Abstract
OBJECTIVE
Studies in sepsis are limited by heterogeneity regarding what constitutes suspicion of infection. We sought to compare potential suspicion criteria using antibiotic and culture order combinations in terms of patient characteristics and outcomes. We further sought to determine the impact of differing criteria on the accuracy of sepsis screening tools and early warning scores.
DESIGN
Observational cohort study
SETTING
Academic center from November 2008 until January 2016
PATIENTS
Hospitalized patients outside the intensive care unit (ICU)
INTERVENTIONS
None
MEASUREMENTS AND MAIN RESULTS
Six criteria were investigated: 1) any culture; 2) blood culture; 3) any culture plus intravenous (IV) antibiotics; 4) blood culture plus IV antibiotics; 5) any culture plus IV antibiotics for at least four of seven days; and 6) blood culture plus IV antibiotics for at least four of seven days. Accuracy of the quick Sepsis-related Organ Failure Assessment (qSOFA) score, SOFA score, systemic inflammatory response system (SIRS) criteria, the National and Modified Early Earning Score (NEWS and MEWS), and the electronic Cardiac Arrest Risk Triage (eCART) score were calculated for predicting ICU transfer or death within 48 hours of meeting suspicion criteria. A total of 53,849 patients met at least one infection criteria. Mortality increased from 3% for group 1 to 9% for group 6 and percentage meeting Angus sepsis criteria increased from 20% to 40%. Across all criteria, score discrimination was lowest for SIRS (median AUC 0.60) and SOFA score (median AUC 0.62), intermediate for qSOFA (median AUC 0.65) and MEWS (median AUC 0.67), and highest for NEWS (median AUC 0.71) and eCART (median AUC 0.73).
CONCLUSIONS
The choice of criteria to define a potentially infected population significantly impacts on prevalence of mortality but has little impact on accuracy. SIRS was the least predictive and eCART the most predictive regardless of how infection was defined.
Keywords: Systemic inflammatory response syndrome, sepsis, multiple organ failure, organ dysfunction scores, early warning scores, SOFA, qSOFA
INTRODUCTION
Sepsis is one of the most deadly and costly medical conditions for hospitalized patients (1, 2). As such, healthcare policy and research have been aimed at early identification and treatment of sepsis in an attempt to improve outcomes (3). Unfortunately, research in this field is limited by the lack of a gold standard for defining sepsis. Historically, many observational studies have relied on billing data, in the form of ICD-9 codes, to identify patients with sepsis (4, 5). However, this method has been shown to have low sensitivity (5, 6). Seymour and colleagues, in their recently published derivation of the quick Sepsis-related Organ Failure Assessment (qSOFA) score, used one specific combination of culture orders and antibiotic administration within a predefined time window to define patients with suspicion for infection (7). However, there are many possible combinations of culture orders and antibiotic use that likely represent different degrees of suspicion of infection, from culture orders without subsequent antibiotics to antibiotics that are continued for several days. Furthermore, the criteria used for infection suspicion are, on the one hand, subjective and, on the other hand, only reflect iatrogenic enthusiasm and not objective evidence of infection. These various combinations may result in important differences in patient outcomes in the study cohort as well as measured accuracy of tools designed to risk stratify septic patients.
Therefore, we aimed to compare the accuracy of qSOFA, SOFA, the systemic inflammatory response syndrome (SIRS) criteria, and general early warning scores for predicting outcomes across different suspicion of infection criteria in hospitalized patients outside the intensive care unit (ICU). We also sought to compare patient characteristics and outcomes across these different criteria.
MATERIALS AND METHODS
Data collection
In this secondary analysis of prospectively collected data, all adult patients admitted to the University of Chicago Medicine from November 2008 to January 2016 were eligible for study inclusion. Vital signs, laboratory values, orders (e.g., blood cultures) and demographic data were collected by the University of Chicago’s Clinical Research Data Warehouse from the electronic health record (EHR) (Epic; Verona, WI). These data were then de-identified and made available on a secure SQL server for analysis. The University of Chicago Institutional Review Board approved the study protocol for this project as non-human subject research (IRB #15-1705).
