ABSTRACT
Circular RNAs are a subclass of noncoding RNAs in mammalian cells; however, whether these RNAs are involved in the regulation of astrocyte activation is largely unknown. Here, we have shown that the circular RNA HIPK2 (circHIPK2) functions as an endogenous microRNA-124 (MIR124–2HG) sponge to sequester MIR124–2HG and inhibit its activity, resulting in increased sigma non-opioid intracellular receptor 1 (SIGMAR1/OPRS1) expression. Knockdown of circHIPK2 expression significantly inhibited astrocyte activation via the regulation of autophagy and endoplasmic reticulum (ER) stress through the targeting of MIR124–2HG and SIGMAR1. These findings were confirmed in vivo in mouse models, as microinjection of a circHIPK2 siRNA lentivirus into mouse hippocampi inhibited astrocyte activation induced by methamphetamine or lipopolysaccharide (LPS). These findings provide novel insights regarding the specific contribution of circHIPK2 to astrocyte activation in the context of drug abuse as well as for the treatment of a broad range of neuroinflammatory disorders.
KEYWORDS: astrocyte, autophagy, circular RNA HIPK2, endoplasmic reticulum, lipopolysaccharide, methamphetamine, MIR124–2HG, sigma non-opioid intracellular receptor 1
Introduction
Astrocytes, the most abundant cell type in the central nervous system (CNS), play critical roles in regulating and maintaining CNS homeostasis in normal physiological situations.1,2 In pathological conditions, astrocytes become activated and are characterized by abnormal morphology with reactive astrogliosis.3-7 Astrocyte activation plays a detrimental role in various neurological pathologies, including stroke,8 Parkinson disease,9,10 Alzheimer disease,11,12 and drug abuse.13 With respect to drug abuse, methamphetamine abuse is a major social and health concern. Astrocyte activation is associated with neuronal damage,14,15 as there is a close relationship between methamphetamine-induced degeneration of dopaminergic neurons in the striatum and concomitant reactive astrogliosis. Methamphetamine-induced neurotoxicity is associated with astrocyte activation in the striata of methamphetamine-treated mice and rats in vivo,16,17 and in vitro.18,19 Moreover, astrocyte activation contributes to the leakage of the blood-brain barrier (BBB) and facilitates the entry of molecules and immune cells into the CNS.20 A detrimental role for astrocyte activation in the BBB damage is supported by a study showing that vascular endothelial growth factor from astrocytes increased the vascular permeability and CNS damage in acute inflammatory lesions.21 Together, evidence demonstrates that astrocyte activation plays a critical role in the recruitment of infiltrating immune cells into the CNS, with subsequent neuroinflammatory cascades resulting in brain tissue injury.
Methamphetamine is known to exhibit moderate affinity for SIGMAR1/OPRS1 (sigma non-opioid intracellular receptor 1), which is a unique drug-binding protein present in the CNS and periphery.22,23 Our previous study demonstrated that methamphetamine-mediated astrocyte activation involves the upregulation of SIGMAR1 expression through a positive feedback mechanism.13 Interestingly, our current study revealed that both methamphetamine and lipopolysaccharide (LPS) induce astrocyte activation via regulation of SIGMAR1 expression, suggesting that SIGMAR1 is a critical player in astrocyte activation. Although SIGMAR1 plays a crucial role in astrocyte activation, regulation of its expression by noncoding RNAs in astrocytes has not yet been explored. Computational algorithms, such as TargetScan, have been used to identify microRNAs (miRNAs) that target evolutionarily conserved sequences in the SIGMAR1-associated gene microRNA-124 (MIR124–2HG, the acronym formatting for humans is MIR124–2HG and for mice is Mir124–2hg, but we henceforth will use MIR124–2HG in both cases for the sake of simplicity), which is conserved among vertebrates and has been predicted to target SIGMAR1. It is the most abundant brain-specific miR that regulates neuronal differentiation during CNS development and adult neurogenesis.24-27 MIR124–2HG promotes microglial quiescence and suppresses experimental autoimmune encephalomyelitis by deactivating macrophages via the CEBPA/C/EBP-a-SPI1/PU.1 pathway.28 Furthermore, a recent study indicated that MIR124–2HG inhibits microglial activation after focal cerebral ischemia.29 However, whether MIR124–2HG regulates SIGMAR1 expression and astrocyte activation remains to be elucidated.
Genome-wide bioinformatic analysis revealed that the circular RNA (circRNA) HIPK2 (circHIPK2, the formatting for humans is circHIPK2 and for mice is circHipk2, but we henceforth will use cirkHIPK2 in both cases for the sake of simplicity), derived from exon 2 of the HIPK2 gene, acts as a sponge for MIR124–2HG. CircRNAs, generated from back-spliced exons, have recently been identified as a naturally occurring family of noncoding RNAs that is highly represented in the eukaryotic transcriptome.30 These endogenous RNAs are characterized by a stable structure and high tissue-specific expression.31 circRNAs are highly homologous to but generally more stable than their linear counterparts because they lack accessible ends and thus are resistant to exonucleases.31 While most of the circular RNAs reported so far have been exonic circular RNAs, a class of intron-containing exonic circRNAs is found predominantly in the nucleus,32 where they promote transcription of their parental genes. Several classes of noncoding RNAs have been shown to be involved in the regulation of physiological and pathophysiological processes including development and heart senescence, hypertrophy and failure, as well as cell growth.33-37 However, whether circHIPK2 is involved in astrocyte activation remains largely unknown, and more extensive study is required.
In this study, we show that circHIPK2 directly binds to MIR124–2HG and acts as an endogenous sponge for MIR124–2HG to inhibit its activity. Knockdown of circHIPK2 expression significantly inhibited astrocyte activation via the regulation of autophagy and endoplasmic reticulum (ER) stress through the targeting of MIR124–2HG. These findings provide the first evidence that the circHIPK2-MIR124–2HG axis mediates a regulatory pathway critical for the regulation of astrocyte activation. Thus, specific blockage of circHIPK2 could be a potential therapeutic target for inhibition of astrocyte activation in the context of drug abuse as well as the treatment of a broad range of neuroinflammatory disorders.
Results
MIR124–2HG participates in the regulation of SIGMAR1
Our previous study indicated that SIGMAR1 upregulation is involved in methamphetamine-induced astrocyte activation. Interestingly, in the current study, we also demonstrated that LPS induced astrocyte activation via SIGMAR1. Treatment of astrocytes with LPS (100 ng/ml) significantly increased the expression of the astrocyte marker glial fibrillary acidic protein (GFAP) (Fig. S1A), with concomitant upregulation of SIGMAR1 expression (Fig. S1B). These findings were further confirmed in an in vivo experiment showing that LPS treatment increased the expression of both GFAP and SIGMAR1 in wild-type (WT) mice and that the expression of these proteins was significantly inhibited in sigmar1 knockout (KO) mice (Fig. S1C).
Given that SIGMAR1 plays a critical role in astrocyte activation, we examined the mechanisms underlying SIGMAR1 expression. MiRNAs are a class of small noncoding RNAs that act as negative regulators of gene expression. To determine whether SIGMAR1 is regulated by miRs, we first predicted the presence of a consensus-binding site of MIR124–2HG in the 3′-untranslated region (3′-UTR) of SIGMAR1 (the gene encoding SIGMAR1) using the TargetScan algorithm. As shown in Fig. 1A, SIGMAR1 has a conserved MIR124–2HG binding site within its 3′-UTR in most species. Intriguingly, cotransfection of a MIR124–2HG-overexpressing vector and pmiR-GLO plasmid with the SIGMAR1 WT 3′-UTR resulted in the downregulation of luciferase activity, and this effect was reversed in HEK293T cells transfected with a mutated SIGMAR1 3′-UTR (Fig. 1B and Table 1A). Next, we aimed to determine whether methamphetamine mediates its effects via the induction of MIR124–2HG and to assess the kinetics of the methamphetamine response. Methamphetamine treatment of the human astrocyte cell line A172 and primary mouse astrocytes resulted in decreased MIR124–2HG expression (Fig. 1C and D). Interestingly and as expected, the methamphetamine-induced modulation of MIR124–2HG was inversely correlated with SIGMAR1 expression (Fig. 1E and F). In line with this finding, MIR124–2HG decreased SIGMAR1 expression, whereas Anti-MIR124–2HG increased its expression in both A172 cells (Fig. 1G) and primary mouse astrocytes (Fig. 1H) at the mRNA level. This finding was further confirmed at the protein level (Fig. 1 I and J).
Figure 1.
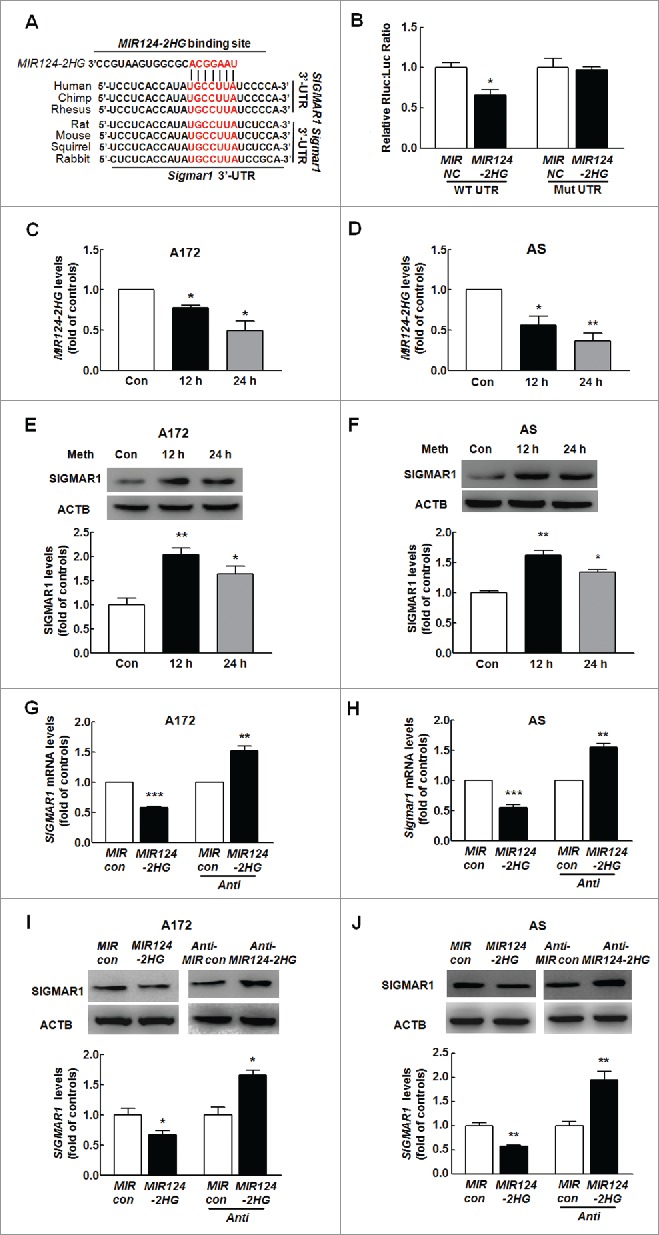
MIR124–2HG regulates SIGMAR1 expression at the post-transcriptional level in astrocytes. (A) Putative MIR124–2HG binding sites in the 3′-UTR of SIGMAR1 and Sigmar1. The potential complementary residues are shown in red. (B) Relative luciferase activity of wild-type and 3′-UTR mutant constructs of SIGMAR1 cotransfected with a MIR124–2HG overexpression vector and pmiR-GLO plasmid. All data are presented as the means ± SD of 3 individual experiments. *P < 0.05 vs. the MIR control cotransfected with the WT construct by one-way ANOVA, followed by the Holm-Sidak test. (C and D) Effect of methamphetamine on MIR124–2HG expression at the mRNA level in A172 cells (C) and primary mouse astrocytes (D) as determined by real-time PCR. Cells were incubated with methamphetamine (100 μM) for 12 h and 24 h, followed by isolation of RNA for measurement of MIR124–2HG expression. All data are presented as the means ± SD of 3 independent experiments. *P < 0.05 and **P < 0.01 vs. the control group by the Student t test. (E and F) Effect of methamphetamine on SIGMAR1 expression in A172 cells (E) and primary mouse astrocytes (F) as determined by western blotting. Cells were incubated with methamphetamine (100 μM) for 12 h and 24 h, followed by measurement of SIGMAR1 expression. Densitometric data of SIGMAR1 expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 and **P < 0.01 vs. the control group by the Student t test. (G and H) Cells were transduced with the MIR control or MIR124–2HG and Anti-MIR control or Anti-MIR124–2HG lentivirus for 24 h, and the mRNA expression of SIGMAR1 was then measured by real-time PCR in A172 cells (G) and primary mouse astrocytes (H). (I and J) Cells were transduced with MIR control or MIR124–2HG and Anti-MIR control or Anti-MIR124–2HG lentivirus for 24 h, followed by measurement of SIGMAR1 expression in A172 cells (I) and primary mouse astrocytes (J). Densitometric analysis of SIGMAR1 expression using ImageJ is presented. All data are presented as the means ± SD of 3 independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. the Mir control or Anti-MIR control group by the Student t test. Meth, methamphetamine; AS, primary mouse astrocytes.
Table 1.
Succinct description of mutated or modified molecules.
| Original molecules | Mutated or modified molecules | Succinct descriptions |
|---|---|---|
| A. pmiR-RB-SIGMAR1 3′-UTR vector | pmiR-RB-SIGMAR1 3′-UTR mutant vector | A vector containing mutated SIGMAR1 3′-UTR that does not target MIR124–2HG |
| B.circHIPK2 probe | biotin-labeled circHIPK2 probe | A single strand DNA probe capable of combining circHIPK2 |
| C.MIR124–2HG | 3′-end biotinylated MIR124–2HG | A 3′-end biotinylated microRNA mimic |
| D.MIR124–2HG | MIR124–2HGΔ | A mutated sequence that does not target the 3′-UTR of SIGMAR1 |
| E.MIR124–2HG probe | digoxigenin-labeled MIR124–2HG probe | A single strand DNA probe capable of combining MIR124–2HG |
MIR124–2HG inhibits astrocyte activation by targeting SIGMAR1 in vitro
Having determined that MIR124–2HG regulates SIGMAR1 expression, we next examined the role of this miRNA in astrocyte activation. Cells were transduced with a lentivirus expressing a MIR124–2HG precursor, and GFAP expression was assessed. Transduction of cells with the MIR124–2HG precursor-expressing lentivirus resulted in significant inhibition of methamphetamine-induced astrocyte activation, as indicated by the GFAP expression levels in both A172 cells (Fig. 2A) and primary mouse astrocytes (Fig. 2B). Next, to determine whether the MIR124–2HG-mediated functional effects were specifically dependent on the upregulation of SIGMAR1 expression, A172 cells were cotransduced with lentiviral vectors expressing Anti-MIR124–2HG and SIGMAR1 siRNA. Transduction of cells with the Anti-MIR124–2HG lentivirus enhanced GFAP expression, and this effect was significantly inhibited in cells cotransduced with the lentiviral vectors expressing Anti-MIR124–2HG and SIGMAR1 siRNA (Fig. 2C). Moreover, transduction of cells with the Anti-MIR124–2HG lentivirus resulted in enhanced GFAP expression (Fig. 2D), and this effect was inhibited in primary mouse astrocytes isolated from sigmar1 KO mice. These rescue experiments were further confirmed in astrocytes treated with methamphetamine (Fig. S2A and B).
