ABSTRACT
The WRKY proteins belong to a superfamily of TFs that play pivotal roles in responses to a wide range of biotic, abiotic, developmental and physiologic cues. Here, we assayed the accumulation of basal WRKY27 transcripts in diverse tissue including root, shoot, leaf and flowers. We demonstrated that plants over-expressing WRKY27 transcript levels exhibit growth aberrations and fertility defects. Scanning electron microscopic data suggest that WRKY27 overexpressor plants exhibit pollen dehiscence defects. Our fluorescein diacetate hydrolysis assay showed that flowers of plants overexpressing WRKY27 display significantly decreased pollen viability. These sterility-related phenotypes were not rescued by the exogenous applications of different phytohormones. Our results indicate the involvement of WRKY27 in particular for proper plant biomass accumulation and male fertility.
KEYWORDS: Anther dehiscence, plant biomass, pollen viability, stunted growth, Transcription factor
Introduction
The regulation of transcription is a fundamental process in all living organisms and eukaryotes have evolved a complex mode of controlling expression of their genes. It is orchestrated by transcription factors (TFs) and other molecules working in concert to fine tune the levels of transcripts produced through a variety of mechanisms.1,2 TFs are essential components of the cellular machinery that regulate constitutively in some instances as well as modulate the spatiotemporal expression of downstream target genes in response to both internal and external stimuli.1 Coordinated transcriptional regulation ensures the precise functions of an organism. All aspects of transcription and its regulation involve dynamic interactions of TFs with chromatin; this fundamental property is known as an important modulator of biologic processes.3,4 Given that TFs are central to the regulation of gene expression, elucidating the molecular mechanisms of their actions is a major focus of research particularly since the complete genome sequence of Arabidopsis thaliana (hereafter Arabidopsis) became available in 2000.5
WRKY proteins are classified into one of the major families of TFs comprising 74 members in Arabidopsis.6-8 WRKY proteins contain one or two domains with a conserved peptide stretch of about 60 amino acids (termed WRKY domain) encompassing a novel zinc-finger motif. The WRKY domain shows a high binding affinity to a distinct cis-acting DNA element termed the W box (TTGACC/T).6,9 Research over the past 15 y demonstrated that WRKY factors are involved in regulating the expression patterns of a plethora of genes, by acting as positive or negative factors.10-15 Mounting evidence suggests that the WRKY proteins actively participate in a wide range of biologic processes. While the roles of numerous WRKY family members in plant biotic and abiotic stresses have been characterized,3,11,15 the underlying molecular mechanisms by which WRKY proteins participate in plant growth, development and flowering are not fully elucidated. Notable examples include WRKY10, also known as MINISEED3 (MINI3), and WRKY44, also referred to as TRANSPARENT TESTA GLABRA2 (TTG2) that play essential roles in seed and trichome development, respectively.16-18 Additionally, in Arabidopsis, WRKY71 was shown to accelerate flowering via direct activation of FLOWERING LOCUS T (FT) and the TF gene LEAFY (LFY),19 as well as affect shoot branching.20 In addition, WRKY6 and WRKY41 were demonstrated to be involved in seed germination and early seed development21 as well as in seed dormancy,22 respectively. Another pair of WRKY factors, WRKY2 and WRKY34, was shown to be required for male gametogenesis.23 Moreover, WRKY23 appears to control the maintenance of the root stem cell niche and to negatively regulate auxin transport.24,25 Finally, OsWRKY78 regulates stem elongation and seed development26 in rice. Here, we demonstrated the involvement of WRKY27 in male sterility. WRKY27, with a single WRKY domain within the C-terminus, belongs to group-IIe in the phylogenetic tree.6,9 WRKY22 and WRKY29 are the closest sequence homologs of WRKY27 that share 54% and 62% identity and 64% and 73% similarity, respectively.
