Abstract
The 2015 Nobel Prize in Physiology or Medicine has been awarded to avermectins and artemisinin, respectively. Avermectins produced by Streptomyces avermitilis are excellent anthelmintic and potential antibiotic agents. Because wild-type strains only produce low levels of avermectins, much research effort has focused on improvements in avermectin production to meet the ever increasing demand for such compounds. This review describes the strategies that have been widely employed and the future prospects of synthetic biology applications in avermectin yield improvement. With the help of genome sequencing of S. avermitilis and an understanding of the avermectin biosynthetic/regulatory pathways, synthetic and systems biotechnology approaches have been applied for precision engineering. We focus on the design and synthesis of biological chassis, parts, devices, and modules from diverse microbes to reconstruct and optimize their dynamic processes, as well as predict favorable effective overproduction of avermectins by a 4Ms strategy (Mine, Model, Manipulation, and Measurement).
Keywords: Avermectins, Metabolic engineering, Streptomyces avermitilis, Synthetic biology
Abbreviations: MDR-TB, multidrug-resistant tuberculosis; XDR-TB, extensively drug-resistant tuberculosis; MRSA, methicillin-resistant Staphylococcus aureus; PBD, Plackett–Burman design; DO, dissolved oxygen; OUR, oxygen uptake rate; EER, ethanol evolution rate; RF, radio frequency; APGD, atmospheric pressure glow discharge; HMGE, high-magnet gravitational environment; UV, ultraviolet rays; MMS, methyl methanesulphonate; NTG, N-methyl-N-nitro-N-nitrosoguanidine; NA, nitrous acid; MTP, microtiter plates; MB-CoA, 2-methybutyryl-CoA; IB-CoA, isobutyryl-CoA; MM-CoA, methylmalonyl- CoA; BCDH, branched-chain alpha-keto acid dehydrogenase; SAM, S-adenosylmethionine; RRF, ribosome recycling factor; GBL, gamma-butyrolactone; STPK, serine-threonine protein kinases; ChIP, chromatin immunoprecipitation; TAR, transformation-assisted recombination
Introduction
Microbial natural products are valuable compounds used in agricultural, pharmaceutical, and food industries. However, there has been an industrial challenge in that wild-type strains isolated from nature usually produce low levels of these compounds that can never meet commercial demands. Avermectin and its analogs, a series of eight major 16-membered macrocyclic polyketides produced by Streptomyces avermitilis, are widely used in the fields of animal health and agriculture, according to their activities against a variety of nematodes and arthropod parasites, with low levels of side effects on humans.1 Because the derivatives of avermectins lowered the incidence of River Blindness and other parasitic diseases, half of the 2015 Nobel Prize in Physiology or Medicine was awarded to avermectin discoverer, William C. Campbell and Satoshi Ōmura.2 Avermectins contain four major (80–90%) components A1a, A2a, B1a, and B2a in varying proportions and four minor (10–20%) components A1b, A2b, B1b, and B2b,3 among which the B1a component has the most effective anthelmintic activity.4 Recently, the pharmaceutical potential of avermectins has been extended against Mycobacterium tuberculosis, including multidrug-resistant tuberculosis (MDR-TB) and extensively drug-resistant tuberculosis (XDR-TB),5 as well as the synergistic effect of avermectin B1a with methicillin (MET) against methicillin-resistant Staphylococcus aureus (MRSA).6, 7
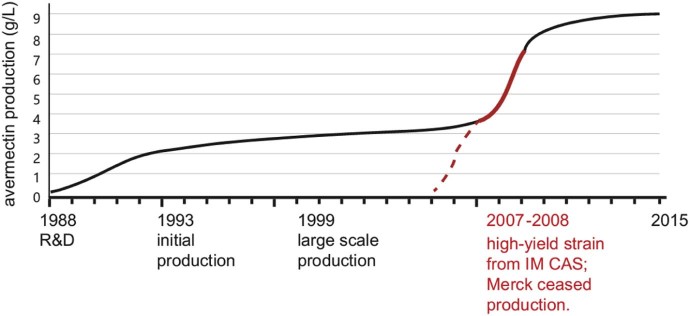
The efforts to produce improved avermectins have never stopped since the discovery of avermectin by Ōmura and co-workers in 1975.1 Avermectins were first commercialized by Merck Sharp & Dohme Research Laboratories, Kitasato Institute, and Kitasato University in 1985. China joined this campaign in 1988 and succeeded in production in 1993. Four companies for avermectin production went public during 1988–2007. The Institute of Microbiology Chinese Academy of Sciences and other institutes, with strong support from those companies, significantly increased the production of avermectin B1a with a titer from 0.009 to 9 g/L (Fig. 1). Now, China is the only avermectin producing country in the world. Avermectin is the only bio-pesticide that has an annual sale above 3 billion RMB, creating great social and economic benefits. Thus, in this review, we summarize the various strategies used to improve production of avermectins.