Defining potentially infected patients
Six criteria meant to signify varying levels of suspicion of infection were created using EHR data based on culture orders and antibiotic administration: 1) any culture; 2) blood culture; 3) any culture plus intravenous (IV) antibiotics; 4) blood culture plus IV antibiotics; 5) any culture plus IV antibiotics for at least four out of seven days; and 6) blood culture plus IV antibiotics for at least four out of seven days. This final criterion is meant to identify a patient subgroup highly likely to be infected but also allowing for “skip days” in antibiotic therapy that might occur, for example, in patients with renal failure. For criteria 5 and 6, if a patient received antibiotics but died before the seven-day period was completed then they were still included in these definitions. These combinations of cultures and antibiotics were chosen to represent a range of suspicion of infection, from low (1 and 2) to high (5 and 6). The time window for culture and antibiotic orders used in this study was the same as in the original qSOFA paper, with either a culture order followed by an antibiotic within 72 hours or an antibiotic followed by a culture order within 24 hours (7). The time of first culture or IV antibiotic was denoted as the time of suspicion of infection, and only admissions first meeting the criteria in the emergency department or wards were included. Two pharmacists reviewed the antibiotics to ensure that they were treatment dose, and prophylactic dose antibiotics were excluded, as previously described (8).
Score calculation
Data from admission until the time that a patient first met each of the suspicion of infection criteria were used to calculate SIRS, SOFA, and qSOFA (7, 9), as well as two general early warning scores, the Modified Early Warning Score (MEWS) (10), the National Early Warning Score (NEWS) (11), and the electronic Cardiac Arrest Risk Triage (eCART) score (12). The MEWS and NEWS are commonly used vital sign based scores and the eCART score is a statistically derived random forest algorithm that includes vital signs, laboratory values, and patient demographics. Only scores calculated in the emergency department and wards were included in the analyses. In addition, non-physiologic values were changed to missing, as previously described (13).
Statistical Analysis
Descriptive statistics for patients meeting the different infection criteria were presented, including the proportion of patients who met Angus sepsis criteria using ICD-9 billing codes (4). Accuracy was calculated for predicting an ICU stay or death within 48 hours of the time of first suspicion of infection using the highest value of each score calculated outside the ICU from admission until the time of first suspicion. Scores were compared using the area under the receiver operating characteristic curve (AUC), sensitivity, and specificity. Two-tailed p-values <0.05 were considered statistically significant, and all analyses were performed using Stata version 14.1 (StataCorps; College Station, Texas).
RESULTS
Study population
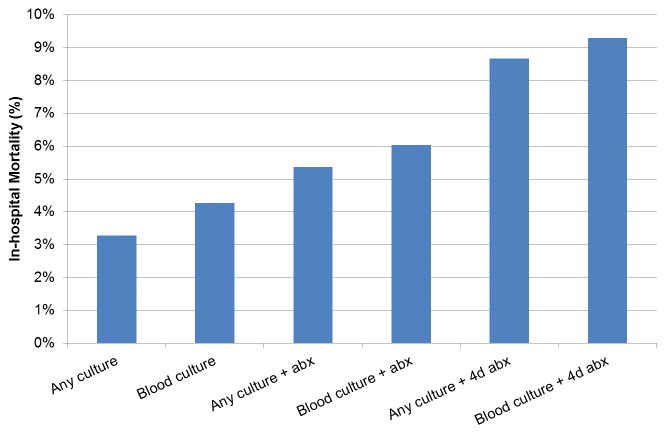
In total, 150,288 patients were hospitalized during the study period and therefore eligible for inclusion. The final study cohort included a total of 53,849 patients who met at least one infection criteria in the emergency department (ED) or wards. Of this cohort, 57% (n=30,677) received at least one dose of IV antibiotics and 31% (n=16,596) received at least 4 days of IV antibiotics in a 7-day period. The median time prior to the combined outcome was 10 hours for any culture order (IQR 4–51 hours), 8 hours for antibiotics (IQR 2–33 hours), and 7 hours for blood culture orders (IQR 2–49 hours). Distributions of age, sex, and race were similar across the different criteria (Table 1). Moving from criteria 1 (any culture) to 6 (blood cultures plus at least four days of antibiotics), the proportion of patients who met Angus sepsis billing criteria, were transferred to the ICU, received a vasopressor, or died increased. For example, 20% (n=10,527) of patients who had any culture met Angus ICD-9 sepsis criteria, compared to 40% (n=5,877) of patients who had a blood culture and at least four days of antibiotics. Seventeen percent of patients with any culture were transferred to the ICU, 6% received a vasopressor, and 3% died. In comparison, of patients with a blood culture and at least 4 days of antibiotics, 33% were transferred to the ICU, 14% received a vasopressor, and 9% died. Mortality steadily increased from criteria 1 (any culture) to criteria 6 (blood cultures plus at least four days of antibiotics) (Figure 1).
