ABSTRACT
Impairment in macroautophagy/autophagy flux and inflammasome activation are common characteristics of nonalcoholic steatohepatitis (NASH). Considering the lack of approved agents for treating NASH, drugs that can enhance autophagy and modulate inflammasome pathways may be beneficial. Here, we investigated the novel mechanism of ezetimibe, a widely prescribed drug for hypercholesterolemia, as a therapeutic option for ameliorating NASH. Human liver samples with steatosis and NASH were analyzed. For in vitro studies of autophagy and inflammasomes, primary mouse hepatocytes, human hepatoma cells, mouse embryonic fibroblasts with Ampk or Tsc2 knockout, and human or primary mouse macrophages were treated with ezetimibe and palmitate. Steatohepatitis and fibrosis were induced by feeding Atg7 wild-type, haploinsufficient, and knockout mice a methionine- and choline-deficient diet with ezetimibe (10 mg/kg) for 4 wk. Human livers with steatosis or NASH presented impaired autophagy with decreased nuclear TFEB and increased SQSTM1, MAP1LC3-II, and NLRP3 expression. Ezetimibe increased autophagy flux and concomitantly ameliorated lipid accumulation and apoptosis in palmitate-exposed hepatocytes. Ezetimibe induced AMPK phosphorylation and subsequent TFEB nuclear translocation, related to MAPK/ERK. In macrophages, ezetimibe blocked the NLRP3 inflammasome-IL1B pathway in an autophagy-dependent manner and modulated hepatocyte-macrophage interaction via extracellular vesicles. Ezetimibe attenuated lipid accumulation, inflammation, and fibrosis in liver-specific Atg7 wild-type and haploinsufficient mice, but not in knockout mice. Ezetimibe ameliorates steatohepatitis by autophagy induction through AMPK activation and TFEB nuclear translocation, related to an independent MTOR ameliorative effect and the MAPK/ERK pathway. Ezetimibe dampens NLRP3 inflammasome activation in macrophages by modulating autophagy and a hepatocyte-driven exosome pathway.
KEYWORDS: autophagy, exosome, inflammasome, inflammation, NASH
Introduction
With the rapid increase in obesity rates, nonalcoholic fatty liver disease (NAFLD) has become one of the most common metabolic liver diseases, affecting up to 30% of the adult population and 70−80% of subjects with obesity and diabetes.1,2 Concomitantly, the increasing prevalence of nonalcoholic steatohepatitis (NASH), which can progress to liver cirrhosis and hepatocellular carcinoma, has caused serious health problems worldwide. NASH is significantly associated with higher mortality not only from complications of liver disorders but also cardiovascular disease.2 However, there is no U.S. Food and Drug Administration (FDA)-approved pharmacological treatment of hepatic NASH in humans.
One hypothesis for the pathogenesis of NASH is the “2-hit theory,” which consists of a first hit of hepatic triglyceride accumulation due to obesity and insulin resistance, and a subsequent second hit of hepatocyte inflammation, necrosis, and fibrosis due to reactive oxidative stress and lipid peroxidation.3 This theory has been modified to a ‘multiple parallel hits’ hypothesis for the pathogenesis of NASH, indicating liver injury by diverse pathogenic events occurring in parallel.4 In the progression of NASH, activated macrophages play a central role in the release of pro-inflammatory M1 mediators (IL1B [interleukin 1 β], IL18 [interleukin 18], and CCL2-CCL5) and liver matrix remodeling, which results in hepatic fibrosis.5 Recent studies reported that autophagy also plays a crucial role in the development of NAFLD and NASH.6 While suppression of autophagy leads to aggravated hepatic steatosis and NASH,7 an increase in autophagy ameliorates fibrosis in a mouse experimental model.8 Furthermore, decline in autophagy is linked to activation of inflammasomes, key signaling platforms for danger signal recognition, leading to inflammatory processes.9,10 Because human NAFLD and NASH are associated with lipotoxicity-induced autophagy defects11 and significantly increase inflammasome expression,12 autophagy induction or inflammasome inhibition have been proposed as a therapeutic option for metabolic pathologies. In this context, autophagy inducers and NLRP3 (NLR family, pyrin domain containing 3) inflammasome inhibitors are currently targets for NASH drug development.1
Ezetimibe is a FDA-approved lipid-lowering agent that blocks NPC1L1 (NPC1 like intracellular cholesterol transporter 1)-dependent cholesterol transport at the border of the intestine and inhibits absorption of dietary and biliary cholesterols.13,14 While NPC1L1 is primarily expressed in the intestine of rodents, in humans NPC1L1 is abundantly expressed in the liver as well as the small intestine.13 In a human clinical trial, 24-mo ezetimibe treatment significantly improved hepatic steatosis and ballooning score.15 Ezetimibe administration to NASH patients also led to improvement in histological features of fibrotic change.16 The proposed explanation for these improvements is the inhibition of cholesterol absorption from the intestine and liver, but this issue remains unclear. Recently, ezetimibe treatment was reported to activate autophagy in human hepatocytes, and increased autophagy flux may be related to ezetimibe-mediated improvement in hepatic steatosis.17 Therefore, in the present study, we investigated whether ezetimibe ameliorates steatohepatitis and fibrosis via induction of autophagic pathways.
Results
Human NASH presents reduced autophagy with increased NLRP3 inflammasome activity
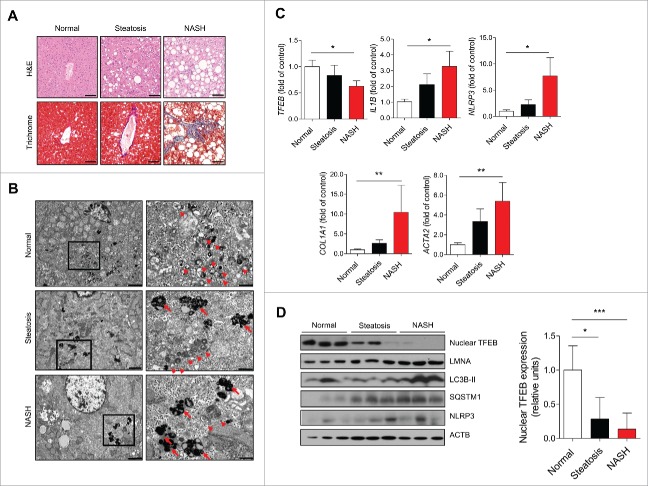
Human livers were histologically evaluated to assess the fatty liver status of patients with normal livers, steatosis, and NASH (Fig. 1A). Masson's trichrome staining revealed marked pericellular fibrosis in the livers of NASH patients. Electron microscopy detected significantly fewer autophagic vacuoles in the livers of patients with steatosis and NASH compared with normal samples (Fig. 1B). In addition, liver specimens from patients with steatosis or NASH had an increased number of lipofuscin granules, a marker of lysosomal damage, composed of the oxidation products of unsaturated fatty acids. Consistently, TFEB (transcription factor EB), a master gene for autophagic and lysosomal biogenesis,18 significantly decreased in steatotic and NASH liver samples compared with normal samples. mRNA expression of IL1B and NLRP3 was markedly increased in steatotic and NASH liver samples, and mRNA expression of fibrosis-related genes (COL1A1 [collagen type I α 1 chain] and ACTA2 [actin, α 2, smooth muscle, aorta]) was significantly increased in steatosis and NASH (Fig. 1C). Parallel decreased protein levels of nuclear TFEB and increased MAP1LC3B/LC3B (microtubule associated protein 1 light chain 3 β)-II and SQSTM1 (sequestosome 1) levels were observed in steatosis and NASH samples (Fig. 1D). NLRP3 levels were also increased in the livers of steatosis and NASH patients compared with normal livers.
Figure 1.
Human NASH demonstrating reduced autophagy with increased NLRP3 inflammasome activity. (A) Representative histological images of H&E- and Masson's trichrome-stained human liver section. Scale bars: 50 μm. (B) Representative transmission electron microscopy images from each type of human liver section. High magnification of boxed areas is presented on the right. Scale bars: 2.5 μm (left), 1 μm (right). Arrowhead, autophagic vacuoles; arrow, lipofuscin granules. (C) qPCR analysis of human liver tissues. Normal (n = 12), simple steatosis (n = 11), and NASH (n = 9). *P < 0.05, **P < 0.01, and ***P < 0.001. (D) Representative immunoblot analyses in normal, steatotic, and NASH liver tissues. Densitometry analysis of nuclear TFEB expression. *P < 0.05 and ***P < 0.001.
