Abstract
Protonated ferryl (or iron(IV)hydroxide) intermediates have been characterized in several thiolate-ligated heme proteins that are known to catalyze C-H bond activation. The basicity of the ferryl intermediates in these species has been proposed to play a critical role in facilitating this chemistry, allowing hydrogen abstraction at reduction potentials below those that would otherwise lead to oxidative degradation of the enzyme. In this contribution, we discuss the events that led to the assignment and characterization of the unusual iron(IV)hydroxide species, highlighting experiments that provided a quantitative measure of the ferryl basicity, the iron(IV)hydroxide pKa. We then turn to the importance of the iron(IV)hydroxide state, presenting a new way of looking at the role of thiolate ligation in these systems.
Introduction
This article examines the factors that govern the reactivity of ferryl centers in heme enzymes. Special emphasis is placed on understanding the role of the ligand bound trans to the ferryl oxygen. Evidence suggests that an electron donating ligand bound at this position can tune the reactivity of the ferryl moiety, making it more reactive towards C-H bonds.1–4 This concept of axial ligand tuning has its origins in a simple observation: Only thiolate-ligated heme proteins, such as cytochrome P450, are known to activate C-H bonds.5 Histidine-ligated peroxidases, which function through sequential one-electron oxidations, typically do not perform the two-electron chemistry required for C-H bond activation.6 Interestingly, there are peroxidases that are known to cleave C-H bonds, but these peroxidases also possess an axial thiolate ligand.7–11 Thus, there appears to be a real connection between axial thiolate ligation and C-H bond activation. We will explore this connection.
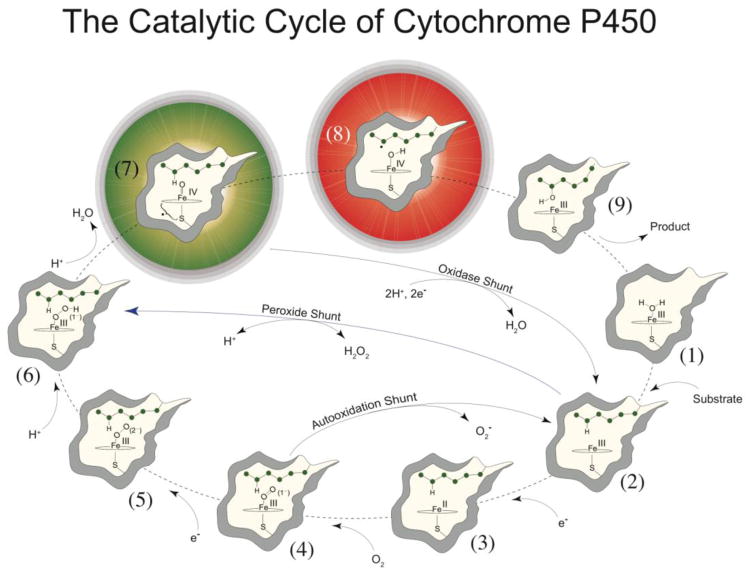
Interest in P450s stems not only from their biological importance but also from a desire to harness their synthetic potential. P450s utilize molecular oxygen and the formal equivalents of dihydrogen (2H+ + 2e−) to catalyze the controlled activation of inert C-H bonds (Figure 1).12–14 The principal oxidant in these demanding transformations is thought to be a short-lived, highly reactive ferryl radical species, called compound I.15–17
Figure 1. General paradigm for P450-catalyzed hydroxylations.
The first step involves the binding of substrate to the resting low-spin ferric enzyme (1). This binding induces structural changes, which often, but not always, manifest themselves in the dissociation of the distally coordinated water and the conversion of the heme from low to high spin (2). These substrate-induced structural changes facilitate reduction of the ferric enzyme, allowing delivery of the first electron to generate the ferrous substrate-bound form of the enzyme (3). Dioxygen then binds to the ferrous heme, forming a species that is best described as a ferric superoxide complex (4). The subsequent reduction of this species forms a ferric peroxo species (5), which is protonated at the distal oxygen to generate a ferric hydroperoxo complex (6). The delivery of an additional proton to the distal oxygen cleaves the O–O bond, yielding compound I (7) and a water molecule. Compound I then abstracts hydrogen from substrate to yield compound II (8) and a substrate radical, which rapidly recombine to yield hydroxylated product and ferric enzyme (9). Hydroxylated product then dissociates, and water coordinates to the heme to regenerate the resting ferric enzyme (1). This figure and caption first appeared in J. Biol. Chem. 2013, 288, 17074–17081.
Ferryl species were first observed in heme peroxidases in the 1930s,18,19 but they were not fully characterized (structurally and electronically) until the 1980s.6,20–26 By the end of the last century, however, it was well understood that the reactive intermediates found in heme peroxidases—known as compound I and compound II—were best described as iron(IV)oxo porphyrin-radical and iron(IV)oxo porphyrin complexes, respectively.27 The S=1 ferryl moiety at the center of these intermediates has two unpaired electrons in orthogonal FeO π* orbitals.28 This electronic configuration results in a double bond with a distance of ~1.65 Å between the iron and oxygen atoms.
Since P450s could be turned over with hydrogen peroxide (via the peroxide shunt pathway, Fig. 1),29,30 it was assumed that similar iron(IV)oxo species existed in P450s. However, due to their highly reactive nature, early insights into these intermediates had to be obtained from calculations and experiments on model systems.28,31–43 Density functional calculations revealed a significant trans-influence between the thiolate and oxo ligands.35 The thiolate ligand is a good π donor, and it can donate electron density into Fe-O orbitals that are π* in character.28 Thus Fe-S bonding is gained at the expense of Fe-O bonding and vice versa. It was speculated that this trans-influence could play an important role in dictating the chemistry of P450s.
To investigate this hypothesis, experiments were performed on chloroperoxidase (CPO).35 CPO is a thiolate-ligated heme peroxidase that is known to cleave activated C-H bonds.11 CPO-I does not perform the more difficult reactions displayed by P450, but, importantly at that time, both CPO compounds I and II could be readily prepared using standard rapid mixing and freezing procedures.44,45
The iron(IV)hydroxide complex
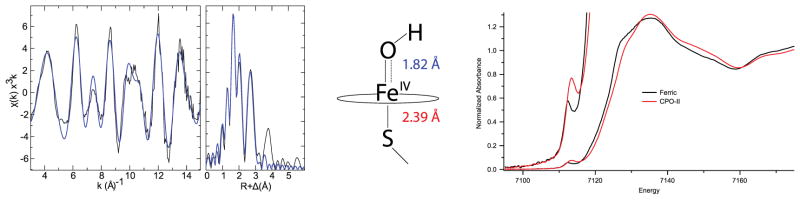
To characterize the extent of the trans-influence in thiolate-ligated ferryl species, x-ray absorption (XAS) measurements were performed to determine metal-ligand bond metrics in CPO-II.35 These experiments revealed an Fe-O bond distance of 1.82 Å, a distance too long to be associated with an iron(IV)oxo species. This result was puzzling, as it was clear from the XAS measurements that the iron center was in a oxidation state that was higher than that of ferric enzyme (Figure 2).