Table 1.
Comparison of patient characteristics across criteria for suspicion of infection
| Characteristic | Any culture (n=53,849) | Blood culture (n=38,438) | Any culture + antibiotics (n=30,677) | Blood culture + antibiotics (n=25,958) | Any culture + 4d antibiotics (n=16,596) | Blood culture + 4d antibiotics (n=14,772) |
|---|---|---|---|---|---|---|
| Infection first suspected in the ED, n (%) | 25,404 (47) | 20,451 (53) | 18,523 (60) | 16,171 (62) | 9,461 (57) | 8,575 (58) |
| Age, mean (SD), years | 57 (18) | 58 (18) | 58 (18) | 58 (18) | 58 (18) | 58 (18) |
| Female sex, n (%) | 29,130 (54) | 19,826 (52) | 16,2116 (53) | 13,307 (51) | 8,297 (50) | 7,248 (49) |
| Race, n (%) | ||||||
| Black | 30,706 (57) | 22,416 (58) | 17,813 (58) | 15,277 (59) | 9,101 (55) | 8,193 (55) |
| White | 19,420 (36) | 13,447 (35) | 10,685 (35) | 8,901 (34) | 6,208 (37) | 5,472 (37) |
| Other | 2,061 (4) | 1,516 (4) | 1,253 (4) | 1,079 (4) | 737 (4) | 651 (4) |
| Unknown | 1,662 (3) | 1,059 (3) | 926 (3) | 701 (3) | 550 (3) | 456 (3) |
| Met Angus sepsis criteria, n (%) | 10,527 (20) | 9,299 (24) | 8,744 (29) | 8,077 (31) | 6,192 (37) | 5,877 (40) |
| Ever ICU transfer, n (%) | 9,117 (17) | 7,788 (20) | 7,258 (24) | 6,578 (25) | 5,151 (31) | 4,830 (33) |
| Ever received vasopressor, n (%) | 3,163 (6) | 2,810 (7) | 2,724 (9) | 2,546 (10) | 2,226 (13) | 2,140 (14) |
| In-hospital mortality, n (%) | 1,772 (3) | 1,644 (4) | 1,649 (5) | 1,568 (6) | 1,440 (9) | 1,374 (9) |
Abbreviations: ED = emergency department, LOS = length of stay, ICU = intensive care unit
Figure 1.