Ezetimibe-induced autophagy ameliorates lipid accumulation and cell apoptosis in hepatocytes
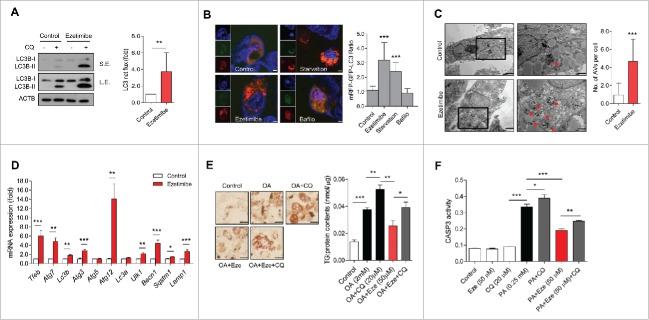
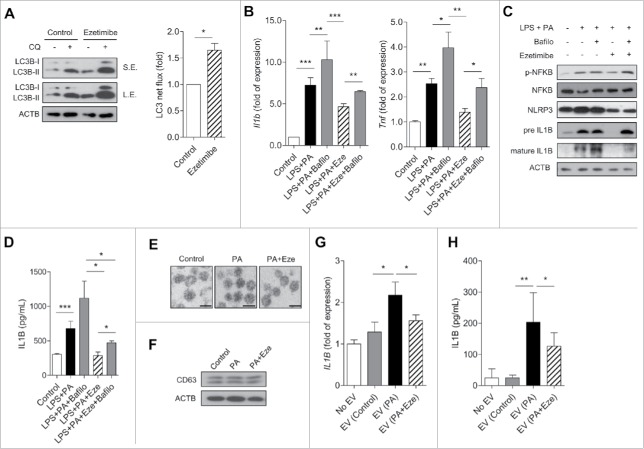
To focus on the impaired autophagy flux and concomitant increase in inflammasome activity in NAFLD and NASH, we investigated the therapeutic effect of ezetimibe and its mechanism in the liver. Ezetimibe significantly increased LC3B-II accumulation, representing LC3 net flux, in hepatocytes when co-treated with chloroquine compared with controls (Fig. 2A). Next, we examined the vesicle formation step of autophagy using the autophagy marker, monomeric red fluorescent protein (mRFP)-green fluorescent protein (GFP)-LC3 in primary hepatocytes in response to glucose starvation, ezetimibe, or ezetimibe + bafilomycin A1 treatment by using fluorescence microscopy. The number of autolysosomes labeled with a red signal was markedly increased in response to glucose starvation or ezetimibe treatment, and decreased by bafilomycin A1 (Fig. 2B). Electron microscopy also demonstrated that ezetimibe significantly increased the number of autophagic vacuoles in primary hepatocytes (Fig. 2C). We assessed the levels of autophagy-related genes including Tfeb, Atg7, Lc3b, Atg3, Atg5, Atg12, Lc3a, Ulk1, Becn1, Sqstm1, and Lamp1 with quantitative polymerase chain reaction (qPCR) analysis and observed significantly increased expression of most of these genes in ezetimibe-treated primary hepatocytes (Fig. 2D). To evaluate the lipophagic effect of ezetimibe on hepatocytes, oleate-treated primary hepatocytes were subjected to Oil red O (ORO) staining and triglyceride content analyses. Ezetimibe significantly attenuated oleate-induced lipid accumulation in hepatocytes (Fig. 2E). Furthermore, to assess whether the effect of ezetimibe against lipotoxicity-mediated apoptosis is related to autophagy, CASP3 (caspase 3) assay was conducted to evaluate the viability or apoptosis of primary hepatocytes. Ezetimibe treatment alone was not cytotoxic to primary hepatocytes and significantly increased viability compared with palmitate treatment alone (Fig. 2F). However, when co-treated with chloroquine (CQ), the cytoprotective effect of ezetimibe was attenuated.
Figure 2.
Ezetimibe-induced autophagy ameliorates lipid accumulation and cell apoptosis in hepatocytes. (A) Primary hepatocytes after 50 μM ezetimibe or control solvent (dimethyl sulfoxide, DMSO) treatment of 16 h in the absence or presence of 20 μM chloroquine (CQ) for the last 2 h were subjected to immunoblotting. Top panel, short exposure (SE). Middle panel, long exposure (L.E.). LC3 net flux was assessed by subtracting the amount of LC3B-II in the absence of CQ from the amount of LC3B-II in the presence of CQ for each of the conditions, as autophagy flux and graphically displayed. **P < 0.01. n = 3–5. (B) Fluorescence microscopy images of primary hepatocytes using the autophagy marker mRFP-GFP-LC3 in response to glucose starvation, 50 μM ezetimibe, or 50 nM bafilomycin A1 (Bafilo) for 2 h. Scale bars: 5 μm. The number of autolysosomes (red puncta) per cell (n = 30) was counted. ***P < 0.001. (C) Representative transmission electron microscopy images of 16 h of 50 μM ezetimibe-treated primary hepatocytes. High magnification of boxed areas is presented on the right. Scale bars: 2.5 μm (left), 1 μm (right). Arrowhead, autophagic vacuole. The number of autophagic vacuoles (AVs) per cell (n = 20) was counted. (D) qPCR analysis of ezetimibe- or vehicle-treated primary hepatocytes for autophagy-related genes and lysosomal genes. n = 5–7. *P < 0.05, **P < 0.01, and ***P < 0.001. (E) Primary hepatocytes were stained with Oil red O and triglyceride content was quantitatively analyzed. Primary hepatocytes were treated with 2 mM oleate (OA) for 16 h and pre-treatment with 50 μM ezetimibe for 2 h before 2 mM OA exposure in the absence or presence of 20 μM chloroquine (CQ) for 2 h. Scale bars: 50 μm. n = 5–7 per group. *P < 0.05, **P < 0.01, and ***P < 0.001. (F) CASP3 assay for relative cell survival of primary hepatocytes exposed to 0.25 mM palmitate (PA) for 24 h or pre-treatment with 50 μM ezetimibe for 2 h before 0.25 mM PA exposure in the absence or presence of 20 μM chloroquine (CQ) for 2 h. n = 5–7 per group. *P < 0.05, **P < 0.01, and ***P < 0.001.
Ezetimibe induces autophagy via the AMPK-TFEB pathway
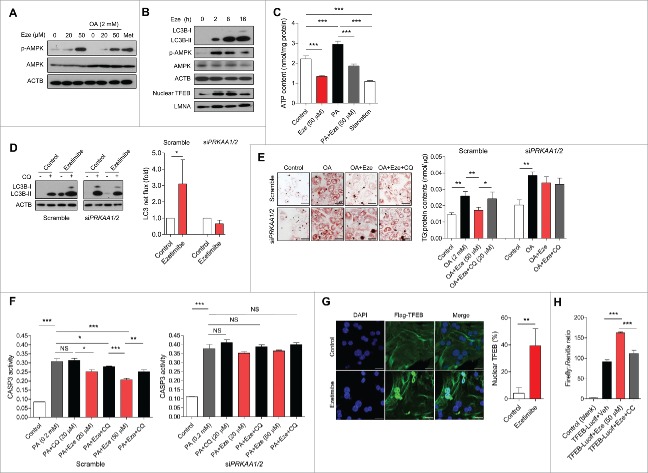
Since the AMP-activated protein kinase (AMPK) plays an essential role in the transcriptional regulation of autophagy,19 PRKA phosphorylation status was evaluated in ezetimibe-treated hepatocytes. Ezetimibe markedly increased phosphorylated (p)-AMPK expression dose-dependently, regardless of nutrient (oleate) overloading condition (Fig. 3A). As TFEB is a key positive regulator of autophagy and lysosome biogenesis,18 we also evaluated whether ezetimibe affects TFEB nuclear translocation. To investigate a time course for the regulation of autophagy by ezetimibe, expression of LC3B-II, p-AMPK, and nuclear TFEB was examined in primary hepatocytes at 0, 2, 8, and 16 h of treatment, and it was increased during 2 to 16 h of treatment (Fig. 3B). Moreover, we evaluated the mechanism of ezetimibe-induced AMPK activation and found that a decrease in cellular ATP contents was observed after treatment with ezetimibe in primary hepatocytes (Fig. 3C) and after HepG2 cells were treated with palmitate (Fig. S1A). To verify that AMPK is required for ezetimibe-induced autophagy we performed an LC3 turnover assay, which indicated that ezetimibe could not induce autophagy after siRNA (small interfering RNA)-mediated knockdown of PRKAA1 (protein kinase AMP-activated catalytic subunit α 1) and PRKAA2 (Fig. S1B and Fig. 3D), whereas p-AMPK was induced by ezetimibe in the AMPK-sufficient condition (Fig. S1C). The formation of autolysosomes labeled with a red signal and expression of Lc3b, Tfeb, and Atg3 were also increased by ezetimibe in the AMPK-sufficient condition, but not in the AMPK-deficient condition (Fig. S1D and S1E).
Figure 3.
Ezetimibe induces autophagy via the AMPK-TFEB pathway. (A) Representative immunoblot analysis of primary hepatocytes treated with 20 or 50 μM ezetimibe for 16 h, pre-treated with ezetimibe for 2 h followed by 2 mM oleate (OA), or treated with 2 mM metformin (Met) for 16 h. n = 3–5. (B) Representative immunoblots of primary hepatocytes treated with 50 μM ezetimibe at 0, 2, 8, and 16 hours. n = 3–5. (C) Cellular ATP levels in primary hepatocytes in the presence or absence of 50 μM ezetimibe for 2 h. PA, 0.2 mM palmitate, n = 3. ***P < 0.001. (D) Representative immunoblots of primary hepatocytes transfected with siRNAs against PRKAA1/2 (siPRKAA1/2) and scramble control incubated with 50 μM ezetimibe or DMSO (control) for 16 h in the absence or presence of 20 μM chloroquine (CQ) for 2 h. n = 3–5. *P < 0.05. (E) Representative staining of scramble control and siPRKAA1/2 primary hepatocytes with Oil red O and triglyceride analysis after treatment with 2 mM oleate (OA) for 16 h or 50 μM ezetimibe pre-treatment of 2 h followed by 2 mM OA in the absence or presence of 20 μM chloroquine (CQ) for 2 h. Scale bars: 50 μm. n = 3–5 per group. *P < 0.05 and **P < 0.01. (F) CASP3 assay for relative survival of scramble control and siPRKAA1/2 primary hepatocytes exposed to 0.2 mM palmitate (PA) for 24 h or pre-treatment with 20 or 50 μM ezetimibe for 2 h before 0.2 mM PA exposure in the absence or presence of 20 μM chloroquine (CQ) for 2 h. n = 3–5 per group. *P < 0.05, **P < 0.01, and ***P < 0.001. (G) Fluorescence microscopy images of Flag-TFEB-transfected primary hepatocytes treated with ezetimibe or DMSO (control) and subjected to nuclear stain (DAPI) and Flag immunostaining (Flag-TFEB). The percentage of cells with nuclear TFEB is presented. Scale bars: 20 μm. n = 3–5. **P < 0.01. (H) Luciferase assay of primary hepatocytes transfected with TFEB-luciferase reporter or TFEB-empty and treated with DMSO (vehicle), 20 μM ezetimibe, 20 μM ezetimibe and 10 μM compound C (CC) for 24 h. n = 5–7 per group. NS, not significant. ***P < 0.001.