Results and discussions
To decipher the potential functions of WRKY27 in plant development and reproduction as well as plant immune responses, we performed transcript analyses of WRKY27 in diverse tissues and upon treatments with pathogens or pathogen-mimic stimuli. We also included WRKY22 and WRKY29, 2 WRKY family group IIe members most closely related to WRKY27,6 in our experiment to delineate the unique and overlapping expression patterns of these WRKY members under diverse physiologic conditions. Firstly, we performed a series of semi-quantitative and quantitative reverse transcription polymerase chain reaction (RT-PCRs) using mRNAs from root, shoot, leaf, flower and silique tissue in 21-day-old wild-type Col-0 plants. Overall, we detected relatively weak but differential transcript levels of WRKY27 in the root, shoot, leaf and flower tissues (Fig. 1A; Supplementary Figure 1A and 1B). Intriguingly, none of the three tested WRKY family members showed expression in siliques under our experimental conditions (Supplementary Figure 1A). In our qRT-PCR analyses, we also detected relatively weak expression levels of WRKY27 compared with WRKY22 in all tissue types (Fig. 1A). In contrast, the expression levels of WRKY29 were lower in leaf and flower compared with WRKY27. Moreover, we also detected relatively weak expression levels of senescence-induced receptor-like kinase 1 (SIRK1), also known as flagellin-induced receptor-like kinase 1, FRK1; a downstream transcriptional target of diverse WRKY members including WRKY22 and WRKY293,27,28 in root, leaf and flower tissues (Fig. 1A). Our comparative organ-specific transcript analysis of WRKY27 with other players is in concordance with the publically available transcriptomic data in The Bio-Analytic Resource (BAR) database29 (Supplementary Figure 1B). These data suggest common and unique functions of these three WRKY factors in plant development and reproduction. Finally, tissue-specific WRKY27 expression patterns are supported by the GUS activity staining revealed in different parts of the plant including anthers and stigmatic papillae (Supplementary Figure 1C-1G) in our previously characterized wrky27–1, an exon trap mutant line, as well as in PWRKY27:GUS transgenic plants.30 These tissue-specific expression patterns of WRKY27 prompted us to consider the potential role of WRKY27 in plant reproduction, in particular anther development and pollen viability.
Figure 1.

Measurement of WRKY27 transcript levels in different tissue (A) Steady state expression levels of WRKY22, WRKY29 and WRKY27 in root, leaf, flower and silique-derived tissues were detected using quantitative real-time PCR. (B) Examination of WRKY27, WRKY22, WRKY29 and SIRK1 (Senescence-induced receptor-like kinase1) expression levels in leaf tissue upon elf18 treatment at indicated time points. The magnified view of expression data-related to WRKY27 and WRKY29 is shown (inset).
In our previous study, we demonstrated that wrky27–1, which lacks a functional WRKY27 transcription factor, exhibited delayed symptoms development in response to the bacterial wilt pathogen Ralstonia solanacearum.30 These data corroborated with the basal and/or induced expression of WRKY 27 in root and shoot vasculature. To investigate whether the transcript levels of WRKY27 can be induced in the leaf, we performed an extensive qRT-PCR analysis using WRKY27-specific primers on cDNAs derived from leaf tissue subjected to a range of biotic stresses. This includes 10 µM elf18 (a peptide derived from the N-terminal region of bacterial elongation factor Tu), syringe-infiltration of avirulent strains of Pseudomonas syringae pv. maculicola (Psm) ES4326 expressing avrRpm1, spray inoculation of P. syringae strain DC3000 and P. syringae strain DC3118 (coronatine toxin-deficient mutant bacterial strain). We also included WRKY22, WRKY29 and SIRK1 as controls since these key immune players were demonstrated to be induced by several pathogens and pathogen-associated molecular patterns (PAMPs).3,27,28,31,32, As expected, we observed a strong induction of WRKY22, WRKY29 and SIRK1 at different time points upon treatment with elf18 (Fig. 1B). Intriguingly, we observed a moderate but significant induction of WRKY27 at 1-hour, 2-hour and 3-hour post treatments with elf18 compared with mock at these specified time points. A similar observation was previously made in a publically available transcriptomics data set that used an unrelated PAMP, flagellin 22 (flg22; TAIR submission ME00332).33 Although WRKY27 has a relatively low basal expression level, it can be strongly induced upon avirulent pathogen (Psm ES4326/avrRpm1) challenge, especially 6h post inoculation, akin to the expression pattern of SIRK1 (Fig. 2A). Finally, we also detected various degrees of induction of WRKY27, WRKY22, WRKY29 and SIRK upon infection with virulent pathogens DC3000 and DC3118 in our time course qRT-PCR assay (Fig. 2B, C). These data corroborate with the previously published microarray data that used DC3000 and DC3000-avrRpm1 for infections (TAIR submission ME00331).33 It's important to note that WRKY27, unlike WRKY22 and WRKY29, failed to induce SIRK1 expression in transient assays30 suggesting the potential involvement of WRKY27 in PAMPs-triggered immunity as well as basal and effector-triggered immunity through a yet-to-be characterized mechanism.
