteins, LC3, one of the Atg8 homologs in mammals, is established as an autophagosome marker and many of the ‘core’ Atg genes have been knocked out for understanding the physiological role of autophagy in mammals.
GFP-LC3 transgenic mice
Mammals have several counterparts of yeast ATG8, one of the core ATG genes; MAP1LC3A, MAP1LC3B, MAP1LC3B2, MAP1LC3C and GABARAP, GABARAPL1, GABARAPL2. The LC3 and GABARAP family proteins are particularly useful for monitoring autophagy. Among the proteins in this family, LC3B is the most widely used marker. Precursor forms of the LC3 and GABARAP family proteins are specifically cleaved at the carboxyl terminus by ATG4 family proteins to form LC3-I (or GABARAP-I, etc.; we will refer exclusively to LC3 hereafter), which has an exposed carboxyl terminal glycine that is conjugated to phosphatidylethanolamine to form LC3-II. LC3-II is tightly bound to both the inner and outer surface of the phagophore and autophagosomal membrane. When the autophagosome fuses with the lysosome, LC3-II on the outer membrane is again cleaved by ATG4 proteins and released from the membranes. By contrast, LC3-II on the inner membrane is degraded by lysosomal enzymes. Therefore, the autophagosome is monitored by tracking LC3.
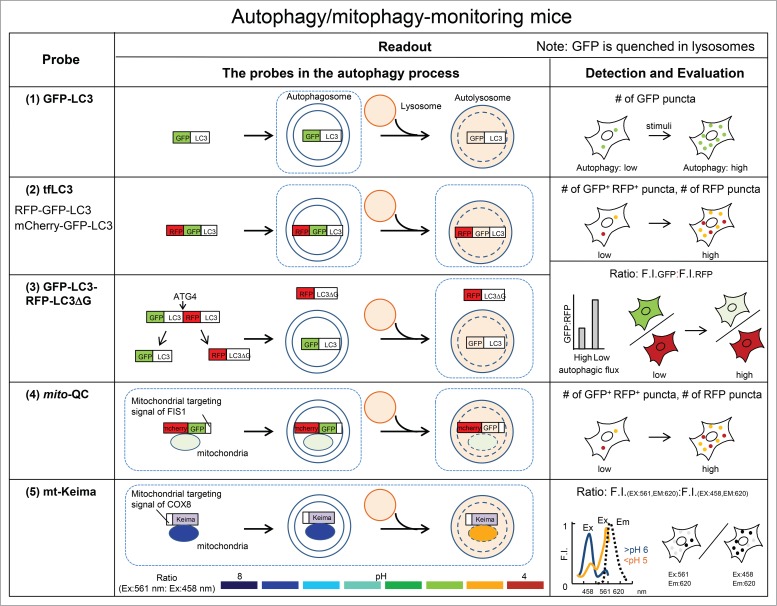
Immunostaining or fluorescent tagging is used for detection of LC3. EGFP-tagged LC3 was the first probe used to monitor autophagy (Fig. 1).2 Because LC3 is cleaved at the carboxyl terminus, EGFP should be fused at the N terminus of LC3. With this probe, autophagosomes are easily detected as fluorescent puncta or sometimes as ring-shaped structures. Although monitoring GFP-LC3 puncta is the most popular assay, it has a limitation. The fluorescent signal of GFP is quenched quickly under the low pH conditions in lysosomes. Therefore, it is difficult to determine how much GFP-LC3 is delivered to lysosomes unless quenching of GFP is inhibited by lysosomotropic reagents. Due to this limitation, a simple increase in the number of GFP-LC3 puncta is not sufficient to conclude that autophagy is activated. The increased number of autophagosomes could represent either induction of autophagy or impairment of autophagosome-lysosome fusion or lysosomal degradation. Therefore, it is critical to distinguish between these options by treatment with lysosomal inhibitors.
Figure 1.
Transgenic mice expressing autophagy probes for monitoring autophagy. (1) GFP-LC3 mice express EGFP-LC3 for labeling phagophores and autophagosomes. The number of GFP puncta is counted before and after stimulation. (2) tfLC3 mice express RFP-EGFP-LC3 or mCherry-EGFP-LC3 for measuring autophagy flux. The numbers of GFP+ RFP+ puncta (yellow) and RFP puncta (red) are counted. Yellow puncta represent phagophores and autophagosomes, and red puncta represent autolysosomes. (3) GFP-LC3-RFP-LC3ΔG mice express GFP-LC3-RFP-LC3ΔG for measuring autophagy flux. The reduction of the fluorescence intensity of GFP indicates autophagy flux because GFP-LC3 is degraded though autophagy (and quenched) as a substrate. The reduction of the fluorescence intensity of GFP-LC3 in the total cells is measured and corrected by the fluorescence intensity of RFP-LC3∆G in the total cells as an internal control. (4) Mito-QC mice express mCherry-EGFP fused with the FIS1 mitochondrial targeting sequence for monitoring mitophagy. The numbers of GFP+ RFP+ (yellow) puncta and RFP puncta (red) are counted. Yellow puncta represent mitochondria, and red puncta represent mitochondria in the lysosomes. (5) Mt-Keima mice express Keima (pH-dependent fluorescent protein) fused with the COX8 mitochondrial-targeting sequence for monitoring mitophagy. mt-Keima is excited predominantly by 458-nm light in a neutral environment (mitochondria), and by 561-nm light in an acidic environment (lysosome). The ratio of the 561-nm:458-nm excited Keima fluorescence intensity indicates the delivery of mitochondria to the lysosome. Squares with a blue dotted-line show the stages detected with the probes. F.I., fluorescence intensity.
The use of EGFP-LC3 has also been applied to mice. Transgenic mice expressing GFP-LC3 in all tissues under the control of the constitutive Cag promoter have been widely used.3 The presence of autophagosomes in mouse tissues can be directly monitored by fluorescence microscopy analysis of cryosections. GFP-LC3 mice were validated by intercrossing with Atg-deficient mice.4 However, in contrast to cells in culture, it is more challenging to measure autophagy flux with transgenic mice. For example, chloroquine has been used for monitoring autophagic flux in mice, but chloroquine treatment is not as robust as has been demonstrated in culture cells, and the efficiency is altered among organs,5 probably due to detoxification of intraperitoneally administered inhibitors, barriers for their access into the tissues such as the blood-brain barrier, and other factors. Therefore, reporter mice expressing probes that could measure autophagic flux without using pharmacological inhibitors of lysosome function have been anticipated (see below).