Figure 2.
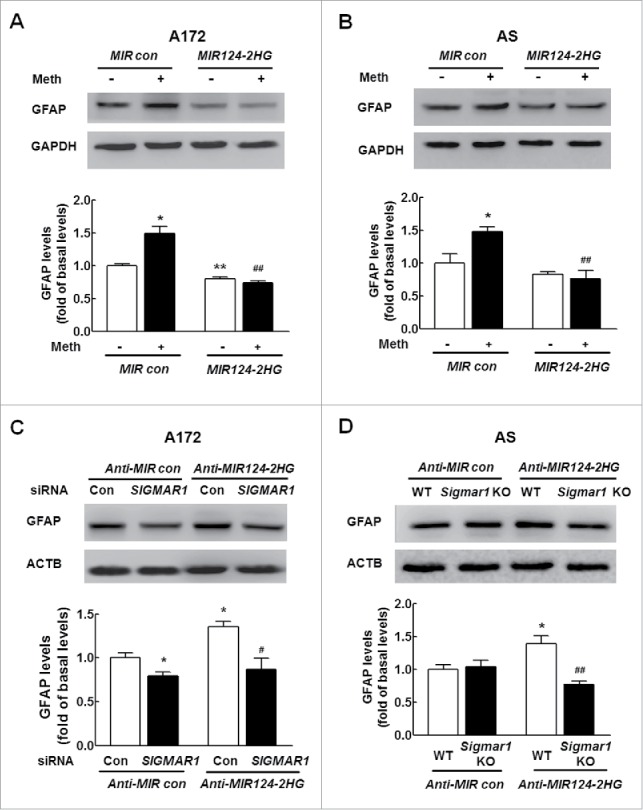
MIR124–2HG inhibits astrocyte activation by targeting SIGMAR1 in vitro. (A and B) Transduction of cells with MIR124–2HG attenuated methamphetamine-induced astrocyte activation in A172 cells (A) and primary mouse astrocytes (B), as determined by western blotting for GFAP expression. Cells were transduced with a MIR124–2HG lentivirus for 24 h, treated with methamphetamine (100 μM) for 12 h followed by measurement of GFAP expression. AS, primary mouse astrocytes. Densitometric data of GFAP expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 and **P < 0.01 vs. the MIR control group; ##P < 0.01 vs. the methamphetamine-treated MIR control group by one-way ANOVA, followed by the Holm-Sidak test. (C) Transfection of A172 cells with SIGMAR1 siRNA significantly inhibited Anti-MIR124–2HG-induced astrocyte activation, as determined by western blotting. Densitometric data of GFAP expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P<0.05 vs. the Anti-MIR control group; #P<0.05 vs. the Anti-MIR124–2HG cotransfected with siRNA control, by one-way ANOVA, followed by the Holm-Sidak test. (D) Transduction of Anti-MIR124–2HG induced astrocyte activation in primary mouse astrocytes isolated from WT mice but not in sigmar1 KO mice, as determined by western blotting. Densitometric data of GFAP expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 vs. Anti-MIR control transfected into primary mouse astrocytes isolated from WT mice; ##P < 0.01 vs. Anti-MIR124–2HG transduced into primary mouse astrocytes isolated from WT mice by one-way ANOVA, followed by the Holm-Sidak test. Meth, methamphetamine; AS, primary mouse astrocytes; MIR124, MIR124–2HG.
MIR124–2HG inhibits astrocyte activation by targeting SIGMAR1 in vivo
Next, to determine whether the Anti-MIR124–2HG-mediated functional effects specifically depend on the upregulation of SIGMAR1 expression, the hippocampi of WT and sigmar1 KO mice were microinjected bilaterally with either an Anti-MIR control-RFP lentivirus or an Anti-MIR124–2HG-RFP lentivirus. It was important to first determine the efficacy of Anti-MIR124–2HG-RFP lentivirus transduction in vivo. As shown in Fig. 3A, Anti-MIR control-RFP-LV (lentivirus) or Anti-MIR control-LV-RFP or Anti-MIR124–2HG-LV-RFP expression was largely restricted to the hippocampus. As expected, increased SIGMAR1 expression was observed on the Anti-MIR124–2HG-RFP-injected side compared with the Anti-MIR control-LV-RFP-injected side (Fig. 3B and C). Moreover, as shown in Fig. 3D and E, the Anti-MIR124–2HG lentivirus induced astrocyte activation, as indicated by the GFAP expression levels, in WT mice, and this effect was inhibited in the sigmar1 KO mice injected with the lentiviral vector expressing Anti-MIR124–2HG. These findings thus support a role for MIR124–2HG in the regulation of methamphetamine-mediated astrocyte activation through the targeting of SIGMAR1 in vivo. These results were further confirmed by immunostaining for GFAP, as shown in Fig. 3F and G. In WT mice, the Anti-MIR124–2HG lentivirus resulted in astrocyte activation in the hippocampus, as demonstrated by a significant increase in the relative integrated optical density (IOD) of GFAP immunoreactivity in astrocytes. However, these effects were inhibited in the sigmar1 KO mice.
Figure 3.
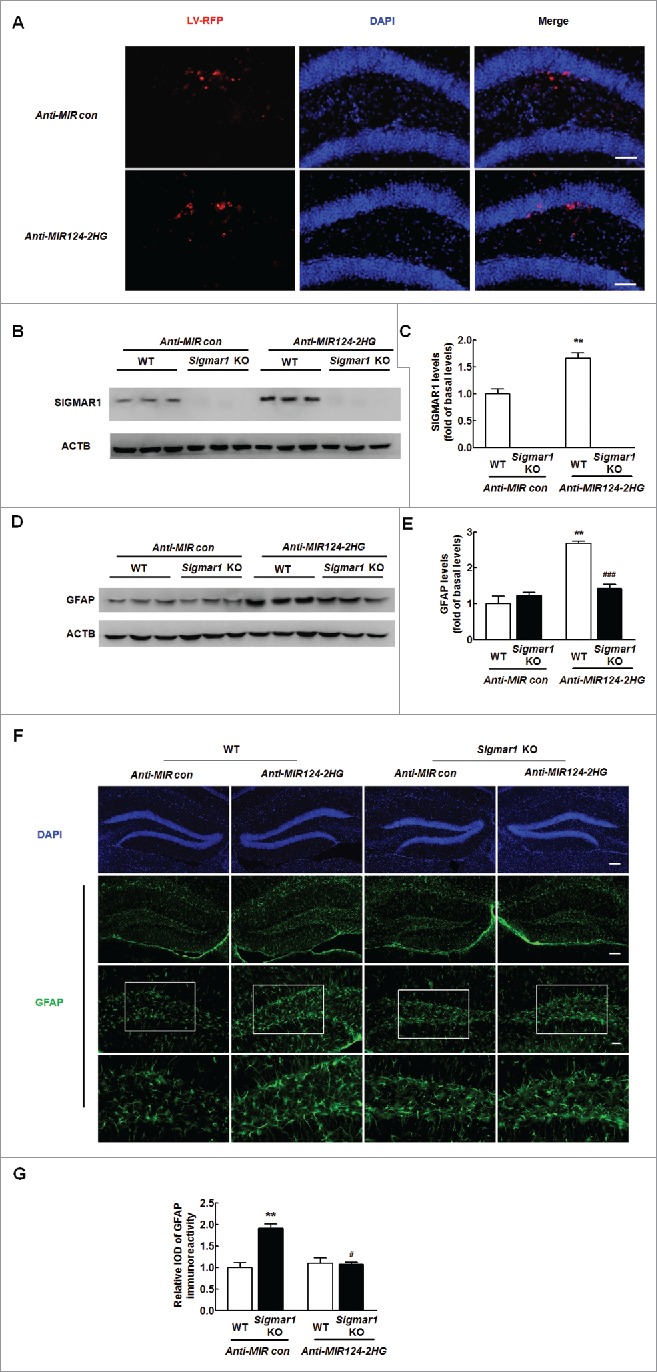
MIR124–2HG inhibits astrocyte activation by targeting SIGMAR1 in vitro. (A) Representative images of C57BL/6 mice microinjected with either Anti-MIR control-RFP or Anti-MIR124–2HG-RFP lentivirus into the hippocampus. Scale bar: 20 μm. The hippocampi of mice were microinjected bilaterally with an Anti-MIR-RFP lentivirus (2 μl of 109 viral genomes µl−1); the mice were killed 2 weeks after microinjection, and RFP expression was measured. (B and C) Anti-MIR124–2HG lentivirus injection successfully increased SIGMAR1 expression, as determined by western blotting. Two wk after microinjection, the mice were killed, and SIGMAR1 expression was measured (B). Densitometric analysis of SIGMAR1 expression using ImageJ (C). (D and E) GFAP expression in the hippocampus was determined in WT and sigmar1 KO mice. WT and sigmar1 KO mice were microinjected with either an Anti-MIR control-RFP or Anti-MIR124–2HG-RFP lentivirus. Two wk after microinjection, the mice were killed, and GFAP expression was determined by western blotting (D). Densitometric analysis of GFAP expression using ImageJ (E). N = 6 animals/group. **P < 0.01 vs. the Anti-MIR control-microinjected WT group; and ###P < 0.001 vs. the Anti-MIR124–2HG-RFP-microinjected WT group by one-way ANOVA, followed by the Holm-Sidak test. (F) Representative images of GFAP immunostaining in the hippocampi of the mice, as described above. Scale bars: 50 μm (upper panel), 50 μm (middle panel) and 20 μm (lower panel). The bottom panels depict outlines of portions of the astrocytes from the corresponding images above. (G) Quantification of GFAP immunofluorescence intensity using ImageJ software. IOD, integrated optical density. N = 6 animals/group. **P < 0.01 vs. the Anti-MIR control-microinjected WT group; #P<0.05 vs. the Anti-MIR124–2HG-RFP-microinjected WT group by one-way ANOVA, followed by the Holm-Sidak test.
circHIPK2 binds MIR124–2HG
Having determined the role of MIR124–2HG-SIGMAR1 axis in the astrocyte activation, we next get insight into the mechanisms of MIR124–2HG regulation. Since circRNAs act as competing endogenous RNA (ceRNA) sponges to interact with miRs and influence their activity, we sought to examine which circRNA can bind MIR124–2HG. Using a bioinformatics program, RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid), we found that circHIPK2 contains one MIR124–2HG target site (Fig. 4A). Consistent with the prediction, circHIPK2 was expressed in primary mouse astrocytes as determined by in situ hybridization analysis using a circHIPK2-specific probe (Fig. 4B and Table. 1B). We then assessed whether circHIPK2 is involved in astrocyte activation induced by different stimuli by qRT-PCR using divergent primers (Fig. 4C). Treatment of astrocytes with methamphetamine significantly increased circHIPK2 expression in both A172 cells and primary mouse astrocytes (Fig. 4D). Furthermore, we applied a biotin-coupled MIR124–2HG mimic (Table 1C) to test whether MIR124–2HG was able to pull down circHIPK2. We observed enrichment of circHIPK2 in the MIR124–2HG-captured fraction compared with that observed following the introduction of mutations that disrupt base paring between circHIPK2 and MIR124–2HG (Fig. 4E). Furthermore, double in situ hybridization indicated that there was colocalization of circHIPK2 and MIR124–2HG in primary mouse astrocytes (Fig. 4F).
Figure 4.
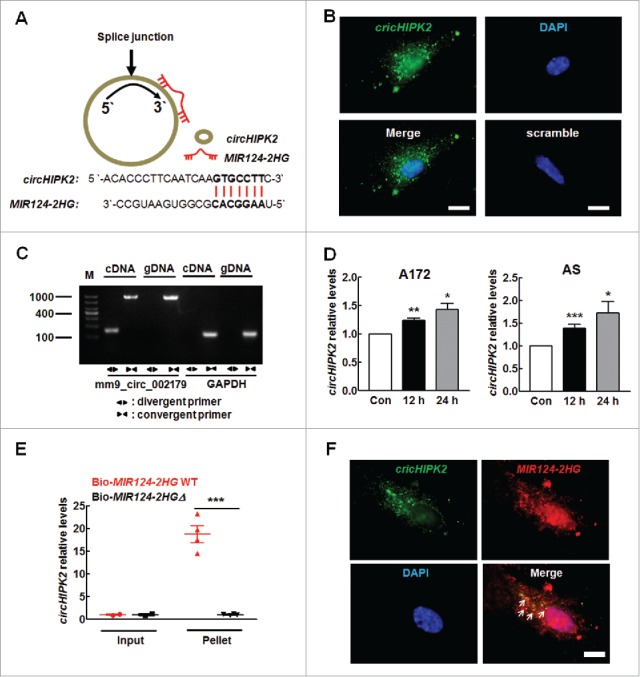
circHIPK2 binds MIR124–2HG. (A) circHIPK2 contains one site complementary to MIR124–2HG, as analyzed using the RNAhybrid bioinformatics program (upper panel). A biotin-coupled MIR124–2HG mutant is shown in the lower panel. (B) Fluorescence in situ hybridization of circHIPK2 in primary mouse astrocytes. Green, circHIPK2; Blue, DAPI. Scale bars: 5 µm. (C) Divergent primers amplified circRNAs in cDNA but not in genomic DNA (gDNA). GAPDH, linear control. (D) Methamphetamine increased the circHIPK2 levels in A172 cells (left panel) and primary mouse astrocytes (right panel), as determined by real-time PCR. All data are presented as the means ± SD of 3 independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. the control group by the Student t test. (E) Biotinylated WT MIR124–2HG (Bio-MIR124–2HG WT) or its mutant (Bio-MIR124–2HGΔ) was transfected into HEK293T cells. After streptavidin capture, the circHIPK2 and GAPDH mRNA levels were quantified by real-time PCR, and the relative immunoprecipitate (IP)/input ratios were plotted. All data are presented as the mean ± SD of 3 independent experiments. ***P < 0.001 vs. the Bio-124-WT group by the Student t test. (F) Fluorescence in situ hybridization of circHIPK2 and MIR124–2HG in primary mouse astrocytes. Green, circHIPK2; Red, MIR124–2HG; Blue, DAPI. Scale bars: 5 µm. Arrows indicate the colocalization of circHIPK2 and MIR124–2HG. Meth, methamphetamine; AS, primary mouse astrocytes.