Figure 2.
XAS experiments on chloroperoxidase compound II. EXAFS data for CPO-II (left). X-ray absorption edges for ferric CPO and CPO-II, showing that the CPO-II sample is in a higher oxidation state than ferric (right). Model of iron(IV)hydroxide center showing metal-ligand bond distances obtained from EXAFS data (center). Samples were prepared and data were analyzed as described in Science 2004, 304, 1653–1656.
The long Fe-O bond distance observed for CPO-II suggested that its ferryl center was protonated. Protonation of the ferryl moiety to yield an iron(IV)hydroxide state would decrease the Fe-O formal bond order from 2 to 1.5, lengthening the Fe-O bond distance from ~1.65 Å to the 1.82 Å measured for CPO-II. The driving force for protonation of the ferryl species could be rationalized by a reduction in the strong trans influence between the oxo and thiolate ligands. When the Fe-O bond lengthens upon protonation, the Fe-S bond shortens, strengthening the interaction between iron and sulfur. Additionally, density functional calculations on a thiolate-ligated iron(IV)hydroxide porphyrin complex yielded an Fe-O distance of 1.81 Å, in good agreement with the XAS measurements.35
It was also known that efforts to locate and measure an Fe=O stretching frequency in CPO-II using resonance Raman spectroscopy had been unsuccessful,46 even though the ferryl stretching frequency of the more reactive CPO-I could be readily observed.46–48 The lack of a signal in CPO-II led researchers to conclude that the Fe-O stretch was not enhanced in this system, presumably due to the unique electronic structure of the thiolate-ligated ferryl intermediate.46 However, the lack of a ferryl stretching frequency was clearly consistent with the EXAFS results. As the Fe-O bond lengthened from 1.65 Å to 1.82Å upon protonation, the Fe-O stretching frequency would shift outside the range observed for ferryl species ~700–850 nm, making it a challenge to locate if one were focused on locating an iron(IV)oxo species.
Thus, the available data seemed to point to a protonated compound II. However, it must be remembered that protons cannot be “seen” in X-ray absorption measurements, and there was a lack of signal in the resonance Raman experiments. The existence of the iron(IV)hydroxide state could only be inferred from the points outlined above.
The assignment of the iron(IV)hydroxide state in CPO-II was not without controversy. At the time, there were no synthetic or mineral iron-hydroxide complexes known above the ferric state. Even today, a definitive characterization of a synthetic iron(IV)hydroxide complex remains elusive.49,50 Ferryl species are typically electrophilic in nature, and the report of a nucleophilic/basic iron(IV)oxo in CPO-II was somewhat unexpected. There were some suggestions that the CPO-II sample may have been photoreduced in the synchrotron x-ray beam, and that the long 1.82 Å Fe-O bond obtained from XAS measurements was representative of ferric enzyme in an unusual coordination environment.
To complicate the matter, a number of histidine-ligated compound II intermediates were suggested to be protonated as well.51 Crystallographic studies reported Fe-O bond distances of 1.84, 1.92, and 1.87 Å for horseradish peroxidase compound II (HRP-II), myoglobin compound II (Mb-II), and cytochrome c peroxidase compound I (CCP-I, also called compound ES, which is like compound II, as it contains a ferryl center with a distant tryptophan radical).51–54 However, the distances obtained from crystallography were at odds with the results from spectroscopic measurements. Resonance Raman experiments at varying pH yielded Fe-O stretching frequencies between 753–805 cm−1 for these intermediates, indicating they were authentic iron(IV)oxo species.55–58 XAS measurements on these intermediates yielded Fe-O bond distance of ~1.65 Å, in agreement with the resonance Raman measurements.22,59,60
It is important to recall that the opposite was true for CPO-II. Spectroscopic measurements suggested that CPO-II was not an iron(IV)oxo species: a ferryl stretch could not be observed and XAS measurement indicated a long Fe-O bond. Thus, there seemed to be something different about the thiolate-ligated system. However, questions about the photoreduction of CPO-II in the synchrotron X-ray beam as well as the assignment of the long Fe-O bonds obtained from crystallographic studies of peroxidases were confounding the issue. To clarify things, two lines of investigation were employed. The first involved the use of Mössbauer spectroscopy to simultaneously determine the oxidation and protonation states of the iron center in compound II samples.61 The second focused on establishing an empirical relationship, known as Badger’s rule, between Fe-O bond distance and stretching frequency for a series of heme and non-heme complexes.62,63
57Fe Mössbauer spectroscopy is a nuclear resonance technique that can be used to assign the oxidation and spin states of iron-containing samples. The technique is particularly suited for investigating the protonation state of the compound II species. Ferryl porphyrins have two unpaired electrons that couple together to yield an S=1 ground state. Integer spin systems, such as this, have distinctive Mössbauer spectra called quadrupole doublets. These spectra consist of two sharp lines separated by a quantity known as the quadrupole splitting. This splitting is known to range from ~ 1.2–1.5 mm/s for ferryl porphyrins. Density functional calculations predicted that protonation of the ferryl moiety would increase the quadrupole splitting to over 2.0 mm/s.61 To test this prediction, cryogenic reduction and annealing techniques were used to generate CPO-II in its iron(IV)oxo and iron(IV)hydroxide forms.61 CPO-I was radiolytically reduced at 77K to produce CPO-II in its unprotonated form. The sample was then annealed at 180K (the lowest temperature at which protons were found to move) allowing protonation of the ferryl moiety. Mössbauer measurements taken before and after annealing were consistent with the theoretical predictions. The quadrupole splitting of the ferryl moiety increased from 1.41 to 2.08 mm/s upon protonation. These experiments seemed to confirm the XAS assignment of the iron(IV)hydroxide center in CPO-II. However, it is important to remember that, just like with XAS, protons cannot be seen with Mössbauer spectroscopy, and the iron(IV)hydroxide state could only be inferred from the changes observed upon annealing of the cryoreduced CPO-I sample and the agreement with the results obtained from DFT calculations. To obtain direct evidence for the existence of the iron(IV)hydroxide state, the Fe-OH stretching frequency would need to be located.
