Abstract
Cell polarization is a key step in the migration, development, and organization of eukaryotic cells, both at the single cell and multicellular level. Research on the mechanisms that give rise to polarization of a given cell, and organization of polarity within a tissue has led to new understanding across cellular and developmental biology. In this review, we describe some of the history of theoretical and experimental aspects of the field, as well as some interesting questions and challenges for the future.
Introduction
Specifying an axis of directionality is essential for most living cells. In the repertoire of cells that move individually, including Dictyostelium amoebae, neutrophils, keratocytes, and fibroblasts, determining the cell front and back is a prerequisite for organizing the machinery that powers cell motility (Fig. 1A). This includes the actin cytoskeleton, myosin, and, in some cases, microtubules. In organized cell collectives such as epithelia, the apical and basal faces of individual cells are distinct from one another and from the lateral surfaces along which cell-cell contact is maintained (Fig. 1B). Even in plant cells, whose shapes are constrained by rigid cell walls, the axis of polarity forms with growth, and is essential for defining cell division planes, and hence, organizing the shape of the plant tissue that emerges (Fig. 1C).
Figure 1. A–C Examples of cell polarity.
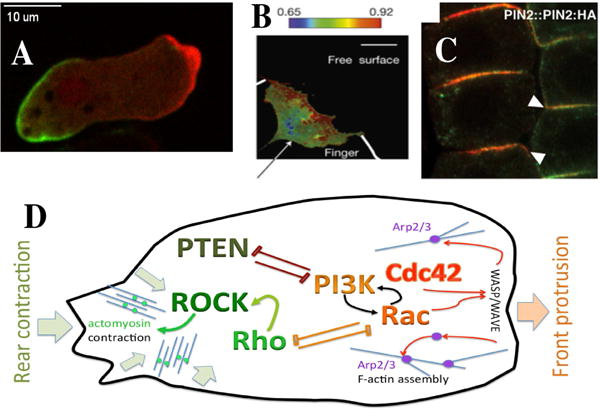
A. A polarized Dictyostelium cell moving to the right and showing an accumulation of myosin at its back and actin enrichment at its front.
B. The distribution of RhoA activity in a leader cell of an invading finger of epithelial Madin-Darby canine kidney cells displays an asymmetric distribution, with the activity being highest at the front edge [154].
C. Polar localizations of the fluorescent marker PIN2:HA at the lower side of root epidermal cells of the plant Arabidopsis thaliana. From [155]. D: A schematic sketch of proteins associated with front (in shades of red) and rear (green) of a polarized cell. The GTPases Rac and Cdc42 promote actin filament assembly (through Wave activating the branching complex Arp2/3). This leads to protrusion of the lamellipod. At the rear, the GTPase Rho activates ROCK which phosphorylates myosin light chain, leading to actomyosin contraction and retraction of the rear. Rac and Rho are often seen to be mutually antagonistic. Other proteins such as PI3K (which catalyzes the reaction PIP2–>PIP3) and PTEN (which catalyzes the reverse reaction) are also associated with the front vs rear of a polarized cell. PI3K participates in a positive feedback loop with Rac.
How cells spontaneously polarize, and how they maintain that polarity (when needed) or adapt it to a changing environment (as required) are fundamental questions in cell biology. For the past few decades, the investigation of such questions has taken place at the intersection of experimental biology, computational biology, physics, and mathematics. Cells can polarize in response to chemical, electrical, mechanical, or other physical stimuli. For example, the link between the direction of an influx of calcium ions and the polarization and direction of growth in the brown algal (fucoid) eggs was made as early as the 1970’s; e.g., [1]. A brief historical account and description of the rapid growth of cell polarization research over the past 40 years is given in [2]. More recently, the study of signaling networks that orchestrate cell polarity has emerged as a subfield of systems biology, with ever-increasing amount of data on the many molecular participants that govern that process. Here we briefly survey the evolution and current state of the field, and suggest some future promising directions.
Several overarching questions have historically focused the research on cell polarity. Among these are (1) What causes the symmetry breaking that polarizes a cell? (2) How can cell polarization be so robust for some cells, such as neutrophils, and very adaptable for others (Dictyostelium). (3) How does cell polarization fit into the broader picture of gradient sensing [3], cell motility [4–11], and chemotaxis [12–16], and more recently, planar cell polarity in tissues [17–19] and collective migration [20,21] (4) When does polarity go wrong, what cellular defects and consequent diseases arise, and how might we target such defects to find cures for the associated diseases [22–24]?
Biological observations that motivated cell polarity research
Early observations of single cells using conventional microscopy defined polarization according to cell shapes with elongated cells being more polarized than round cells. For moving cells, the migration direction is typically in the direction of the polarity axis, defined as the long-axis of the cell. Although these observations were instructive, they did not provide any data about the intracellular distributions of signaling components. This information has become available, however, due to advances in microscopy, and in particular fluorescence microscopy. Immunostaining, for example, allows the visualization of proteins in fixed cells, thus enabling the quantification of asymmetries in intracellular and membrane localizations. Even more informative, however, are methods that fuse signaling components to fluorescent probes, enabling the dynamic visualization of their spatio-temporal distribution.
These experiments have revealed that many signaling components are roughly uniformly distributed within the cytosol in unpolarized cells but have a pronounced asymmetric distribution in polarized cells. In moving cells, for example, the front is associated with high concentrations of actin while the back displays elevated myosin concentrations [25]. Other non-cytoskeleton components that show distinct localization during polarization include the phosphatidylinositol lipid PIP3 (front), along with its phosphatase and kinase (PTEN, back, and PI3K, front) [26,27]. In addition, many small GTPases display asymmetric distributions within a polarized cell. In budding yeast, Cdc42 localizes to the budding site, while in neutrophils and other migrating mammalian cells, Cdc42 is found at the front and Rho is found primarily at the back of the cell [28,29]. These distributions can be measured with high temporal and spatial precision, even under changing conditions. The emergence of such quantitative data has resulted in a surge of interest in the development of models for cell polarity establishment and maintenance, as described below.
What stimuli lead to symmetry breaking and cell polarization?
A universal mechanism for cellular polarization is chemoattraction: chemical stimuli such as Cyclic AMP (CAMP) for Dictyostelium, fMLP for neutrophils, or other chemical attractants that lead to cellular chemotaxis reorient cells and lead to their polarization. Many of the original works in the field, addressed the issue of how a weak chemical gradient of as little as 2% across a cell diameter can give rise to a macroscopic internal “map”, reviewed in [30,31]. But chemical factors are not the only type of cue that elicits polarization. Membrane tension [32] can lead to spontaneous cell polarization on a faster timescale than diffusible signals, especially in elongated cells; see [33]. In fish epidermal keratocytes, Xenopus mesendoderm cells, and Dictyostelium cells, polarization can be induced by mechanical stimuli such as the flow of fluid from a micropipette towards one edge of the cell [34,35] or forces applied locally to cadherins [36]. Electric fields are also known to induce cell polarization and migration in fish keratocytes [37] and Dictyostelium cells [38] and are responsible for cell migration during wound healing with a PI3K-dependent transduction mechanism [39]. Substrate rigidity can also result in polarization of cells, with cells moving toward stiff regions [40]. Furthermore, in plants, where cell walls are rigid, cell polarity depends on cell division planes. The plant hormone auxin is closely linked to cell division, growth, and development, and its distribution is governed by the pattern of localization of transporters (PINs) on the lateral sides of a cell. The precise signaling networks that control PIN localization are still not fully known [41]. See [42] for a computational model and [43] for an example review of work in this area. Finally, we note that some experiments suggest that Dictyostelium cells have a pre-existing polarity and continuously form pseudopods [44]. External cues can then guide these pseudopods, resulting in directed motion.
