Summary
The spindle assembly checkpoint (SAC) prevents premature sister chromatid separation during mitosis. Phosphorylation of unattached kinetochores by the Mps1 kinase promotes recruitment of SAC machinery that catalyzes assembly of the SAC effector mitotic checkpoint complex (MCC). The SAC protein Bub3 is a phospho-amino acid adaptor that forms structurally related stable complexes with functionally distinct paralogs named Bub1 and BubR1. A short motif (“loop”) of Bub1, but not the equivalent loop of BubR1, enhances binding of Bub3 to kinetochore phospho-targets. Here, we asked whether the BubR1 loop directs Bub3 to different phospho-targets. The BubR1 loop is essential for SAC function and cannot be removed or replaced with the Bub1 loop. BubR1 loop mutants bind Bub3 and are normally incorporated in MCC in vitro but have reduced ability to inhibit the MCC target anaphase-promoting complex (APC/C), suggesting that BubR1:Bub3 recognition and inhibition of APC/C requires phosphorylation. Thus, small sequence differences in Bub1 and BubR1 direct Bub3 to different phosphorylated targets in the SAC signaling cascade.
Keywords: gene duplication, sub-functionalization, spindle assembly checkpoint, kinetochore, Bub1, BubR1, Bub3, Knl1, APC/C, MCC
Graphical Abstract
Highlights
-
•
The molecular basis of kinetochore recruitment of Bub1 and BubR1 is dissected
-
•
Bub1 and BubR1 modulate the ability of Bub3 to recognize phosphorylated targets
-
•
A newly identified BubR1 motif targets Bub3 to the anaphase-promoting complex
-
•
The newly identified motif of BubR1 is required for checkpoint signaling
In spindle assembly checkpoint (SAC) signaling, the phospho-amino acid adaptor Bub3 forms complexes with Bub1 and BubR1 paralogs. Whether Bub3-Bub1 and Bub3-BubR1 bind distinct targets has been unclear. Overlack et al. demonstrate that this is the case and identify a motif in BubR1 that directs Bub3 to the SAC target, the anaphase-promoting complex.
Introduction
Bub1 and BubR1 (also known as Mad3 in certain organisms) are paralogous proteins that fulfill different crucial functions in chromosome alignment on the mitotic spindle and in the spindle assembly checkpoint (SAC), a safety mechanism that ensures accurate chromosome segregation during mitosis [1, 2]. Bub1 and BubR1 originated through multiple independent gene-duplication events from a precursor (singleton) surmised to be already present in the hypothetical last eukaryotic common ancestor (LECA). Gene duplication invariably led to sub-functionalization of the resulting gene products [3, 4] (Figure 1A).
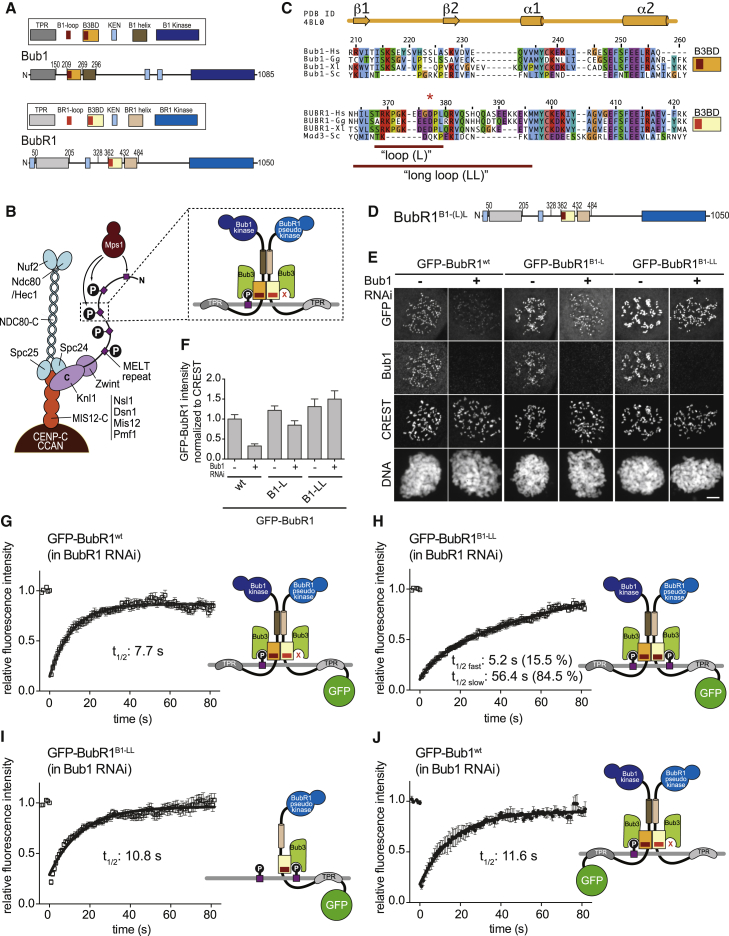
Figure 1.
Kinetochore Localization and Turnover of Long BubR1 Loop Mutants in HeLa Cells
(A) Schematic overview of Bub1 and BubR1 domain organization. B1, Bub1; B3BD, Bub3 binding domain; BR1, BubR1; KEN, lysine-glutamate-asparagine motif; TPR, tetratrico peptide repeat.
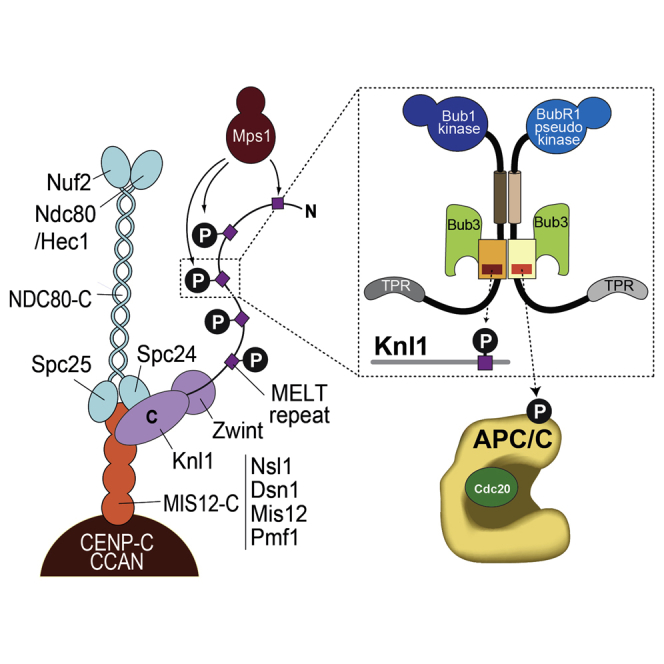
(B) Schematic depiction of the outer kinetochore (KMN network). MELT repeats in Knl1 are phosphorylated by the checkpoint kinase Mps1 and recruit Bub1:Bub3. Bub1:Bub3 in turn recruits BubR1:Bub3 via a pseudo-symmetric interaction, which involves equivalent segments of Bub1 and BubR1 comprising the B3BD and the C-terminal extension whose first part is predicted to form a helix in both proteins. The presence of Bub3 on both proteins seems to be essential for this interaction. The TPR regions of human Bub1 and BubR1 bind to non-conserved short motifs of Knl1 named KI1 and KI2, respectively [5, 6, 7].
(C) Multiple sequence alignments of the B3BDs of Bub1 and BubR1 from four different species: Hs, Homo sapiens; Gg, Gallus gallus; Xl, Xenopus laevis; and Sc, Saccharomyces cerevisiae. Mad3 is the budding yeast BubR1 homolog. The initial loop (L) and the long loop (LL) are indicated by the red lines; exact residue numbers are indicated in the main text.
(D) Domain organization of the BubR1 constructs with the Bub1 loop. BubR1B1-L contains Bub1 residues 214–226; BubR1B1-LL contains Bub1 residues 209–235.
(E) Representative images of HeLa cells transfected with the indicated GFP-BubR1 constructs showing that GFP-BubR1B1-LL localizes better to kinetochores than the short loop mutant (B1-L) in presence and absence of endogenous Bub1. In brief, after transfection, cells were depleted of endogenous Bub1 by RNAi, synchronized with a double thymidine block, and arrested in mitosis with nocodazole. The scale bar represents 10 μm.
(F) Quantification of BubR1 kinetochore levels in cells treated as in (E). The graph shows mean intensity from three independent experiments. Error bars represent SEM. Values for BubR1wt in non-depleted cells are set to 1.
(G–J) FRAP analyses of GFP-tagged BubR1wt (G), BubR1B1-LL (H and I), and GFP-Bub1wt in absence of endogenous BubR1 (G and H) or endogenous Bub1 (I and J). Relevant recovery parameters are shown. The graphs show mean with SEM. The cartoons beside the graphs depict the expected mode of kinetochore localization of each construct.
See also Figures S1–S3.
The molecular mechanism subtending to sub-functionalization of Bub1 and BubR1 is an active area of research. Bub1 is a Ser/Thr kinase [8] whose kinase activity may be strictly required for chromosome alignment, but not for SAC signaling [9, 10, 11, 12, 13, 14, 15, 16, 17]. Bub1 localizes to kinetochores in early prometaphase and is thought to perform its main role in the SAC by acting as a scaffold for the recruitment of downstream checkpoint components, including Mad1, Mad2, BubR1, Bub3, and Cdc20 [11, 14, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28]. Bub1 promotes the incorporation of a subset of these proteins, including BubR1, Bub3, Mad2, and Cdc20, into the mitotic checkpoint complex (MCC), the main checkpoint effector, which directly inhibits the ability of the E3 ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C) to promote exit from mitosis [1, 2]. Promotion of MCC formation by Bub1 likely occurs through a direct interaction with a Mad1:Mad2 template that catalyzes MCC assembly [11, 29, 30, 31, 32, 33, 34, 35].
Unlike Bub1, BubR1 is an inactive pseudokinase [4]. It contributes directly to the SAC and to APC/C inhibition as a subunit of the MCC (together with Bub3, Cdc20, and Mad2) [36, 37, 38]. Two Lys-Glu-Asn (KEN) motifs (also called KEN boxes) in the N-terminal region of BubR1 have been implicated in Cdc20 binding and APC/C inhibition. The first KEN box is essential for stable incorporation of BubR1 in a complex with Cdc20 and Mad2 that represents the core of the MCC [39, 40, 41, 42, 43]. The second KEN box promotes binding to a second Cdc20 molecule, possibly when the latter is already bound to the APC/C [44, 45, 46, 47]. Both KEN motifs are required for effective APC/C inhibition and SAC function (e.g., see [42]). In addition to its role in SAC activation, BubR1 also contributes to the formation of stable kinetochore-microtubule interactions and SAC silencing through kinetochore recruitment of the phosphatase PP2AB56 [48, 49, 50, 51, 52].
In human cells, kinetochore localization of Bub1 and BubR1 requires phosphorylation by the SAC kinase Mps1 of so-called Met-Glu-Leu-Thr (MELT) motifs in the outer kinetochore protein Knl1 (also known as Spc105, Spc7, and Casc5) [53, 54, 55]. It also requires binding to Bub3, which acts as a targeting adaptor [40, 42, 56, 57, 58]. Bub1 and BubR1 bind to Bub3 via conserved Bub3-binding domains (B3BD or GLEBS; Figure 1A) [57, 59]. Bub3, a seven-bladed β propeller, contains an evolutionarily conserved binding pocket that accommodates the phosphorylated Thr residue of the MELT motifs (Figures 1B, S1A, and S1B) [58]. By binding to Bub3, the B3BD of Bub1 is sufficient for kinetochore localization in the absence of other Bub1 regions [5, 57]. We recently reported that a short region of the B3BD of Bub1, which we designated as the “loop” (Figures 1C, S1A, and S1B), markedly increases the binding affinity of Bub3 for phosphorylated MELT motifs (MELTP) through direct contacts [58, 60]. The loop region in the B3BD of BubR1, on the other hand, is unable to increase the binding affinity of Bub3 for Knl1-MELTP sequences, rendering kinetochore localization of BubR1:Bub3 dependent on additional interactions [5, 60]. We reported that kinetochore-bound Bub1:Bub3 provides these additional interactions [60], thus establishing a clear hierarchy in which Bub1:Bub3 is first directly recruited to the kinetochore via interaction with MELTP motifs, after which BubR1:Bub3 is recruited through a direct interaction with Bub1 (Figure 1B). This model agrees with observations that kinetochore recruitment of Bub1 is independent of BubR1, whereas kinetochore localization of BubR1 depends on Bub1 [11, 12, 21, 56, 60, 61, 62, 63]. Hetero-dimerization of Bub1:Bub3 and BubR1:Bub3 requires equivalent domains in Bub1 and BubR1, including the B3BD and a region, directly following the B3BD, predicted to adopt helical conformation. Furthermore, both proteins need to be bound to Bub3 for their effective recruitment to kinetochores (Figure 1B) [60].