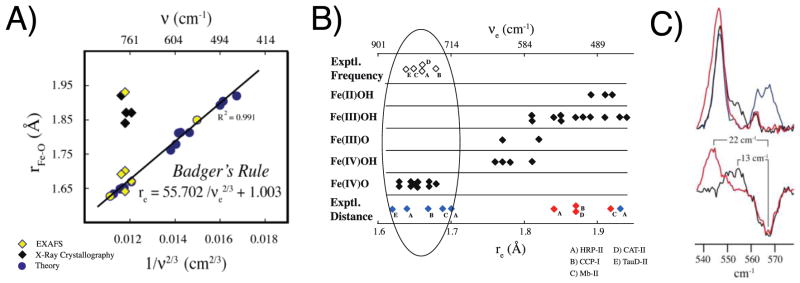
In an effort to locate the Fe-OH vibration in CPO-II and make sense of the structural and vibrational data available for the peroxidase and globin intermediates, researchers turned to Badger’s rule.62 The hope was that this empirical relationship could provide insight into discrepancies in the bond distances obtained from crystallography and spectroscopy, while predicting a location for the hydroxide stretch in CPO-II. Using density functional theory (DFT), the applicability of Badger’s rule to Fe-O bonds in heme and non-heme systems was examined for over 30 compounds. The bond distances and stretching frequencies obtained from DFT calculations were found to be in good agreement with Badger’s rule (R2 = 0.991). Plotting the Fe-O distances and stretching frequencies obtained from experiments revealed an interesting trend. The values obtained from spectroscopy were consistent with Badger’s rule, while the distances obtained from crystallography were not (Figure 3A). The Badger’s rule analysis revealed that the long Fe-O distances obtained from the peroxidase structures were the hallmarks of ferric/ferrous enzyme (Figure 3B). That is, the peroxidase compound II crystals had been reduced in the synchrotron beam. Importantly, Badger’s rule predicted that the Fe-OH stretch of the iron(IV)hydroxide center in CPO-II would be located at 563 cm−1, well outside the region were iron(IV)oxo vibrations are found. Using resonance Raman spectroscopy in conjunction with 18O isotopic labeling, the Fe-OH stretch in CPO-II was identified at 565 cm−1 (Figure 3C).64 This vibrational frequency was found to downshift 12 cm−1 in D2O, in excellent agreement with the value predicted for an Fe-OH diatomic harmonic oscillator (where the OH is treated as a single atom).64 These resonance Raman experiments provided the first direct evidence for the existence of an iron(IV)hydroxide species in any system.
Figure 3.
Badger’s rule analysis and resonance Raman data. A) Fe-O bond length versus 1/νFeO2/3. Yellow and blue circles represent calculated bond distances and frequencies. Best-fit line to theoretical data is given by Badger’s rule. Diamonds represent experimental data. Yellow diamonds are from resonance Raman and EXAFS experiments. Black diamonds are from resonance Raman and X-ray crystallography. This figure first appeared in J. Am. Chem. Soc. 2006, 128, 1902–1906. B) Bottom axis is Fe-O bond distance. Top axis is vibrational frequency obtained by applying Badger’s rule as parametrized in A. Black diamonds represent theoretical results obtained from calculations on a series of compounds. The top and bottom rows contain the results of experimental investigations. Open diamonds represent the experimental vibrational frequencies obtained from resonance Raman measurements. Blue and red diamonds represent the bond distances obtained from EXAFS measurements and X-ray crystallography, respectively. Experimental systems are (A) HRP-II, (B) CCP-I, (C) Mb-II, (D) CAT-II, and (E) the ferryl form of TauD. The ellipse highlights the agreement between theory and EXAFS and resonance Raman experiments. This figure first appeared in J. Am. Chem. Soc. 2006, 128, 1902–1906. C) Low-frequency-resonance Raman data of CPO-II. (Upper) Overlay of CPO-II spectra: 16OH (blue), 16OD (black), and 18OH (red). (Lower) Difference spectra reveal changes in νFe(IV)–OH upon deuterium (black) and 18O (red) substitutions. This figure first appeared in PNAS, 2006, 103, 12307–12310.
The Badger’s rule analysis outlined above inspired follow up investigations of the peroxidase and globin systems. In a dosage-dependent crystallographic study, it was found that the Fe-O bond length in CCP-I correlated well with x-ray exposure time.65 At very low doses it was found that CCP-I did indeed contain an authentic iron(IV)oxo center, but as the exposure increased the ferryl species was reduced and the Fe-O bond lengthened. This result was confirmed by a neutron diffraction study of CCP-I.66 Neutron diffraction does not suffer from photoreduction and perhaps more importantly the technique can see protons. Neutron diffraction of CCP-I revealed that this system is not protonated. It contains an authentic iron(IV)oxo species, consistent with the results of the Badger’s rule analysis. More recently a study which used a combination of EXAFS, Mössbauer, and resonance Raman spectroscopies found that Mb-II was also not protonated.67 Mössbauer measurements suggested that the iron(IV)hydroxide state of Mb-II has a pKa ≤ 3. To date, there has been no evidence to suggest that an iron(IV)hydroxide state can be stabilized in any histidine-ligated heme protein. In contrast, the iron(IV)hydroxide state has been characterized in a number of thiolate-ligated heme proteins.35,51,68,69
The Iron(IV)hydroxide pKa
The importance of the iron(IV)hydroxide state lies in its connection to P450 chemistry. P450s function through the controlled activation of C-H bonds. In the generally accepted “rebound mechanism”, Compound I abstracts hydrogen from the substrate to produce an iron(IV)hydroxide species and a substrate radical, which rapidly recombine to yield hydroxylated product.12,13 This reaction, as shown in Fig. 1 (species 7–9), involves H-atom abstraction by a metal-oxo complex. There is considerable evidence that reactions of this type are controlled by ground state thermodynamics.70,71 That is, the reactivity (or rate of reaction) is related to the driving force, which is given by the difference between the iron(IV)hydroxide D(O-H) and the substrate D(C-H) bond strengths. For unactivated hydrocarbons, D(C-H) is typically around 100 kcal/mol, while the iron(IV)hydroxide bond strength can be determined from Eq. 1.5,71,72 This equation shows that D(O-H) depends upon the one-electron reduction potential of compound I and the pKa of the iron(IV)hydroxide complex (i.e. compound II). These thermodynamic parameters hold the key to understanding C-H bond activation in these systems. To date, only the iron(IV)hydroxide pKa has been measured.69
| Eq. 1 |
As one might imagine, the measurement of thermodynamic quantities can be quite challenging for short-lived reactive species. And, for measurements of the compound II pKa, there is the additional complication of pH stability. Investigations of CPO-II, for example, were hindered not so much by the lifetime of CPO-II but by the enzyme’s pH stability. CPO undergoes an irreversible structural rearrangement above pH 7 that renders the enzyme inactive.73 As a result, only a lower limit (pKa≥8) could be obtained for the iron(IV)hydroxide state in this system.35 Investigations with P450CAM and P450BM3 showed that the compound II forms of these enzymes were also iron(IV)hydroxides at pH 7.74 Reactions with these P450s could be pushed to pH 8, but there was a significant reduction in the already low yields of compound II, making these systems unsuitable for determining the iron(IV)hydroxide pKa. Thus, while various spectroscopic measurements could confirm the basic nature of thiolate ligated ferryls in these systems, low yields and the inability to prepare the intermediates at high pH precluded efforts to determine the pKa of these iron(IV)hydroxide species.