Modeling of single cell polarity — historical development as an offshoot of pattern formation theories
One of the first modeling treatments of cell polarity appeared in an early paper about biological pattern formation [45], some 20 years after the seminal pattern formation theory of Turing [46]. Relying primarily on simulations, Gierer and Meinhardt kindled the interest and curiosity of developmental biologists with their idea of lateral inhibition (local activation to amplify a small signal and long-range inhibition to prevent the activation from spreading throughout the domain unchecked) as a mechanism for pattern formation. Nearly 25 years later, in a follow-up paper [47] a model for cell polarization was revisited and sparked a revitalized interest in formulating theoretical basis to explain how polarity forms.
Meinhardt’s 1999 paper in J Cell Sci [47] was followed closely by seminal work [12,48] that investigated how cells can robustly achieve polarity even though the chemoattractant concentrations and gradient strengths can vary significantly. The basic idea in this work is that adaptation to uniform stimuli could be set up in a system where a stimulus simultaneously triggers activating and inhibitory signals. If the activator is local while the inhibitor acts globally the mechanism results in polarized cells for a wide range of gradient parameters. This local excitation and global inhibition mechanism (LEGI for short) became highly influential both theoretically and experimentally [49–52], accompanied by related models [53]. As in many pattern forming systems, cell polarity networks have both highly localized and quickly-spreading components. For instance, active forms of the protein have relatively slow spatial spread, e.g. by confinement to the membrane or to immobile cellular structures. Inhibitors, or inactive forms of the protein are assumed to be highly mobile or even global and uniform, spreading very rapidly in the cytosol. Typically, rates of diffusion of membrane versus cytoplasmic forms of the same protein (or proteins of comparable molecular weights) differ by 2–3 orders of magnitude.
The idea that cell polarization is a type of pattern formation (with a particularly simple “unimodal” pattern) led to a proliferation of models, many based on Turing-type reaction-diffusion systems of partial differential equations [3,4,54–56]. Popular among these were activator-inhibitor models known for their pattern-forming ability. The inhibitors proposed in some of the early reaction-diffusion or LEGI models have not been identified, despite much experimental effort, and are unlikely to be single agents. We should note, however, that membrane tension has also been suggested to play the role of long-range inhibitor [57].
Some, but not all models, were based on known molecular players such as the antagonists PI3K and PTEN [58,59] or small GTPases such as Cdc42, Rac, and Rho [5,56,60] in either mammalian cells or in yeast [61]. Protein modules that can act as mutually antagonistic “front-back” organizers are now known. These include Rac and Rho whose mutual antagonism can set up polarity or other cell behavior [23,62,63]. It is now accepted that activators of the cell edge cytoskeleton include Rac, or Ras, PI3K, and downstream effectors such as WASP/WAVE Arp2/3 that organize actin branching nucleation. The roles of these are clearly identified as promoting actin assembly and edge protrusion in motile cells. A schematic view of the distribution of signaling molecules in a polarized cells is shown in Fig. 1D. We should point out, however, that in some cells, the pathways directing polarity are slightly different. For example, in Dictyostelium cells, where multiple parallel pathways control directed migration [64], PI3K appears to be not essential for chemotaxis [65,66].
Recent progress in cell polarization research
(A) Progress in Experimental Methods
An important new experimental direction in the field of cell polarization is to quantify cell polarity, including spatio-temporal dynamics of signaling components, in response to carefully controlled external cues or in well-defined geometries. These cues are typically generated using microfluidics which allows the researcher to expose cells to precisely defined external cues [67–71]. These devices are particularly well-suited to generating precise and reproducible gradients of different strengths and profiles and can be used to investigate the chemotactic response of cells [16,72]. Furthermore, they can be used to rapidly change external conditions so as to study how polarity is established, reversed or maintained [67,73].
Other novel studies quantify the response of cells and cell polarity in specific geometric confinements. For example, substrates can be patterned such that cells only adhere to certain well-defined parts. Using this technique, it was found that the behavior of cells crawling on 1D stripes is much more consistent with 3D behavior than cells moving on flat, 2D substrates [74]. These patterned substrates can be used to investigate polarity questions: forcing the cells to crawl on a 1D stripe results in a strongly and consistently polarized cell. Reversal of cell polarity can then be studied by determining how cells behave when they collide “head-on”[75]. These, and other studies, have shown that contact inhibition of locomotion (CIL) plays an important role in cell-cell contacts [76,77]. The same CIL is also believed to play a significant role in the motion of monolayers and might be responsible for the response of cell clusters to chemical gradients [78,79].
Cell polarization can also be induced by geometries that force cells to move within channels with cross sections that are comparable to cell sizes [80,81]. These channel geometries were shown to induce cell polarization and resulted in persistent motion in different cell types, including Dictyostelium cells, cancer cells and human neutrophils [80–82]. In fact, geometry-induced polarity can be so persistent that neutrophils moving in U-shaped microfluidic channels can move against a gradient for extended periods of time (Fig. 2A) [83].
Fig. 2. Examples of recent experimental techniques used in studies on cell polarization.

A. Human neutrophils migrating persistently in a U-shaped microchannel containing [83]. The channel contains a gradient in chemoattractant pointing to the tip of the U and the cell is labeled with PH-Akt-GFP, a polarization marker. Throughout the cell’s trajectory, its cell polarization is persistent, even when the cell moves towards decreasing a chemoattractant concentration.
B. Light induced induction of lamellipodia in live NIH3T3 cells. A red light source was targeted to a small area and moved outwards. The laser light induces the activation of Rac, resulting in a cellular extension of approximately 30 μm.
C. The response of a Dictyostelium cell, expressing the actin marker LimEΔ-GFP (green) with phase contrast in red, to a rapid flow reversal as indicated by the white arrows [70]. The outer cell contour is shown as a white line. The quantitative data obtained from these cell reversal experiments can be used to validate models.
Another exciting new technique involves the use of light to control cellular behavior. First used to regulate neuronal spike timing [84], these optogenetic techniques can now be used to activate certain signaling components [85,86]. Importantly, the activation is achieved with high spatial and temporal resolution and is reversible. Optogenetics can be used to target a single protein, making it possible to determine how a signaling cascade is activated [87]. It can also be used to determine how cell polarity is controlled, both in single cells and in groups of cells [88–91] and promises to be an exciting new experimental tool (Fig. 2B).
(B) Progress in theoretical models and methods
The biological research in polarization has also led to a number of new mathematical findings and methods to explore typical kinds of models that arise: (well-mixed) differential equations or reaction-diffusion systems. Motivated by small GTPase biology, [92] showed that membrane-bound active GTPase with positive feedback (or mutually inhibitory Rac-Rho active forms), cytosolic inactive forms, with the total GTPase conserved in a cell can result in a rapid wave of polarization in response to a super-threshold stimulus. The wave freezes under specific conditions [93], resulting in robust polarization. An approximate bifurcation method (Local Perturbation Analysis, LPA) invented by Marée and Grieneisen [42,94], has proven to be a useful shortcut in analysis of RD systems that comprise both slow and fast rates of diffusion [95], scaling up to larger systems more conveniently than the traditional Turing stability analysis. A way to visualize a multi-parameter space was proposed by [96] and applied in the Rac-Rho cell bistability experiments described in [23].