Knockdown of circHIPK2 expression inhibits astrocyte activation by targeting MIR124–2HG in vitro
To study the effects of circHIPK2 on SIGMAR1 expression and astrocyte activation, astrocytes were transfected with circHIPK2 siRNA. As shown in Fig. S3, circHIPK2 siRNA caused a decrease in circHIPK2 expression compared with the control siRNA, as determined by real-time PCR. We further assessed the effect of circHIPK2 on methamphetamine-induced astrocyte activation. As shown in Fig. 5A, transfection of cells with circHIPK2 siRNA significantly inhibited methamphetamine-induced astrocyte activation, as determined by western blotting (WB) for GFAP expression. We also examined the effect of circHIPK2 on SIGMAR1 expression. The results showed that knockdown of circHIPK2 significantly inhibited the methamphetamine-induced increase in SIGMAR1 expression. Notably, transfection of cells with circHIPK2 siRNA resulted in decreased SIGMAR1 expression (Fig. 5B). Next, to further verify that MIR124–2HG is a mediator of circHIPK2, A172 astrocytes were cotransfected with Anti-MIR124–2HG and circHIPK2. Knockdown of circHIPK2 attenuated the inductive effects of Anti-MIR124–2HG on SIGMAR1 expression (Fig. 5C) and astrocyte activation, as indicated by the GFAP expression levels (Fig. 5D). To claim that these individually observed effects of MIR124–2HG or circHIPK2 on astrocyte activation were induced by methamphetamine, these rescue experiments were performed under methamphetamine treatment. Consistent with the findings, knockdown of circHIPK2 significantly inhibited the expression of SIGMAR1 and activation of astrocytes induced by Anti-MIR124–2HG in primary mouse astrocytes treated with methamphetamine as shown in Fig. S4A and B. These results indicate that circHIPK2 acts as an endogenous MIR124–2HG sponge to regulate SIGMAR1 expression and astrocyte activation.
Figure 5.
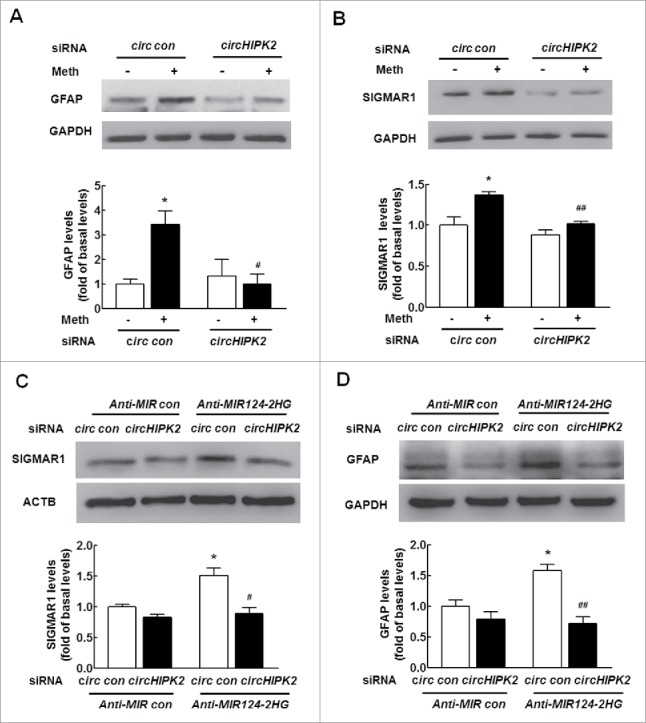
Knockdown of circHIPK2 expression inhibits astrocyte activation by targeting MIR124–2HG in vitro. (A) Transfection of cells with circHIPK2 siRNA attenuated methamphetamine-induced astrocyte activation, as determined by western blotting for GFAP expression. Cells were transfected with circHIPK2 siRNA for 24 h and were then treated with methamphetamine (100 μM) for 12 h, and GFAP expression was measured. (B) Transfection of cells with circHIPK2 siRNA attenuated the methamphetamine-induced increase in SIGMAR1 expression, as determined by western blotting. Densitometric data of GFAP and SIGMAR1 expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 vs. the siRNA control group; #P < 0.05 and ##P<0.01 vs. the methamphetamine-treated siRNA control group by one-way ANOVA, followed by the Holm-Sidak test. (C and D) Transduction of A172 cells with Anti-MIR124–2HG significantly inhibited circHIPK2 siRNA-induced SIGMAR1 expression (C) and astrocyte activation (D), as determined by western blotting. Densitometric data of GFAP and SIGMAR1 expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 vs. the Anti-MIR control cotransduced with siRNA control; #P < 0.05 and ##P < 0.01 vs. the Anti-MIR124–2HG cotransduced with siRNA control, by one-way ANOVA, followed by the Holm-Sidak test. Meth, methamphetamine.
Knockdown of circHIPK2 expression inhibits astrocyte activation in vivo
Having demonstrated that circHIPK2, involved in regulating SIGMAR1 expression, plays a critical role in astrocyte activation, we sought to validate the role of circHIPK2 in vivo by microinjecting a circHIPK2 siRNA lentivirus into the hippocampi of C57BL/6 mice. The hippocampi of C57BL/6 mice were microinjected bilaterally with either a circ siRNA control-GFP lentivirus or a circHIPK2 siRNA-GFP lentivirus and monitored for astrocyte activation in response to methamphetamine. However, it was important to first determine the efficacy of circHIPK2 siRNA-GFP lentivirus transduction in vivo. As shown in Fig. 6A, GFP expression was largely restricted to the hippocampus. As expected, decreased circHIPK2 expression was observed on the circHIPK2 siRNA-injected side compared with the circ siRNA control-GFP lentivirus-injected side (Fig. 6B). Two wk after the administration of the lentivirus, the mice were treated with methamphetamine (30 mg/kg), and GFAP expression in the hippocampus was assessed. As shown in Fig. 6C and D, methamphetamine treatment increased GFAP and SIGMAR1 expression compared with saline treatment, and these effects were significantly attenuated by circHIPK2 siRNA microinjection. Similar to the methamphetamine treatment, circHIPK2 siRNA microinjection also significantly inhibited the LPS-induced increase in GFAP expression, as shown in Fig. 6E and F. These results were further confirmed by immunostaining (Fig. 6G and H).
Figure 6.
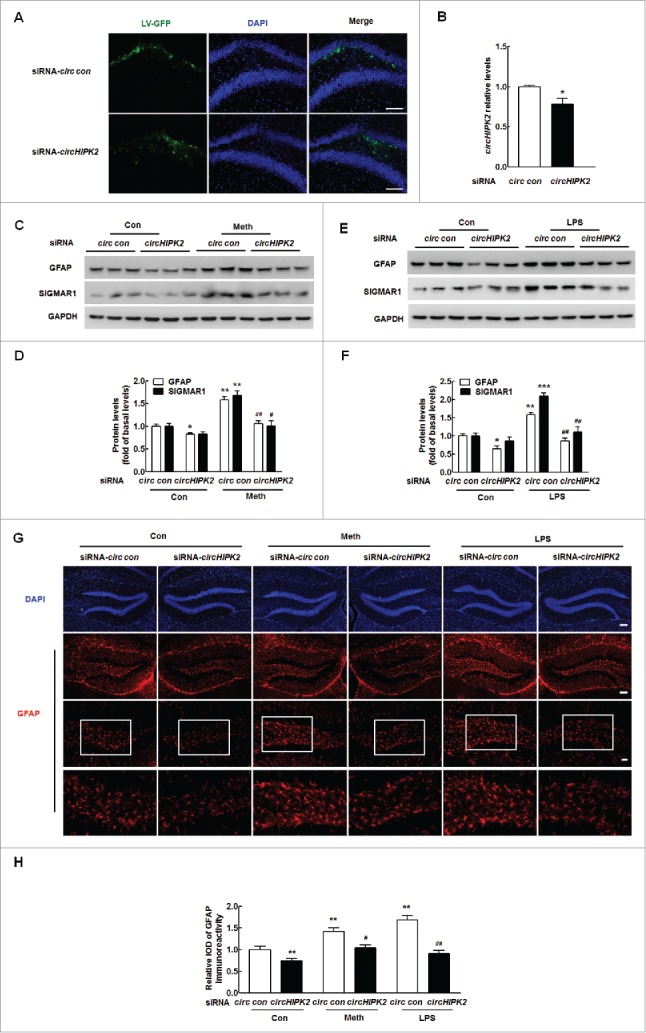
Knockdown of circHIPK2 expression inhibits astrocyte activation. (A) Representative images of hippocampi of C57BL/6 mice microinjected with circ siRNA-GFP lentivirus. Scale bar: 100 μm. The hippocampi of mice were microinjected bilaterally with either circ siRNA control-GFP or circHIPK2 siRNA-GFP lentivirus (2 μl of 109 viral genomes µl−1); the mice were killed 2 wk after microinjection, and GFP expression was measured. (B) circHIPK2 siRNA lentivirus injection successfully decreased circHIPK2 expression, as determined by real-time PCR. Two wk after microinjection, the mice were killed, and circHIPK2 expression was measured. N = 6 animals/group. *P<0.05 vs. the circ siRNA control group by the Student t test. (C and D) Microinjection of the circHIPK2 siRNA lentivirus into the hippocampus inhibited methamphetamine-induced astrocyte activation (C). Densitometric data of GFAP and SIGMAR1 expression using ImageJ (D) are shown MIR124–2HG. (E and F) Microinjection of the circHIPK2 siRNA lentivirus into the hippocampus inhibited astrocyte activation induced by LPS (E). Densitometric data of GFAP and SIGMAR1 expression using ImageJ (F) are shown. (G) Representative images of GFAP immunostaining in the hippocampi of mice treated with methamphetamine or LPS, as described above. Scale bars: 50 μm (upper panel), 50 μm (middle panel) and 20 μm (lower panel). The bottom panels depict outlines of segmented astrocytes from the corresponding images above. (H) Quantification of GFAP immunofluorescence intensity using ImageJ software. IOD, integrated optical density. Two wk after microinjection, the mice were treated with methamphetamine (i.p., 30 mg/kg) for 24 h or with LPS (i.p., 20 mg/kg) for 4 h. N = 6 animals/group. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. the saline-treated circ siRNA control group; #P < 0.05 and ##P < 0.01 vs. the methamphetamine- or LPS-treated circ siRNA control group by one-way ANOVA, followed by the Holm-Sidak test. Meth, methamphetamine.
Knockdown of circHIPK2 expression inhibits astrocyte autophagy by targeting MIR124–2HG
Having determined that circHIPK2 is involved in methamphetamine-induced astrocyte activation, we next assessed the underlying mechanisms. First, we examined the effect of methamphetamine on MAP1LC3B/LC3B-II expression in astrocytes. As shown in Fig. 7A, methamphetamine treatment of A172 cells increased the expression of ATG5 and BECN1/Beclin 1 and induced the production of MAP1LC3B-II, which is a cleaved MAP1LC3B-phosphatidylethanolamine conjugate and a general autophagosomal marker.38 These findings were further confirmed in primary mouse astrocytes (Fig. 7B). During the process of autophagy, SQSTM1/p62 acts as a receptor protein that links MAP1LC3B with ubiquitin moieties on misfolded proteins. Therefore, autophagy mediates the clearance of SQSTM1 along with ubiquitinated proteins. Consistently, SQSTM1 expression was downregulated by methamphetamine treatment in both A172 cells (Fig. 7A) and primary mouse astrocytes (Fig. 7B). This effect was also confirmed by immunofluorescence staining, which showed that methamphetamine treatment resulted in an increased number of vacuoles, as indicated by the endogenous MAP1LC3B-II expression levels (Fig. 7C).
Figure 7.
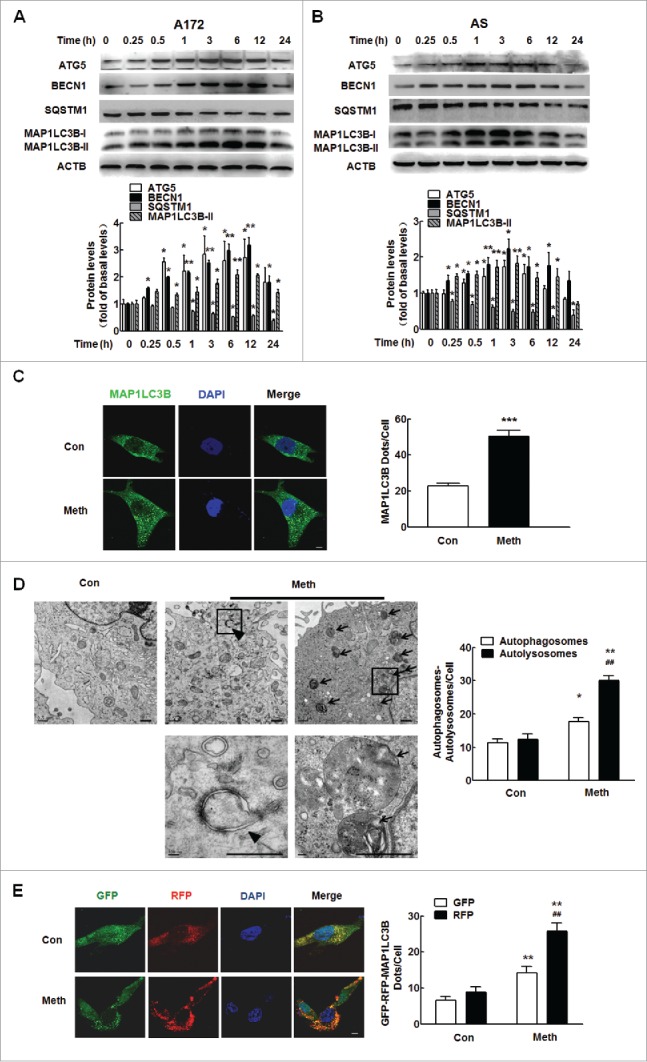
Methamphetamine induces astrocyte autophagy. (A and B) The expression of ATG5, BECN1, MAP1LC3B-II and SQSTM1 in A172 cells (A) and primary mouse astrocytes (B) induced by methamphetamine. Cells were treated with methamphetamine (100 μM) for 0.25 h, 0.5 h, 1 h, 3 h, 6 h, 12 h and 24 h. Densitometric data of ATG5, BECN1, MAP1LC3B-II and SQSTM1 expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 and **P < 0.01 vs. the control group by the Student t test. (C) Effect of methamphetamine on the formation of MAP1LC3B puncta. Cells were treated with methamphetamine (100 μM) for another 12 h. MAP1LC3B puncta were then analyzed by confocal microscopy. Scale bar: 20 μm. All data are presented as the means ± SD of 3 independent experiments. ***P < 0.001 vs. the control group by the Student t test. (D) Transmission electron microscopic imaging showing autolysosomes (arrows) and autophagosomes with double-membraned autophagic vacuoles (arrowheads) in primary microglial cells (left panel) and quantification of autolysosomes and autophagosomes (right panel) in primary microglial cells treated with methamphetamine (100 μM) for 12 h. Scale bar: 200 nm. (E) A172 cells were infected with an RFP-GFP-MAP1LC3B adenovirus and were then treated with 100 μM methamphetamine for 12 h. Effects of methamphetamine on RFP- and GFP-MAP1LC3B puncta (left panel). Scale bar: 5 μm. The numbers of RFP- and GFP-MAP1LC3B puncta per cell were counted, and the results are shown in the right panel. The numbers of puncta per cell are presented as the means ± SD of 3 independent experiments. *P < 0.05 and **P < 0.01 vs. the control group; ##P < 0.01 vs. the methamphetamine-treated group by one-way ANOVA, followed by the Holm-Sidak test. Meth, methamphetamine; AS, primary mouse astrocytes.