In-hospital mortality by criteria for suspicion of infection
Abbreviations: abx = antibiotics
Accuracy comparisons
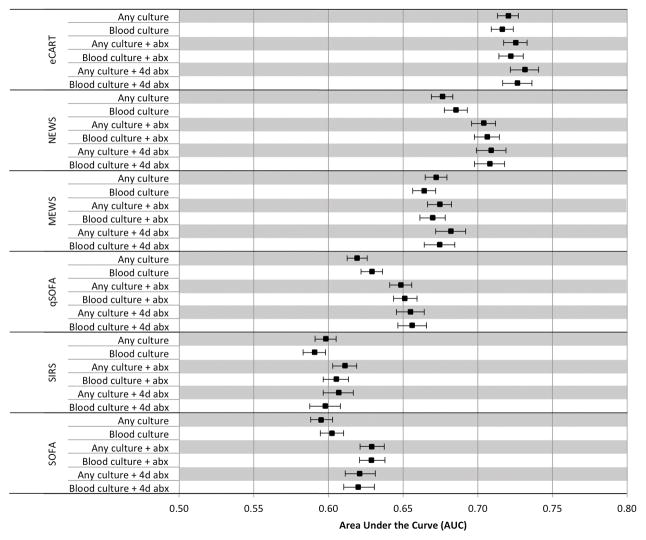
Across all criteria of infection, overall accuracy for predicting the composite outcome in the 48 hours following the time of infection suspicion was lowest for SIRS (median AUC 0.60 (range 0.60–0.61)) and SOFA (median AUC 0.62 (range 0.60–0.63)), intermediate for qSOFA (median AUC 0.65 (range 0.62–0.66)) and MEWS (median AUC 0.67 (range 0.66–0.68)) and highest for NEWS (median AUC 0.71 (range 0.68–0.71)) and eCART (median AUC 0.73 (range 0.72–0.73)) (Figure 2). For the definition with the highest mortality (i.e., blood cultures plus at least four days of antibiotics), the AUC was 0.60 (95% CI 0.59–0.61) for SIRS, 0.66 (95% CI 0.65–0.67) for qSOFA, 0.67 (95% CI 0.66–0.69) for MEWS, 0.71 (95% 0.70–0.72) for NEWS, and 0.73 (0.72–0.74) for eCART. eCART was significantly more accurate than NEWS, with P<0.001. A post-hoc analysis of a model that combined qSOFA and SIRS (i.e., qSOFA plus SIRS) had a median AUC of 0.65 (range 0.64–0.66). As shown in the Figure, accuracy varied little for each score across the varying criteria, although the accuracy of qSOFA, SOFA, and NEWS were lower in the groups that did not require antibiotics for inclusion compared to the criteria used in the original qSOFA study. The sensitivity of qSOFA ≥1 ranged from 69–73% at a 51–57% specificity. At cut-offs with a similar sensitivity to qSOFA ≥1, the specificity of MEWS ranged from 51–57% and the specificity of eCART ranged from 61–65% (Table 2). The sensitivity of SIRS ≥2 ranged from 66–69% at a specificity of 46–52%, compared to the sensitivity of qSOFA ≥2, which ranged from a sensitivity of 18–23% at 91–94% specificity. At a cut-off with a similar specificity to qSOFA ≥2, the sensitivity of MEWS ≥6 ranged from 17–21%, the sensitivity of NEWS ≥9 ranged from 20–25%, and the sensitivity of eCART ≥25 ranged from 25–30% (Table 3).
Figure 2.
Discrimination of scores across different criteria for suspicion of infection
Abbreviations: abx = antibiotics
Table 2.