A breakthrough came in the form of CYP158, a P450 with a large solvent exposed active site from the soil bacterium S. coelicolor.69,75 During efforts to characterize high-valent intermediates in this system, it was found that meta-chloroperbenzoic acid (m-CPBA) could be used to prepare compound II in high yield. Importantly, CYP158 showed excellent pH stability on short (100ms) timescales, and the enzyme could be taken to high pH and returned to neutral (or slightly basic) conditions without degradation. As a result, rapid mixing techniques could be used to prepare CYP158-II over a wide pH range. Mössbauer, UV/Visible, and EXAFS spectroscopies on these samples revealed an iron(IV)hydroxide center with pKa=12. This is a remarkable value. It indicates that the thiolate-ligated ferryl in CYP158-II is at least 9 orders of magnitude more basic than the ferryl center in Mb-II.67
It is reasonable to question how much of this increase in basicity can be attributed to differences in the active site environment versus change in axial ligation. It is known, for example, that the alkaline transition of HRP, which converts the water-bound ferric high-spin form of HRP to the hydroxide-bound low-spin form of the enzyme, is isozyme-dependent. HRP isozymes A and C have pKas of 9.2 and 10.7, respectively, indicating that differences in proximal and distal environments can play a role in modulating ligand basicity.76 However, it seems clear that the lion’s share of the increase in basicity must originate from the difference in axial ligation. An iron(IV)hydroxide has yet to be characterized in a histidine-ligated compound II, while all of the thiolate-ligated species are known to be iron(IV)hydroxides. To investigate variations in the thiolate-ligated systems, efforts were made to measure the compound II pKa in another P450, one with a very different substrate and active site. Key to these efforts were insights gained from CYP158, particularly the understanding of the mechanism by which compound II was generated in such high yield.
The use of m-CPBA, a two-electron oxidant, to prepare CYP158-II, which is only one equivalent above the ferric state, may seem surprising, but it is typical for P450s. For example, the P450CAM-II and P450BM3-II species discussed above were prepared with peracetic acid. The reaction of ferric enzyme with these oxidants produces compound I, and, in the absence of substrate, this highly reactive intermediate can strip an electron from the protein framework to yield compound II and a protein-based radical (resulting in a species similar to CCP-I). The oxidation of tyrosine and tryptophan residues in this manner has proven to be a major obstacle in efforts to isolate P450-I for characterization.77,78 In CYP158, however, this feature of P450 chemistry proved to be advantageous for the preparation of compound II.
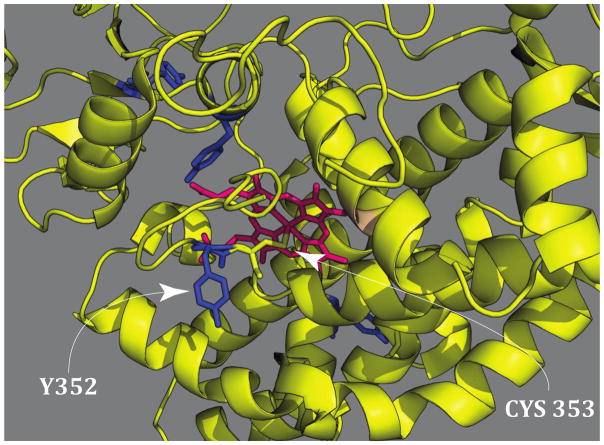
In the generation of CYP158-II from CYP158-I, the protein-derived reducing equivalent comes from a solvent-exposed tyrosine adjacent to the axial cysteine ligand (Fig. 4). This tyrosine is well-coupled to the active site, allowing for the efficient and rapid reduction of compound I to compound II. As a result, very high yields of CYP158-II can be obtained. The tyrosine was confirmed to be the origin of the reducing equivalent through site-directed mutagenesis. Replacing the tyrosine with a phenylalanine resulted in a CYP158 variant that allowed for the preparation of P450-I in high yield (~80%).69
Figure 4.
CYP158 crystal structure. (PDB accession code: 1S1F) The arrow on the left designates the positions of tyrosine 352 which is adjacent to the axial cysteine and ~ 10 Å away from the heme iron atom. Tyrosine 352 was changed to a phenyalanine in the CYP158 variant that generates compound I. This figure was adapted from the supporting information of Science, 2013, 342, 825–829.
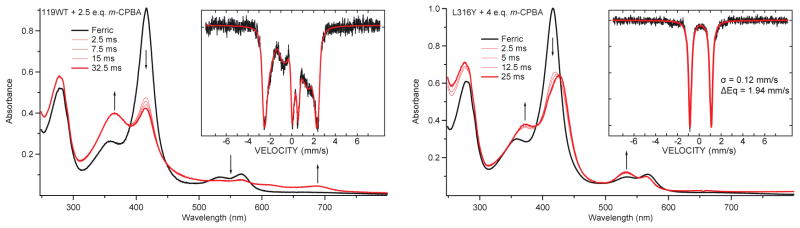
Using this insight, the iron(IV)hydroxide pKa of a second P450 was determined. CYP119 is a P450 that is known to react with m-CPBA to produce compound I in high yield.15 CYP119-I decays rapidly to ferric enzyme, but compound II does not accumulate in the wild type enzyme. The incorporation of a tyrosine residue adjacent to the axial cysteine of CYP119 results in a variant that produces compound II in high yield (Fig. 5).69 Similar pH jump experiments indicate that the iron(IV)hydroxide center of CYP119-II has a pKa=12. Fig. 6 shows the results from these experiments.
Figure 5.
The reaction of m-CPBA with wild type CYP119 generates compound I in high yield (left). The L316Y CYP119 variant incorporates a tyrosine at the same position as Y352 in CYP158. The reaction of this variant with m-CPBA generates compound II in high yield (right). Samples were prepared and analyzed as described in Science, 2013, 342, 825–829.
Figure 6.
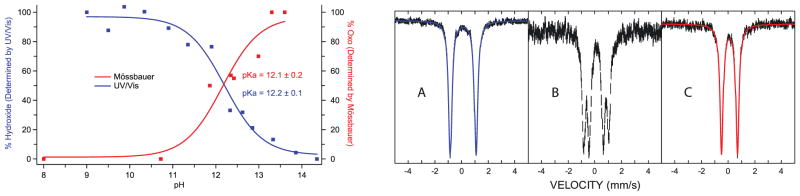
The pKa of the L316Y CYP119-II variant. Relative concentrations of oxo and hydroxo species obtained from UV/Visible and Mössbauer experiments at varying pH. Mossbauer and UV/Visible measurements both yield a pKa=12 (left). Mossbauer spectra of CYP119-II at pH 8.0 (A), pH 11.8 (B), and pH 13.6 (C). As the pH increases the iron(IV)hydroxide state transitions to the oxo species which has a smaller quadrupole splitting. These figures first appeared in the supporting information of Science, 2013, 342, 825–829.