TfLC3 transgenic mice
Because red fluorescent protein (RFP) is more resistant to quenching in the lysosome than EGFP, tandem fluorescent-tagged LC3 (tfLC3) reporters such as mRFP-EGFP-LC3 and mCherry-EGFP-LC3 have been developed as single-molecule probes to detect autophagosomes and autolysosomes specifically.6 In green and red merged images with this probe, autophagosomes are labeled with yellow (mRFP and GFP) and autolysosomes are labeled with red (mRFP only) signals. Therefore, the entire process of autophagy can be traced morphologically with this probe. When autophagy is activated, the numbers of both yellow (autophagosomes) and red (autolysosomes) puncta increase; by contrast, when autophagosome maturation into autolysosomes is blocked, only the number of yellow puncta (autophagosomes) increases.6
These probes have also been applied to in vivo assays by virus-mediated intracranial administration of tfLC3 and have been used to generate transgenic tfLC3 mice.7-9 tfLC3 mice were validated by examining the response to starvation and drugs such as rapamycin, which are established autophagy-inducing conditions. In the heart, autophagy is increased by ischemia-reperfusion injury through an oxidative stress-dependent mechanism.9 Also in renal proximal tubules, autophagy is activated after ischemia-reperfusion injury, which might be for clearance of damaged organelles and remodeling of tubular cells during renal repair.8
GFP-LC3-RFP-LC3ΔG transgenic mice
Another reporter model for assessing autophagic flux is GFP-LC3-RFP-LC3ΔG mice.10 The GFP-LC3-RFP-LC3ΔG probe was developed for accurate quantification of autophagic flux by detecting a reduction of GFP-LC3 signals in the lysosomes. When expressed in cells, GFP-LC3-RFP-LC3ΔG is cleaved by endogenous ATG4, producing equimolar amounts of GFP-LC3 and RFP-LC3ΔG. GFP-LC3 on the autophagosomal inner membrane is delivered into the lysosome and quenched. By contrast, RFP-LC3ΔG is not lipidated due to lack of the carboxyl-terminal glycine, but remains in the cytosol and serves as an internal control. Thus, the GFP:RFP ratio is reduced if autophagic activity is enhanced. In the case of GFP-LC3 and tfLC3 probes, LC3 is used as a marker for labeling autophagosome structures, and the readout is the number of GFP-LC3 and of RFP-LC3 puncta. In contrast, with the GFP-LC3-RFP-LC3ΔG probe, GFP-LC3 is used as a substrate for autophagic degradation (and quenching of the GFP signal), and the readout is the fluorescence intensity of GFP and RFP in the total cell population. The GFP:RFP ratio can be measured using a fluorescence microscope, flow cytometer, or microtiter plate reader.
The GFP-LC3-RFP-LC3ΔG probe can also be used to monitor autophagic flux in vivo. Zebrafish and mice expressing GFP-LC3-RFP-LC3ΔG were generated (the mouse model only expresses an adequate amount of the probe in muscles). Zebrafish expressing GFP-LC3-RFP-LC3ΔG were validated by intercrossing with Atg-deficient animals. Using this probe, it has been successfully observed that autophagy flux is induced by fertilization in both zebrafish and mouse eggs. One notable advantage of this probe is that it is capable of measuring basal autophagy activity. Because basal autophagic activity is usually very low and fluctuates at different time points dependent on nutrient availability and other factors, it is difficult to evaluate by a “snapshot” assay. This probe can overcome these inherent difficulties because it can measure basal autophagic flux as an overall cumulative index. The reporter mouse revealed that the level of autophagy is relatively high in fast-twitch type II fibers compared with slow-twitch type I fibers in the muscle of fed mice. As basal autophagy plays an important role in quality control in vivo, the ability to measure basal autophagy is a strong advantage of this probe.
Although both GFP-LC3-RFP-LC3ΔG and tfLC3 probes are available to measure autophagic flux without requiring the use of lysosomal inhibitors, which is a great advantage especially in mouse experiments, they also have their own limitations. A limitation of GFP-LC3-RFP-LC3ΔG is that the time base for analysis is in hours, because this probe quantifies autophagic flux as the reduction of the GFP signal in a whole cell. Conversely, the tfLC3 probe measures autophagic flux by detecting the number of tfLC3 puncta, which is clearly observed within 30 min after starvation treatment in cultured cells. Therefore, tfLC3 could be more suitable for measuring flux in short-term experiments.
It has been reported that tfLC3 is also available for measuring autophagic flux by detecting the GFP:RFP ratio, similar to GFP-LC3-RFP-LC3ΔG.11,12 However, the lysosome-targeted RFP of tfLC3 was not originally designed to be used as an internal control; RFP is less stable in the lysosome compared with that in the cytosol, especially in long-term experiments. Therefore, RFP-LC3ΔG should be more suitable for an internal control than tfLC3 for measuring the GFP:RFP ratio. It is important to choose an appropriate method and, ideally, a combination of at least 2 methods would be preferred.
Mitophagy-monitoring mice
Selective degradation of mitochondria by autophagy, termed mitophagy, can be a selective pathway to remove damaged mitochondria, and could be a major means of mitochondrial quality control. However, the importance of mitophagy in vivo remains unclear. So far, 2 mouse models have been developed for monitoring mitophagy: mt-Keima transgenic mice and mito-QC transgenic mice.13,14 A mitochondria-targeted form of Keima (mt-Keima), a ratiometric fluorescence probe, is engineered by fusing a tandem repeat of the COX8 pre-sequence with Keima.15 Keima is a lysosome-resistant fluorescent protein that is activated in a unique manner. mt-Keima is excited predominantly by 458-nm light in a neutral environment (mitochondria), and by 561-nm light in an acidic environment (lysosome). The ratio of the 561-nm:458-nm excited Keima fluorescence intensity indicates the delivery of mitochondria to lysosomes, i.e., the mitophagic activity.