The crucial role of the Bub1 loop as enhancer of MELTP binding was emphasized by the fact that grafting the Bub1 loop onto BubR1 promoted Bub1-independent kinetochore recruitment of BubR1 [60]. The observation that this loop swap mutant, however, was unable to support BubR1 SAC function [60] raised the question whether the BubR1 loop performs a specific and so far unidentified function. In this study, we set out to investigate the role of the BubR1 loop in more detail. We demonstrate that the BubR1 loop promotes stable association of the MCC complex with the APC/C.
Results
Definition of Functional Loop Regions of Bub1 and BubR1
The B3BD of Bub1 is sufficient for kinetochore recruitment through Bub3 [5, 57, 60]. In our previous studies, we identified the Bub1 loop region (Bub1L) (residues 214–226 within the Bub1B3BD; Figures 1A and 1C) as a determinant of kinetochore localization of the Bub1:Bub3 complex and showed that Bub1L enhances the affinity of the interaction of Bub3 for Knl1-MELTP motifs at kinetochores [60]. We also showed that, when grafted onto BubR1, Bub1L promotes Bub1-independent kinetochore recruitment of BubR1. This result agreed with observations in vitro that the equivalent BubR1 loop region (BubR1L) (residues 368–379) is unable to promote the interaction of Bub3 with Knl1-MELTP motifs and is therefore functionally distinct from Bub1L [60].
As clarified in Figure S2 (and legend), we discovered that grafting onto BubR1 longer regions of Bub1 (residues 209–235), ranging from the first β sheet (β1) until the beginning of the highly conserved core of the B3BD (Figures 1C and 1D), resulted in more robust kinetochore localization in comparison to that of BubR1B1-L, especially after depletion of endogenous Bub1 (Figures 1E and 1F; to differentiate this longer sequence from that of Bub1L, we denote it as “long loop” [Bub1LL]). BubR11–431/B1-LL, which lacks the predicted helical region required for dimerization with Bub1, localized to kinetochores in presence or absence of endogenous Bub1 (Figures S2E and S2F). Thus, the Bub1LL region is sufficient to mediate kinetochore localization when grafted on a dimerization-deficient BubR1 mutant. When we grafted the equivalent region of BubR1 (residues 363–396; BubR1LL) onto Bub1 (Bub1BR1-LL), we observed that it impaired kinetochore localization of Bub1 significantly more pervasively than when grafting the shorter BubR1L sequence (Bub1BR1-L) (Figures S2G and S2H). Collectively, these experiments identify the LL regions of Bub1 and BubR1 as crucial determinants of their localization and demonstrate that these sequences impart substantial functional divergence to the Bub1 and BubR1 paralogs.
Kinetochore Turnover of BubR1 Loop Mutants
We investigated the effects of grafting Bub1LL on BubR1 kinetochore turnover. The halftime of kinetochore localization of BubR1 measured by fluorescence recovery after photobleaching (FRAP) is relatively fast (t1/2 = 3–20 s) [64, 65]. This rapid turnover of BubR1 likely reflects its dimerization with kinetochore Bub1 and subsequent release, possibly in complex with other MCC subunits. In agreement with the published data [64], we found in FRAP experiments in HeLa cells depleted of endogenous BubR1 that GFP-BubR1WT showed a recovery halftime of 7.7 s (fit with a single exponential curve; Figure 1G; Table S1; the cartoon beside the graph depicts the expected mode of kinetochore localization of the construct).
If grafting of Bub1LL allows BubR1B1-LL to interact with MELTP in addition to dimerizing with Bub1, substantial increases of its kinetochore residence time might be expected if the two binding modes occurred concomitantly. FRAP curves of the GFP-BubR1B1-LL mutant were best fitted with a double exponential curve (Figure 1H; Table S1). A minority of GFP-BubR1B1-LL (15.5%) turned over with a halftime of 5.2 s, likely reflecting single binding events (dimerization with Bub1 or direct binding to MELTP sequences). The majority of GFP-BubR1B1-LL, on the other hand, cycled with a halftime of 56.4 s, indeed suggesting the possibility that the two kinetochore-binding modes, namely the dimerization with Bub1 and the direct interaction of GFP-BubR1B1-LL with MELTP, occur concomitantly and strongly stabilize GFP-BubR1B1-LL at kinetochores. GFP-BubR1LL showed higher kinetochore levels than GFP-BubR1WT in cells depleted of endogenous BubR1 (Figures S3A and S3B), as expected given its increased halftime. We note that this is compatible with previous studies showing that only a subset of all MELTP motifs of Knl1 are occupied with Bub1 at any given time [66].
This binding model predicts that depletion of endogenous Bub1 will ablate dimerization, only allowing a single binding mode of GFP-BubR1B1-LL through MELTP binding. In agreement with this hypothesis, depletion of endogenous Bub1 by RNAi resulted in a recovery curve for GFP-BubR1B1-LL that could be fitted with a single exponential function with rapid turnover (t1/2 = 10.8 s; Figure 1I; Table S1). Like BubR1B1-LL, also Bub1 interacts with kinetochores by binding MELTP sequences, and therefore, it is expected to turn over at kinetochores with a similarly rapid halftime. Whereas previous studies reported that Bub1 turns over relatively slowly at kinetochores [64, 65], our measurements indicate rapid kinetochore turnover of GFP-Bub1WT (t1/2 = 11.6 s; Figure 1J; Table S1), very similar to the recovery halftime of GFP-BubR1B1-LL after Bub1 depletion. Our observations are corroborated by a recent study reporting a recovery halftime of 15 s for kinetochore Bub1 [67].
The Bub1 Loop Cannot Promote the SAC Function of BubR1 In Vivo
Next, we asked whether BubR1B1-LL was able to rescue SAC signaling in HeLa cells depleted of endogenous BubR1. A BubR1 alanine mutant in the first KEN box (KEN1; Figure 2A) was used as a control for checkpoint deficiency in these experiments [42]. Asynchronous cells entering mitosis in presence of low concentrations of the spindle poison nocodazole arrested robustly in mitosis for several hours, indicative of SAC activation (Figure 2B). Depletion of BubR1 prevented mitotic arrest but could be rescued by expression of a wild-type GFP-BubR1 transgene. On the other hand, BubR1B1-LL was unable to rescue the deleterious effects on the SAC caused by depletion of endogenous BubR1 to an extent similar to that of the KEN1 box mutant (Figure 2B). Thus, the Bub1LL region cannot functionally replace the equivalent region of BubR1. These findings were corroborated by immunoprecipitation experiments (IPs) of the GFP-BubR1 species, followed by western blotting (WB) to detect MCC and APC/C subunits. This revealed a substantial decrease in the association of GFP-BubR1B1-LL or GFP-BubR1KEN1/AAA with Mad2 and Cdc20 (but not Bub3, as expected) as well as the APC/C subunits Cdc27, Apc7, and Apc4 (Figure 2C; quantified in Figure S4A). Similar results were obtained when we replaced the sequence of the BubR1 loop with a neutral Gly-Ser-linker sequence (Figures S5A–S5D).
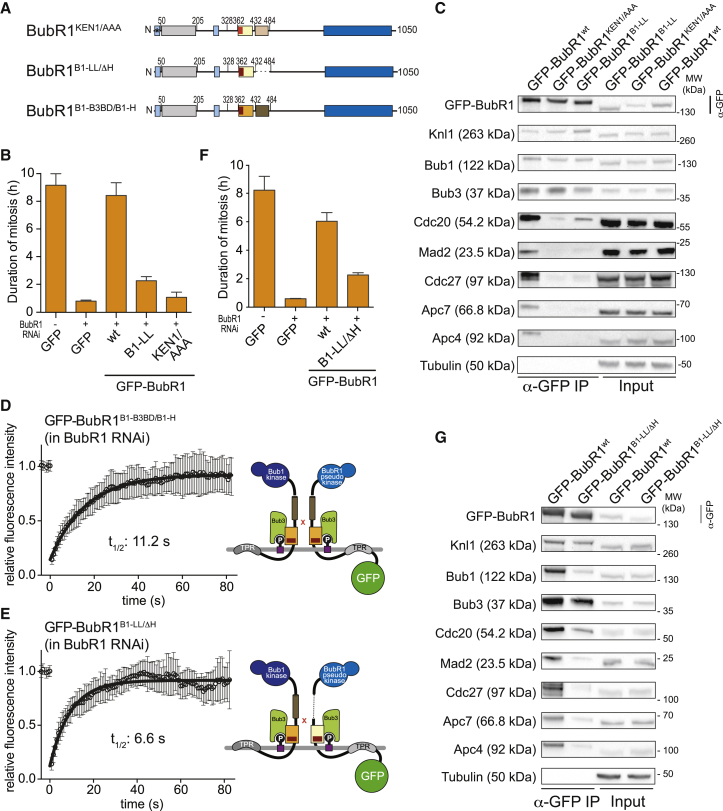
Figure 2.
The Bub1 Loop Cannot Promote the SAC Function of BubR1 In Vivo
(A) Domain organization of the BubR1 constructs.
(B and F) Mean duration of mitosis of Flp-In T-REx stable cell lines expressing the indicated GFP-BubR1 constructs in the absence of endogenous BubR1 and in the presence of 50 nM nocodazole. Cell morphology was used to measure entry into and exit from mitosis by time-lapse microscopy (n > 23 for BubR1B1-LL and BubR1KEN1/AAA [B]; n > 37 for BubR1B1-LL/ΔH [F] per cell line per experiment) from three independent experiments. Error bars depict SEM.
(C and G) Western blot of immunoprecipitates (IPs) from mitotic Flp-In T-REx cell lines expressing the indicated GFP-BubR1 constructs (BubR1B1-LL and BubR1KEN1/AAA [C]; BubR1B1-LL/ΔH [G])showing that the replacement of the BubR1 loop with the Bub1 loop results in strongly impaired APC/C binding. Tubulin was used as loading control.
(D and E) FRAP analyses of GFP-tagged BubR1B1-B3BD/B1-H (D) and BubR1B1-LL/ΔH (E) in absence of endogenous BubR1. Relevant recovery parameters are shown. The graphs show mean and SD. The cartoons beside the graphs depict the expected mode of kinetochore localization of each construct.
See also Figures S4 and S5 and Table S1.
As explained in the previous section, data in Figures 1G–1J suggest that GFP-BubR1B1-LL dimerizes with Bub1 and also binds to phosphorylated MELT motifs at kinetochores. We hypothesized that the SAC defect following grafting of the Bub1LL region into BubR1 might be caused by increased kinetochore residence of this mutant, which also reflected in a strongly increased interaction with the Knl1 kinetochore receptor (Figure S4A). If rapid kinetochore turnover of BubR1 is required for its efficient incorporation into MCC, tighter kinetochore binding might counteract incorporation into MCC. We therefore asked whether we could rescue the SAC deficiency of the GFP-BubR1B1-LL mutant by restoring rapid kinetochore turnover.
To test this hypothesis, we designed two constructs expected to reduce the time of kinetochore residence of GFP-BubR1B1-LL (see Figure 2A for schematics). First, we combined grafting of the Bub1LL region with deletion of the predicted helical region (residues 432–484) involved in BubR1 dimerization with Bub1 (designated as GFP-BubR1B1-LL/ΔH). GFP-BubR1B1-LL/ΔH is expected to localize correctly but exclusively through Bub1LL-mediated recognition of MELTP and not through dimerization. Importantly, deletion of the BubR1 helical region is fully compatible with SAC signaling (see next paragraph). Second, we grafted the entire B3BD and helical region of Bub1 onto GFP-BubR1 (GFP-BubR1B1-B3BD/B1-H), thus again forcing BubR1 to interact with kinetochores exclusively through MELTP and not through an interaction with Bub1 (because neither Bub1 nor BubR1 can form homodimers).
In agreement with our expectations, and more generally with our model of kinetochore recruitment of Bub1 and BubR1, both GFP-BubR1B1-B3BD/B1-H and GFP-BubR1B1-LL/ΔH showed robust localization to kinetochores (Figures S4B and S4C), and FRAP analyses of both mutants demonstrated rapid kinetochore turnover (Figures 2D and 2E), with recovery halftimes that were essentially indistinguishable from those of GFP-BubR1WT (Figure 1G; Table S1). Despite normal kinetochore turnover, however, GFP-BubR1B1-LL/ΔH was unable to rescue the SAC defect caused by BubR1 depletion (Figure 2F) and was correspondingly unable to form stable complexes with MCC and APC/C subunits in IP experiments (Figure 2G; quantified in Figure S4D). Importantly, GFP-BubR1ΔH is fully SAC proficient (Figure S4E) [60], strongly suggesting that the SAC defect is caused by Bub1LL grafting. Also, GFP-BubR1B1-B3BD/B1-H was unable to rescue the SAC in cells depleted of endogenous BubR1 (Figure S4F). In non-depleted cells, GFP-BubR1B1-B3BD/B1-H interacted with BubR1 as expected, but its interaction with MCC and APC/C subunits was reduced (Figure S4G). Collectively, these results indicate that the SAC defect introduced by grafting the Bub1LL region in GFP-BubR1B1-LL is not due to increased kinetochore residency of this mutant.