More recently an iron(IV)hydroxide pKa was measured for the thiolate-ligated aromatic peroxygenase (APO) from Agrocybe aegerita. APO is also a potent catalyst, with a high activity for hydrocarbon oxidations. APO-II was generated from APO-I using nitroxyl radicals, and stopped-flow UV/Visible measurements were used to obtain a pKa=10 for the iron(IV)hydroxide state of this system.68
The Role of the Thiolate
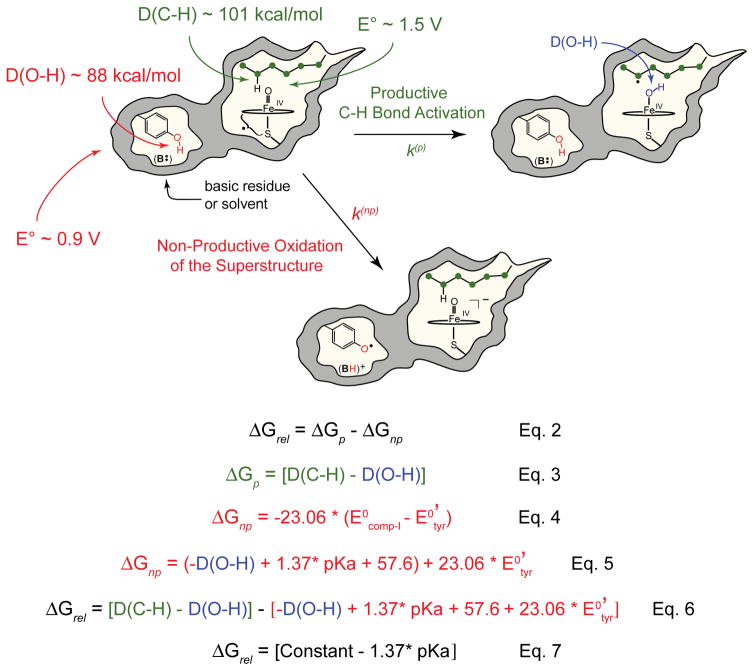
We now return to the importance of the iron(IV)hydroxide pKa and its connection to C-H bond activation. It has been proposed that P450’s axial thiolate ligand facilitates C-H bond activation at biologically viable compound I potentials, promoting substrate oxidation, while minimizing off pathway oxidations of the protein superstructure.35,69 To understand this proposal, it is helpful to consider the relative free energy for the productive and non-productive pathways illustrated in Figure 7.
Figure 7.
Comparison of the productive and non-productive pathways for compound I decay. Equations show how the relative free energy for these pathways is calculated. See text for discussion.
In the productive pathway compound I abstracts hydrogen from substrate to form an iron(IV)hydroxide and a substrate radical. These species subsequently recombine to yield the hydroxylated product. In the non-productive pathway compound I oxidizes a tyrosine (or tryptophan) residue generating an unprotonated compound II species and a protein based radical.
The free energy change for the productive pathway, ΔGp, is the difference between the iron(IV)hydroxide O-H bond strength, D(O-H), and the substrate C-H bond strength, D(C-H) (Fig. 7, Eq. 3), while ΔGnp for the non-productive pathway is determined by the reduction potentials of compound I and the oxidized tyrosine residue (Fig. 7, Eq. 4).
Using Eq. 1 for D(O-H), ΔGnp can be written as Eq. 5 in Fig. 7, allowing the relative free energy, ΔGrel, to be expressed as Eq. 6 in Fig. 7. This latter equation has some interesting features. First, notice the D(O-H) terms cancel, making ΔGrel independent of D(O-H). Second, for a given substrate and tyrosine environment, ΔGrel is a line with a slope of −1.37 with respect to the iron(IV)hydroxide pKa. Thus, for each pKa unit, the relative free energy shifts towards the productive pathway by 1.37 kcal/mol.
Consider the implications for the role of the axial thiolate ligand in P450. Experiments indicate that the difference in pKa between a histidine and thiolate ligated iron(IV)hydroxide species is > 9 units. This means that the thiolate ligand shifts ΔGrel toward the productive pathway by > 12 kcal/mol relative to a histidine ligated system.
It has been argued that an elevated iron(IV)hydroxide pKa affects not only the thermodynamics of the system but also the kinetics.69 For a given ΔGp, a 12 kcal/mol shift of ΔGrel towards the productive pathway corresponds to a drop of > 0.5 V in the compound I reduction potential. This drop in the potential attenuates the rate constants for non-productive oxidations, biasing the system towards C-H bond activation.
The ideas expressed in the preceding paragraphs were originally developed in Yosca et al.,69 and we direct the interested reader there for the full mathematical details. What we would like to present here is a new graphical way of thinking about these ideas, one that makes use of concepts from freshman chemistry (like Hess’s law and the sign of ΔG) that we have found to be effective when discussing the importance of the iron(IV)hydroxide pKa with a general audience.
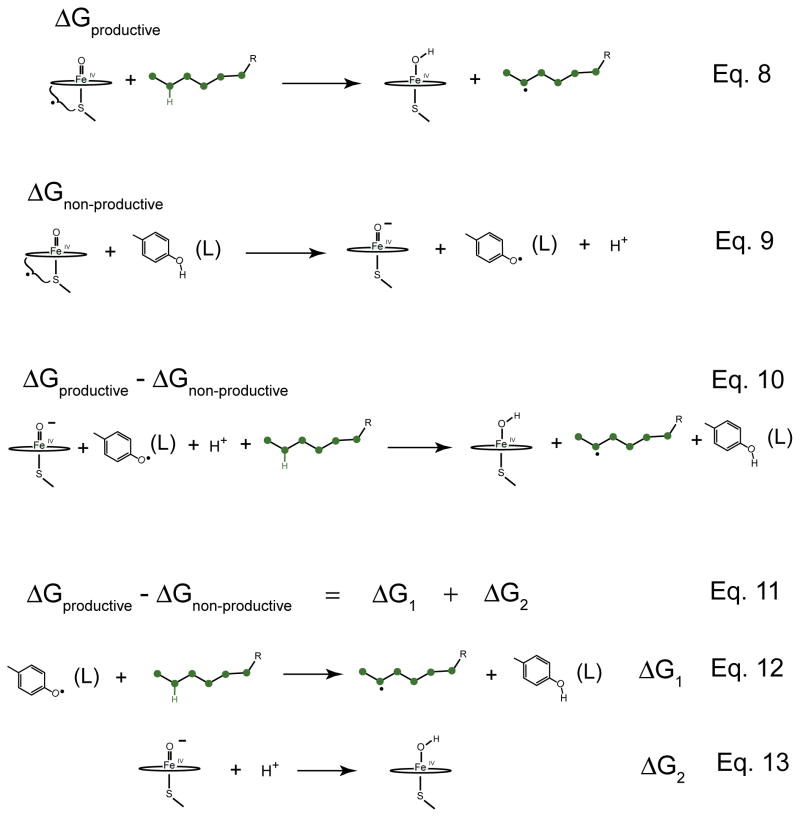
Eq. 8 in Fig. 8 shows the productive reaction. Again, compound I abstracts an H-atom from the substrate, leaving the iron(IV)hydroxide and the substrate radical. The non-productive reaction is illustrated in Eq. 9 of Fig. 8. Compound I oxidizes a tyrosine residue by one electron, generating a ferryl species, a tyrosine radical, and releasing a proton. Subtracting Eq. 9 from Eq. 8 yields ΔGrel, which is given by Eq. 10. It is interesting to note that compound I does not appear in the ΔGrel reaction.
Figure 8.