In addition to the new analyses described above, several new computational techniques are currently pursued. In particular, methods that can describe cells as deformable objects are under development. These models are particularly well-suited if non-homogeneous intracellular regions are of importance or when there exists a feedback between shape, motility, and polarization. An important contribution to this type of research is the work of Mackenzie and coauthors who solved equations proposed by Meinhardt [47] on a moving, deforming 2D cell boundary [97]. Another technique that has been adapted to handle intra-cellular reaction-diffusion models is the cellular Potts model, [98]. In this model, cells are represented by a finite number of sites on a lattice (in 2D or 3D). Each site is assigned a “spin value” which specifies to which cell the site belongs and cell motion is implemented via a Monte Carlo update of spins which attempts to copy the spin from one lattice site to a neighboring site. This model was originally developed to describe cell sorting but has been extended to include polarity terms [99–101] as well as the effect of signaling on actin assembly and cell shape [4,5]. A recent, computationally light phenomenological example inspired by actin dynamics is [102].
Another technique uses elements from pattern formation and introduces an auxiliary field, termed “phase field”, that purely functions as a computational tool. This phase field takes on one value in the interior and another in the exterior of the cell and smoothly varies between these two values in a region that represents the cell’s boundary. A judicial coupling between the phase field and the underlying reaction-diffusion equations can then result in a model that recovers the original model in the limit of vanishing membrane thickness, the so-called sharp interface limit. A major advantage of this technique is that it does not require the explicit tracking of the membrane. For this reason, it has been used extensively in other so-called “free boundary problems”, especially in the field of pattern formation [103–106]. Studies have shown how to incorporate intra-cellular reaction diffusion as well as intra-cellular fluid flow into phase field cell models and have modeled single migratory, deformable cells [8,9,107,108]. More recent work has extended the phase field approach to motion of multiple cells [109–112]. One potential drawback of this methodology is the relatively high computational cost, since each cell requires solving its own set of equations. However, the use of GPU based computational platforms, combined with specialized algorithms, can significantly reduce these costs [109,110].
(C) Progress in linking experiments and theory
The combination of quantitative experiments and modeling can lead to new insights into the mechanisms of cell polarity. For example, experiments that mimic the periodic cAMP waves seen for DictyosteIium cells in aggregates can be generated in microfluidic devices. This allows one to determine the chemotactic response of cells and their polarity as a function of the wave period. It was found that cells have a cellular memory, i.e., they were able to maintain their polarity and ignore the so-called back of the wave, as long as the period was not more than 10 minutes [15]. These results can then be used as input to models for adaptation and memory, resulting in predictions of the topology of signaling networks. A similar approach was also carried out by Bretschneider et al who exposed Dictyostelium cells to alternating gradients of mechanical shear flow [70]. The experiments carefully quantified the spatio-temporal distributions of a fluorescent reporter for actin (Fig. 2C). The experimental findings were then tested against three different reaction-diffusion models for cell polarity. The study found that two of these models, the aforementioned models of Meinhardt and the LEGI model, were able to fit the experimental data in a satisfactory way. Thus, by quantifying the spatio-temporal responses of fluorescent reporters, these studies are able to function as model selectors.
The role of systems biology in driving cell polarization research
An interesting philosophical question is to what extent the methods of systems biology and high throughput biological observations can lead to a better understanding of cell polarity mechanisms and dynamics. Here, our review must of necessity be woefully brief, as this aspect of the research is only now beginning to take shape. To this point, most of the mechanistic modeling in the area has been built around a detailed understanding of very small networks (e.g. Rho GTPase crosstalk [113]) or simplified toy models [92,93,114] that can be fully analyzed mathematically. Such efforts can be expanded to additional layers (e.g. feedback from and to phosphoinositides [5,59]) downstream to the cytoskeleton, protrusion and contraction that generate cell shape changes and motion [5,115]. While we are beginning to study larger networks (described further on), challenges remain in developing techniques to analyze their behavior. A recent example with potential to explore the interactions of genes, mechanical forces, and multicellular interactions is the open source platform MecaGen [116].
Multicellular vs unicellular cell polarity
It is well-known that cell polarity in single migrating cells is dynamic, allowing cells to amplify external gradients and orient in complex environments; see, for example, the model by [117]. While serving a distinct function, dynamic changes in cell polarity are sometimes essential for normal development of a multicellular tissue. One example is the formation of lumens, hollow tubes of tissue consisting of one or more cell layers, as in kidney ducts and other structures. As shown in vitro by [18], such structures are initiated via cell division with the 2-cell stage having a reverse polarity; a marker such as Podocalyxin starts at the extracellular-matrix (ECM) cell surface. As the structure grows and develops, this marker is down-regulated from the “outside” and reassembled at the “interior” (lumen) cell surfaces. This reorientation involves integrins (beta1-integrin/FAK) and the antagonism of RhoA and Rac1. Defects in ECM-signaling, inhibiting beta1-integrin or disrupting the GTPase balance leads to a multicellular front-rear polarity and (aberrant) collective migration of the entire cell cluster, often associated with invasive or metastatic growth.
Collective migration plays a significant role in many normal biological processes and examples include wound healing in epithelial cell layers, specific cell groups during development. A number of papers have proposed the idea of self-generated gradient chemotaxis, wherein a cluster of cells consumes chemokine or ligand so as to form and follow that chemical gradient [118–121]. Collective migration is also a well-known aspect of tumor cell clusters during cancer metastasis. Cells within collectively migrating clusters can have different polarity states. Most notably, cells that are located at the edge of the cluster, termed leader cells, can have a larger size and more active lamellipodium than cells in the interior of the layer [122]. Leader cells are also essential for collective migration of MDCK cells [123] and Rac, PI3K, and Integrin beta 1 are upregulated in a more dramatic polar distribution in leader cells than in follower cells. In [124], the authors showed the connection between the distribution of microtubules and localization of adherens junctions in the posterior lateral line primordium of zebrafish in vivo, and explained the transition between the leader cells in front, and the more epithelial cells that form rosettes in the back.
Within collectively migrating clusters, cell-cell interaction can have a profound effect on cell polarization. Perhaps the best-studied example of this is the previously mentioned CIL, during which a cell repolarizes away from its contact points with other cells. For a review of CIL, including its molecular basis, see [125]. Other possibilities for cell-cell interactions include alignment mechanisms in which the cell’s velocity or polarity is determined by the state of its neighbors [20]. Recent studies have shown that these mechanisms can have profound impacts on the resulting migration of clusters [21,126,127].
A parallel but related realm of research in developmental biology has centered on planar cell polarity (PCP) in tissues [128]. In Drosophila wing development, protein complexes such as Frizzled-Disheveled-Flamingo (FZ-Dsh-Fmi) and Van Gogh- Prickled-Flamingo (Vang-Pk-Fmi) segregate asymmetrically to opposite sides of cells and “hold hands” across cell-cell interfaces to hard-wire polarity across neighboring cells, as reviewed in [129]. The same spontaneous emergence of asymmetry and polarity occurs in Par proteins, first identified in Caenorhabditis elegans: PAR-3 and PAR-6 concentrate at the anterior, whereas PAR-1 and PAR-2 at the posterior end as early as the single-cell stage of the embryo.
Several models have been proposed for the emergence of global organization. The polarization of PAR proteins in the C elegans embryo has been modeled in [130,131]. Similarly, PCP has been an area of active theoretical work for years. Many of the models invoke either cell-cell coupling or cell-cell comparisons, together with innate mechanisms that spawn polarization of a given cell or of the interface between neighboring cells, reviewed in [43,132]. Global signals that provide cues on the scale of many cells are proposed, but the molecular components at work in, for example, the Drosophila wing, are not yet identified. In one of the earliest models of Drosophila wing PCP [133,134], a reaction-diffusion mathematical model for the FZ-Dsh and the Vang-Pk complexes was proposed. In [43], individual cells can polarize spontaneously in absence of external bias, but cell-cell coupling organizes the spatial distribution of the polarity vectors. Another treatment using Hierarchically Colored Petri Nets was described in [135].