Figure 2.

Detection of biotic stress-triggered expression patterns of WRKY27, WRKY22, WRKY29 and SIRK1 in leaf tissue. (A) mRNA levels of WRKY27, WRKY22, WRKY29 and SIRK1 detected using quantitative real-time PCR analysis in leaf tissue upon Pseudomonas syringae pv maculicola (Psm) ES4326 expressing avrRpm1 (Psm ES4326/avrRpm1) challenge. Accumulation of WRKY27, WRKY22, WRKY29 and SIRK1 transcripts in leaf tissue upon infection with P. syringae strain DC3000 (B) and P. syringae strain DC3118 (C), challenge. Time points after pathogen challenge are indicated.
While the roles of WRKY27 in root and plant vasculature as well as the plant immune system were described previously,30 the expression of WRKY27 in different floral tissues including anthers intrigued us to investigate its potential contributions in anther development and male sterility. At first glance, we did not observe any macroscopic floral-related phenotypes in the wrky27–1 plants. This might be due to possible functional redundancy among members of the WRKY family. Potential candidates for such redundancy include the closely-related WRKY22 TF but also WRKY2 and WRKY34 that were previously shown to be required for pollen development,23 as well as WRKY31, WRKY42 and WRKY47 that showed elevated expression in pollen and stamen based on microarray studies.29 Such functional redundancy among TFs and other proteins has been well documented in Arabidopsis.34,35 For instance, plants lacking functional MYC5 TF exhibited no visible phenotypes, while overexpression of MYC5 fused with a repressive domain showed male sterility.35 Thus, overexpression or missexpression of a gene product can provide an alternative and complementary approach to help define the functions of genes that could not be revealed using classical reverse genetics approaches.36 Hence, we generated a series of WRKY27 overexpressing constructs using the full-length cDNA of WRKY27 fused with the 3´ terminator element or in frame to a StrepII sequence allowing epitope tagging of the respective protein. We transformed the resultant constructs, 2 × 35S::WRKY27-terminator and 2 × 35S::WRKY27-StrepII-terminator into Arabidopsis Col-0 wild-type and wrky27–1 mutant plants using Agrobacterium-mediated transformation.37,38 Northern blot and qRT-PCR analyses confirmed the overexpression of WRKY27 in several transgenic lines (Supplementary Figure 2A and 2B). We subjected these transgenic overexpressor (OE) lines for further comprehensive phenotypic analyses to elucidate the roles of WRKY27 in vegetative and reproductive phases of development. Compared with wild-type Col-0, plants overexpressing WRKY27, regardless of the transgene construct type (with or without an epitope tag), exhibited visible phenotypes associated with stunted growth during the entire vegetative growth phase resulting in a smaller plant stature (Fig. 3A, B). In addition, we observed that the leaves of WRKY27 overexpressing plants were smaller in area and more curled when compared with corresponding leaves of wild-type Col-0 plants 4 weeks post germination (Fig. 3C-E). We also measured root and shoot biomass and demonstrated that (OE)-Strep-4, OE-Strep-7 and OE-Strep-8 transgenic lines display significantly reduced biomass in both above- and below-ground plant tissues compared with Col-0 (Fig. 3I). The most evident observation, however, was partial sterility and a delay in senescence in perianth organs. The young siliques of the overexpressor transgenic plants were underdeveloped and empty, and started to eventually elongate at very late stages of flowering (Fig. 3F-H). Subsequently, we quantified the number of underdeveloped empty and underdeveloped partially filled siliques in OE-Strep-4, OE-Strep-7 and OE-Strep-8 transgenic lines. We determined that over 70% of the siliques were underdeveloped and empty for these three representative WRKY27 overexpressing plants, while less than 30% were partially filled (Fig. 3J). In any case, normal silique size was never attained and the seed content was significantly reduced, compared with those of wild-type plants (Fig. 3).