Autophagy is a dynamic process of protein degradation characterized by the formation of double-membraned cytoplasmic vesicles.39 Structural analysis via electron microscopy allows for the visualization of autophagy, characterized by the massive accumulation of autophagic vacuoles (autophagosomes) and autolysosomes in the cytoplasm. As shown in Fig. 7D, methamphetamine treatment resulted in the accumulation of numerous autolysosomes (arrow) and double-membraned autophagic vacuoles (arrowhead) in A172 cells.
Autophagic flux was further monitored in A172 astrocytes that were transduced with a tandem fluorescent-mRFP-GFP-MAP1LC3B-adenovirus, a specific marker of autophagosome formation that relies on the differences in GFP and RFP fluorescence under acidic conditions. GFP fluorescence is sensitive to the acidic conditions of the lysosome lumen, while RFP is relatively stable under acidic conditions. Thus, the colocalization of GFP and RFP signals (yellow dots) indicates the lack of fusion of phagophores or autophagosomes with lysosomes, whereas RFP-only signals (red puncta) indicate the presence of autolysosomes. As shown in Fig. 7E, methamphetamine treatment significantly increased the number of yellow dots per cell, with a concomitant greater increase in RFP-only MAP1LC3B dots in primary mouse microglia transduced with tandem fluorescent-tagged nRFP-GFP-MAP1LC3B.
Next, we elucidated the effect of circHIPK2 on astrocyte autophagy. As shown in Fig. 8A, transfection of cells with circHIPK2 siRNA significantly inhibited methamphetamine-induced astrocyte autophagy, as determined by WB for MAP1LC3B-II expression. Having determined that MIR124–2HG regulates astrocyte activation, we next examined its role in astrocyte autophagy. Transduction of cells with a MIR124–2HG precursor-expressing lentivirus significantly inhibited methamphetamine-induced astrocyte autophagy, as indicated by the MAP1LC3B-II expression levels, in both A172 cells (Fig. 8B) and primary mouse astrocytes (Fig. 8C). Next, to determine whether the MIR124–2HG-mediated functional effects are specifically dependent on the upregulation of SIGMAR1 expression, A172 cells were cotransduced with lentiviral vectors expressing Anti-MIR124–2HG and SIGMAR1 siRNA. Transduction of cells with the Anti-MIR124–2HG lentivirus enhanced GFAP expression, and this effect was significantly inhibited in cells cotransduced with the lentiviral vectors expressing Anti-MIR124–2HG and SIGMAR1 siRNA (Fig. 8D). Moreover, transduction of cells with the Anti-MIR124–2HG lentivirus resulted in enhanced MAP1LC3B-II expression (Fig. 8E), and this effect was inhibited in primary mouse astrocytes isolated from sigmar1 KO mice. In addition, knockdown of circHIPK2 expression significantly attenuated the inductive effect of Anti-MIR124–2HG on astrocyte autophagy (Fig. 8F). These rescue experiments were further confirmed in astrocytes treated with methamphetamine as shown in Fig. S5A-C. These results indicate that circHIPK2 acts as an endogenous MIR124–2HG sponge to regulate astrocyte autophagy.
Figure 8.
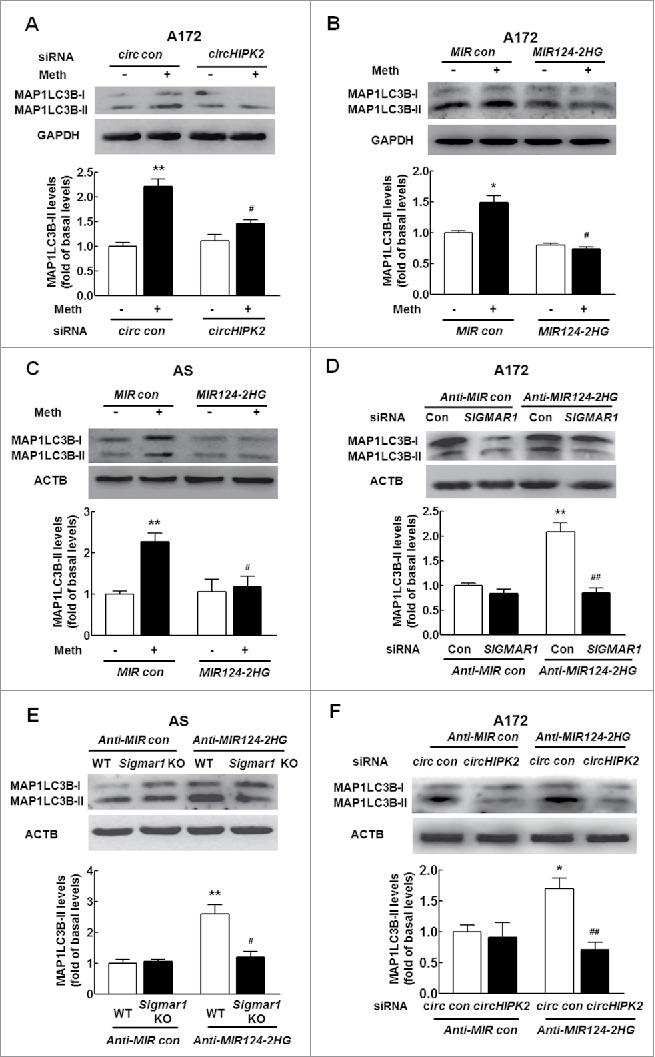
Knockdown of circHIPK2 expression inhibits astrocyte autophagy by targeting MIR124–2HG. (A) Transfection of cells with circHIPK2 siRNA attenuated methamphetamine-induced astrocyte autophagy, as determined by western blotting for MAP1LC3B-II expression. Cells were transfected with circHIPK2 siRNA for 24 h and were then treated with methamphetamine (100 μM) for 12 h, followed by measurement of MAP1LC3B-II expression. Densitometric data of MAP1LC3B-II expression using ImageJ are presented as the means ± SD of 3 independent experiments. **P < 0.01 vs. the siRNA control group; #P<0.05 vs. the methamphetamine-treated siRNA control group by one-way ANOVA, followed by the Holm-Sidak test. (B and C) Transduction of cells with MIR124–2HG attenuated methamphetamine-induced astrocyte autophagy in A172 cells (B) and primary mouse astrocytes (C), as determined by western blotting for MAP1LC3B-II expression. Densitometric data of MAP1LC3B-II expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 and **P < 0.01 vs. the MIR control group; #P < 0.05 vs. the methamphetamine-treated MIR control group by one-way ANOVA, followed by the Holm-Sidak test. (D) Transfection of A172 cells with SIGMAR1 siRNA significantly inhibited Anti-MIR124–2HG-induced astrocyte autophagy, as determined by western blotting. Densitometric data of MAP1LC3B-II expression using ImageJ are presented as the means ± SD of 3 independent experiments. **P < 0.01 vs. the Anti-MIR control group; ##P < 0.01 vs. the Anti-MIR124–2HG cotransduced with siRNA control, by one-way ANOVA, followed by the Holm-Sidak test. (E) Transduction of Anti-MIR124–2HG induced astrocyte activation in primary mouse astrocytes isolated from WT mice but not in sigmar1 KO mice, as determined by western blotting. Densitometric data of MAP1LC3B-II expression using ImageJ are presented as the means ± SD of 3 independent experiments. **P < 0.01 vs. Anti-MIR control transduced into primary mouse astrocytes isolated from WT mice; #P < 0.05 vs. Anti-MIR124–2HG transduced into primary mouse astrocytes isolated from WT mice by one-way ANOVA, followed by the Holm-Sidak test. (F) Transduction of A172 cells with Anti-MIR124–2HG significantly inhibited the circHIPK2 siRNA-induced increase in MAP1LC3B-II expression, as determined by western blotting. Densitometric data of MAP1LC3B-II expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 vs. Anti-MIR control cotransduced with siRNA control; ##P<0.01 vs. the Anti-MIR124–2HG cotransduced with siRNA control, by one-way ANOVA, followed by the Holm-Sidak test. Meth, methamphetamine; AS, primary mouse astrocytes.
Knockdown of circHIPK2 expression inhibits ER stress in astrocytes by targeting MIR124–2HG
Having determined that circHIPK2 is involved in methamphetamine-induced astrocyte activation and autophagy, we next investigated the role of circHIPK2 in ER stress in astrocytes. First, we examined the effect of methamphetamine on ER stress in astrocytes. As shown in Fig. S6, methamphetamine treatment of A172 astrocytes increased the expression of EIF2AK3/PERK, EIF2S1/EIF-2α, DDIT3/CHOP, ERN1/IRE1 and ATF6.
Next, we assessed the effect of circHIPK2 on ER stress in astrocytes. As shown in Fig. 9A, transfection of astrocytes with circHIPK2 siRNA significantly inhibited methamphetamine-induced ER stress, as demonstrated by WB for DDIT3 expression. Having determined that MIR124–2HG regulates astrocyte activation and autophagy, we next examined its role in ER stress in astrocytes. Transduction of cells with a MIR124–2HG precursor-expressing lentivirus significantly inhibited the methamphetamine-induced ER stress associated with astrocyte autophagy, as indicated by the DDIT3 expression levels, in both A172 cells (Fig. 9B) and primary mouse astrocytes (Fig. 9C). Next, to determine whether the MIR124–2HG-mediated functional effects are specifically dependent on the upregulation of SIGMAR1 expression, A172 cells were cotransduced with lentiviral vectors expressing Anti-MIR124–2HG and SIGMAR1 siRNA. Transduction of cells with the Anti-MIR124–2HG lentivirus enhanced DDIT3 expression, and this effect was significantly inhibited in cells cotransduced with the lentiviral vectors expressing Anti-MIR124–2HG and SIGMAR1 siRNA (Fig. 9D). Moreover, transduction of cells with the Anti-MIR124–2HG lentivirus resulted in enhanced DDIT3 expression (Fig. 9E), and this effect was inhibited in primary mouse astrocytes isolated from sigmar1 KO mice. Next, we further assessed whether MIR124–2HG is a mediator of circHIPK2. A172 astrocytes were cotransfected with the Anti-MIR124–2HG lentivirus and circHIPK2 siRNA, and DDIT3 expression was measured. As shown in Fig. 9F, knockdown of circHIPK2 expression significantly counteracted the inductive effect of Anti-MIR124–2HG on ER stress in astrocytes. These rescue experiments were further confirmed in astrocytes treated with methamphetamine, as shown in Fig. S7A to C. These results indicate that circHIPK2 acts as an endogenous MIR124–2HG sponge to regulate ER stress in astrocytes.
Figure 9.
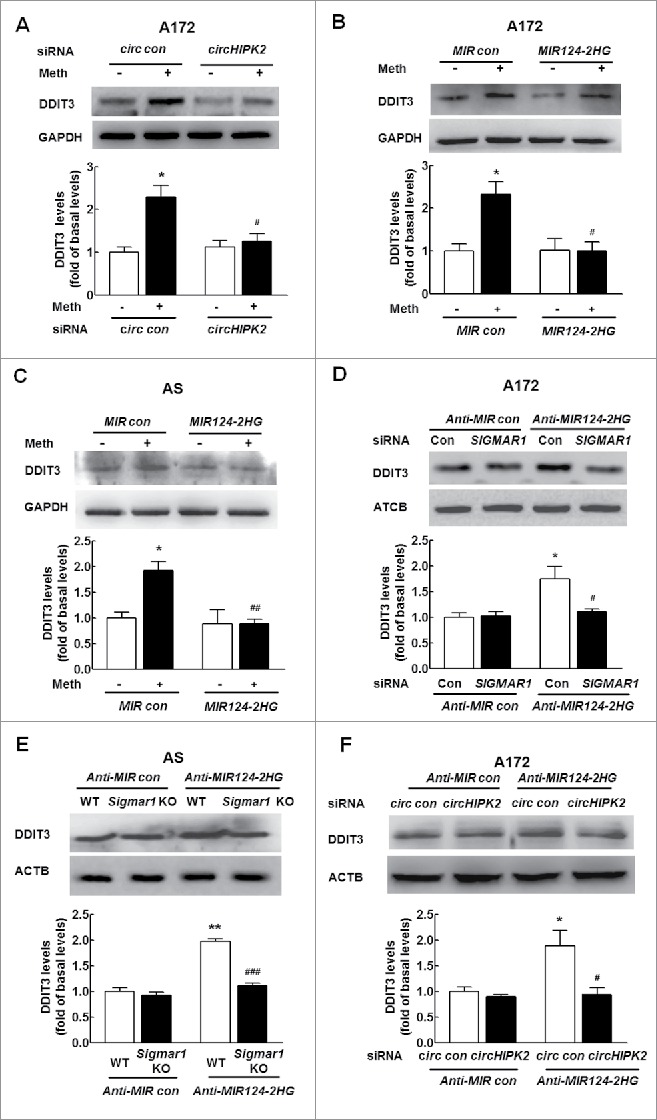
Knockdown of circHIPK2 expression inhibits ER stress in astrocytes by targeting MIR124–2HG. (A) Transfection of cells with circHIPK2 siRNA attenuated methamphetamine-induced ER stress in astrocytes, as determined by western blotting for DDIT3 expression. Densitometric data of DDIT3 expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 vs. the siRNA control group; and #P<0.05 vs. the methamphetamine-treated siRNA control group by one-way ANOVA, followed by the Holm-Sidak test. (B and C) Transduction of cells with MIR124–2HG attenuated methamphetamine-induced ER stress in A172 cells (B) and primary mouse astrocytes (C), as determined by western blotting for DDIT3 expression. Densitometric data of DDIT3 expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P<0.0 vs. the MIR control group; #P < 0.05 and ##P < 0.01 vs. the methamphetamine-treated MIR control group by one-way ANOVA, followed by the Holm-Sidak test. (D) Transfection of A172 cells with SIGMAR1 siRNA significantly inhibited Anti-MIR124–2HG-induced ER stress in astrocytes, as determined by western blotting. Densitometric data of DDIT3 expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 vs. Anti-MIR control cotransduced with siRNA control; #P < 0.05 vs. Anti-MIR124–2HG cotransduced with siRNA control, by one-way ANOVA, followed by the Holm-Sidak test. (E) Transduction of Anti-MIR124–2HG induced ER stress in primary mouse astrocytes isolated from WT mice but not in sigmar1 KO mice, as determined by western blotting. Densitometric data of DDIT3 expression using ImageJ are presented as the means ± SD of 3 independent experiments. **P < 0.01 vs. Anti-MIR control transduced into primary mouse astrocytes isolated from WT mice; ###P<0.001 vs. Anti-MIR124–2HG transduced into primary mouse astrocytes isolated from WT mice by one-way ANOVA, followed by the Holm-Sidak test. (F) Transduction of A172 cells with Anti-MIR124–2HG significantly inhibited the circHIPK2 siRNA-induced increase in DDIT3 expression, as determined by western blotting. Densitometric data of DDIT3 expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 vs. Anti-MIR control cotransduced with siRNA control; #P < 0.05 vs. Anti-MIR124–2HG cotransduced with siRNA control, by one-way ANOVA, followed by the Holm-Sidak test. Meth, methamphetamine; AS, primary mouse astrocytes.