Transgenic mice expressing mt-Keima have been generated.13 mt-Keima mice were validated by intercrossing Atg7-deficient mice and checking colocalization with organelles such as mitochondria and lysosomes in mouse embryonic fibroblasts and tissues. As lysosomal activity should be maintained to produce correct acidic signals, it is important to maintain tissue viability during microscopy observation. Using mice expressing mt-Keima, it has been shown that mitophagy can be induced by several stimuli/stresses such as hypoxia, mitochondrial DNA mutations, and tumors, and it can be reduced by high-fat diet, aging, and HTT (huntingtin) expression. In heart, mitophagy is transiently activated and then suppressed in response to pressure overload.16
Another probe for monitoring mitophagy is mito-QC.17 Mito-QC is engineered by fusing the mitochondrial targeting sequence of the outer mitochondrial membrane proteins FIS1 to a tandem mCherry-GFP tag (mCherry-GFP-FIS1; mito-QC). As with autophagosomes labeled with tfLC3, the cytoplasmic mitochondria display both red and green signals. Upon mitophagy, mitochondria are delivered to lysosomes and mito-QC produces only red signals. Transgenic mice harboring mito-QC in the Rosa26 locus are generated.14 mito-QC mice were also validated by checking colocalization with organelles such as mitochondria, lysosomes, and autophagosomes in tissues. Validity was further confirmed by electron microscopy analysis in tissues. Using mice expressing mito-QC, it has been shown that basal mitophagic activity differs among tissues and cell types. The kidneys are a major site of mitophagy, and cardiomitophagy is activated during development in mice.14
Although both mt-Keima and mito-QC mouse models are valuable resources and their utility is based on a similar concept that mitophagy is detected by a change in pH, they have different advantages and limitations. The limitation of the mt-Keima mouse is that the Keima protein is incompatible with fixation, and freshly excised organs are required for observation. Therefore, this probe is incompatible with performing immunohistochemical experiments. Another limitation of mt-Keima is that the excitation spectra between acidic and neutral environments are not completely separated, but this problem will be improved by genetic alterations in the structure of Keima in the future.
There is a growing interest in the physiological and pathological role of mitophagy. Assessing mitophagic activity in diseased states in which mitophagy is thought to be involved (e.g., neurodegenerative diseases and heart diseases) by crossing the mitophagy monitoring-mice with pathological mouse models may provide valuable insights into the role of mitophagy in vivo.
Analysis of the role of autophagy using autophagy-deficient mice
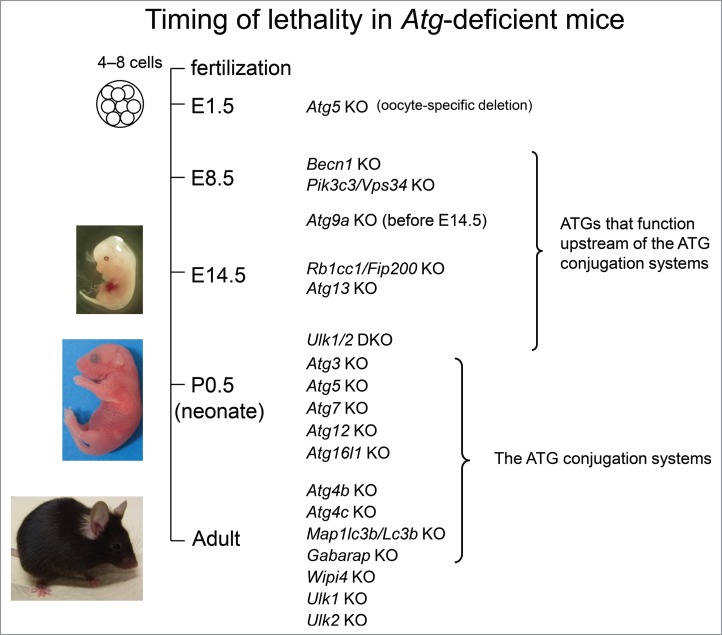
Analysis of Atg gene knockout mice has greatly contributed to understanding the physiological roles of autophagy in vivo. There are approximately 20 core autophagy-related genes involved in autophagosome formation in mammals. So far, 14 of them have been knocked out in mice (Table 1, Fig. 2). These Atg knockout mice die at different stages: (1) some die in utero, (2) some are born alive but die within 1 d of birth, and (3) some show no obvious abnormalities. atg4b−/−, atg4c−/−, lc3b−/−, gabarap−/−, ulk1−/−, and ulk2−/− mice show no obvious (or weak) phenotypic effects. This is probably because these Atg genes have homologs that function redundantly. Mice deficient in nonredundant genes involved in the ATG conjugation systems and in ulk1−/- ulk2−/− (double-knockout mice) were neonatal lethal, and mice deficient in nonredundant genes not involved in the ATG conjugation systems were embryonic lethal. The reasons for the phenotypic differences between Atg mutant mice are not well understood. Some of the autophagy receptors involved in selective autophagy have also been knocked out in mice.
Table 1.
Mouse defects associated with Atg gene knockouts (systemic knockout mouse).
| Atg gene | Survival time | Phenotypes | Ref. of the phenotypes |
|---|---|---|---|
| Becn1 | E8.5 | Defects in proamniotic canal closure, (spontaneous tumors in heterozygotes) | (28,29) |
| Pik3c3/Vps34 | E8.5 | Fail to form mesoderm, reduced cell proliferation | (30) |
| Atg9a | Embryonic lethal | Growth retardation | (31) |
| Rb1cc1 | E14.5-E16.5 | Defective heart and liver development | (27) |
| Atg13 | E17.5 | Growth retardation | (26) |
| Defective heart development | |||
| Ulk1/2 | Neonatal lethal | Impaired lung function | (25) |
| Atg3 | Neonatal lethal | Die within one d after birth, | (17) |
| Atg5 | Neonatal lethal | morphologically normal, | (18) |
| reduced amino acids levels, suckling defect | |||
| Atg7 | Neonatal lethal | (19) | |
| Atg12 | Neonatal lethal | (20) | |
| Atg16l1 | Neonatal lethal | (21) | |
| Atg4b | Viable | Balance dysfunction | (54) |
| Atg4c | Viable | Increased susceptibility to carcinogen-induced fibrosarcomas | (16) |
| Map1lc3b | Viable | No obvious abnormality | (14) |
| Gabarap | Viable | No obvious abnormality | (15) |
| Ulk1 | Viable | Increased reticulocyte number, delayed mitochondrial clearance | (55) |
| Ulk2 | Viable | No obvious abnormality | (56) |
Figure 2.
Timing of lethality in autophagy-deficient mice. Mice deficient in genes involved in the ATG conjugation system (atg3−/−, atg5−/−, atg7−/−, atg12−/−, atg16l1−/−) and ulk1/2 DKO mice are born with normal morphology but die within 1 d after birth. Mice deficient in genes functioning upstream of the ATG conjugation system (rb1cc1−/−, atg13−/−, becn1−/−, uvrag−/−, pik3c3/vps34−/−, atg9a−/−) die in utero. Others (atg4b−/−, atg4c−/−, lc3b−/−, gabarap−/−, ulk1−/−, ulk2−/−) show no obvious (or weak) phenotypic effects.