Accuracy of the different scores across criteria for suspicion of infection, with cut-offs matched to the sensitivity of qSOFA ≥1
| Algorithm | Score | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|
| SOFA ≥1 | Any culture | 68% | 45% | 15% | 91% |
| Blood culture | 69% | 46% | 18% | 89% | |
| Any culture + abx | 68% | 53% | 23% | 89% | |
| Blood culture + abx | 69% | 52% | 25% | 88% | |
| Any culture + 4d abx | 67% | 53% | 28% | 85% | |
| Blood culture + 4d abx | 69% | 51% | 30% | 84% | |
| SIRS ≥2 | Any culture | 66% | 50% | 15% | 91% |
| Blood culture | 68% | 46% | 18% | 89% | |
| Any culture + abx | 66% | 52% | 22% | 88% | |
| Blood culture + abx | 68% | 49% | 23% | 87% | |
| Any culture + 4d abx | 66% | 51% | 29% | 83% | |
| Blood culture + 4d abx | 69% | 47% | 25% | 84% | |
| qSOFA ≥1 | Any culture | 69% | 52% | 17% | 92% |
| Blood culture | 71% | 51% | 20% | 91% | |
| Any culture + abx | 69% | 57% | 25% | 90% | |
| Blood culture + abx | 72% | 55% | 27% | 90% | |
| Any culture + 4d abx | 71% | 56% | 33% | 86% | |
| Blood culture + 4d abx | 73% | 54% | 30% | 88% | |
| MEWS ≥3 | Any culture | 69% | 57% | 18% | 93% |
| Blood culture | 72% | 51% | 20% | 91% | |
| Any culture + abx | 69% | 56% | 24% | 90% | |
| Blood culture + abx | 72% | 52% | 26% | 89% | |
| Any culture + 4d abx | 71% | 55% | 32% | 86% | |
| Blood culture + 4d abx | 74% | 51% | 29% | 88% | |
| NEWS ≥5 | Any culture | 63% | 64% | 19% | 93% |
| Blood culture | 66% | 62% | 23% | 91% | |
| Any culture + abx | 64% | 67% | 28% | 90% | |
| Blood culture + abx | 67% | 65% | 31% | 90% | |
| Any culture + 4d abx | 65% | 67% | 32% | 86% | |
| Blood culture + 4d abx | 68% | 64% | 34% | 88% | |
| eCART ≥10 | Any culture | 69% | 64% | 21% | 94% |
| Blood culture | 71% | 61% | 24% | 92% | |
| Any culture + abx | 69% | 65% | 29% | 91% | |
| Blood culture + abx | 72% | 62% | 30% | 91% | |
| Any culture + 4d abx | 71% | 64% | 38% | 88% | |
| Blood culture + 4d abx | 73% | 61% | 34% | 89% |
Table 3.
Accuracy of the different scores across criteria for suspicion of infection, with cut-offs matched to the sensitivity of qSOFA ≥2
| Algorithm | Score | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|
| SOFA ≥4 | Any culture | 20% | 88% | 19% | 89% |
| Blood culture | 22% | 87% | 23% | 86% | |
| Any culture + abx | 19% | 90% | 28% | 84% | |
| Blood culture + abx | 21% | 89% | 31% | 83% | |
| Any culture + 4d abx | 19% | 89% | 32% | 80% | |
| Blood culture + 4d abx | 22% | 88% | 36% | 79% | |
| qSOFA ≥2 | Any culture | 18% | 92% | 24% | 89% |
| Blood culture | 21% | 91% | 29% | 87% | |
| Any culture + abx | 19% | 94% | 39% | 85% | |
| Blood culture + abx | 21% | 93% | 41% | 84% | |
| Any culture + 4d abx | 21% | 93% | 48% | 79% | |
| Blood culture + 4d abx | 23% | 92% | 44% | 81% | |
| MEWS ≥6 | Any culture | 17% | 96% | 37% | 89% |
| Blood culture | 19% | 94% | 36% | 87% | |
| Any culture + abx | 18% | 95% | 42% | 85% | |
| Blood culture + abx | 20% | 94% | 43% | 84% | |
| Any culture + 4d abx | 19% | 95% | 54% | 79% | |
| Blood culture + 4d abx | 21% | 93% | 45% | 81% | |
| NEWS ≥9 | Any culture | 20% | 94% | 31% | 90% |
| Blood culture | 22% | 94% | 39% | 87% | |
| Any culture + abx | 21% | 95% | 46% | 85% | |
| Blood culture + abx | 22% | 94% | 46% | 84% | |
| Any culture + 4d abx | 23% | 95% | 58% | 80% | |
| Blood culture + 4d abx | 24% | 94% | 52% | 82% | |
| eCART ≥25 | Any culture | 25% | 94% | 36% | 90% |
| Blood culture | 28% | 93% | 41% | 88% | |
| Any culture + abx | 27% | 94% | 48% | 86% | |
| Blood culture + abx | 28% | 93% | 48% | 85% | |
| Any culture + 4d abx | 29% | 93% | 56% | 81% | |
| Blood culture + 4d abx | 30% | 92% | 51% | 83% |
DISCUSSION
In this study comparing different methods of identifying potentially infected patients using EHR data, we found that patient outcomes varied widely depending on the criteria used. Patients identified using the strictest criteria (blood cultures plus four days of antibiotics) had a threefold higher in-hospital mortality compared to those using any culture as the criteria of infection. However, the accuracy of risk stratification algorithms was not impacted appreciably. Specifically, we found both eCART to be the most accurate algorithm studied, followed by NEWS, MEWS, and qSOFA, with the SIRS criteria being the least accurate, regardless of how suspicion of infection was defined. These findings extend our prior work (8), where we found that NEWS had higher discrimination than qSOFA when using the same definition of suspicion of infection as in the original paper by Seymour et al.