Distinct roles of autophagy and ER stress in methamphetamine-induced astrocyte activation
Having determined that methamphetamine induces astrocyte activation, we next sought to examine the roles of autophagy and ER stress in this process. Pretreatment of cells with an ER stress inhibitor, salubrinal, significantly inhibited methamphetamine-induced astrocyte activation (Fig. 10A and B), suggesting that ER stress enhances activation of these cells. Salubrinal acts as a phosphatase inhibitor and increases EIF2S1 phosphorylation, leading to a global decrease in translation and a subsequent reduction in ER stress. Moreover, both A172 cells and primary mouse astrocytes were treated with an autophagy inhibitor, 3-methyladenine (3-MA), which is a PtdIns3K inhibitor that prevents autophagy at an early stage of autophagosome formation. As shown in Fig. 10C and D, pretreatment of cells with 3-MA enhanced methamphetamine-induced GFAP expression in both A172 astrocytes (Fig. 10C) and primary mouse astrocytes (Fig. 10D). However, pretreatment of cells with an autophagy inducer, rapamycin significantly lessened methamphetamine-induced astrocyte activation in both A172 astrocytes (Fig. 10E) and primary mouse astrocytes (Fig. 10F). This finding was further confirmed in vivo, as rapamycin pretreatment significantly inhibited activation of astrocytes induced by methamphetamine, with concomitant enhancement of autophagy (Fig. S8). These findings suggest that autophagy inhibits astrocyte activation, while the inhibition of autophagy results in enhanced astrocyte activation.
Figure 10.
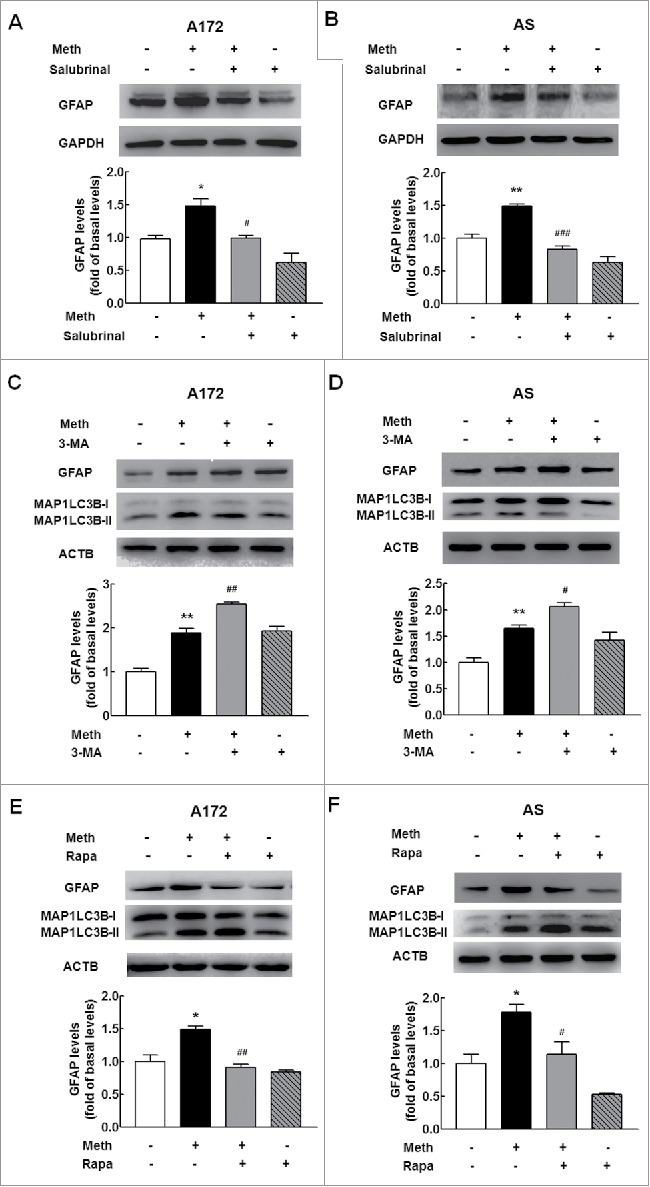
Distinct roles of autophagy and ER stress in methamphetamine-induced astrocyte activation. (A and B) Pretreatment of cells with an ER stress inhibitor, salubrinal, significantly inhibited methamphetamine-induced astrocyte activation in A172 cells (A) and primary mouse astrocytes (B). Cells were pretreated with salubrinal (20 μM) for 1 h and were then treated with methamphetamine (100 μM) for 12 h. (C and D) Pretreatment of cells with an autophagy inhibitor, 3-MA (3 mM), enhanced methamphetamine-induced astrocyte activation in A172 cells (C) and primary mouse astrocytes (D). Cells were pretreated with 3-MA (3 mM) for 1 h and were then treated with methamphetamine (100 μM) for 3 h. (E and F) Pretreatment of cells with an autophagy inducer, rapamycin, attenuated methamphetamine-induced astrocyte activation in A172 cells (E) and primary mouse astrocytes (F). Cells were pretreated with rapamycin (0.1 μM) for 1 h and were then treated with methamphetamine (100 μM) for 12 h. All densitometric data of GFAP expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 and **P < 0.01 vs. the control group; #P < 0.05, ##P < 0.01 and ###P < 0.001 vs. the methamphetamine-treated group by one-way ANOVA, followed by the Holm-Sidak test. Meth, methamphetamine; AS, primary mouse astrocytes; 3-MA, 3-methyladenine; Rapa, rapamycin.
Interplay between autophagy and ER stress induced by methamphetamine
Having determined that methamphetamine induces autophagy and ER stress in astrocytes, further study was undertaken to elucidate the role of ER stress in the autophagy induced by methamphetamine using the ER stress inhibitor salubrinal. As shown in Fig. 11A and B, pretreatment of cells with salubrinal resulted in abrogation of the increased autophagy induced by methamphetamine as evidenced by decreased expression of MAP1LC3B-II, thereby confirming that ER stress response is upstream of autophagy in methamphetamine-treated astrocytes. We next assessed the effect of autophagy on the ER stress response. Pretreatment of cells with the autophagy inducer rapamycin resulted in abrogation of the increased ER stress induced by methamphetamine as evidenced by decreased expression of DDIT3 in both A172 cells (Fig. 11C) and primary mouse astrocytes (Fig. 11D). Consistent with this finding, pretreatment of cells with the autophagy inhibitor 3-MA further enhanced the increased ER stress in both A172 cells (Fig. 11E) and primary mouse astrocytes (Fig. 11F), suggesting an interplay between autophagy and the ER stress response in astrocytes induced by methamphetamine.
Figure 11.
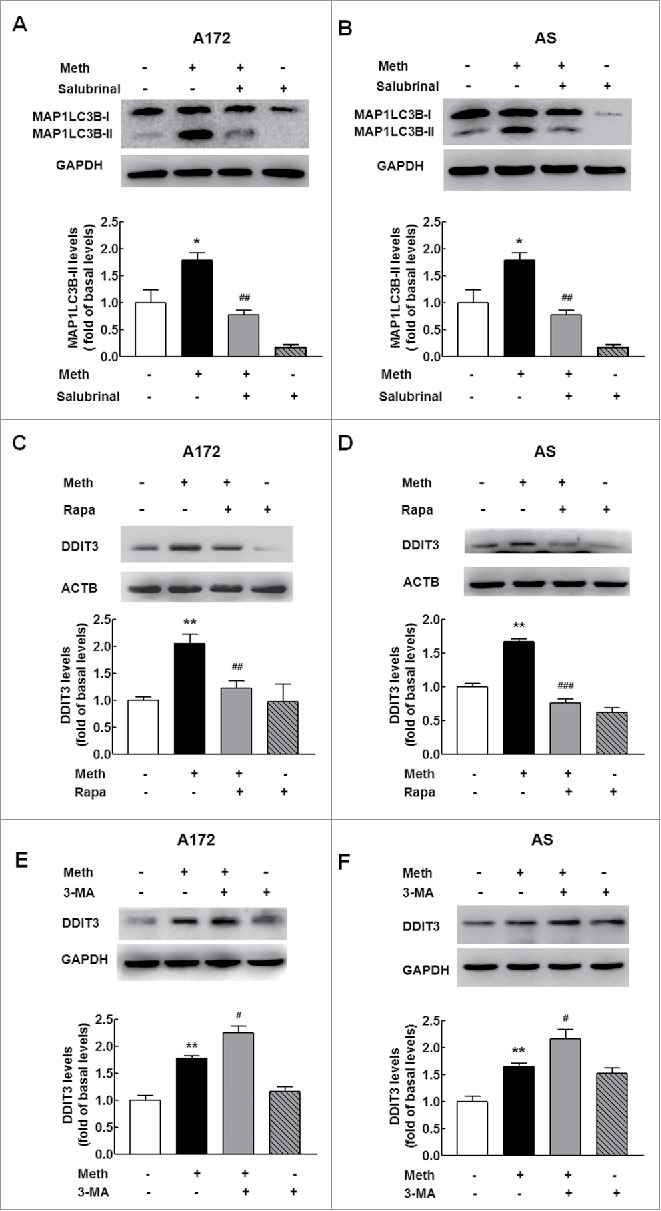
Interplay between autophagy and ER stress induced by methamphetamine. (A and B) Pretreatment of cells with an ER stress inhibitor, salubrinal, significantly inhibited methamphetamine-induced astrocyte autophagy in A172 cells (A) and primary mouse astrocytes (B). Cells were pretreated with salubrinal (20 μM) for 1 h and were then treated with methamphetamine (100 μM) for 12 h. (C and D) Pretreatment of cells with an autophagy inducer, rapamycin, attenuated methamphetamine-induced astrocyte ER stress in A172 cells (C) and primary mouse astrocytes (D). Cells were pretreated with rapamycin (0.1 μM) for 1 h and were then treated with methamphetamine (100 μM) for 12 h. (E and F) Pretreatment of cells with an autophagy inhibitor, 3-MA (3 mM), enhanced methamphetamine-induced astrocyte ER stress in A172 cells (E) and primary mouse astrocytes (F). Cells were pretreated with 3-MA (3 mM) for 1 h and were then treated with methamphetamine (100 μM) for 3 h. All densitometric data of MAP1LC3B-II and DDIT3 expression using ImageJ are presented as the means ± SD of 3 independent experiments. *P < 0.05 and **P < 0.01 vs. the control group; #P < 0.05, ##P < 0.01 and ###P < 0.001 vs. the methamphetamine-treated group by one-way ANOVA, followed by the Holm-Sidak test. Meth, methamphetamine; AS, primary mouse astrocytes; 3-MA, 3-methyladenine; Rapa, rapamycin.
Discussion
Our study provides new insights into the function of circHIPK2, which was found to significantly inhibit astrocyte activation via the regulation of autophagy and ER stress via the targeting of MIR124–2HG and subsequent targeting of MIR124–2HG-SIGMAR1 (Fig. 12). These findings provide the first evidence that the circHIPK2-MIR124–2HG axis mediates a regulatory pathway critical for the regulation of astrocyte activation. Specific blockage of circHIPK2 could be envisioned as a potential therapeutic target for the inhibition of astrocyte activation in the context of drug abuse as well as in the treatment of a broad range of neuroinflammatory disorders.
Figure 12.
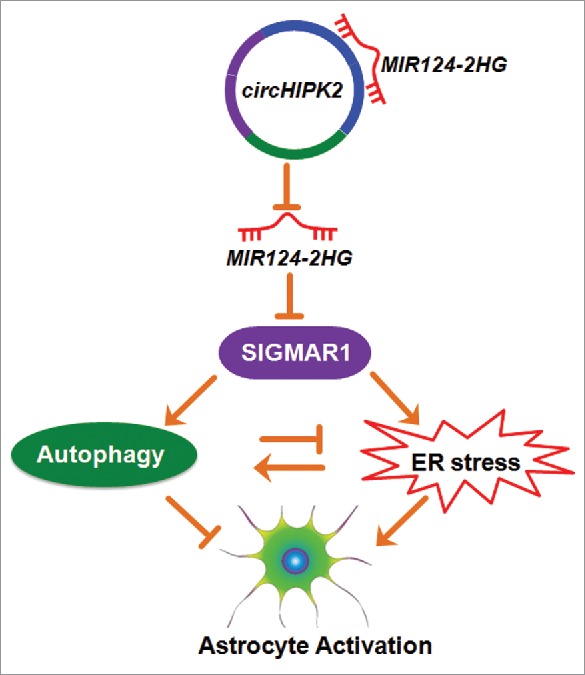
Role of circHIPK2 in astrocyte activation. circHIPK2, identified as a ceRNA, functions as an endogenous MIR124–2HG sponge to sequester MIR124–2HG and inhibit its activity, resulting in increased SIGMAR1 expression and consequent astrocyte activation via the interplay between autophagy and endoplasmic reticulum stress.
Astrocyte activity may exacerbate inflammatory reactions and neuronal tissue damage or promote immune suppression and tissue repair depending on the timing and context.20 However, clinical and experimental findings have indicated that astrocyte activation is critically involved in the pathogenesis of neurotoxicity induced by methamphetamine,40 strongly suggesting that the inflammatory status might critically determine the outcome and prognosis of neuronal tissue injury. Strategies aimed at mitigation of astrocyte activation have been shown to have therapeutic potential in experimental models of neuroinflammation induced by methamphetamine. Thus, researchers have recently focused on developing anti-inflammatory strategies for the treatment of astrocyte activation induced by methamphetamine. For example, our previous studies and other studies indicate that inhibition of SIGMAR1 significantly lessens astrocyte activation.17,41 In this study, we explored molecular players with pivotal roles in astrocyte activation.
Accumulating evidence indicates that circRNAs are not simply the by-products of mis-splicing or splicing errors; in fact, they have important functions in development and heart senescence, hypertrophy and failure, as well as cell growth.33-36 Despite the biological importance of circRNAs, it has not yet been clarified whether they are involved in the regulation of astrocyte activation in the brain. Our present study revealed a novel function of circHIPK2 in the regulation of astrocyte activation via the interplay between autophagy and ER stress. A better understanding of the role of this circRNA in astrocyte activation may facilitate the identification of a novel therapeutic target for the treatment or control of related diseases.