The BubR1 Loop Is Required for SAC Function In Vivo
As an alternative explanation for why GFP-BubR1B1-LL is SAC defective, we considered the hypothesis that the substitution of the BubR1LL region with the Bub1LL region may interfere with the interaction of the BubR1:Bub3 complex with a crucial SAC target. To test this idea, we created two additional BubR1 mutants in which parts of the loop region were deleted (ΔL and ΔLL mutant, corresponding to deletions of residues 368–379 and 363–396, respectively; Figure 3A). GFP-BubR1ΔL and GFP-BubR1ΔLL localized to kinetochores at levels that were indistinguishable from those of GFP-BubR1WT, and their localization depended on Bub1 (Figures 3B and 3C). In FRAP experiments, GFP-BubR1ΔLL appeared to have a recovery halftime of 9.9 s (Figure 3D; Table S1), essentially identical to that of GFP-BubR1WT (Figure 1G). These observations are consistent with our model that the BubR1 loop region is not required for kinetochore recruitment [60] (and this study).
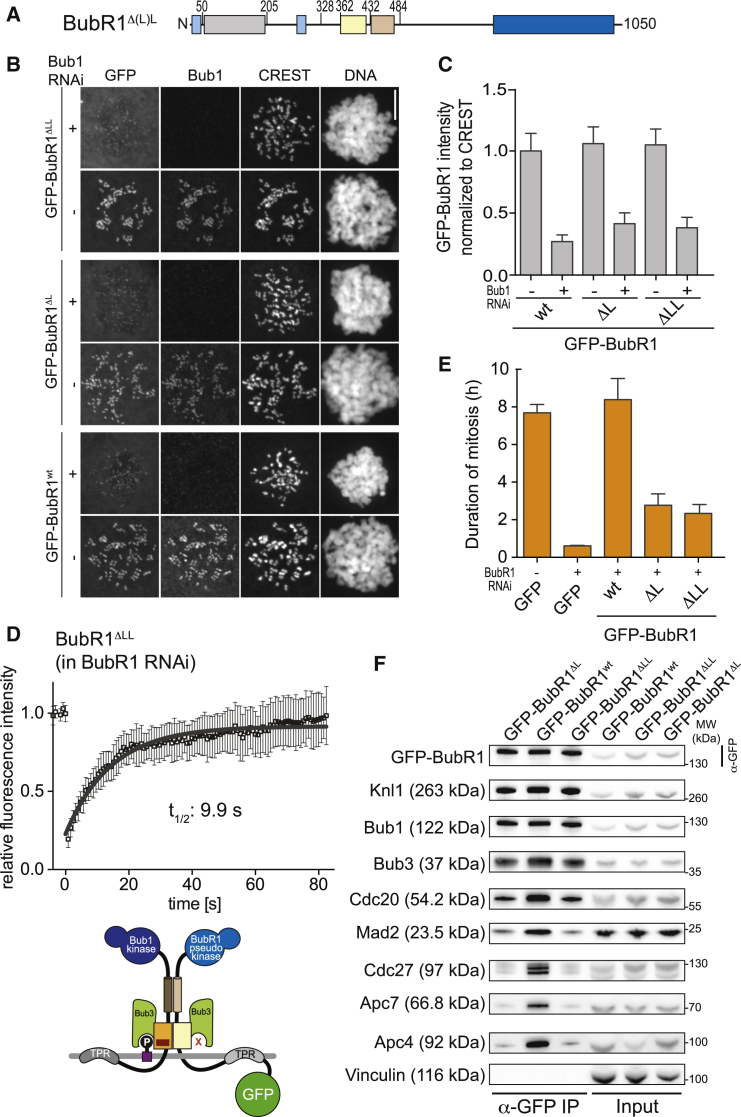
Figure 3.
The BubR1 Loop Is Required for SAC Function In Vivo
(A) Domain organization of the BubR1 constructs used. Constructs lacking the short (L) and long loop (LL) versions were created by deleting residues 368–379 or 363–396 of BubR1, respectively.
(B) Representative images of HeLa cells transfected with the indicated GFP-BubR1 constructs in presence or absence of endogenous Bub1, showing that the lack of the loop does not influence kinetochore localization, as expected. Cells were treated as in Figure 1E. The scale bar represents 10 μm.
(C) Quantification of BubR1 kinetochore levels in cells treated as in (B). The graph shows mean intensity from three independent experiments. Error bars represent SEM. Values for BubR1wt in non-depleted cells are set to 1.
(D) FRAP analysis of BubR1ΔLL in the absence of endogenous BubR1. Relevant recovery parameters are shown. The graph shows mean and SD. The cartoon depicts the expected mode of kinetochore localization of the construct.
(E) Mean duration of mitosis of Flp-In T-REx stable cell lines expressing the indicated GFP-BubR1 constructs in the absence of endogenous BubR1 and in the presence of 50 nM nocodazole. Cell morphology was used to measure entry into and exit from mitosis by time-lapse microscopy (n > 32 for BubR1Δ(L)L per cell line per experiment) from two independent experiments. Error bars depict SEM.
(F) Western blot of IPs from mitotic Flp-In T-REx cell lines expressing the indicated GFP-BubR1 constructs showing that the lack of the BubR1 loop results in strongly impaired APC/C binding. Vinculin was used as loading control.
Next, we asked whether GFP-BubR1ΔL and GFP-BubR1ΔLL were able to support the SAC in cells depleted of endogenous BubR1. Both mutants were largely unable to restore the SAC function of BubR1 (Figure 3E). Furthermore, when we combined deletion of the loop with deletion of the helical region (GFP-BubR1ΔL/ΔH; Figure S4H), we observed impairment of the SAC (Figure S4E), accompanied by reduced interaction with the APC/C (Figure S4I). In IP experiments, GFP-BubR1ΔL and GFP-BubR1ΔLL were shown to bind to the MCC subunits Mad2 and Cdc20, although at reduced levels in comparison to GFP-BubR1WT, but were also largely impaired in their ability to interact with the APC/C, likely explaining why these mutants cannot support the SAC (Figure 3F; quantified in Figure S4J).
The BubR1 Loop Promotes APC/C Binding
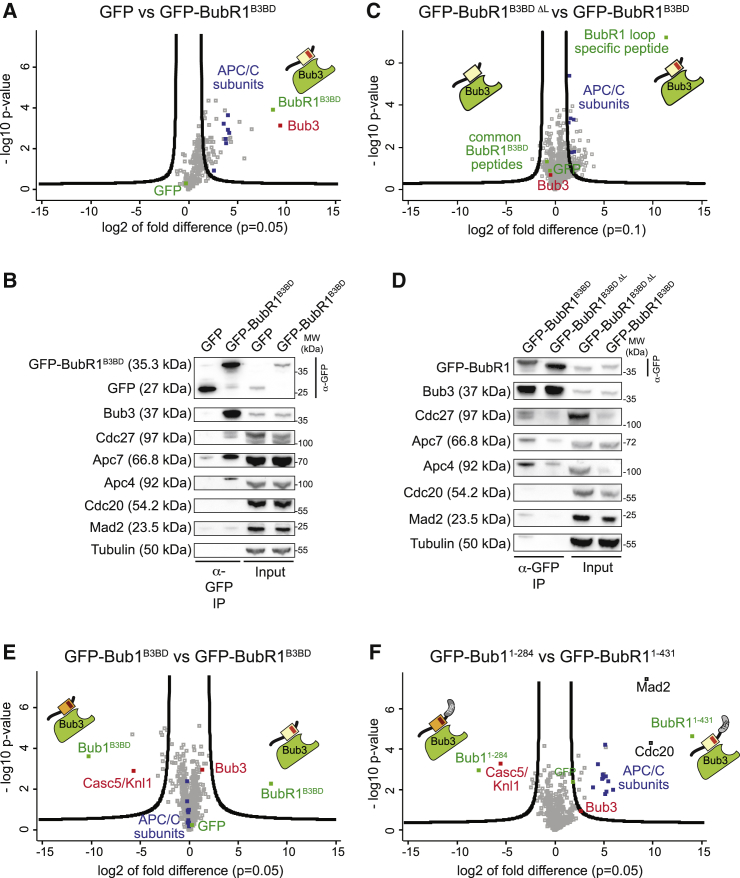
Collectively, these observations support the idea that the loop region of BubR1 is required for the recognition of crucial SAC target(s). To identify these targets, we used the SILAC (stable isotope labeling with amino acids in cell culture) [68] approach to perform quantitative IP and mass spectrometry identification of proteins bound to minimal reporter constructs in mitotic lysates of HeLa cells. Because the B3BD of Bub1 is sufficient for Bub3 binding and for recognition of Knl1-MELTP repeats [60], we started our analysis with a construct corresponding to GFP-BubR1B3BD (residues 362–431). We found that several of the APC/C subunits (blue squares, Figure 4A) were specifically enriched in the GFP-BubR1B3BD precipitates in comparison to those in the GFP control. We validated these interactions by western blotting against the APC/C subunits Cdc27/Apc3, Apc7, and Apc4 on GFP-BubR1B3BD precipitates (Figure 4B). In reciprocal IP experiments, we detected GFP-BubR1B3BD in complex with the APC/C subunit Cdc27 (Figure S6A).
Figure 4.
The BubR1 Loop Promotes APC/C Binding
(A and C) Volcano plot showing the results from three independent SILAC experiments using GFP and GFP-BubR1B3BD (residues 362–431) (A) or GFP-BubR1B3BD ΔL and GFP-BubR1B3BD (C) as affinity resins to identify specific interaction partners in mitotic lysates showing that the BubR1 B3BD binds to the APC/C and that this depends on the BubR1 loop. A p value of 0.05 and 0.1 was used as cutoff for significance, respectively.
(B and D) Western blot of IPs from mitotic Flp-In T-REx cell lines expressing the indicated GFP-BubR1 constructs showing that the B3BD of BubR1 is able to pull down APC/C subunits (B) and that this interaction is impaired if the loop is deleted (D). Tubulin was used as loading control.
(E and F) Volcano plot showing the results from two (E) or three (F) independent SILAC experiments using GFP-Bub1B3BD (residues 209–270) and GFP-BubR1B3BD (E) or GFP-Bub11–284 and GFP-BubR11–431 (F) as affinity resins to identify specific interaction partners in mitotic lysates, showing that the BubR1 N-terminal region increases the specificity of the BubR1-APC/C interaction. A p value of 0.05 was used as cutoff for significance.
See also Figures S6 and S7.
Next, we compared the interactome of GFP-BubR1B3BD with and without the loop region (Figure 4C). This showed that APC/C was enriched in precipitates of intact GFP-BubR1B3BD, indicating that the loop promotes binding of the APC/C (Figure 4C). The role of the BubR1 loop on APC/C binding was recapitulated in a GFP-IP experiment followed by western blotting (Figure 4D; quantified in Figure S6B). Collectively, these results indicate that the BubR1 loop promotes an interaction of the GFP-BubR1B3BD:Bub3 complex with the APC/C.
Our results also indicate that the B3BDs of Bub1 and BubR1 direct Bub3 to Knl1 and the APC/C, respectively. To corroborate this idea, we compared precipitates of GFP-Bub1B3BD (residues 209–270) and GFP-BubR1B3BD in another SILAC experiment (Figure 4E). In agreement with the hypothesis, Knl1 was clearly enriched in precipitates of GFP-Bub1B3BD. On the other hand, we did not observe an enrichment of APC/C subunits with GFP-BubR1B3BD in comparison to GFP-Bub1B3BD. Instead, we found APC/C subunits to be strongly enriched in precipitates of both constructs (and therefore appearing in the middle of the volcano plot, together with Bub3). This result suggests that the B3BD of Bub1 can, in principle, interact both with Knl1 and with the APC/C, whereas that of BubR1 can only bind the APC/C.
Based on these observations, we asked whether the presence of longer segments of Bub1 and BubR1 would increase the selectivity for these substrates. GFP-Bub11–284 and GFP-BubR11–431 encompass the entire N-terminal region in addition to the B3BD, including the TPR repeats and, in case of BubR1, also its two KEN boxes, the first of which is essential for binding to Cdc20 and therefore incorporation into the MCC [42] (Figure 1A). In SILAC IP experiments, we observed that GFP-Bub11–284 and GFP-BubR11–431 have exquisite specificity for Knl1 and the APC/C, respectively. Furthermore, we also detected Mad2 and Cdc20 in the BubR11–431 IPs, likely because this BubR1 segment, which contains the KEN boxes, is sufficient for an interaction with these MCC subunits (Figure 4F). Thus, the selectivity of BubR1 for the APC/C results from a combination of factors, including, in addition to the KEN boxes, the BubR1 loop as a direct APC/C binder. This was further confirmed by examining IPs of constructs in which only the loop regions were swapped. GFP-Bub1BR1-LL and GFP-BubR1B1-LL were both unable to interact effectively with the APC/C (Figure S7A).