Despite the hallmarks of sepsis being first recognized over two millennia ago, there is still no gold standard for its diagnosis (14, 15). This problem is further compounded when the goal is to study sepsis epidemiology, risk factors, and outcomes in large patient cohorts. For example, although ICD-9 codes have been widely used to study sepsis trends over time, they have been found to have low sensitivity, missing over one-third of patients on the wards who develop severe sepsis or septic shock (5, 6). Investigating combinations of antibiotics and culture orders, as was done in the original qSOFA paper, has the advantage of identifying a specific time point where infection was first suspected (7). It is also possible that these orders might be more accurate than billing codes for identifying septic patients retrospectively. However there are a large number of potential combinations of antibiotics and culture orders that are possible, in addition to choosing a time window in which these orders need to occur. Our data illustrate the importance of determining which of these combinations most accurately identifies infected patients for large-scale studies investigating the incidence or outcomes of septic patients due to the wide variability in outcomes depending on the criteria used.
The proliferation of EHRs across the country has also spurred research using large datasets to develop models to identify high-risk patients with sepsis (7, 16, 17). Although ICD-9 codes can be used to identify patients for these studies, they lack a specific time component regarding the onset of sepsis. Therefore, it is likely that studies that utilize combinations of infection-related orders to identify these patients will become more common over time. Our study clearly demonstrated that the accuracy of several tools varied little based on the criteria used. The fact that these differing criteria resulted in similar model accuracy suggests that the vital sign abnormalities that predict adverse outcomes in infected patients are similar to those that predict outcomes in other conditions. Although this idea deserves further research, it is bolstered by the fact that NEWS, MEWS and eCART, which are early warning scores designed for use on the general wards in all patients, were as accurate or more accurate than both SIRS and qSOFA. SOFA was less accurate than these scores, likely because it includes several laboratory parameters, which may not be readily available early on in a patient’s admission, and only data up to the time of initial suspicion of infection was included in this study. Our findings also suggest that non-specific early warning scores can play an important role in sepsis risk stratification.
The greater accuracy of eCART compared to the other scores in this study illustrates the potential for statistically developed scores that utilize advanced statistical methods to improve the detection of high risk infected patients (12, 18). As access to EHR data becomes ubiquitous around the country and other parts of the world, a shift toward more complex and accurate clinical decision support tools will occur. Furthermore, when deciding between SIRS and qSOFA the decision has to be made regarding selecting a score with high sensitivity (SIRS) or high specificity (qSOFA), without the ability to select a threshold that has both characteristics at the time of infection suspicion. The ability of the EHR to both collect and then calculate more complex scores makes simplifying models unnecessary (19). Several groups have already developed complex models for accurately identifying septic patients in the emergency department and on the wards (16, 17), with other studies showing improved processes of care due to early detection through EHR alerting (20–22). However, hospitals need to consider the relative improvements in accuracy of these complex models when compared to more standard scores, such as NEWS, in light of the challenges and costs that come with implementing machine learning models in practice.