Then we examined whether the AMPK-dependent pathway is involved in the protective effect of ezetimibe on lipid accumulation and lipotoxicity-mediated apoptosis via autophagy. Ezetimibe did not inhibit oleate-induced intracellular lipid accumulation in primary hepatocytes transfected with PRKAA1/2 siRNA, and triglyceride accumulation was not affected by ezetimibe in the presence of CQ in both AMPK-sufficient and -deficient conditions (Fig. 3E). Ezetimibe ameliorated lipotoxicity-related cell death dose-dependently in the scramble controls, but the effects were not maintained upon CQ treatment or PRKAA1/2 knockdown (Fig. 3F). After transfection of a plasmid encoding Flag-tagged TFEB into primary hepatocytes, TFEB nuclear translocation was observed in ezetimibe-treated cells compared with controls (Fig. 3G). By contrast, ezetimibe did not promote nuclear TFEB translocation in the PRKAA1/2-knockout condition (Fig. S1F). To determine whether ezetimibe influences TFEB transcriptional activity via AMPK-dependent pathways, TFEB promoter activity in primary hepatocytes was measured by luciferase assay. TFEB transcriptional activity was significantly upregulated by ezetimibe, and it was attenuated by co-treatment with compound C, an AMPK inhibitor (Fig. 3H).
Ezetimibe induces autophagy via MAPK/ERK-related pathway
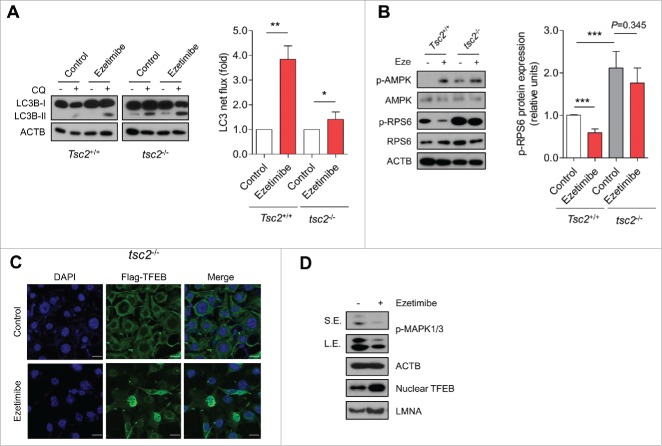
A previous study demonstrated that ezetimibe induced autophagy via inhibition of MTOR (mechanistic target of rapamycin),17 a crucial mediator of autophagy pathway regulation including TFEB nuclear translocation. To verify whether ezetimibe-induced autophagy is dependent on MTOR pathway inhibition, we used mouse embryonic fibroblasts (MEFs) lacking Tsc2 (tuberous sclerosis 2), which encodes an important negative regulator of MTOR, which can lead to constitutive MTOR activation. Autophagy flux determined by LC3 turnover assay demonstrated that ezetimibe could still induce autophagy under this MTOR-activated condition (Fig. 4A). Furthermore, ezetimibe treatment resulted in increased p-AMPK expression in tsc2−/− MEFs, with a slight, but not significant, decrease in levels of p-RPS6 (ribosomal protein S6), a substrate downstream of MTOR (Fig. 4B). In Tsc2+/+ MEFs, ezetimibe inhibited RPS6 phosphorylation, consistent with a previous report.17 Immunofluorescence microscopy showed that ezetimibe induced nuclear TFEB translocation in tsc2−/− MEFs (Fig. 4C).
Figure 4.
Ezetimibe induces autophagy related to the MAPK/ERK pathway. (A) Tsc2+/+ and tsc2−/− MEFs after 20 μM ezetimibe or control solvent (dimethyl sulfoxide, DMSO) treatment of 16 h in the absence or presence of 20 μM chloroquine (CQ) for the last 2 h were subjected to immunoblotting. LC3 net flux was assessed by subtracting the amount of LC3B-II in the absence of CQ from the amount of LC3B-II in the presence of CQ for each of the conditions, and graphically displayed. n = 6–8. *P < 0.05 and **P < 0.01. (B) Representative immunoblot analysis in Tsc2+/+ and tsc2−/− MEFs treated with or without 20 μM ezetimibe for 16 h. Expression of phosphorylation of RPS6 was graphically displayed from the optical density-based data of immunoblots. n = 6–8. ***P < 0.001. (C) Fluorescence microscopy images of tsc2−/− MEFs transfected with Flag-TFEB and treated with 20 μM ezetimibe or DMSO (control) for 2 h. Scale bars: 20 μm. n = 3–5. (D) Immunoblot analysis for p-MAPK1/ERK2-MAPK3/ERK1, ACTB, nuclear TFEB, and LMNA in primary hepatocytes treated with or without 50 μM ezetimibe for 2 h. Top panel, short exposure (SE). Middle panel, long exposure (L.E.). n = 3–5.
Next, we investigated the downstream mechanism of ezetimibe on nuclear TFEB translocation under MTOR-activated conditions. Other kinases such as MAPK1 (mitogen-activated protein kinase)/ERK2 and PRKC (protein kinase C) regulate TFEB phosphorylation and determine its subcellular localization.20,21 We found that ezetimibe treatment decreased phosphorylation of MAPK1/ERK2-MAPK3/ERK1 at threonine 202/tyrosine 204 and increased nuclear TFEB in primary hepatocytes, suggesting that ezetimibe induces nuclear TFEB translocation by inhibiting MAPK1/ERK2-MAPK3/ERK1 (Fig. 4D). However, ezetimibe-induced nuclear TFEB translocation was not blocked by a PRKC inhibitor (Fig. S2). Collectively, these data indicate that ezetimibe induces autophagy under MTOR-activated condition via a MAPK/ERK-related pathway, but not a PRKC pathway.
Ezetimibe ameliorates inflammation via an autophagy-mediated NLRP3 inflammasome-IL1B pathway in macrophages and modulates cell-cell interaction via extracellular vesicles
Next, we sought to determine the role of ezetimibe in macrophages, which play an essential role in the inflammatory processes that occur in NASH. The mRNA expression of NPC1L1, the target of ezetimibe, were confirmed in macrophages including human leukemia cells (THP-1), a human monocytic cell line (Fig. S3). Ezetimibe increased autophagy in both THP-1 and primary peritoneal macrophages (Fig. 5A and Fig. S4). mRNA expression of NFKB (nuclear factor kappa B)-dependent cytokines Il1b and Tnf (tumor necrosis factor) was suppressed by ezetimibe, whereas this effect was blocked by co-treatment with bafilomycin A1 (Fig. 5B). Bafilomycin A1 co-treatment also attenuated the protective effect of ezetimibe on inflammation mediated by the NLRP3 inflammasone-IL1B pathway in macrophages (Fig. 5C). In addition, decreased IL1B release from macrophages with ezetimibe treatment significantly increased with bafilomycin A1 co-treatment (Fig. 5D and Fig. S5).
Figure 5.
Ezetimibe ameliorates inflammation via the autophagy-mediated NLRP3 inflammasome-IL1B pathway in macrophages and modulation of cell-cell interaction via extracellular vesicles. (A) Expression of genes related to LC3B net flux in THP-1 cells treated with 25 μM ezetimibe or vehicle for 16 h in the absence or presence of 20 μM chloroquine (CQ) for 2 h. Top panel, short exposure (SE). Middle panel, long exposure (L.E). Quantified LC3 net flux was graphically displayed. *P < 0.05. n = 3–5. (B) Gene expression of Il1b and Tnf by THP-1 cells in response to 0.1 μg/mL lipopolysaccharide (LPS) and 0.2 mM palmitate (PA) in the presence or absence of 25 μM ezetimibe or 50 nM bafilomycin A1 (Bafilo) for 16 h. n = 5–7 per group. *P < 0.05, **P < 0.01, and ***P < 0.001. (C) THP-1 cells were incubated with 0.1 μg/mL lipopolysaccharide (LPS) and 0.2 mM palmitate (PA) for 16 h with or without 25 μM ezetimibe or 50 nM bafilomycin A1 (Bafilo) for 2 h and subjected to immunoblot analysis for expression of inflammasome-related proteins. n = 3–5. (D) ELISA assays for IL1B levels of THP-1 cells cultured with 0.1 μg/mL lipopolysaccharide (LPS) and 0.2 mM palmitate (PA) for 16 h with or without pre-treatment with 25 μM ezetimibe or 50 nM bafilomycin A1 (Bafilo) for 2 h. n = 5–7 per group. *P < 0.05 and ***P < 0.001. (E) Representative transmission electron microscopy images of isolated extracellular vesicles from HepG2 cells treated with DMSO (control), 0.2 mM palmitate (PA), or 0.2 mM PA + 50 μM ezetimibe for 24 h. Scale bars: 100 nm. n = 3–5 per group. (F) Immunoblot assay for exosome-enriched CD63 protein and ACTB in primary hepatocytes. n = 3–5 per group. (G and H) RT-PCR and ELISA analysis of IL1B from THP-1 cells cultured with extracellular vesicles released from HepG2 cells under treatment with DMSO (control), 0.2 mM palmitate (PA), or 0.2 mM PA + 25 μM ezetimibe for 16 h. n = 5–7 per group. *P < 0.05 and **P < 0.01.
Inflammatory extracellular vesicles (EVs) were recently identified as intercellular mediators between hepatocytes and macrophages in NASH.22 To confirm whether ezetimibe is involved in the inflammasome pathway via hepatocyte-derived EVs, EVs isolated from HepG2 cells were visualized by electron microscopy (Fig. 5E), with confirmation of CD63 protein expression, an exosomal marker, by immunoblots (Fig. 5F). EV-exposure experiments were conducted (Fig. S6), and macrophage expression of both Il1b mRNA and IL1B protein was suppressed by ezetimibe-treated EVs (Fig. 5G and H). Thus, ezetimibe treatment improved inflammation by a hepatocyte-driven exosome pathway.