Deletion of Atg genes in the conjugation systems
Systemic knockout mice of Atg genes in the conjugation system
The Atg12 and Atg8 conjugation systems consist of 8 Atg proteins in yeast (Atg3, Atg4, Atg5, Atg7, Atg8, Atg10, Atg12, Atg16). Mice deficient in each of these proteins (except Atg10) have been generated. Among them, lc3b−/−, gabarap−/−, atg4b−/−, and atg4c−/— mice show no obvious (or weak) phenotypic effects.18-20 Mice deficient in nonredundant genes involved in the 2 conjugation systems (atg3−/−, atg5−/−, atg7−/−, atg12−/−, atg16l1−/−) are born developmentally normal but die within 1 d of birth.4,21-24 Autophagy is massively induced immediately after birth as an adaptation response to starvation due to the sudden interruption of the transplacental nutrient supply.4 During this neonatal starvation period, amino acid supply through autophagic degradation of ‘self’ proteins is important for the maintenance of amino acid levels.4,21,22 However, the exact cause of death had remained unknown. Recently, it was reported that neuronal dysfunction caused by autophagy deficiency, resulting in suckling failure, is the primary cause of the neonatal lethality of Atg5-null mice; neuron-specific expression of ATG5 can rescue these mice.25 Neonates defective in autophagy suffer from and die as a result of a combination of serious malnutrition due to a lack of self-derived nutrients and suckling failure. Therefore, autophagy plays 2 critical roles in neonates: prevention of neuronal dysfunction and adaptation to starvation.
It has been considered that neonatal lethality is the phenotype of autophagy-deficient mice because it is commonly observed in mice deficient in Atg genes encoding proteins involved in the conjugation systems; however, another possibility has emerged. Recently, it was reported that autophagosomes could be formed even in the absence of the ATG conjugation systems, although at a reduced rate.26 The autophagosomes in ATG conjugation system-deficient cells could fuse with lysosomes; however, degradation of the inner membrane of autophagosomes was significantly delayed. Thus, even though autophagic activity in ATG conjugation system-deficient cells is strongly suppressed, a very low level of autophagic activity might remain, leading to a milder phenotype observed in ATG conjugation system-deficient mice compared with those with deletion of upstream Atg genes, such as Rb1cc1 or Atg13 (Fig. 2).26
Systemic Atg7 and Atg5 knockout in adult mice
For physiological analysis, conditional tissue-specific Atg gene knockout mice have been mainly used, because embryonic or neonatal lethality is observed in conventional Atg gene knockout mice. However, it is also possible to investigate autophagy in the whole body in adult mice by using conditional Atg-deficient mice in combination with Ubc-CreERT2 or atg5−/−; Eno2-Atg5 mice.25,27 Ubc-CreERT2 mice are used for tamoxifen (TAM)-inducible Cre-mediated deletion of floxed genes throughout the body. By administration of TAM to Atg7F/F;CreERT2 mice at 8 wk of age, a near-complete and sustained loss of ATG7 protein is accomplished in 2 wk. By 6–12 wk after deletion at 8 wk of age, many histological abnormalities are observed in atg7Δ/Δ mice, and more than 90% of these mice die within 12 wk after deletion. The majority of atg7Δ/Δ mice die because of neurodegeneration or bacterial infection. atg7Δ/Δ mice have reduced body size and body weight with less fat and lean body mass. Liver abnormalities, splenic enlargement, testicular degeneration, low white adipose tissue, degenerative changes in muscle, small myofibers, and pancreatic damage are observed in these animals. These abnormalities are also observed in atg5−/−; Eno2/NSE-Atg5 mice (Eno2 is a neuron-specific promoter) and other tissue-specific Atg knockout mice. In the brain, the numbers of pyramidal neurons and Purkinje cells are decreased, and progressive motor and behavioral deficits are observed. In addition, atg7Δ/Δ mice cannot survive 24-h fasting due to severe hypoglycemia. Therefore, both adult and neonatal mice require autophagy for adaptation to starvation.
Another mouse model deleting Atg in the whole body in adult mice corresponds to the atg5−/−; Eno2-Atg5 mice referred to above. atg5−/−; Eno2-Atg5 mice were generated to rescue Atg5-null mice from neuronal damage and neonatal lethality.25 Expression of exogenous ATG5 only in the brain under control of the Eno2 promoter is sufficient to rescue Atg5-null mice. The majority of these rescued mice survive for between 8 wk and 8 mo. Although the cause of lethality in atg5−/−; Eno2-Atg5 mice has not been established, the wide window of death indicates that these mice might die due to several reasons besides abnormalities in the brain, which is the main cause of death of atg7Δ/Δ mice. In histological analyses, atg5−/−; Eno2-Atg5 mice show similar abnormalities to atg7Δ/Δ mice. Additional previously uncharacterized abnormalities observed in atg5−/−; Eno2-Atg5 mice are morphological changes related to inflammation in many tissues (liver, small intestine, kidney, lung, adipose tissues, spleen, lymph nodes), atrophy in organs regulated by sex hormones (testis, seminal vesicle, ovary, uterus, pituitary gland, salivary gland), and iron-deficiency anemia. SQSTM1/p62 (sequestosome 1), a selective substrate of autophagy, accumulated in all organs examined at 8 wk of age. Accumulation of phosphorylated SQSTM1 causes hyperactivation of the transcription factor NFE2L2/NRF2 (nuclear factor, erythroid derived 2, like 2); however, significant increases in phosphorylated SQSTM1 and NFE2L2 are observed only in the liver and skeletal muscle. Ubiquitinated proteins clearly accumulate only in the liver, heart, skeletal muscle, and pancreas, suggesting that autophagy-dependent protein quality control is important in these organs. This finding also indicates that there may be causes, other than accumulation of phospho-SQSTM1 and ubiquitinated proteins, for the abnormalities observed in other organs. Utilization of these mouse models will provide new insights into the physiological roles of autophagy.