Understanding the role of the thiolate ligand in shifting the relative free energy of the productive and non-productive pathways. Eq. 8 represents the productive pathway. It involves the oxidation of substrate by compound I. Eq. 9 represents the non-productive pathway. It involves the oxidation of tyrosine residue by compound I. Subtracting Eq. 9 from Eq. 8 gives Eq. 10 for ΔGrel. To keep the system on the productive pathway, we would like to push this equation to the right, minimizing ΔGrel. Using Hess’s law Eq. 10 can be split into two new reactions (Eqs. 12 and 13). The C-H bond of the substrate is stronger than the O-H bond of the tyrosine. Thus, ΔG1 is positive. To minimize ΔGrel, we must rely on ΔG2. With increasing pKa, ΔG2 decreases, pushing the system towards the productive pathway.
To keep the system on the productive pathway, we would like to push this reaction to the right, finding conditions to minimize ΔGrel. Here it helps to use Hess’s law to break ΔGrel into two new reactions, Eq. 11. The first of these is shown in Eq. 12 of Fig. 8. In this reaction a tyrosine radical abstracts hydrogen from the substrate to yield a tyrosine and a substrate radical. It is clear that this reaction will be unfavorable. The C-H bond is much stronger than the O-H bond. So, the driving force for this reaction, ΔG1, is positive and, for a given substrate and tyrosine environment, a constant. Therefore, to minimize ΔGrel, we must rely upon the second equation (Eq. 13 in Fig. 8). This equation involves the protonation of a ferryl species to make an iron(IV)hydroxide complex. With increasing pKa, ΔG2 decreases, and it is only by this mechanism that we can push ΔGrel towards the productive pathway.
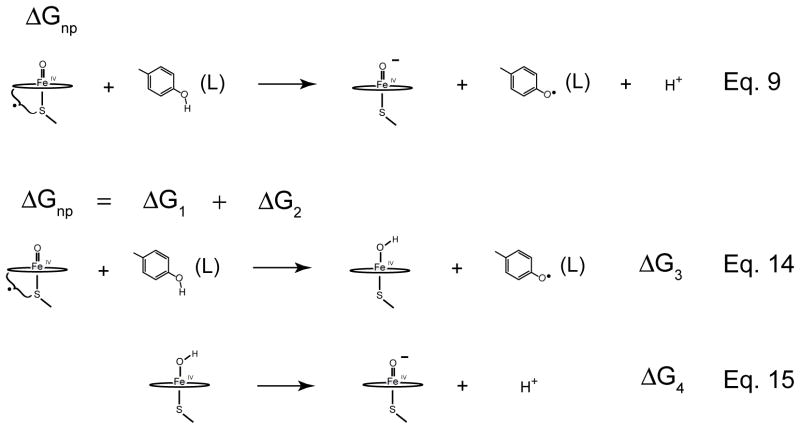
We can use a similar analysis to understand how the thiolate ligand attenuates the rate constants for non-productive oxidations (Fig. 9). Here we want to maximize ΔGnp, pushing the reaction towards the left. Again it helps to uses Hess’s law to generate two new equations. The first of these reactions is the oxidation of tyrosine by compound I (Eq. 14 in Fig. 9). The driving force for this reaction, ΔG3, is clearly negative. In the absence of substrate, compound I readily oxidizes tyrosine residues. Again we turn to the second reaction to achieve our goal. Eq. 15 in Fig. 9 is the deprotonation of the iron(IV)hydroxide state. With increasing pKa, ΔG4 increases, maximizing ΔGnp and minimizing the driving force for non-productive oxidations.
Figure 9.
Understanding the role of the thiolate ligand in attenuating non-productive rate constants. Eq. 9 represents the non-productive pathway. Using Marcus theory the rate constants for the non-productive oxidations can be related to the driving force for these processes. To attenuate the rate constant for non-productive oxidations we would like maximize ΔGnp. Using Hess’s law, ΔGnp can be split into two new reactions (Eqs. 14 and 15). The first of these involves the oxidation of a tyrosine by compound I to yield and iron(IV)hydroxide and a tyrosine radical. ΔG3 is clearly negative. In the absence of substrate compound I readily oxidizes tyrosine and tryptophan residues. Again we must rely on the second equation to accomplish our goal. Increasing the pKa increases ΔG4, pushing Eq. 9 to the left and minimizing the non-productive rate constants.
Conclusion
In closing, we thought it would be of interest to briefly touch upon some of the questions that remain concerning the role of the thiolate ligand in C-H bond activation and its effect upon compound I and the iron(IV)hydroxide state. We have discussed a role for the thiolate ligand in promoting C-H bond activation by minimizing off pathway chemistry. But it is possible that strong electron donation from the axial thiolate ligand also makes compound I more reactive with respect to C-H bond activation. Work on synthetic model systems has shown that reactivity with respect to oxygen atom transfer and C-H bond activation increases with electron donation trans to the oxo ligand.1–3 However, this connection has proven difficult to test in P450s, and to date a direct link between electron donation and reactivity has yet to be established.
The ability to prepare compound I in high yields in CPO and P450 has allowed for detailed comparisons of the electronic and geometric structures of these intermediates. EXAFS measurements revealed a significantly shorter (0.1 Å) Fe-S bond in P450-I than in CPO-I. The shorter Fe-S bond in P450-I suggests increased electron donation in P450, consistent with its greater reactivity towards C-H bonds. But, if electron donation does increase the reactivity of compound I with respect to C-H bond activation, how does it do it? Does it weaken the Fe-O bond of compound I, priming the ferryl oxygen for H atom abstraction?79 Does it increase the D(O-H) bond strength of the iron(IV)hydroxide state?80 Density functional calculations suggest that the oxidation of unactivated hydrocarbons by compound I is about ~6 kcal/mol uphill.81 Is this correct?
With respect to the characterization of P450 compound I, some high value targets remain that could provide considerable insight into the role of the axial thiolate ligand. Principal among these is the reduction potential of compound I. Obtaining this quantity for an enzyme intermediate with a half-life of ~100 ms is obviously a challenging prospect. However, the determination of this parameter would yield D(O-H), Eq.1, allowing insight into the protective role of the thiolate and the importance of ground state thermodynamics in this system. Hopefully future investigations will be able to address this issue as well as the questions outlined above, providing further insight into fundamental aspects of C-H bond activation in this important class of enzymes.
Acknowledgments
This work was supported by NIH (R01-GM101390).