Ezetimibe ameliorates hepatic steatosis and fibrosis via autophagy induction
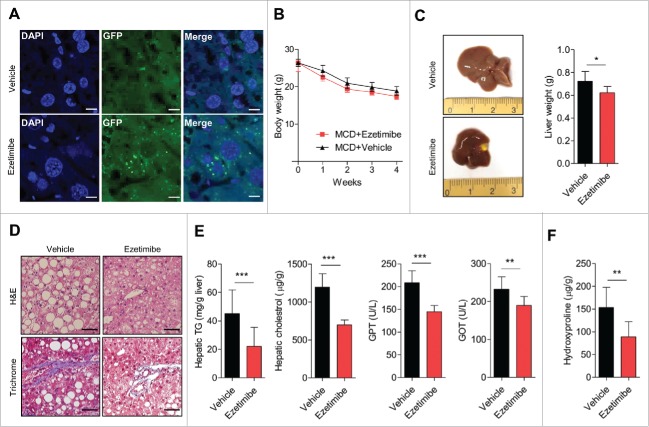
After administration of ezetimibe, the number of GFP-LC3 puncta indicating autophagosomes significantly increased in the liver tissue of chow-fed and methionine- and choline-deficient (MCD) diet-fed GFP-LC3 transgenic mice compared with controls, indicating in vivo autophagy induction by ezetimibe (Fig. 6A and Fig. S7, respectively). On the MCD diet, body weight was comparably reduced in ezetimibe- and vehicle-treated liver-specific Atg7 wild-type mice (Fig. 6B); however, liver size and weights were markedly decreased in ezetimibe-treated mice compared with vehicle-treated mice (Fig. 6C). The administration of ezetimibe markedly decreased multiple lipid droplets and ameliorated fibrotic change in liver sections stained with H&E and Masson's trichrome, respectively (Fig. 6D). Hepatic triglyceride and cholesterol levels and serum GPT/ALT (glutamic puruvic transaminase, soluble) and GOT/AST (glutamic-oxaloacetic transaminase) levels were significantly attenuated by ezetimibe (Fig. 6E). Hepatic hydroxyproline content, representing hepatic fibrosis, was markedly decreased in ezetimibe-treated mice compared with vehicle-treated mice (Fig. 6F).
Figure 6.
Ezetimibe ameliorates hepatic steatosis and fibrosis via autophagy induction. (A) Representative sections of the liver from MCD diet-fed GFP-LC3 transgenic mice treated with vehicle or ezetimibe (10 mg/kg) daily for 4 wk. Scale bars: 10 μm. n = 3 per group. (B) Changes in body weight of liver-specific Atg7 wild-type mice on the MCD diet during 4-wk treatment with vehicle or ezetimibe (10 mg/kg/d). n = 10 per group. (C) Gross images of liver tissue and liver weights from liver-specific Atg7 wild-type mice. *P < 0.05. (D) H&E and Masson's trichrome staining of liver sections from liver-specific Atg7 wild-type mice. Scale bars: 50 μm. (E) Hepatic triglycerides (TG) and cholesterol, and serum GPT and GOT levels. (F) Hepatic hydroxyproline. **P < 0.01, and ***P < 0.001.
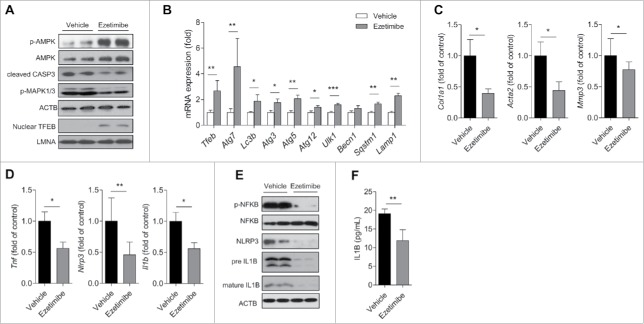
Consistent with our in vitro findings, ezetimibe markedly increased the expression of p-AMPK and nuclear TFEB and decreased cleaved CASP3 (Fig. 7A). The levels of autophagy-related genes including Tfeb, Atg7, Lc3b, Atg3, Atg5, Atg12, Ulk1, Sqstm1, and Lamp1 were significantly increased in the liver from ezetimibe-treated mice (Fig. 7B). Hepatic gene expression of fibrosis markers Col1a1, Acta2, and Mmp3 and inflammatory markers Tnf, Nlrp3, and Il1b was significantly decreased in ezetimibe-treated mice (Fig. 7C and D). p-NFKB, NLRP3, and mature IL1B expression was dramatically attenuated by ezetimibe, and, consistently, ezetimibe resulted in decreased serum IL1B levels (Fig. 7E and F). These results suggest that ezetimibe ameliorates hepatic inflammation and fibrosis.
Figure 7.
Effects of ezetimibe on inflammation and fibrosis-related genes. (A) Liver tissues from liver-specific Atg7 wild-type mice treated with MCD diet or MCD + ezetimibe (10 mg/kg/d) were subjected to immunoblot analysis. n = 10 per group. (B) qPCR analysis of Atg7 wild-type mouse liver tissue treated with ezetimibe or vehicle for autophagy-related genes and lysosomal genes. *P < 0.05, **P < 0.01, and ***P < 0.001. (C) Hepatic gene expression of fibrosis and (D) inflammatory markers in Atg7 wild-type mouse liver tissue. *P < 0.05 and **P< 0.01. (E) The expression of inflammasome-related proteins in Atg7 wild-type mouse liver was determined by immunoblot. n = 10 per group. (F) ELISA assays of serum IL1B in Atg7 wild-type mice. n = 10 per group. **P < 0.01.
Ezetimibe ameliorates steatohepatitis in liver-specific Atg7 haploinsufficient mice but not in atg7 knockout mice
We next investigated whether ezetimibe exerts its protective effects against steatohepatitis in autophagy-insufficient or -absent mouse models. After 4-wk ezetimibe or vehicle administration with MCD diet, liver weights were lower in ezetimibe-treated liver-specific Atg7 haploinsufficient mice (Fig. S8A). Liver tissue sections stained with H&E and Masson's trichrome from liver-specific Atg7 haploinsufficient mice exhibited decreased hepatic lipid droplets and fibrosis by ezetimibe, respectively (Fig. S8B). Hepatic triglyceride and hydroxyproline contents and serum levels of GPT and GOT were significantly decreased in ezetimibe-treated mice relative to vehicle-treated mice (Fig. S8C). Similar to Atg7 wild-type mice, phosphorylation of AMPK and nuclear TFEB were slightly upregulated by ezetimibe in liver-specific Atg7 haploinsufficient mice (Fig. S8D). Ezetimibe also significantly lowered Il1b mRNA expression in the Atg7 haploinsufficient liver (Fig. S8E). p-NFKB and NLRP3 inflammasome-IL1B pathways were consistently downregulated by ezetimibe (Fig. S8F). Consequently, circulating levels of IL1B were markedly decreased in ezetimibe-treated liver-specific Atg7 haploinsufficient mice (Fig. S8G).
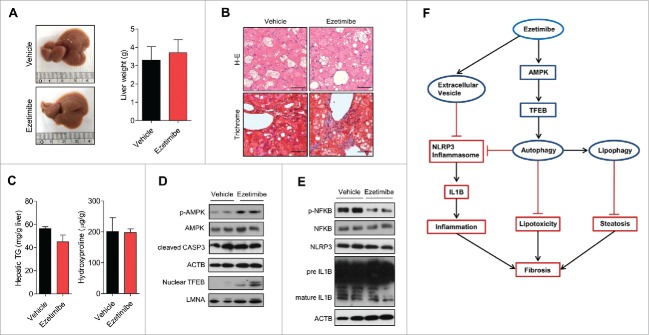
To confirm the effect of ezetimibe on liver steatosis and fibrosis via autophagy induction, we investigated metabolic changes in liver-specific atg7 knockout (atg7−/−) mice treated with vehicle or ezetimibe. The gross morphology of liver tissues and liver weights were similar between vehicle and ezetimibe-treated mice under autophagy-absent conditions (liver-specific atg7−/− mice) (Fig. 8A). Lipid accumulation and fibrosis were not ameliorated by ezetimibe in liver-specific atg7−/− mice (Fig. 8B). Consistently, hepatic triglyceride and hydroxyproline contents did not differ between the 2 groups of liver-specific atg7−/− mice (Fig. 8C). Although the protective effect of ezetimibe was not observed in the blocked-autophagy condition, nuclear TFEB and AMPK phosphorylation were still upregulated by ezetimibe (Fig. 8D). Mature IL1B expression was similar in the presence or absence of ezetimibe, whereas p-NFKB was decreased by ezetimibe in the atg7−/− livers (Fig. 8E). These findings indicate that the effect of ezetimibe on attenuating liver steatosis, inflammation, and fibrosis is autophagy pathway-dependent.
Figure 8.
Ezetimibe could not ameliorate lipid accumulation and fibrosis in liver-specific atg7−/− mice. (A) Representative gross images of liver tissues and liver weights from liver-specific atg7−/− mice treated with vehicle or ezetimibe (10 mg/kg/d) for 4 wk. n = 10 per group. (B) H&E and Masson's trichrome staining of liver sections from liver-specific atg7−/− mice. Scale bars: 50 μm. (C) Hepatic triglycerides (TG) and hydroxyproline. (D) Western blot analysis of liver-specific atg7−/− mouse liver tissue. (E) Inflammasome-related protein expression in liver-specific atg7−/− mouse liver tissue was determined by immunoblot. (F) A schematic illustration showing a plausible mechanism by which ezetimibe ameliorates steatohepatitis and fibrosis.
Discussion
In this study, we report the beneficial effects of ezetimibe for ameliorating hepatic steatosis, inflammation, and fibrosis by inducing AMPK-mediated autophagic activation. We first demonstrated that ezetimibe regulates the inflammasome pathway by modulating autophagy and EV-mediated hepatocyte-macrophage communication. Ezetimibe ameliorated lipid accumulation and lipotoxicity in conjunction with induced autophagy. In the same context, human NAFLD or NASH liver presented decreased autophagic vacuole formation and autophagy pathway impairment with decreased expression of nuclear TFEB. Ezetimibe upregulated AMPK activation, which leads to TFEB nuclear translocation and autophagy induction, independent of the MTOR pathway. Ezetimibe-induced autophagy significantly blocked NLRP3 inflammasome activation and subsequent IL1B release in macrophages. In line with this, human liver with NASH demonstrated increased NLRP3 and IL1B expression. Decrease in IL1B in macrophages cultured with EVs released from palmitate-treated hepatocytes indicated that ezetimibe regulated inflammation via a hepatocyte-driven EV pathway. In vivo experiments using liver-specific Atg7 wild-type, haploinsufficient, and atg7-/− mice verified that ezetimibe can exert immunometabolic effects via autophagy induction.