The BubR1 Loop Is Required for APC/C Inhibition In Vitro
We expressed recombinant versions of Bub3 complexes of a BubR1 segment (residues 1–571) that is larger than the minimal segment of BubR1 that can restore SAC function in BubR1-depleted cells [42, 69]. In the same scaffold, we also created the chimeric mutants BubR1B1-LL and BubR1ΔLL. We then purified the resulting protein complexes to homogeneity. In isolation, BubR1WT:Bub3, BubR1B1-LL:Bub3, and BubR1ΔLL:Bub3 shared identical retention volumes in size exclusion chromatography experiments, attesting to their stability (Figure S7B). BubR1WT:Bub3, BubR1B1-LL:Bub3, and BubR1ΔLL:Bub3 also formed stoichiometric MCC complexes with Cdc20 and Mad2 with indistinguishable retention volumes (Figures 5A and S7B). We used a recently described in vitro assay to monitor assembly kinetics of MCC complexes containing BubR1B1-LL:Bub3 and BubR1ΔLL:Bub3 [31] and found them to be identical to those of BubR1WT:Bub3 (Figure S6C). Collectively, these observations indicate that the BubR1 loop is dispensable for MCC stability and assembly kinetics.
Figure 5.
The BubR1 Loop Is Required for APC/C Inhibition In Vitro
(A) BubR1wt:Bub3 as well as the two loop mutants BubR1B1-LL:Bub3 and BubR1ΔLL:Bub3 interact in size exclusion chromatography with the other two MCC components Cdc20 and Mad2. In the chromatogram, the height of elution curves for the three different MCC complexes were rescaled to match that of MCC containing BubR1ΔLL, which emphasizes the remarkable similarity of the elution profiles, an indication that the different MCC complexes are structurally stable and virtually identical. Vertical dashed lines indicate the elution volumes of the individual constituents of the three MCC complexes. The corresponding elution profiles and SDS-PAGE analyses are shown in Figure S7B.
(B) Ubiquitination reactions in the presence of recombinant APC/C-pE (carrying 68 phospho-mimicking mutations) [70] and the fluorescently labeled N-terminal domain of cyclin B were analyzed by SDS-PAGE and fluorescence scanning. MCC containing BubR1ΔLL:Bub3 is less efficient in inhibiting APC/C cyclin B ubiquitination activity in comparison to MCC containing BubR1wt:Bub3. Omitting Bub3 from MCC also reduces APC/C inhibition. NTD, N-terminal domain; Ub, ubiquitin.
See also Figures S6 and S7.
Therefore, we next tested the ability of MCC containing either BubR1WT:Bub3 or BubR1ΔLL:Bub3 to inhibit APC/C activity in vitro by evaluating ubiquitination of the APC/C substrate cyclin B. For these assays, we used concentrations of MCC (30 nM), APC/C (20 nM), and Cdc20 (100 nM) that are considered physiological [31]. To obtain active APC/C, we used APC/C-pE, an APC/C mutant carrying 68 phosphomimetic mutations that activate APC/C [70]. Wild-type and mutant MCC complexes were allowed to assemble for 15 hr and were purified to homogeneity (Figure S6D). They were then added to APC/CCdc20 (besides being an MCC subunit, Cdc20 also acts as an activator of APC/C), and the cyclin B ubiquitination reaction was initiated by addition of the E1 ubiquitin mix. Under these conditions, we observed near complete ubiquitination of cyclin B already within 30 min (Figure 5B, lanes 2–4). There was substantial inhibition of this reaction upon addition of wild-type MCC, which continued well into the 120 min time point (Figure 5B, lanes 5–7). MCC containing BubR1ΔLL:Bub3, on the other hand, clearly retained partial functionality but inhibited the cyclin B ubiquitination activity of APC/C less efficiently than the wild-type complex (Figure 5B; compare lanes 5–7 with lanes 8–10).
These observations argue that removal of the loop region of BubR1 causes a partial impairment of the ability of MCC to inhibit APC/C. Because our previous studies argue the loop region of Bub1 acts to modulate the binding affinity of Bub3 for phosphorylated MELT repeats [60] and our data so far suggest that this may hold true also for the BubR1 loop, we assembled MCCs with BubR1WT or BubR1ΔLL that lacked bound Bub3 and purified them to homogeneity. Importantly, the absence of Bub3 did not overtly impair MCC formation or stability (Figure S6D). Both MCC versions lacking Bub3 were less efficient in inhibiting APC/C cyclin B ubiquitination activity than MCC containing BubR1WT bound to Bub3 and showed an even stronger defect than that of MCC containing BubR1ΔLL:Bub3 (Figure 5B, lanes 11–16).
Bub3 Is Required for Robust SAC Signaling
Collectively, these results show that Bub3 plays a role in APC/C inhibition and support the hypothesis that the BubR1 loop contributes to the regulation of this process. To test directly the role of Bub3 in MCC function, we examined the effects on the SAC of expressing BubR1 mutants impaired in their interaction with Bub3. Specifically, we deleted the complete B3BD (BubR1ΔB3BD) or introduced two point mutations in the B3BD that are known to prevent Bub3 binding (BubR1E409K+E413K; Figure 6A) [40, 42]. Both mutants were unable to support the SAC in the absence of endogenous BubR1 (Figure 6B). Furthermore, BubR1 mutants defective in Bub3 binding were impaired in binding to APC/C subunits in IPs, similar to the BubR1ΔLL mutant (Figure 6C; quantified in Figure 6D). Collectively, our data provide a strong indication that Bub3, in complex with BubR1, plays a role in the SAC and that the BubR1 loop region works by modulating the interaction of Bub3 with the APC/C.
Figure 6.
Bub3 Is Required for Robust SAC Signaling
(A) Domain organization of the BubR1 constructs used.
(B) Mean duration of mitosis of Flp-In T-REx stable cell lines expressing the indicated GFP-BubR1 constructs in the absence of endogenous BubR1 and in the presence of 50 nM nocodazole. Cell morphology was used to measure entry into and exit from mitosis by time-lapse microscopy (n > 55 per cell line per experiment) from two independent experiments. Error bars depict SEM.
(C) Western blot of IPs from mitotic Flp-In T-REx cell lines expressing the indicated GFP-BubR1 constructs showing a defect especially in binding to APC/C subunits if BubR1 cannot bind to Bub3. Tubulin was used as loading control. ∗, band resulting from previous incubation with Bub1 antibody; ∗∗, unspecific band recognized by the Bub1 antibody.
(D) Quantification of the western blot in Figure 6C. The amounts of co-precipitating proteins were normalized to the amount of GFP-BubR1 bait present in the IPs. Values for GFP-BubR1wt are set to 1. The graph shows mean intensity of two independent experiments. Error bars represent SEM.
See also Figure S4.
Discussion
BubR1 is an essential component of the SAC effector, the MCC. Our understanding of the function of BubR1 in the SAC has been greatly facilitated by functional and structural studies on the MCC and of its interaction with APC/C. High-resolution structures of the APC/CMCC complex [43, 46, 47] have offered an ideal framework to understand the molecular basis of the interaction of MCC with the APC/C, including the role of specific BubR1 sequences identified in previous studies [71], such as the KEN1 and KEN2 boxes and the more recently identified ABBA motifs [3, 39, 41, 42, 43, 44, 72, 73, 74, 75, 76, 77]. The overall picture emerging from these analyses is that a MCC core complex containing one copy each of BubR1, Bub3, Cdc20, and Mad2 binds a second Cdc20 molecule, possibly already bound to the APC/C [2, 44, 45, 46, 47]. In the APC/CMCC complex, BubR1 binds extensively to both Cdc20 subunits, stabilizes the interaction of Mad2 with one of the two Cdc20 subunits, and provides extensive contacts with the APC/C that reinforce its interaction with MCC [46, 47].
The SAC role of Bub3, a constitutive binding partner of BubR1 and Bub1, has remained more elusive. In most organisms, Bub3 forms constitutive complexes both with Bub1 and with BubR1 [57]. An exception is Schizosaccharomyces pombe, where the BubR1 ortholog Mad3 does not interact with Bub3 [43, 78]. The Bub1 and BubR1 paralogs perform distinct functions and have distinct localization patterns. Ablation of Bub3 affects the function of both paralogs, introducing a significant complication in the interpretation of the resulting mitotic defect. On the other hand, preventing the selective interaction of Bub1 or BubR1 with Bub3 by introducing point mutations in the Bub3 binding domains of Bub1 or BubR1 lends itself to the objection that these mutations, by disrupting an interaction with Bub3, might destabilize MCC or its interaction with the APC/C. To overcome these limitations, we took advantage of our previous structural analysis of the Bub1:Bub3:MELTP ternary complex, which identified Bub3 as a phospho-amino acid adaptor and suggested that a region of Bub1, the loop, contributes to the binding affinity for phosphorylated targets [58]. Modifications of the Bub1 loop predictably alter the function of associated Bub3 [60], providing for the first time a clean handle to distinguish the mitotic functions of Bub1-associated and BubR1-associated Bub3 without a need to ablate Bub3 or its interactions with Bub1 or BubR1. In this study, we have taken advantage of this recent progress to test the role of the BubR1 loop and discovered that it is essential for SAC function. Thus, our analysis identifies another short, crucial sequence determinant of BubR1 function in the SAC response.
Previously, it was proposed that Bub3 acts as a catalytic enhancer of the BubR1:Cdc20 interaction at kinetochores and in the cytosol [77]. The moderate stimulation by Bub3 of MCC-dependent APC/C inhibition was interpreted in light of this possible catalytic effect of Bub3 [77]. This alternative hypothesis, however, is at odds with our recent demonstration that the interaction of Mad2 with Cdc20 is the sole rate-limiting step of MCC assembly [31].
Our interpretation of the role of Bub3 is rather that it contributes, thanks to modulation by the BubR1 loop motif, to the binding affinity of the MCC for the APC/C. Recombinant loop mutants (BubR1B1-LL and BubR1ΔLL) engaged in stable MCC complexes with Mad2, Cdc20, and Bub3 in vitro, which assembled with rates identical to those observed with wild-type BubR1. Thus, it is unlikely that the SAC defect observed with these mutants reflects a problem in MCC assembly. In vitro cyclin B ubiquitination assays showed that BubR1 lacking the loop is less efficient in inhibiting APC/C than BubR1WT, and this correlates with a loss of APC/C binding affinity in vivo. Two Bub3-binding defective BubR1 mutants (BubR1ΔB3BD and BubR1E409K+E413K) showed the same SAC defect phenotype described for the loop mutants. Their binding to APC/C subunits in coIPs was also impaired.
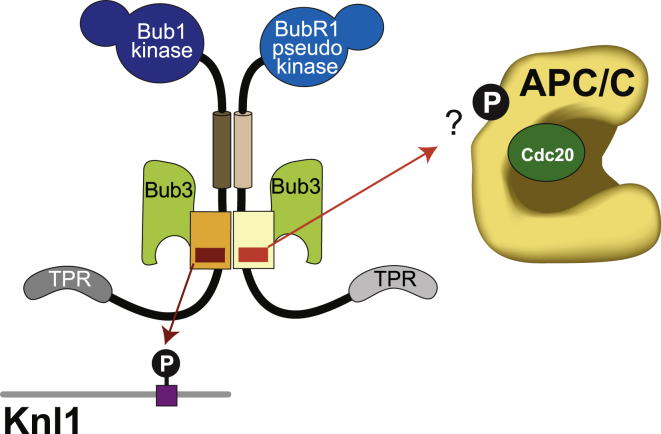
Phosphorylation of a loop of Apc1 promotes binding of Cdc20 and APC/C activation [70, 79, 80, 81, 82, 83, 84]. Similarly, phosphorylation may regulate SAC-dependent inhibition of APC/C. Bub3 is an adaptor protein for phosphorylated motifs, and in complex with Bub1 (through the contribution of the Bub1 loop), it binds to phosphorylated MELT repeats of Knl1 [58, 60]. In analogy to the Bub1 loop, we suspect that the BubR1 loop contributes to the recognition of a phosphorylated motif on the APC/C by Bub3 (Figure 7) [2]. Bub3 (with the help of the BubR1 loop) might be expected to bind sequences related to the MELTP motifs of Knl1. Two such motifs, with sequences MEVT and MELT, exist, respectively, in the Apc4 (residues 116–119) and Apc5 (residues 175–178) subunits of the APC/C, and at least T178Apc5 is phosphorylated during mitosis [70]. Furthermore, T178Apc5 and S179Apc5 are part of the pool of putative phosphomimetic mutations in the APC/C pE sample. We created a new APC/C mutant (APC/C pE-3A; Figure S6D) carrying alanine mutations at T119Apc4, T178Apc5, and S179Apc5 and asked whether mutation of these putative Bub3 target motifs in APC/C weakened the sensitivity of APC/C to wild-type MCC, phenocopying the removal of the BubR1 loop. However, APC/C pE-3A remained as sensitive to MCC as APC/C pE, suggesting that phosphorylation of these residues is not important for MCC inhibition or that the penetrance of the phosphomimetic mutations is limited (Figure S6E).