Fig. 1.
Avermectin production improvement in China. The solid line indicates the avermectin production level in industry in China. The red line indicates contribution by the Institute of Microbiology, CAS. The red dashed line indicates starting from a wild type strain.
Improving the production of avermectin by traditional mutagenesis methods
Microbial fermentation and random mutagenesis are conventionally applied industrially to produce natural products, displaying the advantages of production improvement of the natural product by strains with little genetic information. These approaches have been applied to improve avermectin production in the fermentation industry and increased the titer to 0.5 g/L by strain selection from ultraviolet (UV) light radiation, Methyl methanesulphonate (MMS) and N-methyl-N-nitro-N-nitrosoguanidine (NTG) treatment and media modifications.8, 9
Optimization of media and the fermentation process
A low-cost medium was developed through optimization of nitrogen and carbon sources, as well as supplementation with 0.2 mM Co2+.10 The production of avermectin B1a has increased to a titer of 0.46 g/L, which is 48.8% higher than that of the production in the original medium. Then, statistical experimental designs were used in consideration of the interactions between different factors.11 Out of nine components, corn starch and yeast extract were found to significantly affect the production of avermectin B1a by Plackett–Burman design (PBD). The optimum values of medium composition of 149.57 g/L corn starch and 8.92 g/L yeast extract were determined.11
Aside from the optimization of the fermentation medium, the addition of possible precursors or stimulators of avermectin also plays an important role during the fermentation process. The influence of the addition of the possible precursors of avermectin, acetate and propionate, were investigated on two different strains.12 The addition of 0.8% (w/w) propionate at 24 h of cultivation resulted in a 12.8–13.8% improvement in the production of avermectin B1a after 5 days of incubation. However, there was no change when propionate was added at the beginning of cultivation. Additionally, the proportion of B1a is not affected by propionate supplementation. In the case of acetate, the avermectin yield improvement was not observed when the acetate was added either at the beginning of or 24 h into cultivation.12
The above evidence indicates that glucose metabolism affects avermectin biosynthesis.10, 12, 13 Indeed, avermectin fermentation and 6-phosphoglu-conate dehydrogenase in the pentose phosphate pathway are significantly suppressed by the addition of glucose at the early stage of fermentation.10 Even though the involvement of the pentose phosphate pathway in avermectin production is still unclear, it may help to supply NADPH in avermectin biosynthesis (data unpublished). Avermectin production can be further increased when glucose is fed at a late stage of fermentation in the flask, bench-top, and pilot-plant scales.10, 12, 13 Moreover, a B1a ratio increase by glucose feeding was observed, and the stimulation is further enhanced by controlled glucose feeding.7, 9 It has been suggested that glucose affects avermectin formation by providing additional dTDP-oleandrose, an immediate precursor of avermectin.14, 15 The B1a ratio might be changed due to the feeding of glucose, which could regulate the activity of aveD, a crucial gene that is responsible for the conversion between avermectin B and A types.16, 17, 18 However, the genetic mechanism of this phenomenon is still under investigation.