Deletion of Atg genes not involved in the conjugation systems
ulk1/2 DKO mice die during the neonatal period similar to the ATG conjugation system-deficient mice.28 Lung function is impaired in ulk1/2 DKO neonates. All mice deficient in Atgs that are not part of the ATG conjugation systems (atg13−/−, rb1cc1−/−, atg9a−/−, becn1−/−, vps34−/−), except ulk1/2 DKO animals as noted above, die at different stages of embryonic development. atg13−/− mice die at embryonic d 17.5 (E17.5) of development with myocardial growth defects, which are similar to those observed in rb1cc1−/− mice.29 rb1cc1−/− mice die at E14.5 with abnormalities of the heart and liver.30 In cultured fibroblasts, sensitivity to TNF (tumor necrosis factor)-induced apoptosis is enhanced by deletion of Atg13 or Rb1cc1. becn1−/− mice die at E8.5 and becn1+/− mice show a high incidence of spontaneous tumors such as lymphomas, hepatocellular carcinomas, and adenocarcinoma of the lung.31,32 pik3c3/vps34−/− mice die at E8.5 with severely reduced cell proliferation.33 atg9a−/− mice also die in utero.34 These findings show that timing of lethality and phenotypes are non-uniform among these mutants and clearly different from the phenotypes observed in mice lacking the ATG conjugation systems.
One explanation for these phenotypic differences could be that these proteins are multifunctional. ULK1/Atg1 has important roles in neuronal function.35 In Caenorhabditis elegans, the unc-51 (atg1)-mutant displays defects in axonal elongation and transport, and uncoordinated movement.36-39 These defects are considered to be independent of autophagy as they are not observed in other atg mutants or following silencing of Atgs. The importance in neuronal function of ULK has also been reported in Drosophila melanogaster and mammals.40-42 RB1CC1 is also a multifunctional protein and regulates cell size, proliferation, migration, and apoptosis by interacting with PTK2, PTK2B, TSC1, TP53, MAP3K5, and TRAF2.43-45 BECN1 is involved not only in autophagy as part of a complex with ATG14, PIK3C3/VPS34, and PIK3R4/p150, but also in regulation of the endocytic pathway as part of a complex with UVRAG, PIK3C3/VPS34, and PIK3R4.46 PIK3C3/VPS34 plays roles in membrane trafficking, phagocytosis, cytokinesis, and nutrient sensing.47 These non-autophagic functions would be essential for embryogenesis. Another explanation could be that embryonic lethality is the true phenotype of complete suppression of autophagy by deletion of these Atg genes, which function upstream of the ATG conjugation systems as discussed above.
Deletion of receptors involved in selective autophagy
Although autophagy was originally characterized as a nonselective bulk degradation system, under certain conditions autophagosomes engulf cytosolic materials selectively. This type of autophagy, termed selective autophagy, is classified according to its cargo: aggrephagy, mitophagy, xenophagy, reticulophagy, ferritinophagy, and glycophagy for aggregate proteins, surplus and damaged mitochondria, invasive microbes and viruses, endoplasmic reticulum, ferritin, and glycogen respectively.48 Selective autophagy modulates the quality and quantity of organelles and removes cytotoxic structures, and also regulates the amount of specific proteins such as SRC kinase, a key component of focal adhesions, and KEAP1 (kelch-like ECH-associated protein 1), an adaptor component of a CUL3 (cullin 3)-based ubiquitin E3 ligase, because both are substrates of autophagy. Specific receptors ensure autophagic selectivity. When autophagic cargo such as damaged mitochondria appear in the cytoplasm, they may be tagged with ubiquitin, leading to assembly of receptor proteins that bind to both marker molecules and ATG8 family proteins around the cargo. Some receptors such as SQSTM1 and OPTN are phosphorylated during selective autophagy, which enhances their binding to labeled cargo.49-52 Gene expression of several receptors with a transmembrane region such as BNIP3L/NIX is induced under certain conditions, and the gene products are directly targeted to specific cargo. After the translocation of these receptors to their cargo, some core ATG proteins such as RB1CC1 also recognize the targets—though detailed molecular mechanisms largely remain unclear—beginning the process of autophagosome formation around the targets.49,53 Thus, cargo labeling as well as the transfer of receptors to cargo are essential for selective autophagy. To date, a large number of receptors have been identified in higher eukaryotes, and systemic knockout mice for more than 10 genes encoding receptors have been generated and analyzed. In contrast to Atg gene-null mice, receptor-knockout mice that have been produced to date are viable and exhibit relatively mild phenotypes. The phenotypes of these knockout mice for representative receptors are summarized in Table 2.
Table 2.
Mouse defects in receptors involved in selective autophagy.
| Receptors | Types of selective autophagy | Phenotypes of systemic knockout mice | Reference |
|---|---|---|---|
| SQSTM1/p62 | Aggrephagy, mitophagy, xenophagy, pexophagy, specific proteins (e.g., KEAP1) | Mature-onset obesity | (57,58) |
| NBR1 | Aggrephagy, pexophagy | Age-dependent increase in bone mass and bone mineral density | (59) |
| TOLLIP | Aggrephagy | Less production of the proinflammatory cytokines after low dose of IL1B/IL-1β and LPS treatment | (60) |
| OPTN | Mitophagy, xenophagy | Susceptible to infection with Salmonella, Citrobacter-induced colitis, and E. coli-induced peritonitis | (61,62) |
| CALCOCO2/NDP52 | Mitophagy | — | — |
| SMURF1 | Mitophagy, xenophagy | Age-dependent increase in bone mass accompanied by enhanced osteoblast activity | (63) |
| BCL2L13 | Mitophagy | — | — |
| BNIP3L/NIX | Mitophagy | Anemia with reduced mature erythrocytes and compensatory expansion of erythroid precursors | (64,65) |
| BNIP3 | Mitophagy | Decreased ischemia-induced myocardial apoptosis | (66) |
| FUNDC1 | Mitophagy | — | — |
| FAM134B | Reticulophagy | Neurodegeneration in peripheral sensory nerves | (67) |
| NCOA4 | Feritinophagy | Mild microcytic hypochromic anemia associated with iron overload | (68,69) |
| TAX1BP1 | Xenophagy | Age-dependent inflammatory cardiac valvulitis and skin dermatitis, premature death, and hypersensitivity to low doses of TNF and IL1B | (70) |
| TECPR1 | Aggrephagy, mitophagy, xenophagy | No gross phenotypic abnormalities | (71) |
| STBD1 | Glycophagy | — | |
| CBL | Specific protein (active SRC) | Changes in hemopoietic profiles, splenomegaly, extensive extramedullary hemopoiesis, and mammary duct hyperplasia | (72) |
| TRIMs | Specific proteins | Refer to ref. 73 | (73) |
Future studies
Although autophagy-monitoring and autophagy-deficient mice have led to a huge increase in knowledge about the functions of autophagy in mammals, many aspects of the physiological roles of autophagy in vivo remain unclear. For example, the role of autophagy in nutrient metabolism is still controversial. The concept that autophagy is important for nutrient supply during starvation was proposed long ago; however, the exact role of autophagy has yet to be established. Selective organelle degradation by autophagy such as mitophagy has been considered to be related to disease, but the physiological significance is still largely unknown. Taking advantaged of mouse models will help us to understand these issues.