Figure 7.
Model for the Differential Functions of the BubR1 and Bub1 Loop
Model showing the different functions of the loops in BubR1 and Bub1. Bub1 and BubR1 form a pseudo-symmetric heterodimer through the B3BDs and the helix as well as through the presence of Bub3. The loops are not involved in this interaction but serve different functions. The Bub1 loop enhances binding of the Bub1:Bub3 complex to Knl1-MELTP motifs, which in turn recruits BubR1:Bub3 to kinetochores. The BubR1 loop, however, is not able to enhance such an interaction of Bub3 with Knl1 but instead seems to promote binding of BubR1:Bub3 to the APC/C. This is required for the SAC function of BubR1. We hypothesize that this interaction could work via modulation of Bub3 and be regulated in a phosphorylation-dependent manner, arguing that the BubR1 loop functions in analogy to the Bub1 loop.
The identification of the relevant phospho-epitopes and of the kinase that generates them is therefore an important priority for future studies. Likely, this will be a challenging task, because the 1.2-MDa APC/C particle is highly phosphorylated in mitosis by several kinases, including Plk1 and Cdk1 [81, 85]. Furthermore, the structural analysis of the APC/CMCC complex offers only limited insight into this specific question, because both Bub3 and the segment of BubR1 that binds to it (the B3BD) were invisible in the structures of APC/CMCC [46, 47].
Our analysis of the role of the BubR1 and Bub1 loop led us to revisit the issue of the molecular basis of kinetochore localization and turnover of Bub1 and BubR1. The main conclusions from this analysis are completely consistent with the model that the B3BD of Bub1 is sufficient for kinetochore localization, whereas that of BubR1 is not. Our analysis tested all major predictions of the model, providing a complete account of the mechanism of Bub1 and BubR1 recruitment to kinetochores. It has recently been proposed that a small autonomous pool of BubR1 can localize to kinetochores in a Bub1-independent fashion to perform its function in the SAC [86]. However, we suspect that this pool of kinetochore BubR1 results from the availability of a high concentration of free MELTP motifs after artificial Bub1 depletion [60, 86]. We show here that BubR1 mutants deprived of the helical domain that mediates robust kinetochore recruitment of BubR1 are SAC proficient and that further mutation of the loop disrupts this SAC function. Finally, the BubR1 loop deletion mutants we have tested localize normally to kinetochores but are entirely SAC defective, clearly showing that the deleterious effects of loop mutations on the SAC are uncorrelated with kinetochore localization of BubR1. In certain organisms, like C. elegans and S. cerevisiae, the BubR1 ortholog Mad3 may not even be able to localize to kinetochores [62, 87]. The significance of kinetochore recruitment of BubR1 remains therefore an open question for future studies.
In conclusion, our studies illustrate how the divergence of two paralogs resulted in the emergence of motifs that modulate the binding affinity of a phosphopeptide-recognition module to allow binding to distinct binding partners, a vivid example of sub-functionalization. The evolutionary forces that drove the specific sub-functionalization of Bub and Mad proteins, however, remain uncertain [88] and an interesting subject for future studies.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit polyclonal anti-GFP | generated in-house | N/A |
| rabbit polyclonal anti-Knl1-N | generated in-house | #SI0787 |
| rabbit polyclonal anti-Bub1 | Abcam | Cat#ab9000 |
| mouse monoclonal anti-BubR1 | BD Transduction lab | Cat#612503 |
| sheep polyclonal anti-BubR1 | Stephen S. Taylor, University of Manchester [89] | N/A |
| mouse monoclonal anti-Bub3 | BD Transduction lab | Cat#611731 |
| mouse monoclonal anti-Tubulin | Sigma | Cat#T9026 |
| rabbit polyclonal anti-Apc7 | generated in-house | #SI0651 |
| goat polyclonal anti-Apc4 | Santa Cruz | Cat#sc21414 |
| mouse monoclonal anti-Cdc20 | Santa Cruz | Cat#sc5296 |
| mouse monoclonal anti-Mad2 | generated in-house | #AS55-A12 |
| mouse monoclonal anti-Cdc27 | BD Transduction lab | Cat#610455 |
| mouse monoclonal anti-Vinculin | Sigma | Cat#V9131 |
| sheep anti-mouse HRP | Amersham | Cat#NXA931-1ML |
| donkey anti-rabbit HRP | Amersham | Cat#NXA934-1ML |
| human anti-centromere (CREST) | Antibodies Inc. | Cat#15-234-0001 |
| donkey anti-goat HRP | Santa Cruz | Cat#sc2020 |
| Protein G HRP | Life Technologies | Cat#P21041 |
| mouse monoclonal anti-Bub1 | Abcam | Cat#ab54893 |
| goat anti-human Alexa Fluor-647 | Invitrogen | Cat#A-21445 |
| goat anti-mouse Rhodamine Red | Jackson ImmunoResearch | Cat#115-295-003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| APC/C-pE | Jan-Michael Peters lab [70] | N/A |
| APC/C-pE-3A | this paper | N/A |
| Uba1 | Jan-Michael Peters lab [70] | N/A |
| UbcH10 | Jan-Michael Peters lab [70] | N/A |
| Ube2S | Jan-Michael Peters lab [70] | N/A |
| CycB-NTD | Jan-Michael Peters lab [70] | N/A |
| Ubiquitin | Enzo Life Sciences Inc. | Cat#BML-UW8795-0005 |
| BubR1 1-571 wt/Bub3 | Andrea Musacchio lab [31] | N/A |
| BubR1 1-571 wt | Andrea Musacchio lab [31] | N/A |
| BubR1 1-571 B1-LL/Bub3 | this paper | N/A |
| BubR1 1-571 ΔLL/Bub3 | this paper | N/A |
| BubR1 1-571 ΔLL | this paper | N/A |
| Cdc20 | Andrea Musacchio lab [31] | N/A |
| Mad2 | Andrea Musacchio lab [31] | N/A |
| Lysyl Endopeptidase | Wako | Cat#125-05061 |
| Glu-C Endopeptidase | Promega | Cat#V1651 |
| Protease-inhibitor mix HP Plus | Serva | Cat#39107 |
| PhosSTOP phosphatase inhibitors | Roche | Cat#04906845001 |
| Zeocin | Invitrogen | Cat#R25001 |
| Doxycycline | Sigma | Cat#D9891; CAS:24390-14-5 |
| Nocodazole | Sigma | Cat#M1404; CAS:31430-18-9 |
| DAPI | Serva | Cat#:18860.01 |
| Poly-L-Lysine | Sigma | Cat#:P4832; CAS:25988-63-0 |
| CO2-independent medium | GIBCO | Cat#18045_054 |
| Thymidine | Sigma | Cat#T1895 |
| Lipofectamine2000 | Invitrogen | Cat#11668-019 |
| X-treme Gene | Roche | Cat#06365809001 |
| Poly-D-Lysine | Millipore | Cat#A-003-E |
| Mowiol | Calbiochem | Cat#475904 |
| SILAC DMEM | PAA | Cat#E15-086 |
| dialyzed serum | PAA | Cat#A11-107 |
| Arg-0 | Sigma | Cat#A6969; CAS:1119-34-2 |
| Lys-0 | Sigma | Cat#L8662; CAS:657-27-2 |
| Arg-10 | Silantes | Cat#201604102 |
| Lys-8 | Silantes | Cat#211604102 |
| Urea | Sigma | Cat#U6504; CAS: 57-13-6 |
| Acetonitrile | Fluka | Cat#34967; CAS: 75-05-8 |
| Trypsin | Promega | Cat#V5113 |
| Ammonium Bicarbonate (Ambic) | Fluka | Cat#C990X98; CAS: 1066-33-7 |
| trifluoroacetic acid (TFA) | Sigma | Lot#RB228879; CAS: 76-05-1 |
| iodoacetamide | Sigma | Cat#I6125; CAS: 144-48-9 |
| Experimental Models: Cell Lines | ||
| Trichoplusia ni: BTI-Tnao38 | Garry W. Blissard Lab | N/A |
| S. frugiperda: Sf9 cells in Sf900™ III SFM | Thermo Fisher | Cat#12659017 |
| Human: HeLa, female Cervix Adenocarcinoma Cells | ATCC | Cat#CCL-2™ |
| Human: Flp-In T-Rex HeLa | Stephen S. Taylor, University of Manchester | N/A |
| Human: Flp-In T-Rex HeLa GFP | Andrea Musacchio lab [60] | N/A |
| Human: Flp-In T-Rex HeLa Bub1 wt | Andrea Musacchio lab [60] | N/A |
| Human: Flp-In T-Rex HeLa Bub1 B3BD | Andrea Musacchio lab [60] | N/A |
| Human: Flp-In T-Rex HeLa BubR1 wt | Andrea Musacchio lab [60] | N/A |
| Human: Flp-In T-Rex HeLa BubR1 B1-L | Andrea Musacchio lab [60] | N/A |
| Human: Flp-In T-Rex HeLa BubR1 B1-LL | this paper | N/A |
| Human: Flp-In T-Rex HeLa BubR1 B1-LL/ΔH | this paper | N/A |
| Human: Flp-In T-Rex HeLa BubR1 B1-B3BD/B1-H | this paper | N/A |
| Human: Flp-In T-Rex HeLa BubR1 KEN1/AAA | this paper | N/A |
| Human: Flp-In T-Rex HeLa BubR1 ΔL | this paper | N/A |
| Human: Flp-In T-Rex HeLa BubR1 ΔLL | this paper | N/A |
| Human: Flp-In T-Rex HeLa BubR1 B3BD | Andrea Musacchio lab [60] | N/A |
| Human: Flp-In T-Rex HeLa BubR1 B3BD ΔL | this paper | N/A |
| Human: Flp-In T-Rex HeLa Bub1 1-284 | this paper | N/A |
| Human: Flp-In T-Rex HeLa BubR1 1-431 | this paper | N/A |
| Human: Flp-In T-Rex HeLa BubR1 ΔB3BD | this paper | N/A |
| Human: Flp-In T-Rex HeLa BubR1 E409K+E413K | Andrea Musacchio lab [60] | N/A |
| Human: Flp-In T-Rex HeLa BubR1 ΔH | Andrea Musacchio lab [60] | N/A |
| Human: Flp-In T-Rex HeLa BubR1 ΔL/ΔH | this paper | N/A |
| Experimental Models: Organisms/Strains | ||
| E.coli:One Shot OmniMAX 2 T1R Chemically Competent Cells | Thermo Fisher | Cat#C854003 |
| Oligonucleotides | ||
| siRNA Bub1: 5′-GGUUGCCAACACAAGUUCU-3′ | Dharmacon, custom made | N/A |
| siRNAi BubR1: 5′-CGGGCAUUUGAAUAUGAAA-3′ | Dharmacon, custom made | N/A |
| Recombinant DNA | ||
| MultiBac | Geneva Biotech | N/A |
| pCDNA 5/FRT/TO plasmid | Invitrogen | Cat#V6520-20 |
| pCDNA5/FRT/TO-EGFP-IRES | Andrea Musacchio lab [5] | N/A |
| pCDNA5/FRT/TO-EGFP-Bub1 wt | Andrea Musacchio lab [5] | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 wt | Andrea Musacchio lab [5] | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 B1-LL | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 B1-L | Andrea Musacchio lab [60] | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 KEN1/AAA | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 B1-LL/ΔH | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 B1-B3BD/B1-H | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 ΔLL | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 ΔL | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 B3BD | Andrea Musacchio lab [60] | N/A |
| pCDNA5/FRT/TO-EGFP-Bub1 B3BD | Andrea Musacchio lab [5] | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 B3BD ΔL | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-Bub1 1-284 | Andrea Musacchio lab [5] | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 1-431 | Andrea Musacchio lab [60] | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 ΔB3BD | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 E409K+E413K | Andrea Musacchio lab [60] | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 ΔH | Andrea Musacchio lab [60] | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 B1-L/ΔH | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 1-431 B1-L | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 1-431 B1-LL | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-Bub1 BR1-L | Andrea Musacchio lab [60] | N/A |
| pCDNA5/FRT/TO-EGFP-Bub1 BR1-LL | this paper | N/A |
| pCDNA5/FRT/TO-EGFP-BubR1 ΔL/ΔH | this paper | N/A |
| Software and Algorithms | ||
| ImageJ 1.46 r | NIH | https://imagej.nih.gov/ij/ |
| Imaris 7.3.4 32-bit | Bitplane | http://www.bitplane.com/imaris |
| GraphPad Prism 6.0 | GraphPad software | http://www.graphpad.com |
| Illustrator CS5.1, version 15.1.0 | Adobe | http://www.adobe.com |
| Photoshop CS5.1, version 12.1 | Adobe | http://www.adobe.com |
| MaxQuant, version 1.5.2.18 | [90] | http://www.coxdocs.org/doku.php?id=maxquant:start |
| Perseus, version 1.5.1.5 | [91] | http://www.coxdocs.org/doku.php?id=perseus:start |
| Other | ||
| Roti®garose Protein A beads | Roth | Cat#1278.1 |
| Protein G affinity resin | Amintra | Cat#APG0005 |
| GFP-Trap_A | ChromoTek | Cat#gta-20 |
| ECL Prime western blotting system | GE Healthcare | Cat#RPN 2232 |
| Amicon concentrators (10K/30K) | Millipore | Cat#UFC901024;UFC903024 |
| Nitrocellulose membrane | GE Healthcare | Cat#10600001 |
| Mowiol mounting media | Calbiochem | Cat#475904 |
| 4%–12% NuPAGE Bis-Tris gels | Life Technologies | Cat#NP0321BOX |
| His Trap FF (5ml) | GE Healthcare | Cat#17-5255-01 |
| Superdex 200 Increase 5/150 GL | GE Healthcare | Cat#28-9909-45 |
| Superdex 200 16/60 | GE Healthcare | Cat#28989335 |
| Sep-Pak C18 Vac Cartridge, 50 mg Sorbent | Waters | Cat#WAT054955 |
| 24-well μ-plate | ibidi | Cat#82406 |
| 35 mm glass bottom μ-dishes | ibidi | Cat#81158 |
| Rotilabo® syringe filters | Roth | Cat#P820.1 |
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to and will be fulfilled by Andrea Musacchio (andrea.musacchio@mpi-dortmund.mpg.de).