Some physiological parameters also affect avermectin production by influencing cell growth, such as dissolved oxygen (DO)19 and the oxygen uptake rate (OUR).20 Higher DO tension (usually >20% saturation) is beneficial for pellet formation and avermectin production during submerged cultivation.19 By controlling OUR between 15 and 20 mmol/L/hour, the production of avermectin B1a reaches 5.568 ± 0.111 g/L, which is 21.8% higher than that of the control. This indicates that the stimulatory effects on avermectin B1a production might contribute to improve the precursor supply.20 The OUR parameter is also used to determine the glucose feeding rate in avermectin production, as well as the ethanol evolution rate (EER).21, 22 The EER parameter is mainly affected by O2 supply and the glucose feeding rate and can be considered as another physiological parameter correlated with OUR.22
Improving the production of avermectin by random mutagenesis and screening
The optimization of media and fermentation process plays an important role for industrial overproduction of natural products. Aiming for the same goal by strain improvement by random mutation and follow-up screening is very labor intensive. However, the metabolic capabilities of the production of desired compounds could be enhanced via manipulating and improving microbial strains.
The traditional mutate-and-screen method is typically performed by subjecting the strains to a variety of physical or chemical mutagens and screening. The mutagens used to improve avermectin production are listed in Table 1. These mutagens introduce mutations into strains and, hence, resulted in different production improvements. Wang et al.23 introduced a new mutation tool called radio frequency (RF) atmospheric pressure glow discharge (APGD) plasma jet, which has a high mutagenic capability compared to traditional mutation methods and is an efficient method because of its higher (>30%) and positive (>20%) mutation rate. The mutant colonies displayed different morphologies and colors, which are feasible for initial screening. Among these mutants, G1-1 showed the highest yield of avermectin B1a, which is increased by 40% compared to the wild type strain. Furthermore, the B1a ratio is also increased in G1-1. However, the detailed mutation effect of APGD plasmas in G1-1 still needs to be classified.
Table 1.
Mutagens used for avermectin overproduction.
| Mutagens | |
|---|---|
| Physical mutagenesis | Ultraviolet rays (UV) |
| 12C+6 heavy ion beams | |
| Co60 gamma rays | |
| High-magnet gravitational environment (HMGE) | |
| RF APGD plasma jet | |
| Spaceflight | |
| Chemical mutagenesis | Methyl methanesulphonate (MMS) |
| N-methyl-N-nitro-N-nitrosoguanidine (NTG) | |
| Nitrous acid (NA) | |
| Composite mutagenesis | UV + HNO2 + NTG + L- Ile |
| Co60 gamma rays-Met-5-fluorouracil | |
| UV + LiCl | |
| UV + NTG |
Gao et al.24 assessed a potential induced mutation strategy called high-magnet gravitational environment (HMGE), a space flight-simulated mutation strategy. HMGE was compared with two other conventional strategies, UV and NTG. An algorithm was used to assess the mutation spectrum, and HMGE was approved to enhance the phenotype distribution and diversity better than UV and NTG, even though the positive mutation rate of NTG was the highest. Another technique called diamagnetic levitation has also been used to simulate the space environment for mutagenesis.25 Diamagnetic levitation generates both a varying magnetic field and altered gravity. The individual effects of magnetic field and gravity were investigated for the first time, and the results demonstrate that the magnetic field is a more dominant factor influencing changes in morphology and avermectin production than altered gravity.25
After introducing mutations to production strains, the screening and acquisition of the overproducers from a large number of mutants is crucial. Earlier screening of avermectin overproducers was based on the identification of morphological features, such as the production of aerial mycelia, spore formation, and melanin production.26 However, this method also introduces many negative mutants.23 Thus, a more efficient screening strategy based on UV absorbance using 96 deep-well microtiter plate (MTP) cultivation was introduced.27 This high-throughput screening approach focuses on the culture and avermectin concentration measurement. It uses the UV assay in solid-state MTP cultures, which is much simpler and more rapid than HPLC assays in Erlenmeyer flask cultures. The correlation between the results of the UV assay and the HPLC assay was investigated to test the accuracy of this strategy, and the UV assay was proved to correspond well with the conventional HPLC assay. Subsequently, it was used for high-throughput screening of avermectin over-producing strains, and a 60% increase in avermectin B1a compared to the parent strain in a 360-m3 batch fermentation was observed.