Autophagy-deficient mice show various abnormalities in various organs. For example, apoptosis and neurodegeneration are observed in the brain, whereas proliferation/tumor growth was observed in the liver. Autophagy is important for quality control and elimination of proteins and organelle, but loss of this function might affect different organs in different ways. For example, although ablation of autophagy causes accumulation of SQSTM1 in both liver and brain, the phenotypes observed in the liver, but not in the brain, are largely depends on the accumulation of SQSTM1. Likewise, sensitivity to accumulation of damaged mitochondria and misfolded proteins due to impaired autophagy might be different among organs. These tissue-specific functions of autophagy should be investigated in more detail in the future.
In the meantime, it has become apparent that the autophagy machinery is not as simple as previously thought. The core Atg gene knockout mice show different phenotypes, but the underlying mechanisms are not elucidated. Some, or probably all, ATG proteins may have autophagy-independent functions, whereas some ATG proteins may not be essential even for canonical autophagy. These findings have led to an important notion that we may not be able to determine the specific function of autophagy simply by deletion of one Atg gene. Confirmation of results using another model system would be helpful. Nevertheless, these valuable mouse models are essential for analysis of the role of autophagy.
Abbreviations
- ATG
autophagy-related
- CreERT2
tamoxifen-inducible Cre recombinase
- DKO
double knockout
- EGFP
enhanced green fluorescent protein
- Em
emission
- Ex
excitation
- PtdIns3K
phosphatidylinositol 3-kinase
- RFP
red fluorescent protein
- TAM
tamoxifen
- tfLC3
tandem fluorescent-tagged LC3
- TNF
tumor necrosis factor
Disclosure of potential conflicts of interest
The authors declare that they have no conflicts of interest related to the contents of this article.
Funding
This work was supported by the Japan Society for the Promotion of Science KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas 25111001 (to MK and NM), 25111005 (to NM), 25111006 (to MK), the Takeda Science Foundation (to MK), and Grant-in-Aid for Scientific Research (C) 16K08613 (to AK).
References
- [1].Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728-41. https://doi.org/ 10.1016/j.cell.2011.10.026. PMID:22078875. [DOI] [PubMed] [Google Scholar]
- [2].Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720-8. https://doi.org/ 10.1093/emboj/19.21.5720. PMID:11060023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101-11. https://doi.org/ 10.1091/mbc.E03-09-0704. PMID:14699058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032-6. https://doi.org/ 10.1038/nature03029. PMID:15525940. [DOI] [PubMed] [Google Scholar]
- [5].Vodicka P, Lim J, Williams DT, Kegel KB, Chase K, Park H, Marchionini D, Wilkinson S, Mead T, Birch H, et al. . Assessment of chloroquine treatment for modulating autophagy flux in brain of WT and HD mice. J Huntingtons Dis. 2014;3:159-74. https://doi.org/ 10.4161/auto.4451. PMID:25062859. [DOI] [PubMed] [Google Scholar]
- [6].Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452-60. PMID:17534139. [DOI] [PubMed] [Google Scholar]
- [7].Castillo K, Valenzuela V, Matus S, Nassif M, Onate M, Fuentealba Y, Encina G, Irrazabal T, Parsons G, Court FA, et al. . Measurement of autophagy flux in the nervous system in vivo. Cell Death Dis. 2013;4:e917. https://doi.org/ 10.1038/cddis.2013.421. PMID:24232093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li L, Wang ZV, Hill JA, Lin F. New autophagy reporter mice reveal dynamics of proximal tubular autophagy. J Am Soc Nephrol. 2014;25:305-15. https://doi.org/ 10.1681/ASN.2013040374. PMID:24179166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hariharan N, Zhai P, Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid Redox Signal. 2011;14:2179-90. https://doi.org/ 10.1089/ars.2010.3488. PMID:20812860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kaizuka T, Morishita H, Hama Y, Tsukamoto S, Matsui T, Toyota Y, Kodama A, Ishihara T, Mizushima T, Mizushima N. An autophagic flux probe that releases an internal control. Mol Cell. 2016;64:835-49. https://doi.org/ 10.1016/j.molcel.2016.09.037. PMID:27818143. [DOI] [PubMed] [Google Scholar]
- [11].Gump JM, Thorburn A. Sorting cells for basal and induced autophagic flux by quantitative ratiometric flow cytometry. Autophagy. 2014;10:1327-34. https://doi.org/ 10.4161/auto.29394. PMID:24915460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 2011;9:38. https://doi.org/ 10.1186/1741-7007-9-38. PMID:21635740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Yoo YH, Combs CA, Finkel T. Measuring In Vivo mitophagy. Mol Cell. 2015;60:685-96. https://doi.org/ 10.1016/j.molcel.2015.10.009. PMID:26549682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].McWilliams TG, Prescott AR, Allen GF, Tamjar J, Munson MJ, Thomson C, Muqit MM, Ganley IG. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. 2016;214:333-45. https://doi.org/ 10.1083/jcb.201603039. PMID:27458135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18:1042-52. https://doi.org/ 10.1016/j.chembiol.2011.05.013. PMID:21867919. [DOI] [PubMed] [Google Scholar]
- [16].Shirakabe A, Zhai P, Ikeda Y, Saito T, Maejima Y, Hsu CP, Nomura M, Egashira K, Levine B, Sadoshima J. Drp1-dependent mitochondrial autophagy plays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure. Circulation. 2016;133:1249-63. https://doi.org/ 10.1161/CIRCULATIONAHA.115.020502. PMID:26915633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127-35. https://doi.org/ 10.1038/embor.2013.168. PMID:24176932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cann GM, Guignabert C, Ying L, Deshpande N, Bekker JM, Wang L, Zhou B, Rabinovitch M. Developmental expression of LC3alpha and beta: Absence of fibronectin or autophagy phenotype in LC3beta knockout mice. Dev Dyn. 2008;237:187-95. https://doi.org/ 10.1002/dvdy.21392. PMID:18069693. [DOI] [PubMed] [Google Scholar]
- [19].O'Sullivan GA, Kneussel M, Elazar Z, Betz H. GABARAP is not essential for GABA receptor targeting to the synapse. Eur J Neurosci. 2005;22:2644-8. https://doi.org/ 10.1111/j.1460-9568.2005.04448.x. PMID:16307606. [DOI] [PubMed] [Google Scholar]