The molecular links between PCP and single cell polarization are now more clearly recognized. The involvement of the cytoskeleton, and segregation of actomyosin to specific lateral sides of the cells is needed for such events as cell elongation along an orthogonal axis, driving convergent extension in Drosophila, or asymmetric contractility, in C. elegans. Hence there has been recent interest in the crosstalk between Rho GTPases and PCP proteins, as reviewed in [136]. In one example, it was shown that a complex that includes Cdc42, Rac1, PAR-6 and PAR-3 is required for polarity in neurons [137]. This suggests that the lessons learned in the single and multicellular contexts will be useful in cross-fertilization and deeper understanding of polarization as a multiscale phenomenon.
Future challenges and directions in cell polarity research
It is tempting to speculate on the direction in which the field is moving. One of the challenges, related to the quest of systems-biology is to identify the full signaling pathway in specific cases. Some inroads have been made in this direction. The mutual inhibition between Rac and Rho embedded in the larger EGF-EGF receptor signaling pathway for example has been modeled using a Boolean approach [138]. See also [139] for a similar approach. A second important area is deciphering the role of mechanical forces such as membrane tension, and incorporating these into models for cell polarization. Both these areas are still in their infancy, and methods to understand these are still needed.
Another problem of interest is to what degree mechanisms need to be fine-tuned to create robust, but adaptable cell polarity. Many of the current models only work in relatively small parts of their parameter space. For example, the Meinhardt model we discussed earlier sensitively depends on some of its parameters. Likewise, building amplification into the LEGI model requires a careful fine-tuning of the parameters [15,52] as does the wave-pinning polarization mechanisms of Mori et al. [92] when captured by a single GTPase. (Robustness increases with the number of participating GTPases and feedbacks [140].) Somewhat related is the question of cell variability: how does the degree of polarization vary from cell to cell? Addressing these questions probably requires an integrated approach in which experiments and models go hand in hand.
It is also interesting to consider cell polarization in distinct geometries, for example in in 3D matrices (e.g. on collagen fibers). A recent work describes the formation of special cell appendages, regulated by a balance of Rac Arp2/3 pathway (promotes the formation of “fin-like” structures) and Rho pathways that promote formin-based actin assembly and myosin-based blebbing behavior [141]. Several modeling studies have explored the connections between the shape of a cell and the dynamics of its intracellular signaling for polarity [5,97,142–144], but these have largely focused on 2D cell projections. The computational issues associated with simulating dynamic intracellular signaling inside a deforming 3D cell are still daunting, but developments of new methods is ongoing by several groups (see, e.g. [145]), and progress has been made since the review in [143].
Now that we have data in many types of cells and conditions, it is instructive to compare between species or conditions: what is universal and what is specific? Proteins such as partitioning defective (PAR), and complexes such as Scribble, and Crumbs are responsible for polarity in epithelial cells, and appear to be universal in multicellular organisms from worms to humans. Interactions of these with the small GTPases are reviewed in [136]. Crumbs localizes to the apical domain, Scribble to the lateral and basal cell surfaces in epithelia, but in single migrating cells, all these are found at the cell front [29]. Rho GTPases and their crosstalk and cytoskeletal effectors also appear to be universal, although a variety of types of Rac but no Cdc42 occur in DictyosteIium.
We are also learning to manipulate the polarity machinery and hoping to understand how RNAi or other knockouts affect behavior. Examples of the outcomes of cells treated with RNAi include [146,147]. Some theoretical analysis of the cell shapes was provided in [62]. We have a better understanding of how cell polarity mechanisms evolved in living cells. Evolution of the family of Rho GTPases is described in [148]. Rac is the “founder”. The expression of Rho mRNA in mouse cells of various types indicates that the specialization of Rhos have led them to become expressed more specifically in particular cells and at specific times.
We expect that coupling experiments to modeling efforts will give us more insights into the topology of signaling networks. The first examples of this approach have resulted in promising results [15,149,150] and new experimental techniques, including optogenetics and microfluidics, will undoubtedly be critical. We also expect that studies in which experimental results are systematically compared to model output, as was carried out by Bretscheider et al [70], will be crucial in determining the mechanisms likely involved in cell polarity. Again, new experimental techniques will allow us to generate quantitative data required for these future studies.
We are beginning to link cell polarity and migration defects to cancer and other diseases. For example, the integrity of epithelia is coordinated by cell-cell contacts such as tight junctions and adherens junctions that form, like rivets, between adjacent cells. These are composed of large complexes of molecules that act as scaffolds for signaling, including spatial and temporal control of the activation (by GEFs) and inactivation (by GAPs) of Rho GTPases. Defects in these complexes, whether due to transcription, viral interference, or other causes can lead to disregulation, EMT, and cancer. See reviews in [22,24,151,152]. Downregulation of RhoB has been linked to changes in focal adhesion, reduced beta1 integrin, and weakening of the epithelia that drive prostate cancer, for example [153]. Further work in this direction is likely to provide better insight into factors promoting metastasis, with potential payoff in better treatment and drug targets. In short, while much progress has taken place, a great deal remains to be done.
Acknowledgments
LEK is supported by an NSERC Discovery grant No. 41870. WJR is supported by NIH Grant No. P01 GM078586. We thank Brian Camley for a careful and critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Robinson KR, Jaffe LF. Polarizing fucoid eggs drive a calcium current through themselves. Science (80- ) 1975;187:70–72. doi: 10.1126/science.1167318. [DOI] [PubMed] [Google Scholar]
- 2.Csikász-Nagy A, Sato M, Salas REC. Projecting cell polarity into the next decade. Phil Trans R Soc B. 2013;368:20130001. doi: 10.1098/rstb.2013.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onsum M. A mathematical model for neutrophil gradient sensing and polarization. PLoS Comput Biol. 2007;3:e36. doi: 10.1371/journal.pcbi.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marée AFM, Jilkine A, Dawes A, Grieneisen VA, Edelstein-Keshet L. Polarization and Movement of Keratocytes: A Multiscale Modelling Approach. Bull Math Biol. 2006;68:1169–1211. doi: 10.1007/s11538-006-9131-7. [DOI] [PubMed] [Google Scholar]