Metabolic engineering
Random mutagenesis and screening is widely applied because it is simple and easy to manipulate for efficient strain improvement. However, the method is time-consuming and laborious. Also, mutations may result in strains with undesirable or detrimental traits. Further, the mechanism behind the increase in production of the strains obtained by this method is largely unknown, and thus, it cannot be applied further for the rational design of an overproducing strain. After discovering the avermectin biosynthetic gene cluster28 and determining the genome sequence of S. avermitilis,29 a more rational method called metabolic engineering was introduced and has been widely applied in avermectin improvement research.
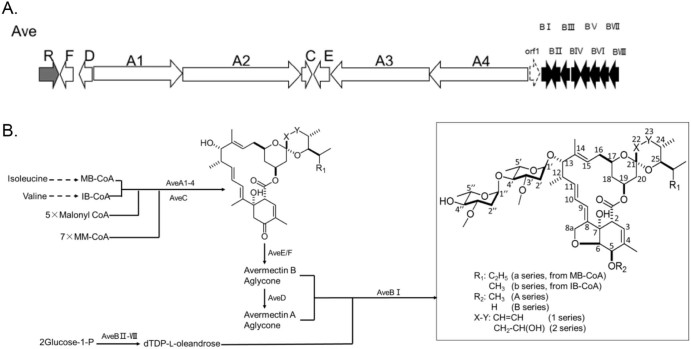
Avermectin biosynthesis consists of the following steps (Fig. 2): (1) the elongation of a polyketide chain by four multi-functional modular polyketide synthase components (AVES 1, 2, 3, and 4), with the addition of five methylmalonyl-CoA (MM-CoA) units and seven malonyl-CoA units to the starter units, 2-methybutyryl-CoA (MB-CoA, “a” components) and isobutyryl-CoA (IB-CoA, “b” components); (2) C22-23 dehydration modification (“1” and “2” components) and spiroketal formation by AveC;30 (3) furan formation and keto reduction by AveE and AveF, respectively; (4) C5 O-methylation by AveD (“A” and “B” components); (5) the biosynthesis of TDP-l-oleandrose by AveBII-VIII; and (6) the glycosylation of aglycones to form the final avermectin compounds.31, 32 In this biosynthetic pathway, AveC performs the dehydration modification function before its spirocyclization formation activity.30 These two functions are independently performed and competed with the same substrate. This dehydration activity can be reduced or increased by mutations in AveC33, 34 without inactivating the spirocyclase. Further elucidation of the AveC structure related to its activity and specificity would take full advantage of this dual function and aid in the development of only “1” components to enhance the production.
Fig. 2.
Biosynthesis of avermectin. (A) The avermectin biosynthesis gene cluster 82 kb. White, the genes involved in the formation of avermectin aglycones; black, the genes involved in glycosylation of the aglycones; gray, the regulator gene; dash line, the gene that is not involved in the biosynthesis of avermectin. (B) The avermectin biosynthetic pathway.
After the complete genome sequencing of S. avermitilis, the study of overall gene expression at the mRNA and protein levels became feasible with the development of genetic manipulation.35 The transcriptome and proteome comparisons between wild-type and avermectin overproducing reveal the possible mechanisms underlying avermectin overproduction and provide new targets for rational yield improvement by using metabolic engineering.36, 37, 38 This method typically involves altering the metabolic flux related to the precursors, regulating the biosynthesis pathway and antibiotic resistance. Here, we present the different genetic approaches used in metabolic engineering for the overproduction of avermectin.