- [20].Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573-83. https://doi.org/ 10.1074/jbc.M701194200. PMID:17442669. [DOI] [PubMed] [Google Scholar]
- [21].Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. . The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762-75. https://doi.org/ 10.1091/mbc.E08-03-0309. PMID:18768753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. . Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425-34. https://doi.org/ 10.1083/jcb.200412022. PMID:15866887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Malhotra R, Warne JP, Salas E, Xu AW, Debnath J. Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy. 2015;11:145-54. https://doi.org/ 10.1038/nature07383. PMID:25585051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. . Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264-8. PMID:18849965. [DOI] [PubMed] [Google Scholar]
- [25].Yoshii SR, Kuma A, Akashi T, Hara T, Yamamoto A, Kurikawa Y, Itakura E, Tsukamoto S, Shitara H, Eishi Y, et al. . Systemic Analysis of Atg5-Null Mice Rescued from Neonatal Lethality by Transgenic ATG5 Expression in Neurons. Dev Cell. 2016;39:116-30. https://doi.org/ 10.1016/j.devcel.2016.09.001. PMID:27693508. [DOI] [PubMed] [Google Scholar]
- [26].Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036-41. https://doi.org/ 10.1126/science.aaf6136. PMID:27885029. [DOI] [PubMed] [Google Scholar]
- [27].Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, et al. . Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914-27. https://doi.org/ 10.1158/2159-8290.CD-14-0363. PMID:24875857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cheong H, Wu J, Gonzales LK, Guttentag SH, Thompson CB, Lindsten T. Analysis of a lung defect in autophagy-deficient mouse strains. Autophagy. 2014;10:45-56. https://doi.org/ 10.4161/auto.26505. PMID:24275123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kaizuka T, Mizushima N. Atg13 is essential for autophagy and cardiac development in mice. Mol Cell Biol. 2015;36:585-95. https://doi.org/ 10.1128/MCB.01005-15. PMID:26644405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol. 2006;175:121-33. https://doi.org/ 10.1083/jcb.200604129. PMID:17015619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Qu X. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809-20. https://doi.org/ 10.1172/JCI20039. PMID:14638851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077-82. https://doi.org/ 10.1073/pnas.2436255100. PMID:14657337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhou X, Takatoh J, Wang F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PloS One. 2011;6:e16358. https://doi.org/ 10.1371/journal.pone.0016358. PMID:21283715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kojima T, Yamada T, Akaishi R, Furuta I, Saitoh T, Nakabayashi K, Nakayama KI, Nakayama K, Akira S, Minakami H. Role of the Atg9a gene in intrauterine growth and survival of fetal mice. Reprod Biol. 2015;15:131-8. https://doi.org/ 10.1016/j.repbio.2015.05.001. PMID:26370455. [DOI] [PubMed] [Google Scholar]
- [35].Ogura K, Wicky C, Magnenat L, Tobler H, Mori I, Muller F, Ohshima Y. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994;8:2389-400. https://doi.org/ 10.1101/gad.8.20.2389. PMID:7958904. [DOI] [PubMed] [Google Scholar]
- [36].Ogura K, Goshima Y. The autophagy-related kinase UNC-51 and its binding partner UNC-14 regulate the subcellular localization of the Netrin receptor UNC-5 in Caenorhabditis elegans. Development. 2006;133:3441-50. https://doi.org/ 10.1242/dev.02503. PMID:16887826. [DOI] [PubMed] [Google Scholar]
- [37].Ogura K, Okada T, Mitani S, Gengyo-Ando K, Baillie DL, Kohara Y, Goshima Y. Protein phosphatase 2A cooperates with the autophagy-related kinase UNC-51 to regulate axon guidance in Caenorhabditis elegans. Development. 2010;137:1657-67. https://doi.org/ 10.1242/dev.050708. PMID:20392746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ogura K, Shirakawa M, Barnes TM, Hekimi S, Ohshima Y. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev. 1997;11:1801-11. https://doi.org/ 10.1101/gad.11.14.1801. PMID:9242488. [DOI] [PubMed] [Google Scholar]
- [39].Lai T, Garriga G. The conserved kinase UNC-51 acts with VAB-8 and UNC-14 to regulate axon outgrowth in C. elegans. Development. 2004;131:5991-6000. [DOI] [PubMed] [Google Scholar]
- [40].Toda H, Mochizuki H, Flores R 3rd, Josowitz R, Krasieva TB, Lamorte VJ, Suzuki E, Gindhart JG, Furukubo-Tokunaga K, et al. . UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev. 2008;22:3292-307. https://doi.org/ 10.1101/gad.1734608. PMID:19056884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mochizuki H, Toda H, Ando M, Kurusu M, Tomoda T, Furukubo-Tokunaga K. Unc-51/ATG1 controls axonal and dendritic development via kinesin-mediated vesicle transport in the Drosophila brain. PloS One. 2011;6:e19632. https://doi.org/ 10.1371/journal.pone.0019632. PMID:21589871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ahantarig A, Chadwell LV, Terrazas IB, Garcia CT, Nazarian JJ, Lee HK, Lundell MJ, Cassill JA. Molecular characterization of Pegarn: A Drosophila homolog of UNC-51 kinase. Mol Biol Rep. 2009;36:1311-21. https://doi.org/ 10.1007/s11033-008-9314-4. PMID:18636236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gan B, Guan JL. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal. 2008;20:787-94. https://doi.org/ 10.1016/j.cellsig.2007.10.021. PMID:18036779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Koinuma D, Shinozaki M, Nagano Y, Ikushima H, Horiguchi K, Goto K, Goto K, Chano T, Saitoh M, Imamura T, et al. . RB1CC1 protein positively regulates transforming growth factor-β signaling through the modulation of arkadia E3 ubiquitin ligase activity. J Biol Chem. 2011;286:32502-12. https://doi.org/ 10.1074/jbc.M111.227561. PMID:21795712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Choi JD, Ryu M, Ae Park M, Jeong G, Lee JS. FIP200 inhibits beta-catenin-mediated transcription by promoting APC-independent beta-catenin ubiquitination. Oncogene. 2013;32:2421-32. https://doi.org/ 10.1038/onc.2012.262. PMID:22751121. [DOI] [PubMed] [Google Scholar]
- [46].Nascimbeni AC, Codogno P, Morel E. Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 2016;284:1267-78. https://doi.org/ 10.1111/febs.13987. PMID:27973739. [DOI] [PubMed] [Google Scholar]