- 5.Marée AFM, Grieneisen VA, Edelstein-Keshet L. How cells integrate complex stimuli: the effect of feedback from phosphoinositides and cell shape on cell polarization and motility. PLoS Comput Biol. 2012;8:e1002402. doi: 10.1371/journal.pcbi.1002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolgemuth CW, Zajac M. The moving boundary node method: A level set-based, finite volume algorithm with application to cell motility. J Comput Phys. 2010;229:7287–7308. doi: 10.1016/j.jcp.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi C, Huang C-HH, Devreotes PN, Iglesias PA. Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells. PLoS Comput Biol. 2013;9:e1003122. doi: 10.1371/journal.pcbi.1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao D, Rappel W-J, Levine H. Computational model for cell morphodynamics. Phys Rev Lett. 2010;105:108104. doi: 10.1103/PhysRevLett.105.108104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shao D, Levine H, Rappel W-JJ. Coupling actin flow, adhesion, and morphology in a computational cell motility model [Internet] Proc Natl Acad Sci U S A. 2012;109:6851–6856. doi: 10.1073/pnas.1203252109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourne HR, Weiner O. A chemical compass [Internet] Nature. 2002;419:21. doi: 10.1038/419021a. [DOI] [PubMed] [Google Scholar]
- 11.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science (80-) 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 12.Levchenko A, Iglesias PA. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys J. 2002;82:50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider IC, Haugh JM. Mechanisms of gradient sensing and chemotaxis: conserved pathways, diverse regulation. Cell Cycle. 2006;5:1130–1134. doi: 10.4161/cc.5.11.2770. [DOI] [PubMed] [Google Scholar]
- 14.King JS, Insall RH. Chemotaxis: finding the way forward with Dictyostelium. Trends Cell Biol. 2009;19:523–530. doi: 10.1016/j.tcb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Skoge M, Yue H, Erickstad M, Bae A, Levine H, Groisman A, Loomis WF, Rappel W-J. Cellular memory in eukaryotic chemotaxis. Proc Natl Acad Sci U S A. 2014;111:14448–14453. doi: 10.1073/pnas.1412197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song L, Nadkarni SM, Bodeker HU, Beta C, Bae A, Franck C, Rappel W-J, Loomis WF, Bodenschatz E. Dictyostelium discoideum chemotaxis: threshold for directed motion. Eur J Cell Biol. 2006;85:981–989. doi: 10.1016/j.ejcb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Lan H, Wang Q, Fernandez-Gonzalez R, Feng JJ. A biomechanical model for cell polarization and intercalation during Drosophila germband extension. Phys Biol. 2015;12:56011. doi: 10.1088/1478-3975/12/5/056011. [DOI] [PubMed] [Google Scholar]
- 18.Bryant DM, Roignot J, Datta A, Overeem AW, Kim M, Yu W, Peng X, Eastburn DJ, Ewald AJ, Werb Z, et al. A molecular switch for the orientation of epithelial cell polarization. 2014;31:171–187. doi: 10.1016/j.devcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gord A, Holmes WR, Dai X, Nie Q. Computational modelling of epidermal stratification highlights the importance of asymmetric cell division for predictable and robust layer formation. J R Soc Interface. 2014;11 doi: 10.1098/rsif.2014.0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarle V, Ravasio A, Hakim V, Gov NS. Modeling the finger instability in an expanding cell monolayer. Integr Biol. 2015;7:1218–1227. doi: 10.1039/c5ib00092k. [DOI] [PubMed] [Google Scholar]
- 21.Camley BA, Zimmermann J, Levine H, Rappel WJ. Collective Signal Processing in Cluster Chemotaxis: Roles of Adaptation, Amplification, and Co-attraction in Collective Guidance. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1005008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halaoui R, McCaffrey L. Rewiring cell polarity signaling in cancer. Oncogene. 2014;34:1–12. doi: 10.1038/onc.2014.59. [DOI] [PubMed] [Google Scholar]
- 23.Byrne KM, Monsefi N, Dawson JC, Degasperi A, Bukowski-Wills J-C, Volinsky N, Dobrzyński M, Birtwistle MR, Tsyganov MA, Kiyatkin A, et al. Bistability in the Rac1, PAK and RhoA signalling network is a feature of cell motility. Cell Syst. 2016;2:38–48. doi: 10.1016/j.cels.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal. 2010;8:23. doi: 10.1186/1478-811X-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franca-Koh J, Devreotes PN. Moving forward: mechanisms of chemoattractant gradient sensing. Physiology. 2004;19:300–308. doi: 10.1152/physiol.00017.2004. [DOI] [PubMed] [Google Scholar]
- 26.Comer FI, Parent CA. Phosphoinositide 3-kinase activity controls the chemoattractant-mediated activation and adaptation of adenylyl cyclase. Mol Biol Cell. 2006;17:357–366. doi: 10.1091/mbc.E05-08-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leslie NR, Batty IH, Maccario H, Davidson L, Downes CP. Understanding PTEN regulation: PIP2, polarity and protein stability. Oncogene. 2008;27:5464–5476. doi: 10.1038/onc.2008.243. [DOI] [PubMed] [Google Scholar]
- 28.Park H, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007 doi: 10.1128/MMBR.00028-06. no volume. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Etienne-Manneville S. Polarity proteins in migration and invasion. Oncogene. 2008;27:6970–6980. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]
- 30.Jilkine A, Edelstein-Keshet L. A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS Comput Biol. 2011;7:e1001121. doi: 10.1371/journal.pcbi.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine H, Rappel W-J. The physics of eukaryotic chemotaxis. Phys Today. 2013;66 doi: 10.1063/PT.3.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsujita K, Takenawa T, Itoh T. Feedback regulation between plasma membrane tension and membrane-bending proteins organizes cell polarity during leading edge formation. Nat Cell Biol. 2015;17:749–758. doi: 10.1038/ncb3162. [DOI] [PubMed] [Google Scholar]
- 33.Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, Weiner OD. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration [Internet] Cell. 2012;148:175–188. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yam PT, Wilson CA, Ji L, Hebert B, Barnhart EL, Dye NA, Wiseman PW, Danuser G, Theriot JA. Actin–myosin network reorganization breaks symmetry at the cell rear to spontaneously initiate polarized cell motility. J Cell Biol. 2007;178:1207–1221. doi: 10.1083/jcb.200706012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalous J, Burghardt E, Muller-Taubenberger A, Bruckert F, Gerisch G, Bretschneider T. Reversal of cell polarity and actin-myosin cytoskeleton reorganization under mechanical and chemical stimulation. Biophys J. 2008;94:1063–1074. doi: 10.1529/biophysj.107.114702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber GF, Bjerke MA, DeSimone DW. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell. 2012;22:104. doi: 10.1016/j.devcel.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen GM, Mogilner A, Theriot JA. Electrophoresis of cellular membrane components creates the directional cue guiding keratocyte galvanotaxis. Curr Biol. 2013;23:560–568. doi: 10.1016/j.cub.2013.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shanley LJ, Walczysko P, Bain M, MacEwan DJ, Zhao M. Influx of extracellular Ca2+ is necessary for electrotaxis in Dictyostelium. J Cell Sci. 2006;119:4741–4748. doi: 10.1242/jcs.03248. [DOI] [PubMed] [Google Scholar]
- 39.Zhao M, Song B, Pu J, Wada T, Reid B, Tai G, Wang F, Guo A, Walczysko P, Gu Y, et al. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature. 2006;442:457–460. doi: 10.1038/nature04925. [DOI] [PubMed] [Google Scholar]
- 40.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Norman JM. Asymmetry and cell polarity in root development. Dev Biol. 2016;419:165–174. doi: 10.1016/j.ydbio.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- 43.Abley K, De Reuille PB, Strutt D, Bangham A, Prusinkiewicz P, Marée AFM, Grieneisen VA, Coen E. An intracellular partitioning-based framework for tissue cell polarity in plants and animals. Development. 2013;140:2061–74. doi: 10.1242/dev.062984. [DOI] [PubMed] [Google Scholar]