Engineering precursors
The sufficiency of biosynthetic precursors is very important for the production of secondary metabolites. These precursors come from primary metabolism, such as the fatty acid, amino acid, and glucose metabolic pathways. Starch is the most important carbon source in the fermentation process of S. avermitilis.11 Starch utilization requires external amylase addition into the medium to form maltose and maltodextrin. The overexpression of malEFG, which encodes a maltose ATP-binding cassette transporter, improves the utilization rate of starch and enhances avermectin production. However, the yields of avermectin are similar when a different copy number of the malEFG is introduced. This may be due to the limitation of the ATPase subunit, which is needed in the ABC transporter.39 This maltose ATP-binding cassette transporter also provides a new method for yield improvement of other natural products that use starch or maltose as a carbon source.
The branched-chain α-keto acid dehydrogenase (BCDH) provides the branched-chain fatty acid starter units 2-methylbutyryl CoA and IB-CoA from the catabolism of L-valine and L-isoleucine, respectively.31, 32 There are two gene clusters encoding the E1α, E1β, and E2 subunits of the BCDH complex.40, 41 Deletion of the 5′ end of bkdF causes complete loss of E1 BCDH activity and the ability to produce natural avermectins, while inactivation of the bkdABC genes does not cause obvious phenotypic changes.41 According to these results, further expression level or enzyme activity enhancement research of BCDH may result in avermectin production improvement.
The loading module of avermectin PKS can also recruit >40 alternative carboxylic acids as the starter units.42 However, the efficiency is much lower than the natural starter unit.43 With structural and specificity analysis of the loading acyltransferase from avermectin PKS, it may be possible to engineer the loading acyltransferase to acquire only the “a” components and enhance avermectin production.44, 45
The overexpression of the S-adenosylmethionine (SAM) synthetase gene (metK) in the wild type strain increases avermectin production.44, 45 The mechanism for this is unclear, but it is hypothesized that it increases intracellular SAM levels, which activates the transcriptional activators responsible for antibiotic biosynthesis or serves as a methyl donor in primary and secondary metabolism.46, 47 This is consistent with the result that the overexpression of the metK gene increases the mRNA levels of metK and the SAM concentration, as well as further upregulates the pathway-specific regulatory gene aveR.45 Further, the overexpression of metK in the avermectin overproducing industrial strains, which already display higher expression levels of metK, aveR, and aveA1, has no effect on avermectin production.45
Engineering regulators
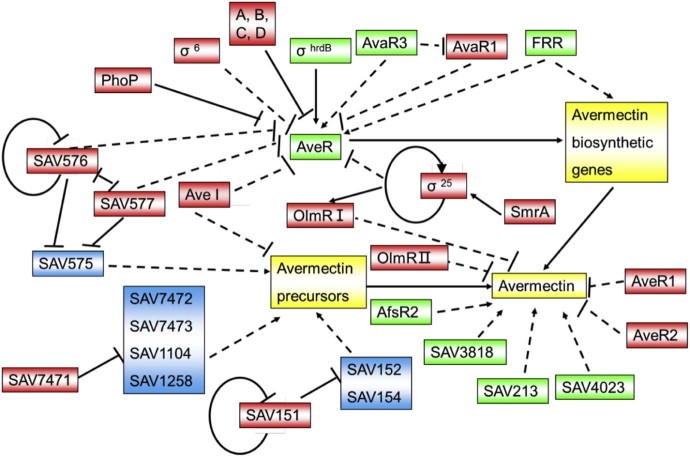
Avermectin biosynthesis is a process under the tight control of multilevel signal transduction mechanisms. There is a putative pathway-specific regulator called aveR located at the far left arm of the avermectin biosynthetic gene cluster, outside of aveF. Tn4560 transposon mutants in the aveR region do not produce avermectins.48 Deletion of aveR results in the complete loss of avermectin production, which can be restored by complementation.49, 50 AveR positively regulates avermectin biosynthesis by specifically binding to the promoter region of the ave structural genes with its C-terminal HTH domain. The overexpression of aveR results in an opposite influence (including improvement and complete loss) on avermectin production in two conflicting reports.49, 50 The discrepancies among these results may come from different wild type strains. However, more research into the binding sequence or the structure of aveR is required to elucidate the regulatory mechanism of avermectin biosynthesis. Aside from the pathway-specific regulator, there are other regulators that have been investigated for avermectin production improvement (Fig. 3).