Figure 3.

Pleiotropic phenotypes of ectopic WRKY27 overexpressor (OE) plants. WT and OE denote wild-type plants and WRKY27 transgenic overexpressor lines, respectively. (A and B) WRKY27 overexpressor lines exhibit dwarf phenotypes compared with wild-type plants. (C–E) Altered leaf shape of WRKY27 overexpressor plants compared with wild-type leaves. (F–H) Partial sterility and delayed senescence in perianth organs detected in plants expressing increased levels of WRKY27 compared with wild-type plants. (I) Root and shoot fresh weight of 5-week-old WT and OE soil grown Arabidopsis. Statistical analysis was performed by 2-way ANOVA followed by Bonferroni test, ***p < 0.001. (J) Quantification of under-developed empty and under-developed partially filled siliques.
Given that concerted actions of plant hormones such as jasmonic acid (JA) and gibberellic acid (GA) are essential for proper stamen development39 and exogenous applications of these hormones or hormone mimics have been shown to rescue sterility-related phenotypes in Arabidopsis,40,41 we subjected the flowers of WRKY27 overexpressor lines to phytohormone treatments. However, the sterility phenotypes of plants overexpressing WRKY27 were not reversible by exogenous application of JA, GA and gamma-amino butyric acid (GABA).
To better understand the nature of the sterility-related phenotypes, we investigated the floral structure of the WRKY27 overexpressor plants in more detail using scanning electron microscopy (SEM). While anther dehiscence in the wild-type flowers normally occurs at the end of flower development, we observed severely delayed or completely inhibited anther dehiscence in the developing flowers of WRKY27 overexpressor plants (Fig. 4A-D). These results indicate that the low fertility of plants expressing WRKY27 might be caused by interrupted anther dehiscence. To search for defects in pollen quality, the pollen grains of WRKY27 overexpressor lines were subjected to fluorescein diacetate (FDA)-based fluorochromatic reaction. This test evaluates the integrity of the plasma membrane of the pollen grains and is an easy, quick and accurate assay to monitor for viability of pollen grains in many plants, as only viable pollen will fluoresce under the microscope.42 Pollen grains from 5 independent 2xCaMV35S::WRKY27 overexpressor lines were stained with FDA. The count of fluorescent pollen grains revealed a significantly lower percentage of viable pollen derived from the overexpressor lines compared with the wild-type (Fig. 4E, F). 43
Figure 4.