Consistent with our findings in human livers, a previous study reported impaired autophagy flux with increased endoplasmic reticulum stress in human liver samples with steatosis and NASH by showing an increase in LC3-II and SQSTM1 expression with the progression of steatosis.11 Since the actual autophagy flux measured by the accumulation of LC3-II in the presence or absence of lysosomal inhibitors is not applicable to human livers, SQSTM1, an autophagy substrate, was used as an indirect marker to assess autophagy, which accumulates under the condition of autophagy inhibition. In addition, we detected a reduced number of autophagic vacuoles with concomitant increase in lipofuscin number, reflecting impaired autophagy with accumulation of undegradable damaged cellular components23,24 in human NAFLD or NASH livers compared with normal livers. Furthermore, we observed for the first time that nuclear expression of TFEB, a master regulator of autophagy, was reduced in human livers with NAFLD or NASH, indicating the inhibition of upstream autophagy regulatory pathways in these tissues. Consistent with our data, previous studies reported that NASH patients had higher hepatic mRNA expression of Il1b and Nlrp3 relative to normal controls.12,25
AMPK has essential roles in autophagy regulation.19 Our data indicated that AMPK was indispensable for the therapeutic effect of ezetimibe on steatosis and lipotoxicity via autophagy induction. Previously, as the canonical mechanisms of activation of AMPK involved increases in AMP and ADP,26 low contents of ATP induced by ezetimibe suggested that ezetimibe may activate AMPK phosphorylation via a depletion of intracellular ATP contents. Moreover, ezetimibe significantly increased the nuclear translocation and transcription of TFEB via an AMPK-dependent mechanism. In previous studies, TFEB nuclear translocation increased the transcription of genes encoding autophagic and lysosomal proteins and consequently promotes autophagosome formation and lysosomal biogenesis to increase autophagy.18 Furthermore, overexpression of TFEB protects against diet-induced steatosis in mouse livers mainly by enhancing lipid degradation and negatively regulating lipid biosynthesis.27 One regulatory mechanism commonly involved in TFEB nuclear translocation is MTOR inhibition.28 Inhibition of MTOR activity is also a well-established mechanism to induce autophagy.29 Previously, ezetimibe was showed to reduce translocation of MTOR to late endosomes/lysosome, wherein free cholesterol accumulated, resulting in the downregulation of MTOR.17 Consistently, we also observed that ezetimibe decreased p-RPS6 levels, indicating the suppression of MTOR activity (Fig. 4B). To determine whether ezetimibe exerts MTOR-independent ameliorative effects on autophagy induction, we used tsc2−/− MEFs and found that ezetimibe continued to induce autophagy, with activation of AMPK and increased translocation of nuclear TFEB. Collectively, these data suggest that the AMPK-TFEB pathway is involved in ezetimibe-induced autophagy, even in MTOR-hyperactivated conditions.
As possible MTOR-independent AMPK-related mechanisms, the MAPK1/ERK2 and PRKC pathways have been reported to phosphorylate TFEB. While MAPK1/ERK2 phosphorylates serine 142 of TFEB and leads to its subcellular localization in the cytosol,20 PRKCB phosphorylates 3 serine residues at the C terminus of TFEB, resulting in its stabilization and increased activity.21 In this regard, we observed that ezetimibe decreased phosphorylated MAPK1/ERK2-MAPK3/ERK1 expression with increased constitutive TFEB nuclear translocation. Because AMPK negatively regulates p-MAPK1/3 by inducing dual-specificity protein phosphatase activity,20 ezetimibe downregulates MAPK1/3 via AMPK activation and consequently leads to nuclear translocation and activation of TFEB. Furthermore, as TFEB has a positive autoregulatory component, binding to its own promoter and modulating its transcription after the initial activation,27 our luciferase assay data also confirmed the autoregulation of TFEB by ezetimibe.
Previous studies demonstrated that autophagy negatively regulates NLRP3 inflammasome activity.30 The inflammasome is a large molecular danger signal-sensing protein complex, primarily induced by NFKB-activating signaling.10 Upon inflammasome activation, matured CASP1 (caspase 1) mediates pre-IL1B cleavage into mature IL1B and secretion of IL1B, a major proinflammatory cytokine involved in the progression of NASH.10 Decline in autophagic activity stimulates the NLRP3 inflammasome, which promotes inflammatory conditions and metabolic disorders.31 Our data clearly demonstrated that ezetimibe not only downregulated NFKB activation but also dampened NLRP3 inflammasome-IL1B signaling via an autophagy-dependent pathway in macrophages and diet-induced NASH models. This anti-inflammatory action of ezetimibe is partially consistent with a previous report showing that ezetimibe inhibited expression of CRP (C-reactive protein), TNF, and NFKB in THP-1 macrophages.32
Recently, cell-cell interaction through exosomes was elucidated as a crucial factor in the development of NAFLD and NASH.33 Exosomes, which contain proteins, mRNAs, and miRNAs, are released by several cell types into biological fluids and taken up by endocytosis into different cell types. This exosome-mediated cell-cell communication can regulate physiological events and modify the microenvironment in target cells, involving mechanisms of hepatocyte regeneration and migration.34,35 It was noted that an increased number of EVs was secreted from macrophages in liver tissues from patients with NAFLD or NASH,36 and secreted EVs from hepatocytes in response to toxic lipids triggered an inflammatory response in macrophages via a TNFSF10/TRAIL (TNF superfamily member 10)-TNFRSF10B/DR5 (TNF receptor superfamily member 10b) ligand signaling pathway in a RIPK1 (receptor interacting serine/threonine kinase 1)-dependent manner, leading to NASH progression.22 Besides the direct inhibitory effect of ezetimibe on NLRP3 inflammasomes in macrophages by autophagy, ezetimibe could also modulate hepatocyte-macrophage interaction via a hepatocyte-driven exosome pathway, as evidenced by the finding of decreased IL1B mRNA and protein in macrophages when cultured with EVs released from ezetimibe/palmitate co-treated hepatocytes. Further investigation of specific structural changes and the detailed mechanism of EV alteration by ezetimibe is needed.
Ezetimibe has been shown to improve NAFLD in several human clinical trials.15,37 Reduction in body weight and visceral fat, decrease in serum triglycerides, and improvement in insulin resistance may indirectly account for this improvement. In addition, ezetimibe has been proven to ameliorate hepatic steatosis in animal models with respect to decrease in hepatic free cholesterol, oxidative stress, and hepatocyte apoptosis.38,39 Despite these multiple effects of ezetimibe, its detailed mechanism needs to be described. Here we newly identify additional mechanisms of ezetimibe for the treatment of NAFLD and NASH, involving roles in autophagic induction, inflammasome inhibition, and exosome-mediated pathway modulation. Ezetimibe was initially discovered from a chemical library in the process of searching novel ACAT (acetyl-CoA acetyltransferase) inhibitors, which mediate cholesterol trafficking.40,41 After confirmation of ezetimibe's cholesterol-lowering effects in the human clinical trials, its putative mechanism related to cholesterol-lowering was elucidated by several papers,13,42 and found to inhibit intestinal cholesterol absorption via the NPC1L1 transporter protein. Therefore, its multiple involvement in pathways protecting against steatohepatitis and fibrosis that we show here, may imply pleiotropic aspects of ezetimibe, outside of a cholesterol absorption inhibitor. These findings open new perspectives of ezetimibe to investigate other possible drug targets, independent of NPC1L1 and suggest drug repositioning of ezetimibe for the treatment of NASH. Because of the lack of FDA-approved treatment modalities for NASH, activation of the AMPK-TFEB pathway should be considered as potential targets for the investigation of therapeutic approaches and discovering new drug candidates for NASH.
In conclusion, the present study demonstrated that ezetimibe ameliorated hepatic steatosis, inflammation, and fibrosis via induction of autophagy through PRKA activation and subsequent TFEB nuclear translocation, related to an MTOR-independent ameliorative effect and the MAPK/ERK pathway (Fig. 8F). Furthermore, autophagy activation by ezetimibe downregulates NLRP3 inflammasome-IL1B-mediated inflammation in macrophages. Moreover, ezetimibe modulates hepatocyte-macrophage communication by EVs released from hepatocytes, consequently suppressing inflammation. Altogether, based on these additional novel implications, ezetimibe is an attractive potential therapy for the treatment of steatohepatitis and fibrosis.
Materials and methods
Antibodies and reagents
The following commercially available antibodies were used: anti-p-PRKAA1/2 (Thr172; Cell Signaling Technology, 2535), anti-PRKAA1/2 (Cell Signaling Technology, 2795), anti-LMNA (Cell Signaling Technology, 2032), anti-phosphorylated MAPK1/ERK2-MAPK3/ERK1 (Thr202/Tyr204; Cell Signaling Technology, 9101), anti-MAPK1/ERK2-MAPK3/ERK1 (Cell Signaling Technology, 9102), anti-CASP3 (Cell Signaling Technology, 9661), anti-p-NFKB (Ser536; Cell Signaling Technology, 3033), anti-NFKB (Cell Signaling Technology, 8242), anti-p-RPS6 (Ser235/236; Cell Signaling Technology, 2211), anti-RPS6 (Cell Signaling Technology, 2217), anti-SQSTM1 (Santa Cruz Biotechnology, sc-28359), anti-NLRP3 (Santa Cruz Biotechnology, sc-66846), anti-IL1B (Santa Cruz Biotechnology, sc-7884), anti-MTOR (Santa Cruz Biotechnology, sc-8319), anti-LC3B (Sigma-Aldrich, L7543), anti-ACTB (Sigma-Aldrich, A5441), anti-Flag (Sigma-Aldrich, F7425), and anti-TFEB (Bethyl Laboratories, Inc., A303–673A). Antibody against CD63 was kindly provided by Myeong Heon Shin (Department of Environmental Medical Biology and Institute of Tropical Medicine, Yonsei University College of Medicine, Seoul, Korea).