Experimental Model and Subject Details
cDNAs used for expression of recombinant proteins were either of human origin, or generated synthetically based on human sequences. HeLa (female Cervix Adenocarcinoma) cells were grown in DMEM (PAN Biotech) supplemented with 10% FBS, penicillin and streptomycin and 2 mM L-glutamine. Cells were grown in a humidified atmosphere of 37°C and 5% CO2.
Method Details
Mammalian plasmids
Plasmids were derived from the pCDNA5/FRT/TO-EGFP-IRES, a previously modified version [5] of the pCDNA5/FRT/TO vector (Invitrogen). To create N-terminally tagged EGFP Bub1 and BubR1 truncation constructs, Bub1 and BubR1 sequences were obtained by PCR amplification from the previously generated pCDNA5/FRT/TO-EGFP-Bub1-IRES and pCDNA5/FRT/TO-EGFP-BubR1-IRES vector, respectively [5] and subcloned in frame with the GFP-tag. Mutations and deletions within the Bub1 and BubR1 constructs were generated by standard site-directed mutagenesis or by a mutagenesis protocol [92]. All Bub1 constructs were RNAi resistant [6]. BubR1-expressing constructs were made siRNA-resistant by changing the sequence targeted by the RNAi oligos to ‘AACGTGCCTTCGAGTACGAGA’. pCDNA5/FRT/TO-based plasmids were used for generation of stable cell lines, as well as for transient transfection. All plasmids were verified by sequencing.
Cell culture and transfection
HeLa cells were grown in DMEM (PAN Biotech) supplemented with 10% FBS (Clontech), penicillin and streptomycin (GIBCO) and 2 mM L-glutamine (PAN Biotech). For all plasmid transfections of HeLa cells X-tremeGENE transfection agent (Roche) was used at a 3:1 ratio with plasmid DNA. Flp-In T-REx HeLa cells used to generate stable doxycycline-inducible cell lines were a gift from S.S. Taylor (University of Manchester, Manchester, England, UK). Flp-In T-REx host cell lines were maintained in DMEM with 10% tetracycline-free FBS (Clontech) supplemented with 50 μg/ml Zeocin (Invitrogen). Flp-In T-REx HeLa expression cell lines were generated as previously described [5]. Briefly, Flp-In T-Rex HeLa host cells were cotransfected with a ratio of 9:1 (w/w) pOG44:pcDNA5/FRT/TO expression plasmid using X-tremeGene transfection agent (Roche). 48 hr after transfection, Flp-In T-Rex HeLa expression cell lines were put under selection for two weeks in DMEM with 10% tetracycline-free FBS (Invitrogen) supplemented with 250 μg/ml Hygromycin (Roche) and 5 μg/ml Blasticidin (ICN Chemicals). The resulting foci were pooled and tested for expression. Gene expression was induced by addition of 0.05-0.5 μg/ml doxycycline (Sigma) for 24 hr. siBUB1 (GE Healthcare Dharmacon; 5′-GGUUGCCAACACAAGUUCU-3′) or siBUBR1 (GE Healthcare Dharmacon; 5′-CGGGCAUUUGAAUAUGAAA-3′) duplexes were transfected with Lipofectamine 2000 (Invitrogen) at 50 nM for 24 hr.
For experiments in HeLa cells, cells were synchronized with a double thymidine arrest 5 hr after transfection with siRNA duplexes. In brief, after washing the cells with PBS they were treated with thymidine for 16 hr and then released into fresh medium. 3 hr after the release, 50 nM siRNA duplexes were transfected for a second time. 5 hr after transfection, cells were treated with thymidine for 16 hr and afterward released in fresh medium. Thymidine (Sigma-Aldrich) was used at 2 mM. Unless differently specified, nocodazole (Sigma-Aldrich) was used at 3.3 μM.
Immunoprecipitation and immunoblotting
To generate mitotic populations for immunoprecipitation experiments, cells were treated with 330 nM nocodazole for 16 hr. Mitotic cells were then harvested by shake off and lysed in lysis buffer [150 mM KCl, 75 mM HEPES, pH 7.5, 1.5 mM EGTA, 1.5 mM MgCl2, 10% glycerol, and 0.075% NP-40 supplemented with protease inhibitor cocktail (Serva) and PhosSTOP phosphatase inhibitors (Roche)]. Extracts were precleared using a mixture of protein A–agarose (Roti®garose Protein A beads; Roth) and protein G-agarose (Protein G affinity resin; Amintra) beads for 1 hr at 4°C. Subsequently, extracts were incubated with GFP-Traps (ChromoTek; 3 μl/mg of extract) for 3 hr at 4°C. Immunoprecipitates were washed with lysis buffer and resuspended in sample buffer, boiled, and analyzed by SDS-PAGE and western blotting using 4%–12% gradient gels (NuPAGE® Bis-Tris Gels, Life technologies). For Cdc27 IPs, cells were lysed in lysis buffer (described above) and extracts were precleared with protein G-agarose beads for 1 hr at 4°C. Afterward, extracts were incubated with 1.5 ug/mg of the anti-Cdc27 primary antibody (mouse monoclonal, BD) for 2 hr at 4°C. Subsequently, protein G-agarose beads were added for 4 hr at 4°C. Immunoprecipitates were washed with a mild wash buffer (lysis buffer without salt) and analyzed as described above. The following antibodies were used: anti-GFP (in house made rabbit polyclonal antibody; 1:1000-3000), anti-Knl1-N (in house made rabbit polyclonal SI0787 antibody; 1:1000), anti-Bub1 (rabbit polyclonal; Abcam; 1:5000), anti-BubR1 (mouse monoclonal; BD; 1:1000), anti-BubR1 (sheep polyclonal, 1:500, a gift from S. S. Taylor (University of Manchester, Manchester, England, UK)), anti-Bub3 (mouse monoclonal; BD; 1:1000), anti-Tubulin (mouse monoclonal; Sigma; 1:8000), anti-Apc7 (in house made rabbit polyclonal antibody SI0651, 1:500), anti-Apc4 (goat polyclonal, Santa-Cruz, 1:100), anti-Cdc20 (mouse monoclonal, Santa Cruz, 1:500), anti-Mad2 (in house made mouse monoclonal antibody, clone AS55-A12, 1:500), anti-Cdc27 (mouse monoclonal, BD; 1:1000-3000), anti-Vinculin (mouse monoclonal, Sigma, 1:20000). Secondary antibodies were anti–mouse (Amersham), anti-goat (Santa-Cruz) and anti–rabbit (Amersham) affinity-purified with horseradish peroxidase conjugate (working dilution 1:10000) or Protein G with horseradish peroxidase conjugate (Life technologies) (working dilution 1:8000). After incubation with ECL western blotting system (GE Healthcare), images were acquired with the ChemiDoc™ MP Imaging System (BioRad) in 16-bit TIFF format. Images were cropped and converted to 8-bit using ImageJ software (NIH). Brightness and contrast were adjusted using Photoshop CS5 (Adobe). Unmodified 16-bit TIFF images were used for quantification with ImageJ software. Measurements were graphed with Excel (Microsoft) and GraphPad Prism version 6.0 for Mac OS X (GraphPad Software).
SILAC and mass spectrometry
For labeling, cells were cultivated for five passages in specialized SILAC medium (DMEM, E15-086, PAA; dialyzed serum, A11-107, PAA) supplemented with either “light” arginine and lysine (referred to as Arg-0 and Lys-0, A6969 and L8662, Sigma) or “heavy” arginine (13C615N4) and lysine (13C615N2) (referred to as Arg-10 and Lys-8) [68, 93]. Afterward, cells were synchronized in prometaphase by the addition of 330 nM nocodazole for 16 hr and harvested by mitotic shake off. During the following anti-GFP IP (described in immunoprecipitation and immunoblotting) the washing steps were performed in a mild wash buffer (75 mM HEPES, pH 7.5, 1.5 mM EGTA, 1.5 mM MgCl2, 10% glycerol) to preserve potentially weak interactions. IPs were usually performed in duplicates swapping the labels (called FOR and REV) and repeated three times to be able to perform statistical analyses of the results. The corresponding heavy and light samples of the forward and reverse experiment were mixed in the last washing step. Afterward, samples were processed for mass spectroscopy. Briefly, samples were reduced, alkylated, digested directly on the beads with LysC/Trypsin and desalted/concentrated on C18-reversed phase stage tips. Samples were then separated on a Thermo Fisher Scientific™ EASY-nLC 1000 HPLC system using a two hour gradient from 5%–60% with 0.1% formic acid and directly sprayed via a nanoelectrospray ion source (Proxeon Biosystems, now Thermo Fisher Scientific) in a quadrupole Orbitrap mass spectrometer (Q Exactive™, Thermo Fisher Scientific). The Q Exactive™ was operated in a data dependent mode acquiring one survey scan and subsequently ten MS/MS scans [94]. Data were analyzed with the quantitative proteomics software MaxQuant (version 1.5.2.18) [90] and further processed in Perseus (version 1.5.1.5) [91]. Contaminants and reverse hits were removed from the protein lists. For t tests and volcano plots, proteins were further filtered to be quantified in at least 2 out of 3 replicates.
Live cell imaging
Cells were plated on a 24-well μ-Plate (Ibidi®). Drugs were diluted in CO2 Independent Medium (GIBCO®) and added to the cells 1 hr before filming. Cells were imaged every 20 to 30 min in a heated chamber (37°C) on a 3i Marianas system (Intelligent Imaging Innovations Inc.) equipped with Axio Observer Z1 microscope (Zeiss), Plan-Apochromat 40x/1.4NA oil objective, M27 with DIC III Prism (Zeiss), Orca Flash 4.0 sCMOS Camera (Hamamatsu) and controlled by Slidebook Software 6.0 (Intelligent Imaging Innovations Inc). For cells expressing the GFP-BubR1 proteins, only cells in which kinetochores were visible (or that were GFP-positive - for constructs that do not localize to kinetochores) were considered for the analysis.