A previous study showed that circRNAs may act as endogenous sponges that interact with miRs and regulate the expression of miRNA target genes. In addition, a recent study demonstrates that the circRNA HRCR acts as a MIR223 sponge to regulate cardiac hypertrophy and heart failure.37 Consistent with previous studies, our current study revealed that circHIPK2 acts a MIR124–2HG sponge to regulate SIGMAR1 expression and astrocyte activation. To improve knockdown efficiency and minimize off-target effects, we further constructed a second circHIPK2 siRNA-GFP lentivirus (circHIPK2 siRNA2) in addition to the first circHIPK2 siRNA-GFP lentivirus (circHIPK2 siRNA1) microinjected in Fig. 6B. Transduction of primary mouse astrocytes with these 2 different circHIPK2 siRNA-GFP lentivirus constructs decreased the expression of circHIPK2 by 41% for circHIPK2 siRNA1 versus 40% for circHIPK2 siRNA2 (Fig. S9A), which are higher than that in A172 cells transfected with human-specific circHIPK2 siRNA (Fig. S3). Furthermore, we also performed circHIPK2 loss-of-function experiments. Knockdown of circHIPK2 expression significantly inhibited activation (Fig. S9B), autophagy (Fig. S9C), ER stress (Fig. S9D) as well as SIGMAR1 expression (Fig. S9E) in astrocytes induced by methamphetamine, which is consistent with the results obtained in A172 cells transfected with human circHIPK2 siRNA as well as that in mice microinjected with circHIPK2 siRNA1 lentivirus.
The interaction between MIR124–2HG and circHIPK2 was further confirmed by a miR affinity-isolation assay. To support the circHIPK2-MIR124–2HG-SIGMAR1 network in our study, we examined the level of MIR124–2HG in astrocytes transduced with circHIPK2 siRNA and found that knockdown of circHIPK2 expression did not significantly affect the level of MIR124–2HG (Fig. S10A and B). However, the level of MIR124–2HG binding with circHIPK2 was significantly decreased in the circHIPK2 siRNA-transduced group compared with the siRNA control-treated group as determined using an affinity-isolation assay (Fig. S10C). Moreover, we examined the effect of circHIPK2-MIR124–2HG axis on the expression of SIGMAR1 in astrocytes without/with methamphetamine. As shown in Fig. S11, knockdown of circHIPK2 significantly inhibited the expression of SIGMAR1 induced by Anti-MIR124–2HG in A172 cells or primary mouse astrocytes treated with methamphetamine. A recent study indicates that another circRNA, circHIPK3, acts as a sponge for MIR124–2HG and regulates cell proliferation.35 circHIPK3 is derived from exon 2 of the HIPK3 gene, and it has been identified in previous reports via the deep sequencing of several human cell lines.31,42,43 To our knowledge, this is the first report demonstrating that knockdown of circHIPK2 expression inhibits astrocyte activation induced by methamphetamine and LPS.
Although the present study showed that MIR124–2HG is regulated by circHIPK2, our results do not exclude the involvement of other circRNAs or molecules in the regulation of MIR124–2HG, either directly or indirectly. On the contrary, circHIPK2 was not specifically targeting MIR124–2HG. Using starBase v2.0 software, we predicted that circHIPK2 is able to bind other miRNAs that are involved in the regulation of autophagy-associated proteins, such as MIR146A, MIR302A, MIR372, MIR373, MIR378A, MIR506 and MIR520A. In our study, we further examined the effect of methamphetamine on the expression of these miRNAs. As shown in Fig. S12, methamphetamine treatment significantly decreased the expression of MIR146A, but not MIR372 or MIR378A. Notably, in primary mouse astrocytes, the levels of Mir302a, Mir373, Mir506 and Mir520a were not detectable. Therefore, it remains to be elucidated whether the binding of other miRNAs to circHIPK2 is involved in astrocyte activation. In terms of the targeting of miRNAs, it is known that one miRNA may have many targets.44 Using TargetScan, we have predicted that, in addition to SIGMAR1, there are other targets for MIR124–2HG including BECN1, RHOG, SNAI2, MAGT1 and PTBP1. Among these targets, only BECN1 is involved in the regulation of autophagy. In our study, methamphetamine treatment increased the expression of BECN1. Consistent with a recent study showing that decreased MIR124–2HG expression enhances the proliferation of breast cancer cells targeting BECN1,45 our current study also indicated that transduction of cells with Anti-MIR124–2HG increased the expression of BECN1, as shown in Fig. S13A. Moreover, loss-of-function experiments knocked down BECN1 expression, suggesting that SIGMAR1 lies upstream of BECN1 (Fig. S13 A and B). Although we could not rule out the possibility that methamphetamine-induced autophagy was due to another target of MIR124–2HG-BECN1, our study provides convincing evidence that the MIR124–2HG-SIGMAR1 axis is involved in the autophagy induced by methamphetamine.
To date, increasing numbers of miRs have been found to be key regulators of different cellular processes, particularly those related to autophagy and ER stress. However, few studies have assessed the regulatory mechanisms of these miRs in astrocytes. MIR124–2HG, the most abundant brain-specific miR, regulates microglial28 and astrocyte29 activation. Consistent with these findings, our study showed that MIR124–2HG expression was decreased in astrocytes treated with methamphetamine and that MIR124–2HG overexpression significantly inhibited methamphetamine-induced astrocyte activation (Fig. 2A and B). To determine whether MIR124–2HG-mediated functional effects depend specifically on SIGMAR1 suppression, a MIR124–2HGΔ lentivirus construct with mutations in the sequence targeting the 3′-UTR of SIGMAR1 was generated (Fig. S14A). Overexpression of a MIR124–2HG, but not the MIR124–2HGΔ construct with mutations in the sequence targeting the 3′-UTR of SIGMAR1, significantly inhibited the increased expression of SIGMAR1, GFAP and MAP1LC3B-II in astrocytes induced by methamphetamine (Fig. S14B). Consistent with this finding, microinjection of a MIR124–2HG lentivirus significantly inhibited the increased expression of GFAP and MAP1LC3B-II induced by methamphetamine (Fig. S15A) or LPS (Fig. S15B). However, the MIR124–2HGΔ construct with mutations in the sequence targeting the 3′-UTR of SIGMAR1 failed to affect these responses. Although previous studies have indicated that MIR124–2HG is involved in glial activation, its detailed function in astrocyte activation and whether it participates in autophagy and ER stress remain an enigma. Our current study focused on elucidation of the role of MIR124–2HG in the regulation of astrocyte activation via the interplay between autophagy and ER stress. Our findings demonstrated that MIR124–2HG inhibits autophagy and ER stress with consequent astrocyte activation through the targeting of SIGMAR1.
Autophagy is a basic cellular process that results in the degradation of defective organelles and misfolded proteins to preserve cellular homeostasis. A previous study indicates that autophagy contributes to astrocyte activation, as evidenced by the finding that the autophagy inhibitor 3-MA decreases astrocyte activation induced by cocaine in astrocytes.46 Surprisingly, our results demonstrated that the autophagy inducer rapamycin significantly decreased astrocyte activation, while the autophagy inhibitor 3-MA had significant inductive effects, suggesting that methamphetamine-induced autophagy inhibits astrocyte activation. To obtain more solid evidence, we further extended the study and confirmed the role of autophagy in astrocyte activation induced by LPS, as evidenced by the finding that rapamycin significantly inhibited the LPS-induced increase in GFAP expression (Fig. S16). Our findings are consistent with a previous study showing that rapamycin decreases astrocyte activation induced by spinal cord injury47 as well as another study demonstrating that conditioned deletion of MTOR in astrocytes with MTOR downregulation significantly ameliorates astrogliosis in the sclerotic hippocampus.45 Controversy exists in the role of autophagy during astrocyte activation, as a recent study indicates that inhibition of autophagy results in the attenuation of astrocyte activation.46 Whether autophagy inhibits or enhances astrocyte activation mainly depends on distinct autophagy-related signaling pathways triggered by different stimuli in specific contexts. In the current study, we provide the first demonstration of a molecular link between circHIPK2 and autophagy. Knockdown of circHIPK2 expression significantly decreased autophagy triggered by methamphetamine via MIR124–2HG and its target SIGMAR1. To the best of our knowledge, this is the first report of a role for this circRNA in the regulation of autophagy. Whether circHIPK2 and MIR124–2HG are also involved in other neurodegenerative diseases is an interesting question that should be addressed in future investigations. Thus, it will be critical to determine whether circHIPK2 and MIR124–2HG function in other astrocyte activation pathways and to identify the relevant substrates.
Given that ER stress induced by numerous stress stimuli, such as hypoxia, infection and nutrient depletion, triggers activation of astrocytes,48,46 we next examined the effect of methamphetamine on ER stress in astrocytes. The results demonstrated that exposure of astrocytes to methamphetamine induced activation of ER stress, ultimately leading to astrocyte activation. Emerging evidence indicates that ER stress plays key roles in the induction of astrocyte activation49 and astrocyte survival.50 Recent studies have shown that ER stress in response to psychostimulants is linked to behavioral sensitization and that the psychostimulant-induced ER stress signaling cascades are closely associated with the pathogenesis of neurodegenerative diseases.46,51 In the current study, we provide the first demonstration of a molecular link between circHIPK2 and ER stress-mediated astrocyte activation. Knockdown of circHIPK2 expression resulted in significantly decreased ER stress triggered by methamphetamine via MIR124–2HG and its target SIGMAR1. To the best of our knowledge, this is the first report of a role for circRNAs in the regulation of ER stress.
Our study has clarified that autophagy has an inhibitory effect on astrocyte activation, while ER stress has an inductive effect, indicating that autophagy and ER stress play distinct roles in this process. Previous studies have demonstrated a central role for ER stress in the induction of autophagy in glioma cells.52,53 To further clarify the relationship between ER and autophagy, we examined the effect of hypoxia (ER stress inducer) and the autophagy inhibitor 3-MA on the expression of GFAP in the same experiment. As shown in Fig. S17, pretreatment of astrocytes with 3-MA significantly inhibited the increased GFAP expression induced by hypoxia, suggesting that ER stress leads to autophagy, which then leads to astrocyte activation. In our current study, methamphetamine treatment also induced ER stress, and the ER stress inhibitor salubrinal inhibited autophagy and the activation of astrocytes induced by methamphetamine (Fig. 11A and B). However, pretreatment of astrocytes with the autophagy inducer rapamycin, but not the autophagy inhibitor 3-MA, inhibited GFAP expression induced by methamphetamine (Fig. 10C to F). While ER stress lies upstream of autophagy, autophagy in turn responds and inhibits the ER stress as evidenced by the fact that rapamycin treatment inhibited the ER stress induced by methamphetamine (Fig. 11C and D), while the autophagy inhibitor 3-MA enhanced ER stress (Fig. 11E and F). Based on our findings, it is possible to interpret the role of autophagy as a “beneficial guardian” that inhibits ER stress through negative-feedback mechanism as well as the activation of astrocytes induced by methamphetamine. Coregulation of methamphetamine-induced astrocyte activation was found to involve autophagy and the ER stress response. Taken together, these findings suggest that autophagy and ER stress function cooperatively in the regulation of astrocyte activation.
Thus, the results of our current study reveal for the first time that circHIPK2 regulates astrocyte activation by targeting the MIR124–2HG-SIGMAR1 pathway via the interplay between autophagy and ER stress. Our data shed new light on the role of circHIPK2 in astrocyte activation. Specific blockage of circHIPK2 shows potential as a therapeutic target for inhibition of astrocyte activation in the context of drug abuse as well as for the treatment of a broad range of neuroinflammatory disorders.
Methods
Reagents
Lentiviral vectors carrying MIR124–2HG, Anti-MIR124–2HG, circHIPK2 siRNA, MIR124–2HGΔ construct with a mutated sequence that does not target the 3′-UTR of SIGMAR1 (MIR124–2HGΔ) and an adenoviral vector carrying mRFP-GFP-MAP1LC3B (HB-AP2100001) were purchased from HanBio (Shanghai, China). Human circHIPK2 siRNA based on the sequence 5′-GUCCAGAUAUUACAGGUAUTT-3′ was purchased from GenePharma (Shanghai, China) and purified by HPLC. Scrambled nontargeting siRNA (GenePharma, Shanghai, China) with the following sequence was used as negative control: 5′-UUCUCCGAACGUGUCACGUTT-3′. For the lentiviral vectors carrying the construct with a mutated MIR124–2HG (MIR124–2HGΔ, Table 1D) that does not target the 3′-UTR of SIGMAR1, the MIR124–2HG sequence (UAAGGCA) targeting the 3′-UTR of SIGMAR1 was changed to UCGACAC, which was obtained from HanBio. Methamphetamine was obtained from the National Institute for the Control of Pharmaceutical and Biological Products. Control siRNA (sc-37007), human SIGMAR1/OPRS1 siRNA (sc-42250), LPS (L2630), rapamycin (R0395) and 3-MA (M9281) were purchased from Sigma-Aldrich. Salubrinal (SML0951) was from Sigma-Aldrich.
Animals
Sigmar1 KO mice were obtained from the Laboratory Animal Center of University of Science and Technology of China (Hefei, China); these mice were backcrossed for 10 generations to a C57BL/6N inbred background. C57BL/6N mice (male, 6 to 8 wk old) were purchased from the Comparative Medicine Center, Yangzhou University (Yangzhou, China). All animals were housed at a constant temperature and humidity with a 12 h light:12 h dark cycle, with lights on at 07:00. Food and water were available ad libitum. The animals were deeply anesthetized by an overdose of isoflurane, followed by pneumothorax, before perfusion. All animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee of the Medical School, Southeast University.
Cell cultures
Primary mouse astrocytes were obtained from postnatal (P1 to P2) C57BL/6N mice. Whole brains were removed from the mice and were then dissected and mechanically dissociated using gauze to remove membranes and large blood vessels. Next, the dissected brain cortices were placed in medium supplemented with phosphate-buffered serum (PBS; 2.96 mM Na2HPO4·7H2O, 1.05 mM KH2PO4, 155.17 mM NaCl, pH 7.4), and the brain tissues were digested with trypsin-EDTA (Gibco, 25200056). Subsequently, the cells were plated on poly-L-lysine precoated cell culture flasks containing Dulbecco's modified Eagle's medium (DMEM; Corning, 32016001) supplemented with fetal bovine serum (10% v/v) and penicillin/streptomycin (1% v/v). The cultures were maintained in a humidified chamber (37 °C, 5% CO2 incubator). After 7 to 10 days, the astrocytes were harvested by trypsinization.
The human astrocytoma cell line A172 (ATCC® CRL1620™) was obtained from the China Center for Type Culture Collection, routinely maintained in DMEM (10% fetal bovine serum and 1% penicillin/streptomycin) and incubated at 37 °C and 5% CO2.