The following chemicals were used: palmitate(Sigma-Aldrich, P9767), oleate (OA; Sigma-Aldrich, O1008), bovine serum albumin (BSA; Sigma-Aldrich, A2058), lipopolysaccharide (LPS; Sigma-Aldrich, L6529), phorbol 12 myristate 13-acetate (PMA; Sigma-Aldrich, P1585), bafilomycin A1 (Sigma-Aldrich, B1793), chloroquine (CQ; Sigma-Aldrich, C6628), Oil red O (ORO; Sigma-Aldrich, O1391), compound C (Sigma-Aldrich, P5499), metformin (Sigma-Aldrich, D15095–9), and PRKC (protein kinase C) inhibitor (Sigma-Aldrich, G1918) and ezetimibe (Cayman Chemical, 16331).
Human subjects
We collected liver tissue samples from a total of 18 subjects who underwent hepatectomy or cholecystectomy at the university-affiliated Severance Hospital, Yonsei University College of Medicine, Republic of Korea, from September 2014 to May 2016. Liver specimens were histologically reviewed by an experienced pathologist and histologically diagnosed as normal (n = 12), simple steatosis (n = 11), or NASH (n = 9). Inclusion criteria for steatosis were based on steatosis with/without necroinflammation and/or fibrosis.43 Exclusion criteria for our protocol were (1) history of alcohol consumption >210 g/wk for men and 140 g/wk for women and (2) any positive serological markers for hepatitis B or hepatitis C virus. All individuals gave informed consent, and the study protocol was approved by the Institutional Review Board at Severance Hospital (IRB No 4–2014–0674).
Isolation of primary hepatocytes and peritoneal macrophages and cell cultures
Hepatocytes were isolated from collagenase-perfused livers of male C57BL/6J wild-type mice (8 wk old, Jackson Laboratory, Bar Harbor, ME, USA) as described previously.44 Hepatocytes (4 × 105 cells/ml) were plated on COL1/collagen type-1-coated wells in 6-well plates and maintained in William's E medium (Gibco, 12551–032) supplemented with 5% fetal bovine serum (Gibco, 16000–044), 1% penicillin, 1% streptomycin (Gibco, 15140–122), 100 nM dexamethasone (Sigma-Aldrich, D1756), and 100 nM insulin (Roche, 11–375–497–001) for 4 h. After 4-h culture, the medium was replaced with serum-containing or serum-free medium (HepatoZYME-SFM; Gibco, 17705–021) for additional time to culture. Peritoneal macrophages were harvested by peritoneal lavage with phosphate-buffered saline (PBS; Gibco, 10010–023), centrifuged, and plated at 106 macrophages per well in 6-well plates containing a defined, RPMI 1640 medium (Gibco, 22400–089). The HepG2 cells and MEF cells were cultured in Dulbecco's modified Eagle's medium (Gibco, 11995–065), whereas THP-1 cells were grown in RPMI 1640 medium (Gibco). Media were supplemented with 10% fetal bovine serum, 1% penicillin, and 1% streptomycin in a 5% CO2 incubator at 37°C.
MEFs with a double KO against Prkaa1/2 subunits (α1−/− and α2−/−) and wild-type controls were generously provided by Benoit Viollet (INSERM U567, CNRS UMR8104, Department of Endocrinology, Metabolism and Cancer, Institut Cochin, Paris, France). Tsc2+/+ and tsc2−/− MEFs were kindly provided by David J. Kwiatkowski (Brigham and Women's Hospital/Harvard Medical School, Boston, MA, USA).
Small interfering RNA (siRNA) transfection
Primary hepatocytes and HepG2 cells were transfected with siRNAs targeting PRKAA1/2 (Santa Cruz Biotechnology, sc-45312) or control siRNA using Lipofectamine 2000 following the manufacturer's instructions (Life Technologies, 11668–019).
Oil Red O staining
Lipid droplets were visualized and quantified by ORO staining. Primary mouse hepatocytes were exposed to 2 mM OA for 16 h, after the treatment with or without 50 μM ezetimibe for 2 h or 20 μM CQ for 2 h, followed by fixation with 10% formalin for 30 min. MEFs were exposed to 0.5 mM OA for 16 h or 20 μM ezetimibe pre-treatment of 2 h followed by 0.5 mM OA in the absence or presence of 20 μM CQ for 2 h, and fixed with 10% formalin. Fixed cells were then stained in ORO solution for 1 h, and stained lipid droplets were observed with a light microscope.
Triglyceride assay
Triglyceride content in hepatocytes was quantified using a Triglyceride Quantification kit (BioAssay Systems, ETGA-200) according to the manufacturer's manual.
ATP assay
Total cellular ATP levels in primary hepatocytes or HepG2 were determined using the ATP Determination Kit (BioVision Inc., K354–100) according to the manufacturer's instructions. The ATP concentration was normalized with the cell protein. The luminescence was measured at 570 nm.
CASP3 assay
Cell viability was assessed by a CASP3 (caspase 3) colorimetric assay kit (BioVision Inc., K106–100) measuring the activity of CASP3-like proteases in the lysate according to the manufacturer's protocol. Primary hepatocytes were incubated in the presence or absence of the indicated concentration of ezetimibe or 20 μM CQ for 2 h and then treated with palmitate (0.25 mM) for 24 h. siRNA-mediated PRKAA1/2 wild-type or silencing primary hepatocytes were incubated with or without various concentrations (20 or 50 μM) of ezetimibe or 20 μM CQ for 2 h and treated with 0.2 mM palmitate for 24 h. Cells were then used to prepare cell lysates in 50 μl and exposed to 5 μl of the CASP3 substrate in assay buffer (total volume 150 μl) for 90 min at 37°C. Optical density was detected using a microplate reader (Molecular Devices, Sunnyvale, CA, USA) at 405 nm.
Luciferase assay
Primary hepatocytes were transiently transfected with luciferase reporter plasmids encoding the TFEB promoter or empty construct using Lipofectamine 2000 (Invitrogen, 11668–019). Cells were treated with DMSO (Sigma-Aldrich, D8414) (vehicle) or ezetimibe, or compound C. Subsequently, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, E1910) on a GloMax Multi detection system (Promega, E7041) according to the manufacturer's instructions. Renilla luciferase signals were normalized to the internal firefly luciferase transfection control. Transfections were performed in at least triplicate for each independent experiment.
Isolation of extracellular vesicles
EVs were isolated from HepG2 cells using the Total Exosome Purification Kit (Invitrogen, 4478359) following the manufacturer's instructions as described previously.22 To remove cellular debris, the culture medium was centrifuged at 2,000 × g for 20 min. The supernatant was transferred to a new microfuge tube, and 0.5 volume of the Invitrogen reagent was added to the sample and vortexed to mix. Samples were incubated overnight at 4°C and subsequently centrifuged at 10,000 × g for 1 h at 4°C. Supernatants were discarded, and pellets were resuspended in 100 µL PBS.
Transmission electron microscopy
Autophagic vacuoles and EVs were detected with an electron microscope (JEM-1011, JEOL/MegaView III, Olympus, Tokyo, Japan). Liver, cell, or pellet specimens were fixed with 2% glutaraldehyde, 2% paraformaldehyde buffered with 0.1 M phosphate buffer, pH 7.2 overnight at 4°C, followed by post-fixation with 1% osmium tetroxide in a 0.1 M sodium cacodylate buffer, pH 7.4 for 1 h at room temperature and graded ethanol dehydration as described previously.45 Samples were embedded in Epon 812 (Electron Microscopy Sciences, 100503–876) and sectioned using an ultramicrotome (Leica, EM UC7, Wetzlar, Germany).
Animal study
Ten-wk-old male C57BL/6J-background liver-specific Atg7 wild-type, haploinsufficient, or homozygous KO mice were fed a MCD as described previously.46 The animals were given free access to diet and water and maintained at a temperature of 23 ± 2°C and humidity of 60 ± 10% on a 12-h light/dark cycle. Food intake and body weight were measured weekly. Randomly assigned mice were orally administered vehicle (distilled water) or ezetimibe (10 mg/kg; Cayman Chemical, 16331) daily for 4 wk. After mice were killed following a 6-h fast, blood was collected via heart puncture, and tissues were harvested. For in vivo autophagy experiments, MCD diet-fed GFP-LC3 transgenic mice were treated with ezetimibe (10 mg/kg) daily for 4 wk, and GFP puncta were identified by fluorescence microscopy. All animal studies were approved by the Animal Care and Use Committee of the Yonsei University College of Medicine.
ELISA for IL1B and biochemical analyses
Serum levels of GPT and GOT were quantified using a colorimetric determination of activity assay kit (BioAssay Systems, EALT-100, EASTR-100). IL1B was measured in THP-1 and primary peritoneal macrophage cultured cell supernatants and serum using enzyme-linked immunosorbent assay (ELISA) kits (eBioscience, human_88–7261–22, mouse_88–7013–22) according to the manufacturer's instructions. Briefly, the supernatant fractions were coated overnight at 4°C with a capture antibody on 96-well ELISA plates. The plates were washed 3 times with wash buffer (0.05% Tween 20 [Sigma-Aldrich, P1379] in PBS, pH 7.4). The supernatant fraction (sample) and standard were added to the well and incubated at room temperature for 2 h, and the human/mouse IL1B detection antibody solution was added. After 2-h incubation at room temperature, the plates were washed 5 times with wash buffer, and streptavidin HRP was added to each well. After a 30-min incubation at room temperature, the plates were visualized in the dark using a substrate solution for reaction with HRP and a microtiter plate reader MAX190 (Molecular Devices, Sunnyvale, CA, USA) calibrated to 450 nm.