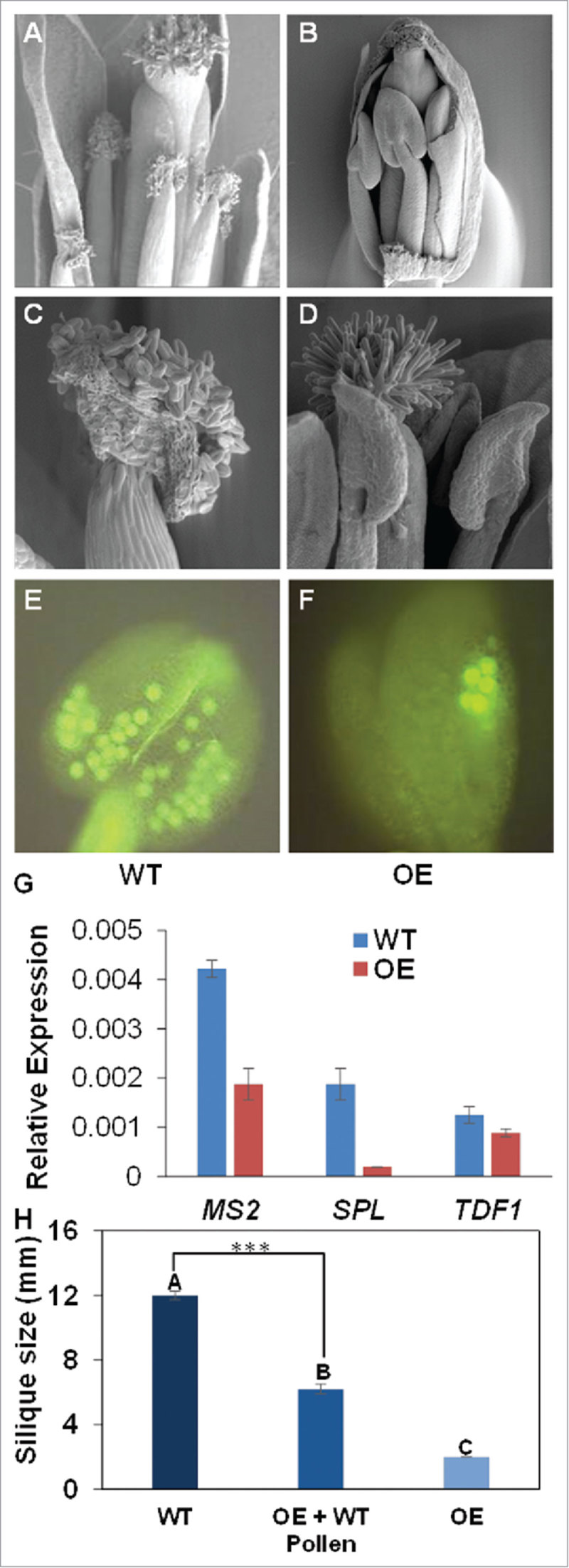
WRKY27 overexpressor lines show reduced fertility. (A–D) Scanning electron microscopy (SEM) analysis of 2 × 35S::WRKY27 flower development. Unopened anthers of WRKY27 transgenic overexpressor plants compared with those of wild-type plants. (E and F) Fluoresceine diacetate (FDA) assay on unopened anthers from 2 × 35S::WRKY27 overexpressor lines showing reduced pollen viability compared with wild-type plants. (G) mRNA levels of SPL/NZZ (SPOROCYTELESS/NOZZLE), TDF1 (tapetal development and function 1) and MS2 (male sterility 2) detected using quantitative real-time Polymerase Chain Reaction (PCR) analysis in floral tissue. (H) Quantification of silique length 7 d after pollination with WT pollens. Self-pollinated OE siliques were used as controls. Statistical analysis was performed by one-way ANOVA, ***p<0.001.
To corroborate our cellular studies, we examined the transcript levels of several well-defined major genetic components of anther and pollen development. This includes SPL/NZZ (SPOROCYTELESS/NOZZLE) that is involved in regulation of anther cell differentiation),44 TDF1 (tapetal development and function 1)45 and MS2 (male sterility 2) 46 that are involved at various phases of pollen and anther development. We demonstrated that WRKY27 overexpressor plants accumulate lower levels of MS2, SPL and TDF1 mRNAs,43 which may explain the deficits in male fertility (Fig. 4G). Finally, we artificially pollinated WRKY27 overexpressor flowers with wild-type pollen at an early flowering stage, which resulted in partial restoration of fertility (Fig. 4H). Taken together, we concluded that overexpression of WRKY27 results in male sterility.
Conclusions
Overall, we discovered that WRKY27 is expressed in diverse tissues including root, shoot, leaf and floral organs. While the requirement of functional WRKY27 in root and leaf vasculature was previously demonstrated, we expanded the potential roles of this important member of the WRKY family in leaf-centered immune responses. This finding will form the basis for future research. In the current manuscript, we focused to decipher the involvement of WRKY27 in floral organs and provide evidence of the dual functions of this key gene in plant defense as well as plant development and male sterility. We showed that the endogenous expression patterns of WRKY27 overlap with the plant organs displaying the aberrant phenotypes. Finally, we also demonstrated that the transgenic plants with increased levels of WRKY27 mRNA displayed several alterations in the morphological phenotype, related to stunted growth, irregular leaf shape, and drastically reduced male fertility. In summary, our results indicate dual roles of WRKY27 in plant immunity and development, in particular proper plant biomass accumulation and male fertility.
