Standfirst
Sequencing methods based on electron tunnelling could lead to breakthroughs in genomics, proteomics and glycomics, but the engineering challenges involved in delivering these devices are formidable.
It is over 20 years since the first proposal was made to sequence DNA by measuring variations in ion current flowing through a nanopore as a single molecule of DNA is driven through the pore by electrophoresis1. Such were the technical challenges involved in this approach that working devices have only recently appeared2: a commercially-engineered version of this technology – the “MinION” from Oxford Nanopore Technologies – uses a protein nanopore and has shipped to hundreds of labs. Users of the MinION have generated reads of up to 100 kilobases in length with per-base read accuracies in excess of 90%3. This is still well below the accuracy of Sanger sequencing but comparable to other single-molecule methods such as the SMRT sequencing from Pacific Biosciences. Given the success of this remarkable low-cost technology, why is there a strong interest in exploring alternative single-molecule DNA sequencing methods and, in particular, developing physical detection methods that use solid-state devices4,5?
One reason for this is that the physics of ion-current flow through a small pore imposes a fundamental limit on resolution. The electric field near a pore of diameter d extends for about a distance d on each side of the pore6. In order to pass single-stranded DNA, the diameter of the pore must be at least 1.4 nm (ref.7), so even pores in atomically-thin membranes (such as those made of graphene, which are discussed in the Review by Stephanie Heerema and Cees Dekker4) will be sensitive to the DNA bases that lie outside the pore on each side (Fig. 1a). However, despite the fact that several bases contribute to the size of the ion current at any one time, a clever analysis can unfold the underlying sequence by analysing the changes in current signals as many bases pass the pore8,9. Graphene membranes would make this process much easier10. Nonetheless, the lack of single-base resolution complicates reads of homopolymer sequences and also the identification of epigenetically-modified bases. In addition, further improvements in accuracy are needed for the reliable detection of single nucleotide polymorphisms in a human genome.
Figure 1.
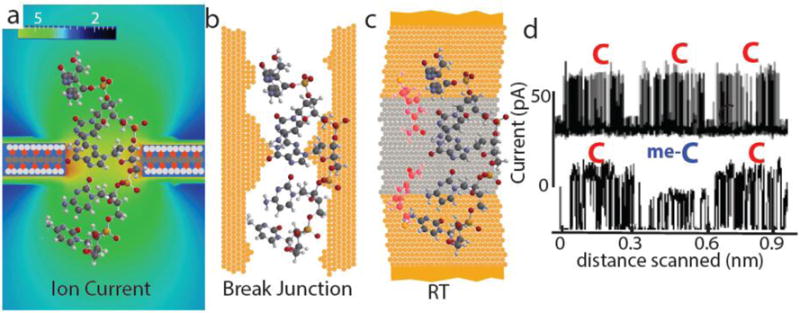
Three physical approaches to sequencing a moving DNA molecule. a, Sequencing by sensing changes in ion current passing through a nanopore as a DNA molecule translocates through the pore. The electric field around a nanopore (colour bar at top - units are MV/m) extends above and below the pore, so even in a thin membrane DNA bases adjacent to the pore lie in regions of relatively high current, limiting resolution. b, A mechanical break junction in which a 0.8 nm gap in a freshly-broken gold wire contacts a DNA base directly, and bases are assigned by means of the tunnel current that flows across them. In this case, the entire DNA cannot be drawn through the gap, so the sequence must be built up statistically by sampling portions of the molecule that land on the gap. c, In recognition tunnelling (RT) only bases contacted simultaneously by recognition molecules (red) attached to the electrodes generate a signal. By choosing recognition molecules with a high electronic conductance, the gap can be increased to 2.5 nm. In one embodiment of RT, a nanopore is drilled through a metal-dielectric-metal stack so the DNA sequence is read by bonding events that bridge the gap between the two electrodes. d, RT can read homopolymer sequences (top) and epigentically-modified bases (me-C is 5-methylcytosine). The signal consists of a series of randomly-timed current spikes, but the transitions from one base to another are clear as dips in the train of spikes.