References
- 1.Prokop KA, de Visser SP, Goldberg DP. Angewandte Chemie-International Edition. 2010;49:5091. doi: 10.1002/anie.201001172. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi A, Kurahashi T, Fujii H. Inorg Chem. 2009;48:2614. doi: 10.1021/ic802123m. [DOI] [PubMed] [Google Scholar]
- 3.Ohno T, Suzuki N, Dokoh T, Urano Y, Kikuchi K, Hirobe M, Higuchi T, Nagano T. J Inorg Biochem. 2000;82:123. doi: 10.1016/s0162-0134(00)00141-0. [DOI] [PubMed] [Google Scholar]
- 4.Krest CM, Silakov A, Rittle J, Yosca TH, Onderko EL, Calixto JC, Green MT. Nat Chem. 2015;7:696. doi: 10.1038/nchem.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green MT. Curr Opin Chem Biol. 2009;13:84. doi: 10.1016/j.cbpa.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 6.Dunford HB. Peroxidases and Catalases: Biochemistry, Biophysics, Biotechnology, and Physiology. 2. John Wiley and Sons; New York: 2010. [Google Scholar]
- 7.Peter S, Kinne M, Wang X, Ullrich R, Kayser G, Groves JT, Hofrichter M. FEBS Journal. 2011;278:3667. doi: 10.1111/j.1742-4658.2011.08285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Peter S, Kinne M, Hofrichter M, Groves JT. J Am Chem Soc. 2012;134:12897. doi: 10.1021/ja3049223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Peter S, Ullrich R, Hofrichter M, Groves JT. Angewandte Chemie-International Edition. 2013;52:9238. doi: 10.1002/anie.201302137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Ullrich R, Hofrichter M, Grove JT. P Natl Acad Sci USA. 2015;112:3686. doi: 10.1073/pnas.1503340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaks A, Dodds DR. J Am Chem Soc. 1995;117:10419. [Google Scholar]
- 12.de montellano pro. cytochrome p450: structure, mechanism, and biochemistry. 3. kluwer academic/plenum publishers; 2005. [Google Scholar]
- 13.Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem Rev. 2005;105:2253. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 14.de Montellano PRO. Chem Rev. 2010;110:932. [Google Scholar]
- 15.Rittle J, Green MT. Science. 2010;330:933. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 16.Egawa T, Shimada H, Ishimura Y. Biochemical and Biophysical Research Communications. 1994;201:1464. doi: 10.1006/bbrc.1994.1868. [DOI] [PubMed] [Google Scholar]
- 17.Kellner DG, Hung SC, Weiss KE, Sligar SG. J Biol Chem. 2002;277:9641. doi: 10.1074/jbc.C100745200. [DOI] [PubMed] [Google Scholar]
- 18.Keilin D, Mann T. Proc R Soc Ser B-Bio. 1937;122:119. [Google Scholar]
- 19.Theorell H. Metal Species in Chemical and Biochemical Oxidations. 1941:303. [Google Scholar]
- 20.Roberts JE, Hoffman BM, Rutter R, Hager LP. J Am Chem Soc. 1981;103:7654. [PubMed] [Google Scholar]
- 21.Roberts JE, Hoffman BM, Rutter R, Hager LP. J Biol Chem. 1981;256:2118. [PubMed] [Google Scholar]
- 22.Pennerhahn JE, Eble KS, McMurry TJ, Renner M, Balch AL, Groves JT, Dawson JH, Hodgson KO. J Am Chem Soc. 1986;108:7819. doi: 10.1021/ja00284a054. [DOI] [PubMed] [Google Scholar]
- 23.Pennerhahn JE, McMurry TJ, Renner M, Latosgrazynsky L, Eble KS, Davis IM, Balch AL, Groves JT, Dawson JH, Hodgson KO. J Biol Chem. 1983;258:2761. [PubMed] [Google Scholar]
- 24.Moss TH, Ehrenber A, Bearden AJ. Biochemistry-Us. 1969;8:4159. doi: 10.1021/bi00838a037. [DOI] [PubMed] [Google Scholar]
- 25.Palaniappan V, Terner J. J Biol Chem. 1989;264:16046. [PubMed] [Google Scholar]
- 26.Kincaid JR, Zheng YH, AlMustafa J, Czarnecki K. J Biol Chem. 1996;271:28805. doi: 10.1074/jbc.271.46.28805. [DOI] [PubMed] [Google Scholar]
- 27.Kuramochi H, Noodleman L, Case DA. J Am Chem Soc. 1997;119:11442. [Google Scholar]
- 28.Green MT. J Am Chem Soc. 1999;121:7939. [Google Scholar]
- 29.Nordblom GD, White RE, Coon MJ. Arch Biochem Biophys. 1976;175:524. doi: 10.1016/0003-9861(76)90541-5. [DOI] [PubMed] [Google Scholar]
- 30.Hrycay EG, Gustafsson JA, Ingelmansundberg M, Ernster L. Biochemical and Biophysical Research Communications. 1975;66:209. doi: 10.1016/s0006-291x(75)80315-9. [DOI] [PubMed] [Google Scholar]
- 31.Groves JT, Mcclusky GA. J Am Chem Soc. 1976;98:859. [Google Scholar]
- 32.Groves JT, Vanderpuy M. J Am Chem Soc. 1976;98:5290. [Google Scholar]
- 33.Gross Z, Nimri S. Inorg Chem. 1994;33:1731. [Google Scholar]
- 34.Weiss R, Mandon D, Wolter T, Trautwein AX, Müther M, Bill E, Gold A, Jayaraj K, Terner J. Journal of Biological Inorganic Chemistry. 1996;1:377. doi: 10.1021/ic951058v. [DOI] [PubMed] [Google Scholar]
- 35.Green MT, Dawson JH, Gray HB. Science. 2004;304:1653. doi: 10.1126/science.1096897. [DOI] [PubMed] [Google Scholar]
- 36.Harris D, Loew G, Waskell L. J Inorg Biochem. 2001;83:309. doi: 10.1016/s0162-0134(00)00177-x. [DOI] [PubMed] [Google Scholar]
- 37.Harris DL, Loew GH. J Am Chem Soc. 1998;120:8941. [Google Scholar]
- 38.Ogliaro F, Cohen S, de Visser SP, Shaik S. J Am Chem Soc. 2000;122:12892. [Google Scholar]
- 39.Ogliaro F, Filatov M, Shaik S. Eur J Inorg Chem. 2000:2455. [Google Scholar]
- 40.Ogliaro F, Harris N, Cohen S, Filatov M, de Visser SP, Shaik S. J Am Chem Soc. 2000;122:8977. [Google Scholar]
- 41.Harris N, Cohen S, Filatov M, Ogliaro F, Shaik S. Angewandte Chemie-International Edition. 2000;39:2003. doi: 10.1002/1521-3773(20000602)39:11<2003::aid-anie2003>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 42.Ogliaro F, Cohen S, Filatov M, Harris N, Shaik S. Angewandte Chemie-International Edition. 2000;39:3851. doi: 10.1002/1521-3773(20001103)39:21<3851::AID-ANIE3851>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 43.Filatov M, Harris N, Shaik S. Angewandte Chemie-International Edition. 1999;38:3510. doi: 10.1002/(sici)1521-3773(19991203)38:23<3510::aid-anie3510>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 44.Lambeir AM, Dunford HB, Pickard MA. Eur J Biochem. 1987;163:123. doi: 10.1111/j.1432-1033.1987.tb10744.x. [DOI] [PubMed] [Google Scholar]