- 44.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 45.Gierer A, Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12:30–39. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 46.Turing AM. The chemical theory of morphogenesis. Phil Trans Roy Soc. 1952;237:37–72. [Google Scholar]
- 47.Meinhardt H. Orientation of chemotactic cells and growth cones: models and mechanisms. J Cell Sci. 1999;112:2867–2874. doi: 10.1242/jcs.112.17.2867. [DOI] [PubMed] [Google Scholar]
- 48.Parent CA, Devreotes PN. A cell’s sense of direction. Science (80-) 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 49.Ma L, Janetopoulos C, Yang L, Devreotes PNN, Iglesias PAA. Two complementary, local excitation, global inhibition mechanisms acting in parallel can explain the chemoattractant-induced regulation of PI(3,4,5)P-3 response in Dictyostelium cells. Biophys J. 2004;87:3764–3774. doi: 10.1529/biophysj.104.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iglesias PA, Devreotes PN. Navigating through models of chemotaxis. Curr Op Cell Biol. 2008;20:35–40. doi: 10.1016/j.ceb.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 51.Iglesias PA, Devreotes PN. Biased excitable networks: how cells direct motion in response to gradients. Curr Opin Cell Biol. 2011;24:245–253. doi: 10.1016/j.ceb.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong Y, Huang CH, Iglesias PA, Devreotes PN. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc Natl Acad Sci U S A. 2010;107:17079–17086. doi: 10.1073/pnas.1011271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levine H, Kessler DA, Rappel WJ. Directional sensing in eukaryotic chemotaxis: A balanced inactivation model. PNAS. 2006;103:9761–9766. doi: 10.1073/pnas.0601302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu J, Wang F, Van Keymeulen A, Herzmark P, Straight A, Kelly K, Takuwa Y, Sugimoto N, Mitchison T, Bourne HR. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;114:201–214. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 55.Wedlich-Soldner R, Altschuler S, Wu L, Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science (80-) 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- 56.Otsuji M, Ishihara S, Co C, Kaibuchi K, Mochizuki A, Kuroda S, et al. A mass conserved reaction-diffusion system captures properties of cell polarity. PLoS Comput Biol. 2007;3(6):e108 1040–1054. doi: 10.1371/journal.pcbi.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, Weiner OD. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration [Internet] Cell. 2012;148:175–188. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin B, Holmes WR, Wang CJ, Ueno T, Harwell A, Edelstein-Keshet L, Inoue T, Levchenko A. Synthetic spatially graded Rac activation drives cell polarization and movement. Proc Natl Acad Sci U S A. 2012;109:E3668–77. doi: 10.1073/pnas.1210295109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dawes AT, Edelstein-Keshet L. Phosphoinositides and Rho proteins spatially regulate actin polymerization to initiate and maintain directed movement in a one-dimensional model of a motile cell. Biophys J. 2007;92:744–768. doi: 10.1529/biophysj.106.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jilkine A, Marée AFM, Edelstein-Keshet L. Mathematical model for spatial segregation of the Rho-family GTPases based on inhibitory crosstalk. Bull Math Biol. 2007;69:1943–1978. doi: 10.1007/s11538-007-9200-6. [DOI] [PubMed] [Google Scholar]
- 61.Goryachev AB. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008;582:1437–1443. doi: 10.1016/j.febslet.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 62.Holmes WR, Edelstein-keshet L. Analysis of a minimal Rho-GTPase circuit regulating cell shape. Phys Biol. 2016;13 doi: 10.1088/1478-3975/13/4/046001. [DOI] [PubMed] [Google Scholar]
- 63.Guilluy C, Garcia-Mata R, Burridge K. Rho protein crosstalk: another social network? Trends Cell Biol. 2011;21:718–726. doi: 10.1016/j.tcb.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bosgraaf L, Van Haastert PJ. {N}avigation of chemotactic cells by parallel signaling to pseudopod persistence and orientation. PLoS One. 2009;4:e6842. doi: 10.1371/journal.pone.0006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 66.Veltman DM, Lemieux MG, Knecht DA, Insall RH. PIP(3)-dependent macropinocytosis is incompatible with chemotaxis. J Cell Biol. 2014;204:497–505. doi: 10.1083/jcb.201309081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skoge M, Adler M, Groisman A, Levine H, Loomis WF, Rappel W-J. Gradient sensing in defined chemotactic fields. Integr Biol. 2010;2:659–668. doi: 10.1039/c0ib00033g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dertinger SK, Chiu DT, Dertinger SK, Whitesides GM. Generation of gradients having complex shapes using microfluidic networks. Anal Chem. 2001;73:1240–1246. [Google Scholar]
- 69.Irimia D, Liu SY, Tharp WG, Samadani A, Toner M, Poznansky MC. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients. Lab Chip. 2006;6:191–198. doi: 10.1039/b511877h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lockley R, Ladds G, Bretschneider T. Image based validation of dynamical models for cell reorientation. Cytom A. 2014 doi: 10.1002/cyto.a.22600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loomis WF, Fuller D, Gutierrez E, Groisman A, Rappel W-J. Innate nonspecific cell substratum adhesion. PLoS One. 2012;7:e42033. doi: 10.1371/journal.pone.0042033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fuller D, Chen W, Adler M, Groisman A, Levine H, Rappel W-J, Loomis WF. External and internal constraints on eukaryotic chemotaxis [Internet] Proc Natl Acad Sci U S A. 2010;107:9656–9659. doi: 10.1073/pnas.0911178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prentice-Mott HV, Meroz Y, Carlson A, Levine MA, Davidson MW, Irimia D, Charras GT, Mahadevan L. Directional memory arises from long-lived cytoskeletal asymmetries in polarized chemotactic cells [Internet] Proc Natl Acad Sci U S A. 2016;113:1267–1272. doi: 10.1073/pnas.1513289113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin B, Yin T, Wu YI, Inoue T, Levchenko A. Interplay between chemotaxis and contact inhibition of locomotion determines exploratory cell migration. Nat Commun. 2015;6:6619. doi: 10.1038/ncomms7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, Stern CD, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abercrombie M, Heaysman JEM. Observations on the social behaviour of cells in tissue culture. I. Speed of movement of chick heart fibroblasts in relation to their mutual contacts. Exp Cell Res. 1953;5:111–131. doi: 10.1016/0014-4827(53)90098-6. [DOI] [PubMed] [Google Scholar]
- 78.Theveneau E, Mayor R. Integrating chemotaxis and contact-inhibition during collective cell migration: Small GTPases at work. Small GTPases. 2010;1:113–117. doi: 10.4161/sgtp.1.2.13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nagel O, Guven C, Theves M, Driscoll M, Losert W, Beta C. Geometry-Driven Polarity in Motile Amoeboid Cells. PLoS One. 2014;9:e113382. doi: 10.1371/journal.pone.0113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Irimia D, Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr Biol. 2009;1:506. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boneschansker L, Yan J, Wong E, Briscoe DM, Irimia D. Microfluidic platform for the quantitative analysis of leukocyte migration signatures [Internet] Nat Commun. 2014;5:4787. doi: 10.1038/ncomms5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hamza B, Wong E, Patel S, Cho H, Martel J, Irimia D. Retrotaxis of human neutrophils during mechanical confinement inside microfluidic channels. Integr Biol. 2014;6:175–183. doi: 10.1039/c3ib40175h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 85.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. LETTERS A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461 doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang X, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol. 2010;12:591. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jost AP, Weiner OD. Probing Yeast Polarity with Acute, Reversible, Optogenetic Inhibition of Protein function. ACS Synth Biol. 2015 doi: 10.1021/acssynbio.5b00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee S, Park H, Kyung T, Kim N, Kim S, Kim J. Reversible protein inactivation by optogenetic trapping in cells. Nature. 2014 doi: 10.1038/nmeth.2940. no volume. [DOI] [PubMed] [Google Scholar]