Fluorescence recovery after photobleaching
For FRAP experiments cells were grown in 35 mm glass bottom μ-dishes (Ibidi®). Experiments were performed in the presence of 3.3 μM nocodazole, the presence of the GFP-tagged wild-type or mutant fusionprotein and in the absence of the endogenous protein. Cells were imaged on a 3i Marianas system (Intelligent Imaging Innovations Inc., described above) using a 100x/1.4NA Oil Objective (Zeiss). Photobleaching was performed as described previously [95]. Briefly, individual kinetochores were bleached with 100% laser power of an Argon-488 laser line. Images were binned 2x2 to increase signal-over-camera noise. At each time point a z stack consisting of 3 sections at 0.27 μm intervals was acquired. The GFP-signal was imaged for 5 time frames before photobleaching. After opening the laser shutter for 5 ms, cells were imaged by time-lapse microscopy, taking a z series every 0.8 s for a total duration of 2 min with an exposure time of 125 ms. Images were converted into maximal intensity projections and exported as 16-bit TIFF files. Measurements of fluorescence intensity were made on the 16-bit maximal intensity projections using ImageJ. Apart from the bleached KT, a non-bleached KT from the same nucleus and a region of the same size outside of the cell were also measured. Afterward, measurements were exported into excel. The relative fluorescence intensity was calculated as RFI = (FROI(t)/FBG(t)) / (FROI(t0)/FBG(t0)), as also described in [96], to correct for background intensity and for photobleaching that occurred during image acquisition. FROI(t) is the intensity of the bleached KT at different time points after bleaching, FBG(t) is the intensity of the control non-bleached KT at the corresponding time points. FROI(t0) is the average intensity of the bleached KT before bleaching, FBG(t0) is the average intensity of the control non-bleached KT before bleaching. A baseline value, calculated from the region outside of the cell, was subtracted from all values before entering the values into the formula shown above. The final data were analyzed using Graph Pad Prism 6.0. Between 5 and 20 cells from at least two independent experiments were analyzed for each investigated construct.
Immunofluorescence
HeLa cells and Flp-In T-REx HeLa cells were grown on coverslips precoated with poly-D-Lysine (Millipore, 15 μg/ml) and poly-L-Lysine (Sigma), respectively. For the experiments with HeLa cells, cells were synchronized with a double thymidine block and after release from that arrested in prometaphase by the addition of 330 nM nocodazole for 3 hr. For all other experiments, asynchronously growing cells were arrested in prometaphase by the addition of nocodazole for 3-4 hr and fixed using 4% paraformaldehyde. Cells were stained for Bub1 (mouse, ab54893, 1:400) and CREST/anti-centromere antibodies (Antibodies, Inc., 1:100), diluted in 2% BSA-PBS for 1.5 hr. Goat anti–human Alexa Fluor 647 (Invitrogen) and goat anti–mouse RRX (Jackson ImmunoResearch Laboratories, Inc.) were used as secondary antibodies. DNA was stained with 0.5 μg/ml DAPI (Serva) and coverslips were mounted with Mowiol mounting media (Calbiochem). Cells were imaged at room temperature using a spinning disk confocal device on the 3i Marianas system equipped with an Axio Observer Z1 microscope (Zeiss), a CSU-X1 confocal scanner unit (Yokogawa Electric Corporation), Plan-Apochromat 63x or 100x/1.4NA Oil Objectives (Zeiss) and Orca Flash 4.0 sCMOS Camera (Hamamatsu). Images were acquired as z sections at 0.27 μm. Images were converted into maximal intensity projections, exported, and converted into 8-bit. Quantification of kinetochore signals was performed on unmodified 16-bit z series images using Imaris 7.3.4 32-bit software (Bitplane). After background subtraction, all signals were normalized to CREST. At least 307 kinetochores were analyzed per condition. Measurements were exported in Excel (Microsoft) and graphed with GraphPad Prism 6.0 (GraphPad Software).
Protein expression and purification
Sequences coding for H6-mTurquoise-BubR11-571-ΔLL and H6-mTurquoise-BubR11-571-B1-LL were sub-cloned into pFLMultiBac vectors and baculoviruses were generated [97]. Expression of H6-mTurquoise-BubR11-571, the BubR1 mutant constructs, untagged Bub3, and Mps1 was performed in TnaO38 insect cells. Expression of H6-3xMyc-and BUB1:BUB3 was carried out in Sf9 cells. Expression of H6-Mad2 was carried out in Escherichia coli [97]. After infection with virus (1:50), cultures were grown at 27°C degrees and harvested after three days, and pellets stored at −20°C. MAD1:C-MAD2 and BUBR1:BUB3 were cultured by mixing individual viruses, each harboring individual genes. MPS1 was expressed in the presence of 2 μM Reversine. Insect cells were harvested by centrifugation at 750 g for 12 min in a Sorvall RC3BP+ (Thermo Scientific) centrifuge with Rotor H6000A. The pellet was resuspended in PBS, centrifuged at 500 g for 5 min, the supernatant was discarded and the pellet was stored at −20°C.
Generally, the cell pellet from 1 l of insect cell culture volume was re-suspended in 250 mL lysis buffer (25 mM HEPES pH 7.5, 300 mM NaCl, 5% glycerol, 10 mM imidazole, 1 mM TCEP, 1 mM PMSF). Cdc20 was purified in lysis buffer with 500 mM NaCl. Cells were lysed by sonication and centrifuged at 108000 g (Rotor JA30.50, Avanti-J30I, Beckman Coulter) for 30 min at 4°C. The supernatant was filtered through 0.8 μm Rotilabo® syringe filters (Carl Roth GmbH). The proteins were isolated from the cleared lysate on a 5 mL HisTrap FF affinity column (GE Healthcare). Peak fractions were pooled, concentrated, and further purified in gelfiltration buffer (10 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM TCEP) by size exclusion chromatography on a S200 16/60 column (GE Healthcare). Peak fractions were pooled, concentrated to typically 3 to 5 mg/ml, flash frozen and stored at −80°C until use.
For the purification of recombinant MCC complexes, individual purified MCC components were mixed to obtain 250 μg of MCC and incubated at 4°C over night. Afterward, gelfiltration was performed using a Superdex 200 increase 5/150 column (GE Healthcare) equilibrated against a buffer containing 20 mM HEPES pH 7.5, 200 mM NaCl and 2 mM DTT. Peak fractions were pooled, concentrated flash frozen and stored at −80°C.
Size-exclusion chromatography mobility shift assay
Proteins tested for interactions were diluted to a final concentration of 5 μM in 50 μl reactions in binding buffer (10 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM TCEP, 1 mM MgCl2) and incubated at 4°C over night. Complex formation was analyzed by size exclusion chromatography on a Superdex 200 increase 5/150 column (GE Healthcare). Eluates were analyzed by SDS-PAGE and Coomassie staining.
MCC assembly kinetics
The assay was performed precisely as described [31]. Fluorimeter scans were performed on a Fluoromax 4 (Jobin Yvon) in a buffer containing fresh 10 mM HEPES (pH 7.5), 150 mM NaCl, 2.5% glycerol, 10 mM beta-mercaptoenthanol and 0.05% Triton X-100. Mixtures were excited at 430 nm and the emissions were scanned from 450 to 650 nm. Single wavelength acceptor fluorescence measurements were carried out at 583 nm. Mixtures of MAD1:C-MAD2 with BUB1:BUB3 and/or MPS1 were pre-incubated at 1 μM for 30 min at 30 degrees. Assays were performed using 100 nM of all proteins, except CDC20, which was added at 500 nM. Curves reporting time-dependent changes in FRET signal report single measurements representative of at least three independent technical replicates.
APC/C-mediated ubiquitination assays
To measure the activity of APC/C, its ability to form poly-ubiquitin chains on its substrate Cyclin B was analyzed. Recombinant APC/C-pE, containing 68 phospho-mimicking mutations [70], was used as mimic of mitotic APC/C. APC/C-pE (20 nM) and Cdc20 (100 nM) were part of a mastermix with 500 nM CycBNTD∗, 500 nM UbcH10, 500 nM Ube2S, 2.5 mM MgATP and 0.5 mg/ml BSA. This was aliquoted and mixed with recombinant MCC versions (30 nM) on ice. Reactions were equilibrated to room temperature for 10 min and afterward started by the addition of the Uba1 (100 nM)/Ub (100 μM) mix. After 30, 60 and 120 min reactions were stopped by the addition of SDS sample buffer. Samples were analyzed by SDS-PAGE and fluorescence scanning.
Quantification and Statistical Analyses
For FRAP experiments, statistical analysis is described in the Figure legends, in the Method Details, and in Table S1. For kinetochore localization experiments and checkpoint assays, quantification and statistical analysis (mean ± SEM) are described in the figure legends. Quantification and statistical analysis of immunoprecipitation experiments (mean ± SEM) are described in the figure legends. Analysis of SILAC data is described in the Methods details.
Data and Software Availability
The full list of interacting proteins identified in the SILAC experiments is available with the online version of the paper as Data S1.
Author Contributions
K.O. carried out all cell-biology-based work, including checkpoint assays, immunoprecipitations, immunofluorescence, and data analyses. K.O. and A.C.F. carried out biochemical analyses and experiments with FRET sensor. I.P. contributed unpublished reagents. K.O., F.M., and T.B. carried out SILAC-mass spectrometry experiments. K.O. and S.M. performed and analyzed FRAP experiments. F.W. and J.-M.P. contributed experiments with purified APC/C. K.O. and A.M. conceived the work, prepared the figures, and wrote the paper with contributions from all authors. A.M. coordinated the working team.
Acknowledgments
We thank the members of the Musacchio laboratory for helpful comments and discussions. A.M. acknowledges funding from the European Research Council (ERC) AdG RECEPIANCE (669686) and the DFG Collaborative Research Centre (CRC) 1093. Research in the lab of J.-M.P. was supported by Boehringer Ingelheim, the Austrian Science Fund (Wittgenstein award Z196-B20) and the Austrian Research Promotion Agency (headquarter grant FFG-852936).
Published: September 21, 2017
Footnotes
Supplemental Information includes seven figures, one table, and one data file and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2017.08.033.
Supplemental Information
The complete list of specific and common/background binders identified in SILAC mass spectrometry experiments discussed in this study.