Aware of these problems, Michael Zwolak and Max Di Ventra11 proposed measuring electron-tunnelling current flowing transverse to the long axis of the DNA (an approach discussed in the Review by Di Ventra and Masateru Taniguchi5). Molecular tunnelling requires good chemical contacts between the molecule and metal electrodes12, and this is difficult to achieve given the rapid accumulation of hydrocarbons on exposed metal surfaces. Taniguchi and colleagues solved this problem by preparing freshly-broken gold gaps that are small enough (0.8 nm) to contact a DNA base directly. (Such a tunnel junction is shown approximately to scale in Fig. 1b.) Clearly, single-stranded DNA, with a diameter of 1.4 nm, cannot enter a 0.8 nm gap. Therefore, it is not possible to pull DNA through the break-junction gap to ensure sequential reading of the bases. Sequencing can be carried out using “stochastic traps”, which means random sampling of the DNA as it lands on, and diffuses over, the junction5. However, it is not clear how this sampling of small fragments could be extended to compete with the 100 kilobase single-molecule reads that are currently achieved using ion-current sensing nanopores.
In order to address this limitation, my group developed recognition tunnelling (RT; the scanning tunnelling microscope implementations of this approach are also described in the Review by Di Ventra and Taniguchi5). In RT, recognition molecules (Fig. 1c, in red), chemically-bound to the electrodes, are used to make good, but non-covalent, contacts to the target molecules. The on-rate for the formation of complexes (such as those shown in Fig.1c) is rapid enough that each base should be captured in turn even with sub-millisecond intervals between bases passing through the junction13. By designing the recognition molecules with the appropriate electronic properties, the size of the reading gap can be increased to 2.5 nm (ref.14): large enough to let single stranded DNA pass and more manufacturable than smaller gaps. In particular, a structure made from metal-dielectric-metal stacks (Fig. 1c) can be readily manufactured on a wafer-scale and can read single nucleotides with high accuracy15.
Using a scanning tunnelling microscope (STM) to achieve directed motion of the molecule with respect to the junction, RT can read homopolymer sequences and epigenetic modifications directly and accurately (Fig. 1d). STM read lengths are, however, short due to the need to track the axis of the polymer through regions of secondary structure where the DNA is folded13. With solid-state tunnel junctions based on the metal-dielectric-metal design, sequencing requires that a small a small diameter (~ 2 nm) aperture be drilled through the layers to direct the DNA over the electrodes in the appropriate geometry (illustrated in Fig. 1c). Drilling these apertures without damaging the electrode structure is a challenge. Large (micron-sized) holes made by selective reactive ion etching (RIE) work very well15 for identifying individual DNA nucleotides. Careful electron-beam lithography has also allowed nanopores as small as 20 nm diameter to be made by RIE in a scalable way16. However, because the geometry of DNA passing through 20 nm apertures is not sufficiently constrained, long DNA molecules produce signals that are reads of the sequence at randomly-selected positions, much like the stochastic sampling described by Di Ventra and Taniguchi5.
In this regard, tunnelling readout is in much the same state that protein-pore based ion-current devices were before Mark Akeson and colleagues introduced molecular control of the motion of DNA into the pore17. In order to follow this approach with solid state pores, it will be necessary to make pores that are smaller than the dimensions of a motor protein (i.e. about 5 nm in diameter) so that an enzyme that controls the DNA motion is trapped against the nanopore. The current methods for making small pores, like electron-beam drilling18 or electrochemical breakdown19, are neither scalable nor gentle enough. Finding ways to make small pores, while leaving the metal-dielectric-metal junction unscathed, is the focus of current research.
Electron-beam drilling can be used to make very small pores in single metal layers that act as models of the electrodes in the tunnel junctions. This single electrode can be chemically-modified in the same way that the tunnelling devices are, so that the effects of transient binding of the DNA bases to the recognition molecules can be explored. We found that DNA translocation is slowed by nearly a factor of 1000 compared to translocation through bare metal pores20. This should be a significant advantage of RT once a reliable and scalable method is found for making small pores.
Any scheme that seeks to confine DNA into a solid-state nanopore will face manufacturing challenges related to the scalability of methods that sculpt fine features into solid state materials; this applies to the several ingenious graphene sensing structures that are currently being explored4 just as much as it does to the pores in metal-based tunnelling devices. This problem has, of course, been solved by nature, and exploited in the protein pores used in the MinION, where the pore channel is defined by the remarkably stable folding of the protein. Even so, the manufacture of devices like the MinION presented its own major engineering challenges. Anyone who has accidently banged a lab-bench on which a nanopore is operating knows how fragile these devices can be, and yet Oxford Nanopore has found a way to mass-produce these devices and ship them worldwide. It is hard to believe that such engineering ingenuity cannot also be applied to solid-state sequencing devices in the future.