Luciferase activity assays
The 3′-UTR of the 945-base pair human SIGMAR1 gene containing the putative MIR124–2HG target site was PCR-amplified from human genomic DNA using forward (5′-GCGCTCGAGCTCACCACCTACCTCT-3′) and reverse (5′-AATGCGGCCGCGGAACAGATCAACAAATCT-3′) primers, and the DNA fragment was cloned into the XhoI and NotI sites on the 3′-end of the luc2 gene in a pmiR-RB-REPORT™ vector (RiboBio). For the pmiR-RB-SIGMAR1–3′-UTR-MIR124–2HG-target-mutant vector, the MIR124–2HG target site (TGCCTTA) within the SIGMAR1 3′-UTR was changed to ACGGAAT by PCR mutagenesis with the primers SIGMAR1-MIR124–2HGT-F (5′-TCACCATAACGGAATTCCCCATTCTACTCCCCT-3′) and SIGMAR1-MIR124–2HGT-R (5′-AATGGGGAATTCCGTTATGGTGAGGACAGGGGA-3′). Briefly, HEK293T cells were transfected with a MIR124–2HG overexpression pLV-[hsa-MIR124–2HG] vector (RiboBio) and a target plasmid, pmiR-RB-SIGMAR1–3′-UTR or pmiR-RB-SIGMAR1–3′-UTR-MIR124–2HG-target-mutant, at a molar ratio of 50:1. A miR control pLV-[MIR control] plasmid was used as a negative control. Luciferase activity was determined 24 h post-transfection, and the reporter assay was performed following the manufacturer's protocol (Promega, E2920). Renilla luciferase activity was normalized to firefly luciferase activity and expressed as a percentage of the control (n>3, determined from 5 wells each sample).
Western blot analysis
Proteins were extracted in RIPA lysis buffer (Beyotime, P0013B). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%) and electrophoretically transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline with Tween-20 (Aladdin, T104863), probed with antibodies overnight at 4°C and then incubated with a horseradish peroxidase-conjugated goat anti-mouse (ZSGB-BIO, ZB5305) or rabbit (ZSGB-BIO, ZB5301) IgG secondary antibody (1:2000). The antibodies used were as follows: anti-GFAP (G3893), obtained from Sigma-Aldrich; anti-SIGMAR1/OPRS1 (sc137075), anti-ACTB/β-actin (sc1616), anti-EIF2AK3 (sc13073), anti-ERN1 (sc20790), anti-ATG5 (sc33210) and anti-ATF6 (sc166659), purchased from Santa Cruz Biotechnology; anti-SQSTM1 (18420), anti-BECN1 (11306), anti-MAP1LC3B (14600), anti-GAPDH (60004) and anti-DDIT3 (15204), acquired from Proteintech; and anti-EIF2S1 (5324s), obtained from Cell Signaling Technology. Detection was performed using a MicroChemi 4.2® (DNR, Israel) digital image scanner. Band intensity was quantified using ImageJ software (NIH).
Real-time PCR
Total RNA was isolated from cells and subjected to reverse transcription using a PrimeScript RT Master Mix Kit (TaKaRa, RR036). Real-time PCR was performed by StepOne™ Real-Time PCR System (Life Technologies, 4376357, Singapore) using the following primers: human divergent circHIPK2 (forward primer: 5′-CTGTGTGCTCCACCTACTTG-3′; reverse primer: 5′-TACCCAGTCATGTCCCAGTTG-3′), human GAPDH (forward primer: 5′-ACCATCTTCCAGGAGCGAGAT-3′; reverse primer: 5′-GGGCAGAGATGATGACCCTTT-3′), mouse divergent circHIPK2 (forward primer: 5′-GACAACCGTACCGAGTGAAG-3′; reverse primer: 5′-GTGTGAGGGGAGAAAACTTGC-3′), mouse circHIPK2 convergent primer (forward: 5′-GCAAGTTTTCTCCCCTCACAC-3′; reverse: 5′-CTTCACTCGGTACGGTTGTC-3′), mouse divergent Gapdh (forward primer: 5′-AGGTCGGTGTGAACGGATTTG-3′; reverse primer: 5′-GGGGTCGTTGATGGCAACA-3′) and mouse Gapdh convergent primer (forward: 5′-AGGTCGGTGTGAACGGATTTG-3′; reverse: 5′-GGGGTCGTTGATGGCAACA-3′). Relative quantification was performed using TaKaRa SYBR® Premix Taq™ (TbI RNase H Plus; TaKaRa, RR420). The sequences for knockdown of circHIPK2 expression: human circHIPK2 siRNA: 5′-GUCCAGAUAUUACAGGUAUTT-3′; mouse circHIPK2 siRNA 1: 5′-UACCGGUAUGGCCUCACAUTT-3′; mouse circHIPK2 siRNA 2: 5′-UCCAGAUACUACCGGUAUGTT-3′. Specific primers and probes for mature MIR124–2HG and RNU6–6P/RNU6B were obtained from RiboBio. All reactions were run in triplicate. The amount of MIR124–2HG was determined by normalizing to the level of RNU6–6P relative to that in the control as previously reported.54
Fluorescence in situ hybridization (FISH)
According to our previous study,55 primary mouse astrocytes cultured in coverslips were fixed with 4% paraformaldehyde for 20 min and incubated in PBS overnight at 4°C followed by the detection of circHIPK2 and MIR124–2HG expression. The cells were permeabilized with 0.25% Triton X-100 (Aladdin, T109027) in PBS for 15 min, prehybridized in hybridization buffer (50% formamide, 10 mM Tris-HCl, pH 8.0, 200 μg/ml yeast tRNA [Sigma-Aldrich, 15401–011], 1X Denhardt solution [Sigma-Aldrich, 30915], 600 mM NaCl, 0.25% SDS [Invitrogen, 15553–035], 1 mM EDTA, 10% dextran sulfate [Sigma-Aldrich, D8906]) for 1 h at 37°C. Then, the coverslips were heated to 65°C for 5 min in hybridization buffer containing 2 nM of a commercially available digoxigenin-labeled MIR124–2HG probe (Exiqon, Table 1E) and 500 nM of a commercially available biotin-labeled circHIPK2 probe (Invitrogen) hybridization occurred at 37°C overnight. The next day, the coverslips were washed 3 times in 2X SSC (10% v/v 20X SSC [Invitrogen, 15557] in DEPC-treated water [Vetec, V900882]) and twice in 0.2X SSC(10% v/v 2X SSC in DEPC-treated water) at 42°C and then blocked with 1% BSA (Biosharp, BS043D), 3% normal goat serum (ZSGB-BIO, ZLI-0956) in PBS for 1 h at room temperature. They were then incubated with a horseradish peroxidase-conjugated anti-digoxigenin antibody (1:200; Roche Diagnostics GmbH, 11207733910) and FITC-streptavidin (1:200; Life Technology, 434311) overnight at 4°C. After the coverslips were washed 3 times with TBS (0.1 M Tris, 0.308 M NaCl, pH 7.4), they were incubated with a TSA Cy5 kit (PerkinElmer, NEL766B001KT) for 10 min at room temperature. Then, the coverslips were washed twice with PBS and mounted with Prolong gold anti-fade reagent containing DAPI (SouthernBiotech, 0100–20). In the FISH experiments, the specificity of the circHIPK2 and MIR124–2HG signal was confirmed via comparison with a scrambled control. Unlike the circHIPK2 and MIR124–2HG probe, the scrambled probe showed no signal in primary mouse astrocytes.
Affinity-isolation assay with biotinylated MIR124–2HG
A total of 2 × 106 HEK 293T cells were seeded the day before transfection. On the following day, the cells were transfected with 3′-end biotinylated MIR124–2HG or MIR124–2HGΔ at a final concentration of 50 nM for 36 h. Then, the cells were washed with PBS, briefly vortexed and incubated in lysis buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.05% Igepal [Sigma, I3021], 60 U/ml Superase-In [Ambion, AM2694], 1 mM DTT [Sigma, 43816] and protease inhibitors [Biotool, B14001]) on ice for 10 min. Lysates were precleared by centrifugation, and 50 μl aliquots of the samples were prepared for input. The remaining lysates were incubated with M-280 streptavidin magnetic beads (Invitrogen, 11205D). The beads were coated with yeast tRNA in advance to prevent the nonspecific binding of RNA and protein complexes. The beads were incubated at 4°C for 2.5 h and were then washed twice with ice-cold lysis buffer, twice with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl) and once with high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 500 mM NaCl). The bound RNAs were purified using Trizol (TaKaRa, 9109) for measurement of circHIPK2 levels. The biotinylated MIR124–2HG at the 3′-end was synthesized by GenePharma (Shanghai, China), and its sequence was 5′-UAAGGCACGCGGUGAAUGCC-3′. The MIR124–2HGΔ sequence was 5′-UCGACACCGCGGUGAAUGCC-3′ and also biotinylated at the 3′-end.
Affinity-isolation assay with DNA probe
The biotinylated DNA probe complementary to circHIPK2 was synthesized and dissolved in 500 μl of wash/binding buffer (0.5 M NaCl, 20 mM Tris-HCl, pH 7.5, 1 mM EDTA). The probes were incubated with streptavidin-coated magnetic beads (Sigma, 08014) at 25°C for 2 h to generate probe-coated magnetic beads. Cardiomyocyte lysates were incubated with probe-coated beads, and after washing with the wash/binding buffer, the RNA complexes bound to the beads were eluted and extracted for realtime PCR. The following primer sequences were used: circHIPK2 affinity-isolation probe, AAAACATGTGAGGCCATACCTGTAATATCTGGACT; and random affinity-isolation probe, AAACAGTACTGGTGTGTAGTACGAGCTGAAGCTAC.
Transduction of astrocytes with lentivirus
Astrocytes were transduced with MIR control, MIR124–2HG, Anti-MIR control, Anti-MIR124–2HG, circ control siRNA and circHIPK2 siRNA lentivirus with multiplicity of infection of 0.5 (primary mouse astrocytes) or 10 (A172 cells), followed by gentle swirling, incubation and replacement of fresh feed medium.
Microinjection of Anti-MIR124–2HG or circHIPK2 siRNA lentivirus
The hippocampi of 8-wk-old C57BL/6 mice were microinjected bilaterally with an Anti-MIR control-RFP, Anti-MIR124–2HG-RFP, circ control siRNA-GFP or circHIPK2 siRNA-GFP lentivirus (2 μl of 109 viral genomes µl−1, HanBio) using the following microinjection coordinates: 2.06 mm behind the bregma and ± 1.5 mm lateral from the sagittal midline at a depth of 2 mm from skull surface. To evaluate the effect of Anti-MIR124–2HG on astrocyte activation in WT and sigmar1 KO mice, 12 mice (n = 6/group; male) were perfused and sectioned 2 wk after lentivirus microinjection, followed by GFAP staining to assess astrocyte activation. To evaluate the effect of circHIPK2 siRNA on methamphetamine/LPS-induced astrocyte activation, 18 mice were divided into 3 groups (n = 6/group; male) 2 wk after lentivirus microinjection and treated with saline, methamphetamine (30 mg/kg, injected every 2 h for a total of 4 times, intraperitoneally [i.p.]) for 24 h or LPS (20 mg/kg, i.p.) for 4 h. Then, the animals were perfused and sectioned, followed by GFAP staining to assess astrocyte activation.
Immunostaining and image analysis
As described in our previous study,55 the sections encompassing the entire hippocampus were cut into 35 μm slices with a cryostat. Then, the sections were incubated with 0.3% Triton X-100 in PBS for 15 min and blocked with 10% normal goat serum in 0.3% Triton X-100 for 1 h at room temperature. Next, the sections were incubated with a mouse anti-GFAP antibody (Sigma-Aldrich, G3893) overnight at 4°C. On the following day, the sections were washed and incubated with Alexa Fluor 488-conjugated anti-mouse IgG (1:250; Invitrogen, A11001) or Alexa Fluor 594 goat anti-mouse IgG (1:250; Invitrogen, A11005) for 1 h to detect GFAP. After a final washing step with PBS, the sections were mounted onto glass slides, and ProLong gold Anti-fade reagent containing DAPI (SouthernBiotech, 0100–20) was applied for visualization of nuclei. Immunofluorescence images were captured by microscopy (Olympus DP73, Olympus, Tokyo, Japan). Average intensities of GFAP were calculated using ImageJ by sampling of a 28 × 28 pixel area, and 36 images were captured from 6 consecutive sections. The values were reported as the average intensity above the background ± SD.
Transmission electron microscopy
Cells were cultured in 24-well plates and treated with 100 μM methamphetamine. After incubation of the cells with methamphetamine for 24 h, the treated cells from each sample were harvested in a 1.5 ml microcentrifuge tube. The cells were washed once with cold PBS and fixed in a mixture of 2% glutaraldehyde and 2% formaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) at 4°C overnight. Then, they were washed 4 times in 0.1 M sodium cacodylate buffer (pH 7.4) containing 264 mM sucrose (Sinopharm, 10021418) before being transferred to 1% osmium tetroxide in 0.1 M sodium cacodylate (pH 7.4) for 2 h. The cells were then washed 3 times in double distilled H2O and stained in 2% aqueous uranyl acetate for 2 h. Next, they were dehydrated in an ethanol series, infiltrated in propylene oxide (Aladdin, R101941) 2 times and embedded in Epon 812 (SPI Science, 90529–77–4). Ultrathin sections (90-nm thick) were cut using a Leica EM UC7 ultramicrotome (Ser. No: 595915, Austria) with a diamond knife. Grids were poststained with 2% saturated uranyl acetate in 50% ethanol and 1% lead citrate (pH 12) and examined using an H-7650 electron microscope (Hitachi, Tokyo, Japan), and images were captured with a Ganton 830 digital camera.
Tandem fluorescent-mRFP-GFP-MAP1LC3B-adenovirus transduction of A172 cells
A172 cells were transfected with a tandem fluorescent-mRFP-GFP-MAP1LC3B-adenovirus (HanBio, HB-AP2100001) that expresses a specific marker of autophagosome formation to detect autophagy, according to the manufacturer's instructions.56 Five fields were chosen from 3 different cell preparations. GFP- and mRFP-expressing spots, which were indicated by fluorescent puncta and DAPI-stained nuclei were counted manually. The number of spots per cell was determined by dividing the total number of spots by the number of nuclei in each field.
Rapamycin administration
Rapamycin (LC Laboratories, R-5000) was dissolved in DMSO (25 mg/ml). For animal injections, the stock solution was diluted immediately before i.p. injection with 100 μl water containing 5% polyethylene glycol 400 (Damas-β, 41713B) and 5% Tween-80 (Aladdin, T104866). Mice received rapamycin (5 mg/kg, i.p.) 24 h before the last methamphetamine injection, and control mice received the drug vehicle.
Statistical analyses
Statistical analysis was performed using the Student t test or one-way analysis of variance (ANOVA) followed by the Holm-Sidak test (SigmaPlot 11.0). The tests used are indicated in the figure legends. The ANOVA results were considered significant at a P < 0.05.