Our study has several limitations. First, it is a single center investigation and thus the findings may not be generalizable to other settings. In addition, we did not investigate all possible combinations of infection related orders or vary the time window for these orders due to the sheer number of possible iterations. Finally, it is unknown which of the patients in our large dataset truly were infected given that manual chart review was not feasible in this large cohort of over 50,000 patients.
CONCLUSIONS
In conclusion, patients identified as infected using different combinations of antibiotic and culture orders had a wide range of outcomes, including in-hospital mortality. However, the accuracy of both sepsis-specific and general early warning scores were similar across these criteria, with eCART being most accurate, followed by NEWS, MEWS and qSOFA, and then the SIRS criteria and SOFA score being least accurate. Future work is needed to determine which suspicion criteria most accurately identifies infected patients before these methods are used in studies on sepsis epidemiology and outcomes in large EHR datasets.
Acknowledgments
Source of Funding: This research was funded in part by an ATS Foundation Recognition Award for Outstanding Early Career Investigators grant (PI: Dr. Matthew Churpek). Dr. Churpek is supported by a career development award from the National Heart, Lung, and Blood Institute (K08 HL121080) and has received honoraria from Chest for invited speaking engagements. Drs. Churpek and Edelson have a patent pending (ARCD. P0535US.P2) for risk stratification algorithms for hospitalized patients. In addition, Dr. Edelson has received research support from Philips Healthcare (Andover, MA) and Early Sense (Tel Aviv, Israel). She has ownership interest in Quant HC (Chicago, IL), which is developing products for risk stratification of hospitalized patients.
We would like to thank Timothy Holper, MS, Julie Johnson, MPH, RN, and Thomas Sutton for assistance with data abstraction, and Nicole Twu, MS for administrative support. Data from this study were provided by the Clinical Research Data Warehouse (CRDW) maintained by the Center for Research Informatics (CRI) at University of Chicago. The Center for Research Informatics is funded by the Biological Sciences Division, the Institute for Translational Medicine/CTSA (NIH UL1 TR000430) at the University of Chicago.
Abbreviations
- AUC
area under the receiver operating characteristic curve
- AVPU
alert, responsive to voice, responsive to pain, and unresponsive
- CI
confidence interval
- EHR
electronic health record
- ICU
intensive care unit
- MEWS
modified early warning score
Footnotes
Copyright form disclosure: Dr. Churpek’s institution received funding from an ATS Foundation Recognition Award for Outstanding Early Career Investigators grant and from the National Heart, Lung, and Blood Institute (K08 HL121080); he received funding from CHEST (honoraria for invited speaking engagements); and he received support for article research from the National Institutes of Health. Drs. Chupek and Edelson disclosed having a patent pending (ARCD. P0535US.P2) for risk stratification algorithms for hospitalized patients. Dr. Edelson’s institution received funding in the form of research support from Philips Healthcare and EarlySense, and she received funding from QuantHC (ownership interest). The remaining authors have disclosed that they do not have any potential conflicts of interest.