So what of a world in which cheap, massively parallel solid-state sequencing devices are ubiquitous? The single most important aspect of this technology is that physical readouts do not depend on the action of a DNA polymerase, and so they are agnostic about the nature of the molecule to be sequenced. The approach has, for example, already been used to identify amino acids and partially sequence peptides5. As Di Ventra and Taniguchi point out5, there is no analogue of the DNA-polymerase chain reaction to amplify tiny amounts of protein in patient biopsies, so a single-molecule analytical technique that uses only tiny amounts of sample will have a profound impact on proteomics. This is turn will set the stage for significant advances in personalized medicine because the proteome provides a detailed, real-time picture of current health status, as well as a record of past pathologies, and does so in massive detail (there are fewer than 20,000 coding genes but millions of gene products in the many forms of protein that are expressed).
Mass spectrometry dominates proteomics at present, but requires large amounts of sample to analyse rare protein variants. Future single molecule detectors should be readily capable of analysing quantities of sample as small as a femtomole. The impact should be even more profound in glycomics where milligrams of sample are currently required to crack the most difficult glycan structures using nuclear magnetic resonance (NMR), an approach which is not feasible in a clinical setting. I expect that RT will, in the future, be able to identify many important compounds using only femtomoles of sample.
Even without nanopores, the current generation of tunnel junctions15 make very sensitive, label-free single molecule detectors, and practical applications can be expected even before the nanopore manufacturing problem is solved.
Acknowledgments
This work was supported in part by the NHGRI (grant HG 006323), F. Hoffman La Roche and Recognition AnalytiX. Useful discussions with P. Pang, P. Zhang, B. Ashcroft and W. Song are acknowledged.
Footnotes
Competing financial interests
S.L. is a co-founder of Recognition AnalytiX, a company that holds the rights to applications of recognition tunnelling technology in the fields of proteomics and glycomics.
References
- 1.Church G, Deamer DW, Branton D, Baldarelli R, Kasianowicz J. 5,795,782. US patent. 1998
- 2.Manrao EA, et al. Nat Biotechnol. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ip CLC, et al. F1000Research. 2015;4:1075. doi: 10.12688/f1000research.7201.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heerema SJ, Dekker C. Nature Nanotech. 2016;X:XXX–XXX. doi: 10.1038/nnano.2015.307. [DOI] [PubMed] [Google Scholar]
- 5.Di Ventra M, Taniguchi Nature Nanotech. 2016;X:XXX–XXX. doi: 10.1038/nnano.2015.320. [DOI] [PubMed] [Google Scholar]
- 6.Hall JE. J Gen Physiol. 1975;66:531–532. doi: 10.1085/jgp.66.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akahori R, et al. Nanotechnology. 2014;25:275501. doi: 10.1088/0957-4484/25/27/275501. [DOI] [PubMed] [Google Scholar]
- 8.Laszlo AH, et al. Nature Biotech. 2014;32:829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain M, et al. Nature Meth. 2015;12:351–356. doi: 10.1038/nmeth.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garaj S, et al. Nature. 2010;467:190–193. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwolak M, Di Ventra M. Nano Lett. 2005;5:421–424. doi: 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]
- 12.Cui XD, et al. Science. 2001;294:571–574. doi: 10.1126/science.1064354. [DOI] [PubMed] [Google Scholar]
- 13.Chang S, et al. Nanotechnology. 2012;23:235101–235115. doi: 10.1088/0957-4484/23/23/235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang S, He J, Zhang P, Gyarfas B, Lindsay S. J Am Chem Soc. 2011;133:14267–14269. doi: 10.1021/ja2067737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang P, et al. ACS Nano. 2014;8:11994–12003. doi: 10.1021/nn505356g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai J, et al. Nanoscale. 2014;6:8900–8906. doi: 10.1039/c3nr06723h. [DOI] [PubMed] [Google Scholar]
- 17.Cherf GM, et al. Nature Biotech. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fologea D, et al. Nano Lett. 2005;5:1905–1909. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briggs K, et al. Nanotechnology. 2015;26:084004. doi: 10.1088/0957-4484/26/8/084004. [DOI] [PubMed] [Google Scholar]
- 20.Krishnakumar P, et al. ACS Nano. 2013;7:10319–10326. doi: 10.1021/nn404743f. [DOI] [PMC free article] [PubMed] [Google Scholar]