- 92.Mori Y, Jilkine A, Edelstein-Keshet L. Wave-pinning and cell polarity from a bistable reaction-diffusion system. Biophys J. 2008;94:3684–3697. doi: 10.1529/biophysj.107.120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mori Y, Jilkine A, Edelstein-Keshet L. Asymptotic and bifurcation analysis of wave-pinning in a reaction-diffusion model of cell polarization. SIAM J Appl Math. 2011;71:1401–1427. doi: 10.1137/10079118X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walther GR, Marée AFM, Edelstein-Keshet L, Grieneisen VA. Deterministic versus stochastic cell polarisation through wave-pinning. Bull Math Biol. 2012;74:2570. doi: 10.1007/s11538-012-9766-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Holmes WR, Mata MA, Edelstein-Keshet L. Local Perturbation analysis: A computational tool for biophysical reaction-diffusion models. Biophys J. 2015;108:230–236. doi: 10.1016/j.bpj.2014.11.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nguyen LK, Degasperi A, Cotter P, Kholodenko BN. DYVIPAC: an integrated analysis and visualisation framework to probe multi-dimensional biological networks. Sci Rep. 2015;5:12569. doi: 10.1038/srep12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Neilson MP, Veltman DM, van Haastert PJM, Webb SD, Mackenzie JA, Insall RH. Chemotaxis: a feedback-based computational model robustly predicts multiple aspects of real cell behaviour. PLoS Biol. 2011;9:e1000618. doi: 10.1371/journal.pbio.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Graner F, Glazier JA. Simulation of biological cell sorting using a two-dimensional extended Potts model [Internet] Phys Rev Lett. 1992;69:2013–2016. doi: 10.1103/PhysRevLett.69.2013. [DOI] [PubMed] [Google Scholar]
- 99.Rappel W-J, Nicol A, Sarkissian A, Levine H, Loomis WF. Self-organized vortex state in two-dimensional Dictyostelium dynamics. Phys Rev Lett. 1999;83:1247–1250. [Google Scholar]
- 100.Szabó A, Ünnep R, Méhes E, Twal WO, Argraves WS, Cao Y, Czirók A. Collective cell motion in endothelial monolayers. Phys Biol. 2010;7:46007. doi: 10.1088/1478-3975/7/4/046007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kabla AJ. Collective cell migration: leadership, invasion and segregation. J R Soc Interface. 2012;9:3268–3278. doi: 10.1098/rsif.2012.0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Niculescu I, Textor J, De Boer RJ. Crawling and gliding: a computational model for shape-driven cell migration. PLoS Comput Biol. 2015;11:e1004280. doi: 10.1371/journal.pcbi.1004280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Karma A, Rappel W-J. Phase-field method for computationally efficient modeling of solidification with arbitrary interface kinetics. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Top. 1996;53:R3017–R3020. doi: 10.1103/physreve.53.r3017. [DOI] [PubMed] [Google Scholar]
- 104.Karma A, Kessler DA, Levine H. Phase-field model of mode III dynamic fracture. Phys Rev Lett. 2001;87:45501. doi: 10.1103/PhysRevLett.87.045501. [DOI] [PubMed] [Google Scholar]
- 105.Fenton FH, Cherry EM, Karma A, Rappel W-J. Modeling wave propagation in realistic heart geometries using the phase-field method. Chaos. 2005;15 doi: 10.1063/1.1840311. [DOI] [PubMed] [Google Scholar]
- 106.Folch R, Casademunt J, Hernandez-Machado A, Ramirez-Piscina L. Phase-field model for Hele-Shaw flows with arbitrary viscosity contrast. I. Theoretical approach. Phys Rev E. 1999;60:1724–1733. doi: 10.1103/physreve.60.1724. [DOI] [PubMed] [Google Scholar]
- 107.Kockelkoren J, Levine H, Rappel W-J. Computational approach for modeling intra- and extracellular dynamics [Internet] Phys Rev E Stat Nonlin Soft Matter Phys. 2003;68:37702. doi: 10.1103/PhysRevE.68.037702. [DOI] [PubMed] [Google Scholar]
- 108.Li X, Lowengrub J, Rätz A, Voigt A. Solving PDEs in complex geometries: a diffuse domain approach. Commun Math Sci. 2009;7:81–107. doi: 10.4310/cms.2009.v7.n1.a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Camley BA, Zhang Y, Zhao Y, Li B, Ben-Jacob E, Levine H, Rappel W-J. Polarity mechanisms such as contact inhibition of locomotion regulate persistent rotational motion of mammalian cells on micropatterns. Proc Natl Acad Sci U S A. 2014;111:14770–14775. doi: 10.1073/pnas.1414498111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kulawiak DA, Camley BA, Rappel W-J. Modeling contact inhibition of locomotion of colliding cells migrating on micropatterned substrates. 2016 doi: 10.1371/journal.pcbi.1005239. submitted. no volume. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nonomura M. Study on Multicellular Systems Using a Phase Field Model. PLoS One. 2012;7:e33501. doi: 10.1371/journal.pone.0033501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marth W, Voigt A. Collective migration under hydrodynamic interactions–a computational approach. Interface Focus. 2016 doi: 10.1098/rsfs.2016.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jilkine A, Marée AFM, Edelstein-Keshet L, Maree AF, Edelstein-Keshet L. Mathematical model for spatial segregation of the Rho-family GTPases based on inhibitory crosstalk. Bull Math Biol. 2007;69:1943–1978. doi: 10.1007/s11538-007-9200-6. [DOI] [PubMed] [Google Scholar]
- 114.Edelstein-Keshet L, Holmes WR, Zajac M, Dutot M, Edelstein-keshet L, Holmes WR, Zajac M, Dutot M. From simple to detailed models for cell polarization From simple to detailed models for cell polarization. 2013;368:20130003. doi: 10.1098/rstb.2013.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Marée AFM, Jilkine A, Dawes A, Grieneisen VA, Edelstein-Keshet L. Polarization and movement of keratocytes: a multiscale modeling approach. Bull Math Biol. 2006;68:1169. doi: 10.1007/s11538-006-9131-7. [DOI] [PubMed] [Google Scholar]
- 116.Delile J, Herrmann M, Peyrie N. A cell-based computational model of early embryogenesis coupling mechanical behaviour and gene regulation. Nat Commun. 2017;8:13929. doi: 10.1038/ncomms13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hecht I, Levine H, Rappel W-J, Ben-Jacob E. “Self-assisted” amoeboid navigation in complex environments. PLoS One. 2011;6:e21955. doi: 10.1371/journal.pone.0021955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dalle Nogare D, Somers K, Rao S, Matsuda M, Reichman-Fried M, Raz E, Chitnis AB. Leading and trailing cells cooperate in collective migration of the zebrafish posterior lateral line primordium. DeveIopment. 2014;141:3188–3196. doi: 10.1242/dev.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tweedy L, Knecht DA, Mackay GM, Insall RH. Self-generated chemoattractant gradients: attractant depletion extends the range and robustness of chemotaxis. PLoS Biol. 2016;14:e1002404. doi: 10.1371/journal.pbio.1002404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Venkiteswaran G, Lewellis SW, Wang J, Reynolds E, Nicholson C, Knaut H. Generation and dynamics of an endogenous, self-generated signaling gradient across a migrating tissue. Cell. 2013;155:674. doi: 10.1016/j.cell.2013.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Streichan SJ, Valentin G, Gilmour D, Hufnagel L. Collective cell migration guided by dynamically maintained gradients. Phys Biol. 2011;8:45004. doi: 10.1088/1478-3975/8/4/045004. [DOI] [PubMed] [Google Scholar]