References
- 1.Lara-Gonzalez P., Westhorpe F.G., Taylor S.S. The spindle assembly checkpoint. Curr. Biol. 2012;22:R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 2015;25:R1002–R1018. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 3.Tromer E., Bade D., Snel B., Kops G.J. Phylogenomics-guided discovery of a novel conserved cassette of short linear motifs in BubR1 essential for the spindle checkpoint. Open Biol. 2016;6:160315. doi: 10.1098/rsob.160315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suijkerbuijk S.J., van Dam T.J., Karagöz G.E., von Castelmur E., Hubner N.C., Duarte A.M., Vleugel M., Perrakis A., Rüdiger S.G., Snel B., Kops G.J. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Dev. Cell. 2012;22:1321–1329. doi: 10.1016/j.devcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Krenn V., Wehenkel A., Li X., Santaguida S., Musacchio A. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J. Cell Biol. 2012;196:451–467. doi: 10.1083/jcb.201110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiyomitsu T., Obuse C., Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Krenn V., Overlack K., Primorac I., van Gerwen S., Musacchio A. KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT repeats. Curr Biol. 2014;24:29–39. doi: 10.1016/j.cub.2013.11.046. [DOI] [PubMed] [Google Scholar]
- 8.Kang J., Yang M., Li B., Qi W., Zhang C., Shokat K.M., Tomchick D.R., Machius M., Yu H. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol. Cell. 2008;32:394–405. doi: 10.1016/j.molcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernius J., Hardwick K.G. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 2007;3:e213. doi: 10.1371/journal.pgen.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawashima S.A., Yamagishi Y., Honda T., Ishiguro K., Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 11.Klebig C., Korinth D., Meraldi P. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 2009;185:841–858. doi: 10.1083/jcb.200902128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perera D., Tilston V., Hopwood J.A., Barchi M., Boot-Handford R.P., Taylor S.S. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev. Cell. 2007;13:566–579. doi: 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Ricke R.M., Jeganathan K.B., Malureanu L., Harrison A.M., van Deursen J.M. Bub1 kinase activity drives error correction and mitotic checkpoint control but not tumor suppression. J. Cell Biol. 2012;199:931–949. doi: 10.1083/jcb.201205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp-Baker H., Chen R.H. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H., Rankin S., Yu H. Phosphorylation-enabled binding of SGO1-PP2A to cohesin protects sororin and centromeric cohesion during mitosis. Nat. Cell Biol. 2013;15:40–49. doi: 10.1038/ncb2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H., Jia L., Yu H. Phospho-H2A and cohesin specify distinct tension-regulated Sgo1 pools at kinetochores and inner centromeres. Curr. Biol. 2013;23:1927–1933. doi: 10.1016/j.cub.2013.07.078. [DOI] [PubMed] [Google Scholar]
- 17.Yamagishi Y., Honda T., Tanno Y., Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- 18.Boyarchuk Y., Salic A., Dasso M., Arnaoutov A. Bub1 is essential for assembly of the functional inner centromere. J. Cell Biol. 2007;176:919–928. doi: 10.1083/jcb.200609044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brady D.M., Hardwick K.G. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- 20.Chen R.H. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 2002;158:487–496. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson V.L., Scott M.I., Holt S.V., Hussein D., Taylor S.S. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 22.Meraldi P., Sorger P.K. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;24:1621–1633. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rischitor P.E., May K.M., Hardwick K.G. Bub1 is a fission yeast kinetochore scaffold protein, and is sufficient to recruit other spindle checkpoint proteins to ectopic sites on chromosomes. PLoS ONE. 2007;2:e1342. doi: 10.1371/journal.pone.0001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vigneron S., Prieto S., Bernis C., Labbé J.C., Castro A., Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruse T., Larsen M.S., Sedgwick G.G., Sigurdsson J.O., Streicher W., Olsen J.V., Nilsson J. A direct role of Mad1 in the spindle assembly checkpoint beyond Mad2 kinetochore recruitment. EMBO Rep. 2014;15:282–290. doi: 10.1002/embr.201338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinrich S., Sewart K., Windecker H., Langegger M., Schmidt N., Hustedt N., Hauf S. Mad1 contribution to spindle assembly checkpoint signalling goes beyond presenting Mad2 at kinetochores. EMBO Rep. 2014;15:291–298. doi: 10.1002/embr.201338114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J.S., Holland A.J., Fachinetti D., Kulukian A., Cetin B., Cleveland D.W. Catalytic assembly of the mitotic checkpoint inhibitor BubR1-Cdc20 by a Mad2-induced functional switch in Cdc20. Mol. Cell. 2013;51:92–104. doi: 10.1016/j.molcel.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mora-Santos M.D., Hervas-Aguilar A., Sewart K., Lancaster T.C., Meadows J.C., Millar J.B. Bub3-Bub1 binding to Spc7/KNL1 toggles the spindle checkpoint switch by licensing the interaction of Bub1 with Mad1-Mad2. Curr. Biol. 2016;26:2642–2650. doi: 10.1016/j.cub.2016.07.040. [DOI] [PubMed] [Google Scholar]
- 29.London N., Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 2014;28:140–152. doi: 10.1101/gad.233700.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., Musacchio A. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 31.Faesen A.C., Thanasoula M., Maffini S., Breit C., Müller F., van Gerwen S., Bange T., Musacchio A. Basis of catalytic assembly of the mitotic checkpoint complex. Nature. 2017;542:498–502. doi: 10.1038/nature21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji Z., Gao H., Jia L., Li B., Yu H. A sequential multi-target Mps1 phosphorylation cascade promotes spindle checkpoint signaling. eLife. 2017;6:e22513. doi: 10.7554/eLife.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overlack K., Krenn V., Musacchio A. When Mad met Bub. EMBO Rep. 2014;15:326–328. doi: 10.1002/embr.201438574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan I., Leontiou I., Amin P., May K.M., Soper Ní Chafraidh S., Zlámalová E., Hardwick K.G. Generation of a spindle checkpoint arrest from synthetic signaling assemblies. Curr. Biol. 2017;27:137–143. doi: 10.1016/j.cub.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyle M.W., Kim T., Hattersley N., Espeut J., Cheerambathur D.K., Oegema K., Desai A. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J. Cell Biol. 2014;204:647–657. doi: 10.1083/jcb.201311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardwick K.G., Johnston R.C., Smith D.L., Murray A.W. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sudakin V., Chan G.K., Yen T.J. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraschini R., Beretta A., Sironi L., Musacchio A., Lucchini G., Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burton J.L., Solomon M.J. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elowe S., Dulla K., Uldschmid A., Li X., Dou Z., Nigg E.A. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. J. Cell Sci. 2010;123:84–94. doi: 10.1242/jcs.056507. [DOI] [PubMed] [Google Scholar]
- 41.King E.M., van der Sar S.J., Hardwick K.G. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lara-Gonzalez P., Scott M.I., Diez M., Sen O., Taylor S.S. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J. Cell Sci. 2011;124:4332–4345. doi: 10.1242/jcs.094763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chao W.C., Kulkarni K., Zhang Z., Kong E.H., Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 44.Izawa D., Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517:631–634. doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Primorac I., Musacchio A. Panta rhei: the APC/C at steady state. J. Cell Biol. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alfieri C., Chang L., Zhang Z., Yang J., Maslen S., Skehel M., Barford D. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature. 2016;536:431–436. doi: 10.1038/nature19083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaguchi M., VanderLinden R., Weissmann F., Qiao R., Dube P., Brown N.G., Haselbach D., Zhang W., Sidhu S.S., Peters J.M. Cryo-EM of mitotic checkpoint complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Mol. Cell. 2016;63:593–607. doi: 10.1016/j.molcel.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foley E.A., Maldonado M., Kapoor T.M. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kruse T., Zhang G., Larsen M.S., Lischetti T., Streicher W., Kragh Nielsen T., Bjørn S.P., Nilsson J. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 2013;126:1086–1092. doi: 10.1242/jcs.122481. [DOI] [PubMed] [Google Scholar]
- 50.Suijkerbuijk S.J., Vleugel M., Teixeira A., Kops G.J. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Xu P., Raetz E.A., Kitagawa M., Virshup D.M., Lee S.H. BUBR1 recruits PP2A via the B56 family of targeting subunits to promote chromosome congression. Biol. Open. 2013;2:479–486. doi: 10.1242/bio.20134051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Espert A., Uluocak P., Bastos R.N., Mangat D., Graab P., Gruneberg U. PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J. Cell Biol. 2014;206:833–842. doi: 10.1083/jcb.201406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., Millar J.B. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 2012;22:891–899. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.London N., Ceto S., Ranish J.A., Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 2012;22:900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamagishi Y., Yang C.H., Tanno Y., Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 2012;14:746–752. doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- 56.Logarinho E., Resende T., Torres C., Bousbaa H. The human spindle assembly checkpoint protein Bub3 is required for the establishment of efficient kinetochore-microtubule attachments. Mol. Biol. Cell. 2008;19:1798–1813. doi: 10.1091/mbc.E07-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor S.S., Ha E., McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Primorac I., Weir J.R., Chiroli E., Gross F., Hoffmann I., van Gerwen S., Ciliberto A., Musacchio A. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife. 2013;2:e01030. doi: 10.7554/eLife.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larsen N.A., Al-Bassam J., Wei R.R., Harrison S.C. Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc. Natl. Acad. Sci. USA. 2007;104:1201–1206. doi: 10.1073/pnas.0610358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Overlack K., Primorac I., Vleugel M., Krenn V., Maffini S., Hoffmann I., Kops G.J., Musacchio A. A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. eLife. 2015;4:e05269. doi: 10.7554/eLife.05269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanoosthuyse V., Valsdottir R., Javerzat J.P., Hardwick K.G. Kinetochore targeting of fission yeast Mad and Bub proteins is essential for spindle checkpoint function but not for all chromosome segregation roles of Bub1p. Mol. Cell. Biol. 2004;24:9786–9801. doi: 10.1128/MCB.24.22.9786-9801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gillett E.S., Espelin C.W., Sorger P.K. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Millband D.N., Hardwick K.G. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 2002;22:2728–2742. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., Salmon E.D. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 65.Shah J.V., Botvinick E., Bonday Z., Furnari F., Berns M., Cleveland D.W. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 66.Vleugel M., Omerzu M., Groenewold V., Hadders M.A., Lens S.M., Kops G.J. Sequential multisite phospho-regulation of KNL1-BUB3 interfaces at mitotic kinetochores. Mol. Cell. 2015;57:824–835. doi: 10.1016/j.molcel.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 67.Asghar A., Lajeunesse A., Dulla K., Combes G., Thebault P., Nigg E.A., Elowe S. Bub1 autophosphorylation feeds back to regulate kinetochore docking and promote localized substrate phosphorylation. Nat. Commun. 2015;6:8364. doi: 10.1038/ncomms9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 69.Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J., van Deursen J.M. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev. Cell. 2009;16:118–131. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiao R., Weissmann F., Yamaguchi M., Brown N.G., VanderLinden R., Imre R., Jarvis M.A., Brunner M.R., Davidson I.F., Litos G. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 2016;113:E2570–E2578. doi: 10.1073/pnas.1604929113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davey N.E., Morgan D.O. Building a regulatory network with short linear sequence motifs: lessons from the degrons of the anaphase-promoting complex. Mol. Cell. 2016;64:12–23. doi: 10.1016/j.molcel.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu D., Hsiao J.Y., Davey N.E., Van Voorhis V.A., Foster S.A., Tang C., Morgan D.O. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J. Cell Biol. 2014;207:23–39. doi: 10.1083/jcb.201402041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diaz-Martinez L.A., Tian W., Li B., Warrington R., Jia L., Brautigam C.A., Luo X., Yu H. The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J. Biol. Chem. 2015;290:2431–2443. doi: 10.1074/jbc.M114.616490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Di Fiore B., Davey N.E., Hagting A., Izawa D., Mansfeld J., Gibson T.J., Pines J. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev. Cell. 2015;32:358–372. doi: 10.1016/j.devcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Fiore B., Wurzenberger C., Davey N.E., Pines J. The mitotic checkpoint complex requires an evolutionary conserved cassette to bind and inhibit active APC/C. Mol. Cell. 2016;64:1144–1153. doi: 10.1016/j.molcel.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lischetti T., Zhang G., Sedgwick G.G., Bolanos-Garcia V.M., Nilsson J. The internal Cdc20 binding site in BubR1 facilitates both spindle assembly checkpoint signalling and silencing. Nat. Commun. 2014;5:5563. doi: 10.1038/ncomms6563. [DOI] [PubMed] [Google Scholar]
- 77.Han J.S., Vitre B., Fachinetti D., Cleveland D.W. Bimodal activation of BubR1 by Bub3 sustains mitotic checkpoint signaling. Proc. Natl. Acad. Sci. USA. 2014;111:E4185–E4193. doi: 10.1073/pnas.1416277111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sczaniecka M., Feoktistova A., May K.M., Chen J.S., Blyth J., Gould K.L., Hardwick K.G. The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-anaphase-promoting complex (APC/C) J. Biol. Chem. 2008;283:23039–23047. doi: 10.1074/jbc.M803594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fujimitsu K., Grimaldi M., Yamano H. Cyclin-dependent kinase 1-dependent activation of APC/C ubiquitin ligase. Science. 2016;352:1121–1124. doi: 10.1126/science.aad3925. [DOI] [PubMed] [Google Scholar]
- 80.Zhang S., Chang L., Alfieri C., Zhang Z., Yang J., Maslen S., Skehel M., Barford D. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature. 2016;533:260–264. doi: 10.1038/nature17973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., Peters J.M. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kramer E.R., Scheuringer N., Podtelejnikov A.V., Mann M., Peters J.M. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Golan A., Yudkovsky Y., Hershko A. The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J. Biol. Chem. 2002;277:15552–15557. doi: 10.1074/jbc.M111476200. [DOI] [PubMed] [Google Scholar]
- 84.Shteinberg M., Protopopov Y., Listovsky T., Brandeis M., Hershko A. Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem. Biophys. Res. Commun. 1999;260:193–198. doi: 10.1006/bbrc.1999.0884. [DOI] [PubMed] [Google Scholar]
- 85.Herzog F., Mechtler K., Peters J.M. Identification of cell cycle-dependent phosphorylation sites on the anaphase-promoting complex/cyclosome by mass spectrometry. Methods Enzymol. 2005;398:231–245. doi: 10.1016/S0076-6879(05)98019-1. [DOI] [PubMed] [Google Scholar]
- 86.Zhang G., Mendez B.L., Sedgwick G.G., Nilsson J. Two functionally distinct kinetochore pools of BubR1 ensure accurate chromosome segregation. Nat. Commun. 2016;7:12256. doi: 10.1038/ncomms12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Essex A., Dammermann A., Lewellyn L., Oegema K., Desai A. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol. Biol. Cell. 2009;20:1252–1267. doi: 10.1091/mbc.E08-10-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen Ba A.N., Strome B., Osman S., Legere E.A., Zarin T., Moses A.M. Parallel reorganization of protein function in the spindle checkpoint pathway through evolutionary paths in the fitness landscape that appear neutral in laboratory experiments. PLoS Genet. 2017;13:e1006735. doi: 10.1371/journal.pgen.1006735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hashimoto Y., Zhang S., Zhang S., Chen Y.R., Blissard G.W. Correction: BTI-Tnao38, a new cell line derived from Trichoplusia ni, is permissive for AcMNPV infection and produces high levels of recombinant proteins. BMC Biotechnol. 2012;12:12. doi: 10.1186/1472-6750-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 91.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 92.Liu H., Naismith J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ong S.E., Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 94.Olsen J.V., Macek B., Lange O., Makarov A., Horning S., Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 95.Maddox P.S., Bloom K.S., Salmon E.D. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen D., Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trowitzsch S., Bieniossek C., Nie Y., Garzoni F., Berger I. New baculovirus expression tools for recombinant protein complex production. J. Struct. Biol. 2010;172:45–54. doi: 10.1016/j.jsb.2010.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The complete list of specific and common/background binders identified in SILAC mass spectrometry experiments discussed in this study.