References
- 1.Liu V, Escobar GJ, Greene JD, et al. Hospital deaths in patients with sepsis from 2 independent cohorts. JAMA. 2014;312(1):90–92. doi: 10.1001/jama.2014.5804. [DOI] [PubMed] [Google Scholar]
- 2.Lagu T, Rothberg MB, Shieh MS, et al. Hospitalizations, costs, and outcomes of severe sepsis in the United States 2003 to 2007. Crit Care Med. 2012;40(3):754–761. doi: 10.1097/CCM.0b013e318232db65. [DOI] [PubMed] [Google Scholar]
- 3.Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41(2):580–637. doi: 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- 4.Angus DC, Linde-Zwirble WT, Lidicker J, et al. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29(7):1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Iwashyna TJ, Odden A, Rohde J, et al. Identifying patients with severe sepsis using administrative claims: patient-level validation of the angus implementation of the international consensus conference definition of severe sepsis. Medical care. 2014;52(6):e39–43. doi: 10.1097/MLR.0b013e318268ac86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whittaker SA, Mikkelsen ME, Gaieski DF, et al. Severe sepsis cohorts derived from claims-based strategies appear to be biased toward a more severely ill patient population. Crit Care Med. 2013;41(4):945–953. doi: 10.1097/CCM.0b013e31827466f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315(8):762–774. doi: 10.1001/jama.2016.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churpek MM, Snyder A, Han X, et al. qSOFA, SIRS, and Early Warning Scores for Detecting Clinical Deterioration in Infected Patients Outside the ICU. Am J Respir Crit Care Med. 2016 doi: 10.1164/rccm.201604-0854OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101(6):1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 10.Subbe CP, Kruger M, Rutherford P, et al. Validation of a modified Early Warning Score in medical admissions. QJM : monthly journal of the Association of Physicians. 2001;94(10):521–526. doi: 10.1093/qjmed/94.10.521. [DOI] [PubMed] [Google Scholar]
- 11.Smith GB, Prytherch DR, Meredith P, et al. The ability of the National Early Warning Score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation. 2013;84(4):465–470. doi: 10.1016/j.resuscitation.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 12.Churpek MM, Yuen TC, Winslow C, et al. Multicenter Comparison of Machine Learning Methods and Conventional Regression for Predicting Clinical Deterioration on the Wards. Crit Care Med. 2016;44(2):368–374. doi: 10.1097/CCM.0000000000001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Churpek MM, Zadravecz FJ, Winslow C, et al. Incidence and Prognostic Value of the Systemic Inflammatory Response Syndrome and Organ Dysfunctions in Ward Patients. Am J Respir Crit Care Med. 2015;192(8):958–964. doi: 10.1164/rccm.201502-0275OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent JL, Opal SM, Marshall JC, et al. Sepsis definitions: time for change. Lancet. 2013;381(9868):774–775. doi: 10.1016/S0140-6736(12)61815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiel SW, Rosini JM, Shannon W, et al. Early prediction of septic shock in hospitalized patients. J Hosp Med. 2010;5(1):19–25. doi: 10.1002/jhm.530. [DOI] [PubMed] [Google Scholar]
- 17.Taylor RA, Pare JR, Venkatesh AK, et al. Prediction of In-hospital Mortality in Emergency Department Patients With Sepsis: A Local Big Data-Driven, Machine Learning Approach. Academic emergency medicine : official journal of the Society for Academic Emergency Medicine. 2016;23(3):269–278. doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churpek MM, Yuen TC, Winslow C, et al. Multicenter development and validation of a risk stratification tool for ward patients. Am J Respir Crit Care Med. 2014;190(6):649–655. doi: 10.1164/rccm.201406-1022OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang MA, Churpek MM, Zadravecz FJ, et al. Real-Time Risk Prediction on the Wards: A Feasibility Study. Crit Care Med. 2016 doi: 10.1097/CCM.0000000000001716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurczewski L, Sweet M, McKnight R, et al. Reduction in time to first action as a result of electronic alerts for early sepsis recognition. Critical care nursing quarterly. 2015;38(2):182–187. doi: 10.1097/CNQ.0000000000000060. [DOI] [PubMed] [Google Scholar]
- 21.Sawyer AM, Deal EN, Labelle AJ, et al. Implementation of a real-time computerized sepsis alert in nonintensive care unit patients. Crit Care Med. 2011;39(3):469–473. doi: 10.1097/CCM.0b013e318205df85. [DOI] [PubMed] [Google Scholar]
- 22.Umscheid CA, Betesh J, VanZandbergen C, et al. Development, implementation, and impact of an automated early warning and response system for sepsis. J Hosp Med. 2015;10(1):26–31. doi: 10.1002/jhm.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]