- 122.Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, Buguin A, Silberzan P. Collective migration of an epithelial monolayer in response to a model wound. Proc Natl Acad Sci. 2007;104:15988. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yamaguchi N, Mizutani T, Kawabata K, Haga H. Leader cells regulate collective cell migration via Rac activation in the downstream signaling of integrin β1 and PI3K. Sci Rep. 2015;5:7656. doi: 10.1038/srep07656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Revenu C, Streichan S, Donà E, Lecaudey V, Hufnagel L, Gilmour D. Quantitative cell polarity imaging defines leader-to-follower transitions during collective migration and the key role of microtubule-dependent adherens junction formation. Development. 2014;141:1282–1291. doi: 10.1242/dev.101675. [DOI] [PubMed] [Google Scholar]
- 125.Stramer B, Mayor R. Mechanisms and in vivo functions of contact inhibition of locomotion. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.118. no volume. [DOI] [PubMed] [Google Scholar]
- 126.Camley BA, Rappel W-J. Velocity alignment leads to high persistence in confined cells. Phys Rev E Stat Nonlin Soft Matter Phys. 2014;89:62705. doi: 10.1103/PhysRevE.89.062705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zimmermann J, Camley BA, Rappel W-J, Levine H. Contact inhibition of locomotion determines cell-cell and cell-substrate forces in tissues. Proc Natl Acad Sci U S A. 2016;113:2660–2665. doi: 10.1073/pnas.1522330113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Devenport D. The cell biology of planar cell polarity. J Cell Biol. 2014;207:171–179. doi: 10.1083/jcb.201408039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zallen JA. Planar polarity and tissue morphogenesis. Cell. 2007;129:1051–1063. doi: 10.1016/j.cell.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 130.Dawes AT, Munro EM. PAR-3 oligomerization may provide an actin-independent mechanism to maintain distinct par protein domains in the early Caenorhabditis elegans embryo. Biophys J. 2011;101:1412–1422. doi: 10.1016/j.bpj.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goehring NW, Hoege C, Grill SW, Hyman AA. PAR proteins diffuse freely across the anterior–posterior boundary in polarized C. elegans embryos. J Cell Biol. 2011;193:583–594. doi: 10.1083/jcb.201011094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Meinhardt H. Computational modelling of epithelial patterning. Curr Opin Genet Dev. 2007;17:272–280. doi: 10.1016/j.gde.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 133.Amonlirdviman K, Khare NA, Tree DRP, Chen W, Axelrod JD, Tomlin CJ. Mathematical modeling of planar cell polarity to understand domineering nonautonomy. Science (80-) 2005;307:423. doi: 10.1126/science.1105471. [DOI] [PubMed] [Google Scholar]
- 134.Axelrod JD. Pattern Formation in Morphogenesis. Springer; 2013. Mathematical Modeling of Planar Cell Polarity Signaling; pp. 27–35. [Google Scholar]
- 135.Gao Q, Gilbert D, Heiner M, Liu F, Maccagnola D, Tree D. Multiscale modeling and analysis of planar cell polarity in the Drosophila wing. IEEE/ACM Trans Comput Biol Bioinforma. 2013;10:337–351. doi: 10.1109/TCBB.2012.101. [DOI] [PubMed] [Google Scholar]
- 136.Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- 137.Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima Y, Ohno S, Hoshino M, Kaibuchi K. PAR-6–PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol. 2005;7:270–277. doi: 10.1038/ncb1227. [DOI] [PubMed] [Google Scholar]
- 138.Hetmanski JHR, Zindy E, Schwartz J-M, Caswell PT. A MAPK-Driven Feedback Loop Suppresses Rac Activity to Promote RhoA-Driven Cancer Cell Invasion. PLoS Comput Biol. 2016;12:e1004909. doi: 10.1371/journal.pcbi.1004909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim TH, Monsefi N, Song JH, Von Kriegsheim A, Vandamme D, Pertz O, Kholodenko BN, Kolch W, Cho KH. Network-based identification of feedback modules that control RhoA activity and cell migration. J Mol Cell Biol. 2015;7:242–252. doi: 10.1093/jmcb/mjv017. [DOI] [PubMed] [Google Scholar]
- 140.Chau AH, Walter JM, Gerardin J, Tang C, Lim WA. Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell. 2012;151:320–332. doi: 10.1016/j.cell.2012.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Guetta-Terrier C, Monzo P, Zhu J, Long H, Venkatraman L, Zhou Y, Wang P, Chew SY, Mogilner A, Ladoux B, et al. Protrusive waves guide 3D cell migration along nanofibers. J Cell Biol. 2015;211:683–701. doi: 10.1083/jcb.201501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Giese W, Eigel M, Westerheide S, Engwer C, Klipp E. Influence of cell shape, inhomogeneities and diffusion barriers in cell polarization models. Phys Biol. 2015;12:66014. doi: 10.1088/1478-3975/12/6/066014. [DOI] [PubMed] [Google Scholar]
- 143.Holmes WR, Edelstein-Keshet L. A Comparison of Computational Models for Eukaryotic Cell Shape and Motility. PLoS Comput Biol. 2012;8:e1002793. doi: 10.1371/journal.pcbi.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Camley BA, Zhao Y, Li B, Levine H, Rappel W-J. Crawling and turning in a minimal reaction-diffusion cell motility model: coupling cell shape and biochemistry. arXiv Prepr arXiv1609.01764. 2016 doi: 10.1103/PhysRevE.95.012401. no volume. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tuncer N, Madzvamuse A. Projected finite elements for systems of reaction-diffusion equations on closed evolving spheroidal surfaces. Commun Comput Physics Accept. 2016 no volume. [Google Scholar]
- 146.Sailem H, Bousgouni V, Cooper S, Bakal C. Cross-talk between Rho and Rac GTPases drives deterministic exploration of cellular shape space and morphological heterogeneity. Open Biol. 2014;4:130132. doi: 10.1098/rsob.130132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cooper S, Sadok A, Bousgouni V, Bakal C. Apolar and polar transitions drive the conversion between amoeboid and mesenchymal shapes in melanoma cells. Mol Biol Cell. 2015;26:4163–4170. doi: 10.1091/mbc.E15-06-0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol. 2007;24:203–216. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Takeda K, Shao D, Adler M, Charest PG, Loomis WF, Levine H, Groisman A, Rappel W-J, Firtel RA. Incoherent feedforward control governs adaptation of activated ras in a eukaryotic chemotaxis pathway. Sci Signal. 2012;5:ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nakajima A, Ishihara S, Imoto D, Sawai S. Rectified directional sensing in long-range cell migration. Nat Commun. 2014;5:5367. doi: 10.1038/ncomms6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zihni C, Terry SJ. RhoGTPase signalling at epithelial tight junctions: Bridging the GAP between polarity and cancer. Int J Biochem Cell Biol. 2015;64:120–125. doi: 10.1016/j.biocel.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McCormack J, Welsh NJ, Braga VMM. Cycling around cell-cell adhesion with Rho GTPase regulators. J Cell Sci. 2013;126:379–91. doi: 10.1242/jcs.097923. [DOI] [PubMed] [Google Scholar]
- 153.Vega FM, Thomas M, Reymond N, Ridley AJ. The Rho GTPase RhoB regulates cadherin expression and epithelial cell-cell interaction. Cell Commun SignaI. 2015;13:1–9. doi: 10.1186/s12964-015-0085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Reffay M, Parrini M-C, Cochet-Escartin Oand Ladoux B, Buguin A, Coscoy S, Amblard Françoisand Camonis J, Silberzan P. Interplay of RhoA and mechanical forces in collective cellmigration driven by leader cells. Nat Cell Biol. 2014;16:217–223. doi: 10.1038/ncb2917. [DOI] [PubMed] [Google Scholar]
- 155.Wisniewska J, Xu J, Seifertová D, Brewer PB, Růžička K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J. Polar PIN Localization Directs Auxin Flow in Plants. Science (80-) 2006;312 doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]