Supplementary Material
Abbreviations
- 3-MA
3-methyladenine
- ACTB
actin β
- ATG5
autophagy-related 5
- BBB
blood-brain barrier
- BECN1
Beclin 1, autophagy related
- CNS
central nervous system
- circRNA
circular RNA
- circHIPK2
circular RNA HIPK2
- DAPI
4′,6-diamidino-2-phenylindole
- DDIT3
DNA damage inducible transcript 3
- ER
endoplasmic reticulum
- EIF2AK3
eukaryotic translation initiation factor 2 α kinase 3
- EIF2S1
eukaryotic translation initiation factor 2 subunit α
- ERN1
endoplasmic reticulum to nucleus signaling 1
- FISH
fluorescence in situ hybridization
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFAP
glial fibrillary acidic protein
- KO
knockout
- LPS
lipopolysaccharide
- LV
lentivirus
- MAP1LC3B
microtubule-associated protein 1 light chain 3 β
- Meth
methamphetamine
- MIR124–2HG
MIR124–2 host gene
- miRNAs
microRNAs
- MTOR
mechanistic target of rapamycin (serine/threonine kinase)
- OE
overexpression
- SIGMAR1/OPRS1
sigma non-opioid intracellular receptor 1
- PCR
polymerase chain reaction
- SQSTM1
sequestosome 1
- UTR
untranslated region
- WT
wild type
Disclosure of potential conflicts of interest
The authors declare that there are no competing financial interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81322048, No. 81673410 and No. 81603090), the Jiangsu Specially Appointed Professor and the Major State Basic Research Development Program of China (973 Program) (2013CB733800 and 2013CB733803).
References
- [1].Shirakawa H, Sakimoto S, Nakao K, Sugishita A, Konno M, Iida S, Kusano A, Hashimoto E, Nakagawa T, Kaneko S. Transient receptor potential canonical 3 (TRPC3) mediates thrombin-induced astrocyte activation and upregulates its own expression in cortical astrocytes. J Neurosci. 2010;30(39):13116-29. doi: 10.1523/JNEUROSCI.1890-10.2010. PMID:20881130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Williams A, Piaton G, Lubetzki C. Astrocytes–friends or foes in multiple sclerosis? Glia. 2007;55(13):1300-12. doi: 10.1002/glia.20546. PMID:17626262 [DOI] [PubMed] [Google Scholar]
- [3].Iadecola C, Nedergaard M. Glial regulation of the cerebral microvasculature. Nat Neurosci. 2007;10(11):1369-76. doi: 10.1038/nn2003. PMID:17965657 [DOI] [PubMed] [Google Scholar]
- [4].Dong Y, Benveniste EN. Immune function of astrocytes. Glia. 2001;36(2):180-90. doi: 10.1002/glia.1107. PMID:11596126 [DOI] [PubMed] [Google Scholar]
- [5].Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32(12):638-47. doi: 10.1016/j.tins.2009.08.002. PMID:19782411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nair A, Frederick TJ, Miller SD. Astrocytes in multiple sclerosis: A product of their environment. Cell Mol Life Sci. 2008;65(17):2702-20. doi: 10.1007/s00018-008-8059-5. PMID:18516496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yan Y, Ding X, Li K, Ciric B, Wu S, Xu H, Gran B, Rostami A, Zhang GX. CNS-specific therapy for ongoing EAE by silencing IL-17 pathway in astrocytes. Mol Ther. 2012;20(7):1338-48. doi: 10.1038/mt.2012.12. PMID:22434134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rossi DJ, Brady JD, Mohr C. Astrocyte metabolism and signaling during brain ischemia. Nat Neurosci. 2007;10(11):1377-86. doi: 10.1038/nn2004. PMID:17965658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shao W, Zhang SZ, Tang M, Zhang XH, Zhou Z, Yin YQ, Zhou QB, Huang YY, Liu YJ, Wawrousek E, et al.. Suppression of neuroinflammation by astrocytic dopamine D2 receptors via alphaB-crystallin. Nature. 2013;494(7435):90-4. doi: 10.1038/nature11748. PMID:23242137 [DOI] [PubMed] [Google Scholar]
- [10].Halliday GM, Stevens CH. Glia: Initiators and progressors of pathology in Parkinson's disease. Mov Disord. 2011;26(1):6-17. doi: 10.1002/mds.23455. PMID:21322014 [DOI] [PubMed] [Google Scholar]
- [11].Verkhratsky A, Olabarria M, Noristani HN, Yeh CY, Rodriguez JJ. Astrocytes in Alzheimer's disease. Neurotherapeutics. 2010;7(4):399-412. doi: 10.1016/j.nurt.2010.05.017. PMID:20880504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rodriguez JJ, Olabarria M, Chvatal A, Verkhratsky A. Astroglia in dementia and Alzheimer's disease. Cell Death Differ. 2009;16(3):378-85. doi: 10.1038/cdd.2008.172. PMID:19057621 [DOI] [PubMed] [Google Scholar]
- [13].Zhang Y, Zhu T, Zhang X, Chao J, Hu G, Yao H. Role of high-mobility group box 1 in methamphetamine-induced activation and migration of astrocytes. J Neuroinflammation. 2015;12:156. doi: 10.1186/s12974-015-0374-9. PMID:26337661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lau JW, Senok S, Stadlin A. Methamphetamine-induced oxidative stress in cultured mouse astrocytes. Ann N Y Acad Sci. 2000;914:146-56. doi: 10.1111/j.1749-6632.2000.tb05192.x. PMID:11085317 [DOI] [PubMed] [Google Scholar]
- [15].Clark KH, Wiley CA, Bradberry CW. Psychostimulant abuse and neuroinflammation: Emerging evidence of their interconnection. Neurotox Res. 2013;23(2):174-88. doi: 10.1007/s12640-012-9334-7. PMID:22714667 [DOI] [PubMed] [Google Scholar]
- [16].Pu C, Vorhees CV. Developmental dissociation of methamphetamine-induced depletion of dopaminergic terminals and astrocyte reaction in rat striatum. Brain Res Dev Brain Res. 1993;72(2):325-8. doi: 10.1016/0165-3806(93)90201-K. PMID:8097974 [DOI] [PubMed] [Google Scholar]
- [17].Robson MJ, Turner RC, Naser ZJ, McCurdy CR, O'Callaghan JP, Huber JD, Matsumoto RR. SN79, a sigma receptor antagonist, attenuates methamphetamine-induced astrogliosis through a blockade of OSMR/gp130 signaling and STAT3 phosphorylation. Exp Neurol. 2014;254:180-9. doi: 10.1016/j.expneurol.2014.01.020. PMID:24508558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shah A, Kumar S, Simon SD, Singh DP, Kumar A. HIV gp120- and methamphetamine-mediated oxidative stress induces astrocyte apoptosis via cytochrome P450 2E1. Cell Death Dis. 2013;4:e850. doi: 10.1038/cddis.2013.374. PMID:24113184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shah A, Silverstein PS, Singh DP, Kumar A. Involvement of metabotropic glutamate receptor 5, AKT/PI3K signaling and NF-kappaB pathway in methamphetamine-mediated increase in IL-6 and IL-8 expression in astrocytes. J Neuroinflammation. 2012;9:52. doi: 10.1186/1742-2094-9-52. PMID:22420994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Colombo E, Farina C. Astrocytes: Key regulators of neuroinflammation. Trends Immunol. 2016;37(9):608-20. doi: 10.1016/j.it.2016.06.006. PMID:27443914 [DOI] [PubMed] [Google Scholar]
- [21].Argaw AT, Asp L, Zhang J, Navrazhina K, Pham T, Mariani JN, Mahase S, Dutta DJ, Seto J, Kramer EG, et al.. Astrocyte-derived VEGF-A drives blood-brain barrier disruption in CNS inflammatory disease. J Clin Invest. 2012;122(7):2454-68. doi: 10.1172/JCI60842. PMID:22653056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sharkey J, Glen KA, Wolfe S, Kuhar MJ. Cocaine binding at sigma receptors. Eur J Pharmacol. 1988;149(1-2):171-4. doi:0014-2999(88)90058-1. PMID:2840298 [DOI] [PubMed] [Google Scholar]
- [23].Su TP, London ED, Jaffe JH. Steroid binding at sigma receptors suggests a link between endocrine, nervous, and immune systems. Science. 1988;240(4849):219-21. doi: 10.1126/science.2832949. PMID:2832949 [DOI] [PubMed] [Google Scholar]
- [24].Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769-73. doi: 10.1038/nature03315. PMID:15685193 [DOI] [PubMed] [Google Scholar]
- [25].Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12(4):399-408. doi: 10.1038/nn.2294. PMID:19287386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27(3):435-48. doi: 10.1016/j.molcel.2007.07.015. PMID:17679093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314(14):2618-33. doi: 10.1016/j.yexcr.2008.06.002. PMID:18619591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011;17(1):64-70. doi: 10.1038/nm.2266. PMID:21131957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hamzei Taj S, Kho W, Riou A, Wiedermann D, Hoehn M. MiRNA-124 induces neuroprotection and functional improvement after focal cerebral ischemia. Biomaterials. 2016;91:151-65. doi: 10.1016/j.biomaterials.2016.03.025. PMID:27031810 [DOI] [PubMed] [Google Scholar]
- [30].Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453-61. doi: 10.1038/nbt.2890. PMID:24811520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333-8. doi: 10.1038/nature11928. PMID:23446348 [DOI] [PubMed] [Google Scholar]
- [32].Li X, Wu Q, Xie Y, Ding Y, Du WW, Sdiri M, Yang BB. Ergosterol purified from medicinal mushroom Amauroderma rude inhibits cancer growth in vitro and in vivo by up-regulating multiple tumor suppressors. Oncotarget. 2015;6(19):17832-46. doi: 10.18632/oncotarget.4026. PMID:26098777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Boeckel JN, Jae N, Heumuller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res. 2015;117(10):884-90. doi: 10.1161/CIRCRESAHA.115.306319. PMID:26377962 [DOI] [PubMed] [Google Scholar]
- [34].Yin TP, Cai L, Xing Y, Yu J, Li XJ, Mei RF, Ding ZT. Alkaloids with antioxidant activities from Aconitum handelianum. J Asian Nat Prod Res. 2016;18(6):603-10. doi: 10.1080/10286020.2015.1114473. PMID:26744060 [DOI] [PubMed] [Google Scholar]
- [35].Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al.. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. PMID:27050392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2016;38(18):1402-12. doi: 10.1093/eurheartj/ehw001. PMID:26873092 [DOI] [PubMed] [Google Scholar]
- [37].Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al.. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur heart J. 2016;37(33):2602-11. doi: 10.1093/eurheartj/ehv713. PMID:26802132 [DOI] [PubMed] [Google Scholar]
- [38].Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3(3):181-206. doi: 10.4161/auto.3678. PMID:17224625 [DOI] [PubMed] [Google Scholar]
- [39].Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, et al.. Classification of cell death: Recommendations of the nomenclature committee on cell death 2009. Cell Death Differ. 2009;16(1):3-11. doi: 10.1038/cdd.2008.150. PMID:18846107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kitamura O, Takeichi T, Wang EL, Tokunaga I, Ishigami A, Kubo S. Microglial and astrocytic changes in the striatum of methamphetamine abusers. Leg Med (Tokyo). 2010;12(2):57-62. doi: 10.1016/j.legalmed.2009.11.001. PMID:20110187 [DOI] [PubMed] [Google Scholar]
- [41].Deng B, Zhang Y, Zhang S, Wen F, Miao Y, Guo K. MicroRNA-142-3p inhibits cell proliferation and invasion of cervical cancer cells by targeting FZD7. Tumour Biol. 2015;36(10):8065-73. doi: 10.1007/s13277-015-3483-2. PMID:25976503 [DOI] [PubMed] [Google Scholar]
- [42].Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141-57. doi: 10.1261/rna.035667.112. PMID:23249747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. PMID:24039610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3-14. doi: 10.1016/j.addr.2015.05.001. PMID:25979468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang F, Wang B, Long H, Yu J, Li F, Hou H, Yang Q. Decreased miR-124-3p expression prompted breast cancer cell progression mainly by targeting beclin-1. Clin Lab. 2016;62(6):1139-45. PMID:27468577 [DOI] [PubMed] [Google Scholar]
- [46].Cai Y, Arikkath J, Yang L, Guo ML, Periyasamy P, Buch S. Interplay of endoplasmic reticulum stress and autophagy in neurodegenerative disorders. Autophagy. 2016;12(2):225-44. doi: 10.1080/15548627.2015.1121360. PMID:26902584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Goldshmit Y, Kanner S, Zacs M, Frisca F, Pinto AR, Currie PD, Pinkas-Kramarski R. Rapamycin increases neuronal survival, reduces inflammation and astrocyte proliferation after spinal cord injury. Mol Cell Neurosci. 2015;68:82-91. doi: 10.1016/j.mcn.2015.04.006. PMID:25936601 [DOI] [PubMed] [Google Scholar]
- [48].Fan Y, He JJ. HIV-1 Tat induces unfolded protein response and endoplasmic reticulum stress in astrocytes and causes neurotoxicity through glial fibrillary acidic protein (GFAP) activation and aggregation. J Biol Chem. 2016;291(43):22819-29. doi: 10.1074/jbc.M116.731828. PMID:27609520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1-222. doi: 10.1080/15548627.2015.1100356. PMID:26799652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Leask A. CCN6: A modulator of breast cancer progression. J Cell Commun Signal. 2016;10(2):163-4. doi: 10.1007/s12079-016-0321-2. PMID:27086280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Go BS, Kim J, Yang JH, Choe ES. Psychostimulant-induced endoplasmic reticulum stress and neurodegeneration. Mol Neurobiol. 2016;54(6):4041-48. doi: 10.1007/s12035-016-9969-0. PMID:27314686 [DOI] [PubMed] [Google Scholar]
- [52].Ciechomska IA, Kaminska B. ER stress and autophagy contribute to CsA-induced death of malignant glioma cells. Autophagy. 2012;8(10):1526-8. doi: 10.4161/auto.21155. PMID:22910018 [DOI] [PubMed] [Google Scholar]
- [53].Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, et al.. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119(5):1359-72. doi: 10.1172/JCI37948. PMID:19425170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hu G, Zhou R, Liu J, Gong AY, Chen XM. MicroRNA-98 and let-7 regulate expression of suppressor of cytokine signaling 4 in biliary epithelial cells in response to Cryptosporidium parvum infection. J Infect Dis. 2010;202(1):125-35. doi: 10.1086/653212. PMID:20486857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yao H, Ma R, Yang L, Hu G, Chen X, Duan M, Kook Y, Niu F, Liao K, Fu M, et al.. MiR-9 promotes microglial activation by targeting MCPIP1. Nat Commun. 2014;5:4386. doi: 10.1038/ncomms5386. PMID:25019481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hariharan N, Zhai P, Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid Redox Signal. 2011;14(11):2179-90. doi: 10.1089/ars.2010.3488. PMID:20812860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