Materials and methods
Plant treatments, RNA and quantitative Real Time PCR (qRT-PCR)
Leaf tissue was collected from 4-week old Arabidopsis plants that were syringe-infiltrated with 10 µM flg22 (Genscript) or 10 µM elf18 (Genscript).
Pathogen treatments were performed using 4-week old plants that were syringe-infiltration with Psm ES4326/avrRpm1 (OD600nm = 0.1), spray inoculated with Pst DC3000 (OD600nm = 0.2, 0.02% Silwet L-77) or Pst DC3118 (OD600nm = 0.2, 0.02% Silwet L-77). Untreated root, shoot leaf, shoot, and flower tissues were collected to determine the basal levels of tarnscripts. We extracted total RNA from the collected samples using RiboZol (AMRESCO). Possible genomic DNA contamination was eliminated using DNase I (Ambion). Formaldehyde agarose gel preparation, quantification, electrophoresis and samples prepration/loading were done following the RNeasy Plant Mini® Kit QIAGEN protocol. SuperScript III first-strand RT-PCR kit (Invitrogen) was used to convert mRNA into cDNA. We used GoTaq qPCR Master Mix (Promega) to perform qRT-PCR using gene-specific primers in a RealPlex S MasterCycler (Eppendorf). Primers used in this study are listed in the Supplementary Table 1.
RNA hybridization and Northern blot analysis
Pre-hybridization and hybridization were performed in hybridization solution in glass tubes (30 cm × 4 cm) at 65°C under continuous rotation in a hybridization oven (Bachofer, Reutlingen, Germany). The pre-hybridization was performed overnight. Upon adding the denatured radio-active probe, the hybridization was performed for at least 16 hours. After hybridization the filter was washed with SSC and SDS solutions. The filter was wrapped in Saran wrap and exposed overnight to a phosphoimager screen (Molecular Dynamics) in a cassette at room temperature. 50–100 ng of gel-purified PCR product for Northern analysis was used. Probe was prepared according to Rediprime II Random Prime Labeling System protocol manual provided by Amersham Biosciences. Probe was later on purified on a Sephadex G25 column.
GUS and FDA staining assays
Plant tissue were stained for GUS activity using a solution containing 2 mM 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc) in 0.1 M Na2HPO4, pH 7.0, 10 mM EDTA, 0.5 mM potassium ferricyanide/ferrocyanide, and 0.06% Triton X-100 at 37°C for 16 hours. The samples were cleared of chlorophyll by sequential washing in 70% ethanol. The fluorochromatic reaction (FCR) procedure was used for determining the viability of pollen. 0.02 g of fluorescein diacetate (FDA) (Sigma-Aldrich GmbH, Munich, Germany), was mixed with 10 ml of acetone. The FDA solution was added drop by drop to 5 ml of 20%sucrose until persistent turbidity was achieved. This solution was used within 30 min of preparation. Each slide containing 10 µl of this solution and pollens were incubated at room temperature for 15 minutes. Viability of pollen grains was examined under a fluorescence microscope (Leica MZ12, excitation filter 450–490 nm).
Phytohormone treatments, biomass and cross-pollination
Diverse concentrations (1 mM to 0.5 M) of Gamma-aminobutyric acid (GABA) soluble in dH2O, 100 µM of Methyl Jasmonate (MeJA) and mock solution 0.2% ethanol, 1 µM GA3 solution (stock solution of 10 mM GA3) were sprayed directly on flowers. Rosettes of individual 5-week-old Arabidopsis plants were removed and weighed as shoot fresh biomass. Root of individual 5-week-old Arabidopsis plants were rinsed in water to remove soil before fresh biomass determination. To achieve cross-pollination, emasculation was performed one day before artificial pollination using pollens from designated plants. Siliques length was measured 7 d after cross-pollination.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the NSF (IOS-1557796) to M.S.M. The authors wish to acknowledge Dr. Peter Huijser and Ms Yali Sun for technical assistance and Dr. K. Mukhtar for critical reading the manuscript.