Immunoblot analyses
Total cell lysates were prepared by lysis of human livers, primary hepatocytes, and HepG2 cells with RIPA buffer (20 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate; Cell Signaling Technology, 9806) and the protein contents were measured using the Bradford assay (Bio-Rad, 500–0006). Equivalent amounts of each protein extract were heat denatured in 5 × sample buffer (2% sodium dodecyl sulfate (Tech&Innovation, BSS-9005), 62.5 mM Tris, pH 6.8, 0.01% bromophenol blue, 1.43 mM mercaptoethanol, and 0.1% glycerol), separated on 10% polyacrylamide gels, and electrophoretically transferred onto a polyvinylidene fluoride membrane (Bio-Rad, 1620175). After blocking, membranes were treated with the appropriate antibodies. Immunostaining was performed using chemiluminescent reagents (SuperSignal West Pico Luminol/Enhancer solution; Thermo Scientific, 34080) and Agfa medical X-ray film (Mortsel, CURIX 60). ACTB protein levels were used as a loading control.
RNA isolation and reverse transcriptase-polymerase chain reaction (RT-PCR) analysis
Total RNA was isolated from cells with TRIzol reagent (Invitrogen, 15596–018) following the manufacturer's instructions, and then 2 μg total RNA was reverse transcribed into cDNA (cDNA) using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, 4368814). The cDNA was then amplified in the ABI 7500 sequence detection system (Applied Biosystems, 4350584) using Power SYBR® Green PCR Master Mix (Applied Biosystems, 4367659) with the following cycling conditions: 40 cycles of 95°C for 5 sec, 58°C for 10 sec, and 72°C for 20 sec. Target gene expression was normalized to that of Gapdh (glyceraldehyde-3-phosphate dehydrogenase) or Actb, and quantitative analyses were conducted using the ΔΔcycle threshold method and StepOne Software version 2.2.2. The primer sets used for RT-PCR are described in Table S1. All reactions were performed in at least triplicate.
Immunofluorescence
For detection of autophagosomes and autolysosomes, primary hepatocytes, HepG2 cells, or MEFs were transfected with the expression vector mRFP-GFP-LC3 (Addgene, plasmid 11546) using Lipofectamine 2000 (Invitrogen, 11668–019) for 48 h. The cells were then incubated with either ezetimibe (50 μM) for 18 h or glucose starvation medium for 18 h or bafilomycin A1 (50 nM) for 2 h. After washing with PBS and fixation in 4% paraformaldehyde, cells were observed under a fluorescence microscope (LSM700; Carl Zeiss Inc., Oberkochen, Germany). To detect nuclear translocation of TFEB, HepG2 cells or MEFs were transfected with blank vector or DNA plasmids pCMV-TFEB-3 × FLAG, kindly provided by Andrea Ballabio (Telethon Institute of Genetics and Medicine, Pozzuoli, Naples). TFEB transfection was detected by probing membranes with anti-FLAG M2 antibody (1:1,000; Sigma, F1804). Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI; Vector Laboratories, H-1200). All experiments were conducted independently at least 3 times.
Histological analysis
Liver tissues were fixed with 10% neutral-buffered formalin and embedded with paraffin. Sections (5 μm) were stained using hematoxylin and eosin (H&E) and Masson's trichrome.
Measurement of hepatic triglyceride, cholesterol, and hydroxyproline
After homogenization, triglyceride and cholesterol contents in liver tissues were measured using a Triglycerides Quantification Kit (Bioassay, ETGA-200) and Total Cholesterol and Cholesteryl Ester Colorimetric/Fluorometric Assay Kit (BioVision, K603–100), respectively, according to the manufacturer's instructions. Hydroxyproline, a determinant of total collagen, was measured by a modification of previously described methods using a Hydroxyproline Colorimetric Assay Kit (BioVision, K555–100).47
Statistical analyses
Data were presented as mean ± standard error of the mean (SEM) from at least 3 independent experiments. Data were analyzed by 2-tailed Student t test for comparisons between 2 groups, or one-way analysis of variance (ANOVA) with post-hoc Bonferroni multiple comparison test for comparisons involving > 2 groups. Statistical analyses were performed using PRISM 5.0 software (GraphPad Software, La Jolla, CA). P values < 0.05 were considered statistically significant.
Supplementary Material
Abbreviations
- ACTA2
actin, α 2, smooth muscle, aorta
- ACTB
actin β
- AMP
activated, α
- ANOVA
analysis of variance
- Bafilo
bafilomycin A1
- BECN1
Beclin 1, autophagy related
- BSA
bovine serum albumin
- CASP1
caspase 1
- CASP3
caspase 3
- COL1A1
collagen, type I, α 1
- DAPI
4',6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- ELISA
enzyme-linked immunosorbent assay
- EV
extracellular vesicles
- FDA
U.S. Food and Drug Administration
- GFP
green fluorescent protein
- GOT/AST
glutamic-oxaloacetic transaminase
- GPT/ALT
glutamic puruvic transaminase, soluble
- H&E
hematoxylin and eosin
- IL1B
interleukin 1 β
- IL18
interleukin 18
- KO
knockout
- LAMP1
lysosomal-associated membrane protein 1
- LMNA
lamin A/C
- LPS
lipopolysaccharide
- MAPK/ERK
mitogen-activated protein kinase
- MAP1LC3B/LC3B
microtubule-associated protein 1 light chain 3 β
- MCD
methionine- and choline-deficient diet
- MEF
mouse embryonic fibroblast
- MMP3
matrix metallopeptidase 3
- MTOR
mechanistic target of rapamycin
- MTT3
3-(4, 5-dimethyldiazol-2-yl)-2, 5-diphenyltetrazolium bromide
- NAFLD
non-alcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- NFKB
nuclear factor kappa B
- NLRP3
NLR family pyrin domain containing 3
- NPC1L1
NPC1 like intracellular cholesterol transporter 1
- ORO
Oil red O
- PRKC
protein kinase C
- PMA
phorbol 12 myristate 13-acetate
- PRKA
protein kinase, AMP-activated
- PRKAA1/2
protein kinase, AMP-activated, catalytic subunit α1/2
- PRKCB
protein kinase C, β
- RPS6
ribosomal protein S6
- PCR
polymerase chain reaction
- siRNA
small interfering RNA
- SQSTM1
sequestosome 1
- TFEB
transcription factor EB
- TNF
tumor necrosis factor
- Tsc2
tuberous sclerosis 2
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Korea Healthcare Technology R&D Project, Ministry of Health and Welfare, Republic of Korea under Grant HI14C2476; Basic Science Research Program through the National Research Foundation of Korea (NRF) by the Ministry of Science, ICT & Future Planning under Grant 2015R1C1A1A01052558; NRF by the Korean Government (MSIP) under Grant NRF-2016R1A5A1010764.
References
- [1].Musso G, Cassader M, Gambino R. Non-alcoholic steatohepatitis: emerging molecular targets and therapeutic strategies. Nat Rev Drug Discovery. 2016;15:249-74. PMID:26794269 [DOI] [PubMed] [Google Scholar]
- [2].Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology (Baltimore, Md). 2012;55:2005-23. doi: 10.1002/hep.25762. PMID:22488764 [DOI] [PubMed] [Google Scholar]
- [3].Day CP, James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114:842-5. doi: 10.1016/S0016-5085(98)70599-2. PMID:9547102 [DOI] [PubMed] [Google Scholar]
- [4].Birkenfeld AL, Shulman GI. Nonalcoholic fatty liver disease, hepatic insulin resistance, and type 2 diabetes. Hepatology (Baltimore, Md). 2014;59:713-23. doi: 10.1002/hep.26672. PMID:23929732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bertola A, Bonnafous S, Anty R, Patouraux S, Saint-Paul MC, Iannelli A, Gugenheim J, Barr J, Mato JM, Le Marchand-Brustel Y, et al.. Hepatic expression patterns of inflammatory and immune response genes associated with obesity and NASH in morbidly obese patients. PloS One. 2010;5:e13577. doi: 10.1371/journal.pone.0013577. PMID:21042596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Quan W, Lee MS. Role of autophagy in the control of body metabolism. Endocrinol Metab (Seoul, Korea). 2013;28:6-11. doi: 10.3803/EnM.2013.28.1.6. PMID:24396643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Amir M, Czaja MJ. Autophagy in nonalcoholic steatohepatitis. Expert Rev Gastroenterol Hepatol. 2011;5:159-66. doi: 10.1586/egh.11.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, et al.. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science (New York, NY). 2010;329:229-32. doi: 10.1126/science.1190354 [DOI] [PubMed] [Google Scholar]
- [9].Wree A, Eguchi A, McGeough MD, Pena CA, Johnson CD, Canbay A, Hoffman HM, Feldstein AE. NLRP3 inflammasome activation results in hepatocyte pyroptosis, liver inflammation, and fibrosis in mice. Hepatology (Baltimore, Md). 2014;59:898-910. doi: 10.1002/hep.26592. PMID:23813842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bauernfeind F, Ablasser A, Bartok E, Kim S, Schmid-Burgk J, Cavlar T, Hornung V. Inflammasomes: current understanding and open questions. Cell Mol Life Sci. 2011;68:765-83. doi: 10.1007/s00018-010-0567-4. PMID:21072676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gonzalez-Rodriguez A, Mayoral R, Agra N, Valdecantos MP, Pardo V, Miquilena-Colina ME, Vargas-Castrillón J, Lo Iacono O, Corazzari M, Fimia GM, et al.. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014;5:e1179. doi: 10.1038/cddis.2014.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wree A, McGeough MD, Pena CA, Schlattjan M, Li H, Inzaugarat ME, Messer K, Canbay A, Hoffman HM, Feldstein AE, et al.. NLRP3 inflammasome activation is required for fibrosis development in NAFLD. J Mol Med (Berlin, Germany). 2014;92:1069-82. doi: 10.1007/s00109-014-1170-1. PMID:24861026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Altmann SW, Davis HR, Jr Zhu LJ, Yao X, Hoos LM, Tetzloff G, et al.. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Sci (New York, NY). 2004;303:1201-4. doi: 10.1126/science.1093131 [DOI] [PubMed] [Google Scholar]