- [47].Backer JM. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem J. 2016;473:2251-71. https://doi.org/ 10.1042/BCJ20160170. PMID:27470591. [DOI] [PubMed] [Google Scholar]
- [48].Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26:6-16. https://doi.org/ 10.1016/j.tcb.2015.08.010. PMID:26437584. [DOI] [PubMed] [Google Scholar]
- [49].Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S, et al. . Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115-28. https://doi.org/ 10.1083/jcb.201304188. PMID:24100292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. . Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228-33. https://doi.org/ 10.1126/science.1205405. PMID:21617041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279-89. https://doi.org/ 10.1016/j.molcel.2011.07.039. PMID:22017874. [DOI] [PubMed] [Google Scholar]
- [52].Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, et al. . TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223-34. https://doi.org/ 10.1016/j.immuni.2012.04.015. PMID:22921120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309-14. https://doi.org/ 10.1038/nature14893. PMID:26266977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Marino G, Fernandez AF, Cabrera S, Lundberg YW, Cabanillas R, Rodriguez F, Salvador-Montoliu N, Vega JA, Germanà A, Fueyo A, et al. . Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331-44. https://doi.org/ 10.1172/JCI42601. PMID:20577052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493-502. https://doi.org/ 10.1182/blood-2008-02-137398. PMID:18539900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A. 2011;108:11121-6. https://doi.org/ 10.1073/pnas.1107969108. PMID:21690395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3:211-22. https://doi.org/ 10.1016/j.cmet.2006.01.011. PMID:16517408. [DOI] [PubMed] [Google Scholar]
- [58].Harada H, Warabi E, Matsuki T, Yanagawa T, Okada K, Uwayama J, Ikeda A, Nakaso K, Kirii K, Noguchi N, et al. . Deficiency of p62/Sequestosome 1 causes hyperphagia due to leptin resistance in the brain. J Neurosci. 2013;33:14767-77. https://doi.org/ 10.1523/JNEUROSCI.2954-12.2013. PMID:24027277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Whitehouse CA, Waters S, Marchbank K, Horner A, McGowan NW, Jovanovic JV, Xavier GM, Kashima TG, Cobourne MT, Richards GO, et al. . Neighbor of Brca1 gene (Nbr1) functions as a negative regulator of postnatal osteoblastic bone formation and p38 MAPK activity. Proc Natl Acad Sci U S A. 2010;107:12913-8. https://doi.org/ 10.1073/pnas.0913058107. PMID:20616007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Didierlaurent A, Brissoni B, Velin D, Aebi N, Tardivel A, Kaslin E, Sirard JC, Angelov G, Tschopp J, Burns K. Tollip regulates proinflammatory responses to interleukin-1 and lipopolysaccharide. Mol Cell Biol. 2006;26:735-42. https://doi.org/ 10.1128/MCB.26.3.735-742.2006. PMID:16428431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chew TS, O'Shea NR, Sewell GW, Oehlers SH, Mulvey CM, Crosier PS, Godovac-Zimmermann J, Bloom SL, Smith AM, Segal AW. Optineurin deficiency in mice contributes to impaired cytokine secretion and neutrophil recruitment in bacteria-driven colitis. Dis Model Mech. 2015;8:817-29. https://doi.org/ 10.1242/dmm.020362. PMID:26044960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Slowicka K, Vereecke L, Mc Guire C, Sze M, Maelfait J, Kolpe A, Saelens X, Beyaert R, van Loo G. Optineurin deficiency in mice is associated with increased sensitivity to Salmonella but does not affect proinflammatory NF-kappaB signaling. Eur J Immunol. 2016;46:971-80. https://doi.org/ 10.1002/eji.201545863. PMID:26677802. [DOI] [PubMed] [Google Scholar]
- [63].Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101-13. https://doi.org/ 10.1016/j.cell.2005.01.035. PMID:15820682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232-5. https://doi.org/ 10.1038/nature07006. PMID:18454133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Diwan A, Koesters AG, Odley AM, Pushkaran S, Baines CP, Spike BT, Spike BT, Daria D, Jegga AG, Geiger H, et al. . Unrestrained erythroblast development in Nix-/- mice reveals a mechanism for apoptotic modulation of erythropoiesis. Proc Natl Acad Sci U S A. 2007;104:6794-9. https://doi.org/ 10.1073/pnas.0610666104. PMID:17420462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, Li H, Kirshenbaum LA, Hahn HS, Robbins J, et al. . Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest. 2007;117:2825-33. https://doi.org/ 10.1172/JCI32490. PMID:17909626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al. . Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354-8. https://doi.org/ 10.1038/nature14498. PMID:26040720. [DOI] [PubMed] [Google Scholar]
- [68].Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, et al. . Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16:1069-79. https://doi.org/ 10.1038/ncb3053. PMID:25327288. [DOI] [PubMed] [Google Scholar]
- [69].Bellelli R, Federico G, Matte A, Colecchia D, Iolascon A, Chiariello M, Santoro M, De Franceschi L, Carlomagno F. NCOA4 Deficiency impairs systemic iron homeostasis. Cell Rep. 2016;14:411-21. https://doi.org/ 10.1016/j.celrep.2015.12.065. PMID:26776506. [DOI] [PubMed] [Google Scholar]
- [70].Iha H, Peloponese JM, Verstrepen L, Zapart G, Ikeda F, Smith CD, Starost MF, Yedavalli V, Heyninck K, Dikic I, et al. . Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-kappaB activation. EMBO J. 2008;27:629-41. https://doi.org/ 10.1038/emboj.2008.5. PMID:18239685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ogawa M, Yoshikawa Y, Kobayashi T, Mimuro H, Fukumatsu M, Kiga K, Piao Z, Ashida H, Yoshida M, Kakuta S, et al. . A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe. 2011;9:376-89. https://doi.org/ 10.1016/j.chom.2011.04.010. PMID:21575909. [DOI] [PubMed] [Google Scholar]
- [72].Murphy MA, Schnall RG, Venter DJ, Barnett L, Bertoncello I, Thien CB, Langdon WY, Bowtell DD. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol Cell Biol. 1998;18:4872-82. https://doi.org/ 10.1128/MCB.18.8.4872. PMID:9671496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kimura T, Mandell M, Deretic V. Precision autophagy directed by receptor regulators - emerging examples within the TRIM family. J Cell Sci. 2016;129:881-91. https://doi.org/ 10.1242/jcs.163758. PMID:26906420. [DOI] [PMC free article] [PubMed] [Google Scholar]