- 45.Rutter R, Hager LP, Dhonau H, Hendrich M, Valentine M, Debrunner P. Biochemistry-Us. 1984;23:6809. doi: 10.1021/bi00321a082. [DOI] [PubMed] [Google Scholar]
- 46.Egawa T, Proshlyakov DA, Miki H, Makino R, Ogura T, Kitagawa T, Ishimura Y. Journal of Biological Inorganic Chemistry. 2001;6:46. doi: 10.1007/s007750000181. [DOI] [PubMed] [Google Scholar]
- 47.Hosten CM, Sullivan AM, Palaniappan V, Fitzgerald MM, Terner J. J Biol Chem. 1994;269:13966. [PubMed] [Google Scholar]
- 48.Egawa T, Miki H, Ogura T, Makino R, Ishimura Y, Kitagawa T. Febs Lett. 1992;305:206. doi: 10.1016/0014-5793(92)80668-7. [DOI] [PubMed] [Google Scholar]
- 49.Hill EA, Weitz AC, Onderko E, Romero-Rivera A, Guo Y, Swart M, Bominaar EL, Green MT, Hendrich MP, Lacy DC, Borovik AS. J Am Chem Soc. 2016;138:13143. doi: 10.1021/jacs.6b07633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boaz NC, Bell SR, Groves JT. J Am Chem Soc. 2015;137:2875. doi: 10.1021/ja508759t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Behan RK, Green MT. J Inorg Biochem. 2006;100:448. doi: 10.1016/j.jinorgbio.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 52.Berglund GI, Carlsson GH, Smith AT, Szoke H, Henriksen A, Hajdu J. Nature. 2002;417:463. doi: 10.1038/417463a. [DOI] [PubMed] [Google Scholar]
- 53.Hersleth HP, Dalhus B, Gorbitz CH, Andersson KK. Journal of Biological Inorganic Chemistry. 2002;7:299. doi: 10.1007/s007750100296. [DOI] [PubMed] [Google Scholar]
- 54.Bonagura CA, Bhaskar B, Shimizu H, Li HY, Sundaramoorthy M, McRee DE, Goodin DB, Poulos TL. Biochemistry-Us. 2003;42:5600. doi: 10.1021/bi034058c. [DOI] [PubMed] [Google Scholar]
- 55.Sitter AJ, Reczek CM, Terner J. J Biol Chem. 1985;260:7515. [PubMed] [Google Scholar]
- 56.Sitter AJ, Reczek CM, Terner J. Biochim Biophys Acta. 1985;828:229. doi: 10.1016/0167-4838(85)90301-2. [DOI] [PubMed] [Google Scholar]
- 57.Reczek CM, Sitter AJ, Terner J. J Mol Struct. 1989;214:27. [Google Scholar]
- 58.Hashimoto S, Teraoka J, Inubushi T, Yonetani T, Kitagawa T. J Biol Chem. 1986;261:1110. [PubMed] [Google Scholar]
- 59.Chance M, Powers L, Kumar C, Chance B. Biochemistry-Us. 1986;25:1259. doi: 10.1021/bi00354a010. [DOI] [PubMed] [Google Scholar]
- 60.Chance M, Powers L, Poulos T, Chance B. Biochemistry-Us. 1986;25:1266. doi: 10.1021/bi00354a011. [DOI] [PubMed] [Google Scholar]
- 61.Stone KL, Hoffart LM, Behan RK, Krebs C, Green MT. J Am Chem Soc. 2006;128:6147. doi: 10.1021/ja057876w. [DOI] [PubMed] [Google Scholar]
- 62.Green M. J Am Chem Soc. 2006;128:1902. doi: 10.1021/ja054074s. [DOI] [PubMed] [Google Scholar]
- 63.Badger RM. J Chem Phys. 1935;3:710. [Google Scholar]
- 64.Stone KL, Behan RK, Green MT. P Natl Acad Sci USA. 2006;103:12307. doi: 10.1073/pnas.0603159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meharenna YT, Doukov T, Li HY, Soltis SM, Poulos TL. Biochemistry-Us. 2010;49:2984. doi: 10.1021/bi100238r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Casadei CM, Gumiero A, Metcalfe CL, Murphy EJ, Basran J, Concilio MG, Teixeira SCM, Schrader TE, Fielding AJ, Ostermann A, Blakeley MP, Raven EL, Moody PCE. Science. 2014;345:193. doi: 10.1126/science.1254398. [DOI] [PubMed] [Google Scholar]
- 67.Yosca TH, Behan RK, Krest CM, Onderko EL, Langston MC, Green MT. J Am Chem Soc. 2014;136:9124. doi: 10.1021/ja503588n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang XS, Ullrich R, Hofrichter M, Groves JT. P Natl Acad Sci USA. 2015;112:3686. doi: 10.1073/pnas.1503340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yosca TH, Rittle J, Krest CM, Onderko EL, Silakov A, Calixto JC, Behan RK, Green MT. Science. 2013;342:825. doi: 10.1126/science.1244373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mayer JM. Annu Rev Phys Chem. 2004;55:363. doi: 10.1146/annurev.physchem.55.091602.094446. [DOI] [PubMed] [Google Scholar]
- 71.Mayer JM. Accounts Chem Res. 1998;31:441. [Google Scholar]
- 72.Goldsmith CR, Cole AP, Stack TDP. J Am Chem Soc. 2005;127:9904. doi: 10.1021/ja039283w. [DOI] [PubMed] [Google Scholar]
- 73.Lambeir AM, Dunford HB. Arch Biochem Biophys. 1983;220:549. doi: 10.1016/0003-9861(83)90446-0. [DOI] [PubMed] [Google Scholar]
- 74.Behan RK, Hoffart LM, Stone KL, Krebs C, Green MT. J Am Chem Soc. 2006;128:11471. doi: 10.1021/ja062428p. [DOI] [PubMed] [Google Scholar]
- 75.Zhao B, Guengerich FP, Bellamine A, Lamb DC, Izumikawa M, Lei L, Podust LM, Sundaramoorthy M, Kalaitzis JA, Reddy LM, Kelly SL, Moore BS, Stec D, Voehler M, Falck JR, Shimada T, Waterman MR. J Biol Chem. 2005;280:11599. doi: 10.1074/jbc.M410933200. [DOI] [PubMed] [Google Scholar]
- 76.Terner J, Reed DE. Biochim Biophys Acta. 1984;789:80. [Google Scholar]
- 77.Jung C, Schunemann V, Lendzian F. Biochemical and Biophysical Research Communications. 2005;338:355. doi: 10.1016/j.bbrc.2005.08.166. [DOI] [PubMed] [Google Scholar]
- 78.Schunemann V, Lendzian F, Jung C, Contzen J, Barra AL, Sligar SG, Trautwein AX. J Biol Chem. 2004;279:10919. doi: 10.1074/jbc.M307884200. [DOI] [PubMed] [Google Scholar]
- 79.Kang Y, Chen H, Jeong YJ, Lai W, Bae EH, Shaik S, Nam W. Chem-Eur J. 2009;15:10039. doi: 10.1002/chem.200901238. [DOI] [PubMed] [Google Scholar]
- 80.de Visser SP. J Am Chem Soc. 2010;132:1087. doi: 10.1021/ja908340j. [DOI] [PubMed] [Google Scholar]
- 81.de Visser SP, Kumar D, Cohen S, Shacham R, Shaik S. J Am Chem Soc. 2004;126:8362. doi: 10.1021/ja048528h. [DOI] [PubMed] [Google Scholar]