- [14].Park SW. Intestinal and hepatic niemann-pick c1-like 1. Diabetes Metab J. 2013;37:240-8. doi: 10.4093/dmj.2013.37.4.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Park H, Shima T, Yamaguchi K, Mitsuyoshi H, Minami M, Yasui K, Itoh Y, Yoshikawa T, Fukui M, Hasegawa G, et al.. Efficacy of long-term ezetimibe therapy in patients with nonalcoholic fatty liver disease. J Gastroenterol. 2011;46:101-7. doi: 10.1007/s00535-010-0291-8. PMID:20658156 [DOI] [PubMed] [Google Scholar]
- [16].Yoneda M, Fujita K, Nozaki Y, Endo H, Takahashi H, Hosono K, Suzuki K, Mawatari H, Kirikoshi H, Inamori M, et al.. Efficacy of ezetimibe for the treatment of non-alcoholic steatohepatitis: an open-label, pilot study. Hepatol Res Official J Japan Society Hepatol. 2010;40:566-73. doi: 10.1111/j.1872-034X.2010.00644.x. PMID:20412324 [DOI] [PubMed] [Google Scholar]
- [17].Yamamura T, Ohsaki Y, Suzuki M, Shinohara Y, Tatematsu T, Cheng J, Okada M, Ohmiya N, Hirooka Y, Goto H, et al.. Inhibition of Niemann-Pick-type C1-like1 by ezetimibe activates autophagy in human hepatocytes and reduces mutant alpha1-antitrypsin Z deposition. Hepatology (Baltimore, Md). 2014;59:1591-9. doi: 10.1002/hep.26930. PMID:24214142 [DOI] [PubMed] [Google Scholar]
- [18].Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al.. TFEB links autophagy to lysosomal biogenesis. Science (New York, NY). 2011;332:1429-33. doi: 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016-23. doi: 10.1038/ncb2329. PMID:21892142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kim HS, Kim MJ, Kim EJ, Yang Y, Lee MS, Lim JS. Berberine-induced AMPK activation inhibits the metastatic potential of melanoma cells via reduction of ERK activity and COX-2 protein expression. Biochemical Pharmacol. 2012;83:385-94. doi: 10.1016/j.bcp.2011.11.008. PMID:22120676 [DOI] [PubMed] [Google Scholar]
- [21].Ferron M, Settembre C, Shimazu J, Lacombe J, Kato S, Rawlings DJ, Ballabio A, Karsenty G. A RANKL-PKCbeta-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27:955-69. doi: 10.1101/gad.213827.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hirsova P, Ibrahim SH, Krishnan A, Verma VK, Bronk SF, Werneburg NW, Charlton MR, Shah VH, Malhi H, Gores GJ, et al.. Lipid-induced signaling causes release of inflammatory extracellular vesicles from hepatocytes. Gastroenterology. 2016;150:956-67. doi: 10.1053/j.gastro.2015.12.037. PMID:26764184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Moore MN, Allen JI, McVeigh A, Shaw J. Lysosomal and autophagic reactions as predictive indicators of environmental impact in aquatic animals. Autophagy. 2006;2:217-20. PMID:16874099. doi: 10.4161/auto.2663 [DOI] [PubMed] [Google Scholar]
- [24].Ahishali E, Demir K, Ahishali B, Akyuz F, Pinarbasi B, Poturoglu S, Ibrisim D, Gulluoglu M, Ozdil S, Besisik F, et al.. Electron microscopic findings in non-alcoholic fatty liver disease: is there a difference between hepatosteatosis and steatohepatitis? J Gastroenterol Hepatol. 2010;25:619-26. PMID:20370732. doi: 10.1111/j.1440-1746.2009.06142.x [DOI] [PubMed] [Google Scholar]
- [25].Csak T, Ganz M, Pespisa J, Kodys K, Dolganiuc A, Szabo G. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology (Baltimore, Md). 2011;54:133-44. PMID:21488066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251-62. doi: 10.1038/nrm3311. PMID:22436748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Settembre C, De Cegli R Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al.. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647-58. doi: 10.1038/ncb2718. PMID:23604321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al.. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095-108. doi: 10.1038/emboj.2012.32. PMID:22343943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim KH, Lee MS. Autophagy–a key player in cellular and body metabolism. Nat Rev Endocrinol. 2014;10:322-37. doi: 10.1038/nrendo.2014.35. PMID:24663220 [DOI] [PubMed] [Google Scholar]
- [30].Salminen A, Kaarniranta K, Kauppinen A. Inflammaging: disturbed interplay between autophagy and inflammasomes. Aging. 2012;4:166-75. doi: 10.18632/aging.100444. PMID:22411934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wen H, Ting JP, O'Neill LA. A role for the NLRP3 inflammasome in metabolic diseases–did Warburg miss inflammation? Nat Immunol. 2012;13:352-7. doi: 10.1038/ni.2228. PMID:22430788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Qin L, Yang YB, Yang YX, Zhu N, Li SX, Liao DF, , Zheng XL. Anti-inflammatory activity of ezetimibe by regulating NF-kappaB/MAPK pathway in THP-1 macrophages. Pharmacology. 2014;93:69-75. doi: 10.1159/000357953. PMID:24557496 [DOI] [PubMed] [Google Scholar]
- [33].Sato K, Meng F, Glaser S, Alpini G. Exosomes in liver pathology. J Hepatol. 2016;65:213-21. doi: 10.1016/j.jhep.2016.03.004. PMID:26988731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology (Baltimore, Md). 2012;56:1946-57. doi: 10.1002/hep.25873. PMID:22684891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Povero D, Feldstein AE. Novel Molecular Mechanisms in the Development of Non-Alcoholic Steatohepatitis. Diabetes Metab J. 2016;40:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kornek M, Lynch M, Mehta SH, Lai M, Exley M, Afdhal NH, Schuppan D. Circulating microparticles as disease-specific biomarkers of severity of inflammation in patients with hepatitis C or nonalcoholic steatohepatitis. Gastroenterology. 2012;143:448-58. doi: 10.1053/j.gastro.2012.04.031. PMID:22537612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Takeshita Y, Takamura T, Honda M, Kita Y, Zen Y, Kato K, Misu H, Ota T, Nakamura M, Yamada K, et al.. The effects of ezetimibe on non-alcoholic fatty liver disease and glucose metabolism: a randomised controlled trial. Diabetologia. 2014;57:878-90. doi: 10.1007/s00125-013-3149-9. PMID:24407920 [DOI] [PubMed] [Google Scholar]
- [38].Van Rooyen DM, Gan LT, Yeh MM, Haigh WG, Larter CZ, Ioannou G, Teoh NC, Farrell GC. Pharmacological cholesterol lowering reverses fibrotic NASH in obese, diabetic mice with metabolic syndrome. Journal of Hepatology. 2013;59:144-52. doi: 10.1016/j.jhep.2013.02.024. PMID:23500152 [DOI] [PubMed] [Google Scholar]
- [39].Lee DH, Han DH, Nam KT, Park JS, Kim SH, Lee M, Kim G, Min BS, Cha BS, Lee YS, et al.. Ezetimibe, an NPC1L1 inhibitor, is a potent Nrf2 activator that protects mice from diet-induced nonalcoholic steatohepatitis. Free Radical Biol Med. 2016;99:520-32. doi: 10.1016/j.freeradbiomed.2016.09.009 [DOI] [PubMed] [Google Scholar]
- [40].Burnett DA, Caplen MA, Davis HR Jr, Burrier RE, Clader JW. 2-Azetidinones as inhibitors of cholesterol absorption. J Med Chem. 1994;37:1733-6. doi: 10.1021/jm00038a001. PMID:8021912 [DOI] [PubMed] [Google Scholar]
- [41].Salisbury BG, Davis HR, Burrier RE, Burnett DA, Bowkow G, Caplen MA, Clemmons AL, Compton DS, Hoos LM, McGregor DG, et al.. Hypocholesterolemic activity of a novel inhibitor of cholesterol absorption, SCH 48461. Atherosclerosis. 1995;115:45-63. doi: 10.1016/0021-9150(94)05499-9. PMID:7669087 [DOI] [PubMed] [Google Scholar]
- [42].Garcia-Calvo M, Lisnock J, Bull HG, Hawes BE, Burnett DA, Braun MP, Crona JH, Davis HR Jr, Dean DC, Detmers PA, et al.. The target of ezetimibe is Niemann-Pick C1-Like 1 (NPC1L1). Proc Natl Acad Sci U S A. 2005;102:8132-7. doi: 10.1073/pnas.0500269102. PMID:15928087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Takahashi Y, Fukusato T. Histopathology of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. 2014;20:15539-48. doi: 10.3748/wjg.v20.i42.15539. PMID:25400438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Seglen PO. Hepatocyte suspensions and cultures as tools in experimental carcinogenesis. J Toxicol Environmental Health. 1979;5:551-60. doi: 10.1080/15287397909529766. PMID:224209 [DOI] [PubMed] [Google Scholar]
- [45].Song YM, Lee YH, Kim JW, Ham DS, Kang ES, Cha BS, Lee HC, Lee BW. Metformin alleviates hepatosteatosis by restoring SIRT1-mediated autophagy induction via an AMP-activated protein kinase-independent pathway. Autophagy. 2015;11:46-59. doi: 10.4161/15548627.2014.984271. PMID:25484077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang HJ, Park JY, Kwon O, Choe EY, Kim CH, Hur KY, Lee MS, Yun M, Cha BS, Kim YB, et al.. Chronic HMGCR/HMG-CoA reductase inhibitor treatment contributes to dysglycemia by upregulating hepatic gluconeogenesis through autophagy induction. Autophagy. 2015;11:2089-101. doi: 10.1080/15548627.2015.1091139. PMID:26389569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ruwart MJ, Rush BD, Snyder KF, Peters KM, Appelman HD, Henley KS. 16,16-Dimethyl prostaglandin E2 delays collagen formation in nutritional injury in rat liver. Hepatol (Baltimore, Md). 1988;8:61-4. doi: 10.1002/hep.1840080112. PMID:3338720 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.