References
- 1.Lenstra TL, Rodriguez J, Chen H, Larson DR. Transcription dynamics in living cells. Annu Rev Biophys. 2016;45:25–47. doi: 10.1146/annurev-biophys-062215-010838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tugrul M, Paixao T, Barton NH, Tkacik G. Dynamics of transcription factor binding site evolution. PLoS Genet. 2015;11:e1005639. doi: 10.1371/journal.pgen.1005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsuda K, Somssich IE. Transcriptional networks in plant immunity. New Phytol. 2015;206:932–47. doi: 10.1111/nph.13286. [DOI] [PubMed] [Google Scholar]
- 4.Tully JP, Hill AE, Ahmed HM, Whitley R, Skjellum A, Mukhtar MS. Expression-based network biology identifies immune-related functional modules involved in plant defense. BMC Genomics. 2014;15:421. doi: 10.1186/1471-2164-15-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muhammad D, Schmittling S, Williams C, Long TA. More than meets the eye: Emergent properties of transcription factors networks in Arabidopsis. Biochim Biophys Acta. 2017;1860(1):64–74. doi: 10.1016/j.bbagrm.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 6.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 7.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–58. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005;12:9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- 9.Rinerson CI, Rabara RC, Tripathi P, Shen QJ, Rushton PJ. The evolution of WRKY transcription factors. BMC Plant Biol. 2015;15:66. doi: 10.1186/s12870-015-0456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birkenbihl RP, Somssich IE. Transcriptional plant responses critical for resistance towards necrotrophic pathogens. Front Plant Sci. 2011;2:76. doi: 10.3389/fpls.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phukan UJ, Jeena GS, Shukla RK. WRKY transcription factors: Molecular regulation and stress responses in plants. Front Plant Sci. 2016;7:760. doi: 10.3389/fpls.2016.00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y, Yu D. WRKY transcription factors: Links between phytohormones and plant processes. Sci China Life Sci. 2015;58:501–2. doi: 10.1007/s11427-015-4849-9. [DOI] [PubMed] [Google Scholar]
- 13.Schluttenhofer C, Yuan L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015;167:295–306. doi: 10.1104/pp.114.251769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripathi P, Rabara RC, Rushton PJ. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta. 2014;239:255–66. doi: 10.1007/s00425-013-1985-y. [DOI] [PubMed] [Google Scholar]
- 15.Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–55. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:17531–6. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Zhang X, Kang X, Zhao X, Zhang X, Ni M. SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell. 2009;21:106–17. doi: 10.1105/tpc.108.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–75. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, Liu Z, Wang L, Kim SG, Seo PJ, Qiao M, Wang N, Li S, Cao X, Park CM, et al.. WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 2016;85:96–106. doi: 10.1111/tpj.13092. [DOI] [PubMed] [Google Scholar]
- 20.Guo D, Zhang J, Wang X, Han X, Wei B, Wang J, Li B, Yu H, Huang Q, Gu H, et al.. The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in arabidopsis. Plant Cell. 2015;27:3112–27. doi: 10.1105/tpc.15.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Feng CZ, Ye Q, Wu WH, Chen YF. Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet. 2016;12:e1005833. doi: 10.1371/journal.pgen.1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding ZJ, Yan JY, Li GX, Wu ZC, Zhang SQ, Zheng SJ. WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 2014;79:810–23. doi: 10.1111/tpj.12597. [DOI] [PubMed] [Google Scholar]
- 23.Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S. Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 2014;10:e1004384. doi: 10.1371/journal.pgen.1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grunewald W, De Smet I, De Rybel B, Robert HS, van de Cotte B, Willemsen V, Gheysen G, Weijers D, Friml J, Beeckman T. Tightly controlled WRKY23 expression mediates Arabidopsis embryo development. EMBO Rep. 2013;14:1136–42. doi: 10.1038/embor.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grunewald W, De Smet I, Lewis DR, Lofke C, Jansen L, Goeminne G, Vanden Bossche R, Karimi M, De Rybel B, Vanholme B, et al.. Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci U S A. 2012;109:1554–9. doi: 10.1073/pnas.1121134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang CQ, Xu Y, Lu Y, Yu HX, Gu MH, Liu QQ. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta. 2011;234:541–54. doi: 10.1007/s00425-011-1423-y. [DOI] [PubMed] [Google Scholar]
- 27.Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–49. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waese J, Provart NJ. The Bio-analytic resource for plant biology. Methods Mol Biol. 2017;1533:119–48. doi: 10.1007/978-1-4939-6658-5_6. [DOI] [PubMed] [Google Scholar]
- 30.Mukhtar MS, Deslandes L, Auriac MC, Marco Y, Somssich IE. The Arabidopsis transcription factor WRKY27 influences wilt disease symptom development caused by Ralstonia solanacearum. Plant J. 2008;56:935–47. doi: 10.1111/j.1365-313X.2008.03651.x. [DOI] [PubMed] [Google Scholar]
- 31.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–83. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 32.Mukhtar MS, McCormack ME, Argueso CT, Pajerowska-Mukhtar KM. Pathogen tactics to manipulate plant cell death. Curr Biol. 2016;26:R608–19. doi: 10.1016/j.cub.2016.02.051. [DOI] [PubMed] [Google Scholar]
- 33.Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The botany array resource: E-Northerns, expression angling, and promoter analyses. Plant J. 2005;43:153–63. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang JZ. Overexpression analysis of plant transcription factors. Curr Opin Plant Biol. 2003;6:430–40. doi: 10.1016/S1369-5266(03)00081-5. [DOI] [PubMed] [Google Scholar]
- 35.Figueroa P, Browse J. Male sterility in Arabidopsis induced by overexpression of a MYC5-SRDX chimeric repressor. Plant J. 2015;81:849–60. doi: 10.1111/tpj.12776. [DOI] [PubMed] [Google Scholar]
- 36.Prelich G. Gene overexpression: Uses, mechanisms, and interpretation. Genetics. 2012;190:841–54. doi: 10.1534/genetics.111.136911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koncz C, Mayerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Redei GP, Schell J. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 1990;9:1337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logemann E, Birkenbihl RP, Ulker B, Somssich IE. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods. 2006;2:16. doi: 10.1186/1746-4811-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song S, Qi T, Huang H, Xie D. Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol Plant. 2013;6:1065–73. doi: 10.1093/mp/sst054. [DOI] [PubMed] [Google Scholar]
- 40.Kwon CT, Paek NC. Gibberellic acid: A key phytohormone for spikelet fertility in rice grain production. Int J Mol Sci. 2016;17:794. doi: 10.3390/ijms17050794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, Chételat A, Haupt I, Kennerley BJ, Hodgens C, et al.. A regulatory network for coordinated flower maturation. PLoS Genet. 2012;8:e1002506. doi: 10.1371/journal.pgen.1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinillos V, Cuevas J. Standardization of the fluorochromatic reaction test to assess pollen viability. Biotech Histochem. 2008;83:15–21. doi: 10.1080/10520290801987204. [DOI] [PubMed] [Google Scholar]
- 43.Ye Q, Zhu W, Li L, Zhang S, Yin Y, Ma H, Wang X. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc Natl Acad Sci U S A. 2010;107:6100–5. doi: 10.1073/pnas.0912333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Huang J, Parameswaran S, Ito T, Seubert B, Auer M, Rymaszewski A, Jia G, Owen HA, Zhao D. The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant physiol. 2009;151:1401–11. doi: 10.1104/pp.109.145896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN. Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008;55:266–77. doi: 10.1111/j.1365-313X.2008.03500.x. [DOI] [PubMed] [Google Scholar]
- 46.Wallace S, Chater CC, Kamisugi Y, Cuming AC, Wellman CH, Beerling DJ, Fleming AJ. Conservation of male sterility 2 function during spore and pollen wall development supports an evolutionarily early recruitment of a core component in the sporopollenin biosynthetic pathway. New phytol. 2015;205:390–401. doi: 10.1111/nph.13012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