Fig. 3.
Networking of regulators that affect avermectin production. Green, positive regulators; red, negative regulators; and blue, the target genes of these regulators; dash lines: indirect effect with unknown mechanism; solid lines: direct effect; arrows: positive effect; bars: negative effect.
Several regulators affect avermectin production through the pathway specific activator aveR. This includes the global regulator σHrdB, which directly recognizes the promoter region of aveR in vitro. The hrdB gene has been engineered in an industrial strain to identify the effect of σHrdB on aveR and avermectin biosynthesis.36 Two high-avermectin producing mutants, A56 and A393, were obtained by high-throughput screening of a hrdB mutant library. The genetically stable mutant A56 was further cultivated in a 180-m3 fermentor, and the production of avermectin B1a reached 6.38 g/L, an increase of 53% over the parent strain. The mutations in the conserved region 1.1 and region 2.4 of hrdB influence the transcriptional levels of aveR and, thus, avermectin production. However, elucidating the mechanism of the hrdB mutant and its effect on the regulatory network or metabolic flux toward avermectin biosynthesis requires further study.36
The extracytoplasmic function (ECF) σ factors σ6 and σ25 inhibit avermectin production by indirectly affecting the transcription of aveR via an unknown mechanism. Through gene-deletion, complementation, and over-expression experiments, the role of σ6 on avermectin production was investigated. The results show that σ6 negatively regulates avermectin production but has no effects on growth, stress responses, or morphology. The avermectin production was increased 2 to 2.7-fold (0.68 g/L) compared to the wild-type strain by deletion of the sig6 gene.51 The deletion of the sig25 gene results in ~1.23-fold higher avermectin production than the wild-type strain.52
σ25 initiates the transcription of olmRI and indirectly activates olmRII expression. olmRI and olmRII are the pathway-specific activator genes of oligomycin biosynthesis, which negatively affect avermectin production.53 The overproduction of avermectins in the olmRI and olmRII deleted mutants may be due to the extended units of competition for polyketide backbone synthesis of the oligomycin and avermectin.54 Thus the effect of σ25 on avermectin production may be induced by the regulation of aveR and the metabolic flux alteration toward avermectin production. Further studies show that σ25 initiates its own transcription, and its expression is directly activated by SmrA, the response regulator of a putative two-component system (TCS) smrAB located upstream of sig25.52 Deletion and complementation experiments with smrAB indicate that smrAB and σ25 function similarly in the regulation of avermectin. However, the exact regulatory mechanism of this ECF σ factor-TCS signal transduction system remains to be clarified.
Avermectin production is positively affected by ribosome recycling factor (RRF), which is involved in the release of ribosomes from the translational post-termination complex for a new round of initiation. The overexpression of frr increases avermectin yield by 3- to 3.7-fold compared to the wild-type strain and exhibits a greater promoting effect with multiple copies of frr. This effect functions by promoting cell growth, as well as the expression of the ave genes (including aveR and the ave structural genes). However, the exact targets of RRF remain a subject for further investigation.55
Some regulators that are involved in autoregulatory signaling also regulate aveR transcription and affect avermectin production. Avenolide is a class of Streptomyces autoregulators essential for eliciting avermectin production. The aco gene (encoding an acyl-CoA oxidase) that is involved in avenolide biosynthesis is clustered at the same locus with three homologs of the γ-butyrolactone autoregulator receptor proteins AvaR1, AvaR2, and AvaR3.56 Deletion of avaR3 results in a great decrease in avermectin production compared to the wild type strain. AvaR3 indirectly controls the expression of aveR and thus activates avermectin production. AvaR3 also negatively regulates the transcription of both avaR1 and avaR2.57
Deletion of avaR1 in a high-producing strain increases the production of avermectin B1a ~1.75-fold compared to the parent strain.58 AvaR1 represses avenolide production by binding to the promoter of the aco gene, and this interaction is inhibited by avenolide.56 AvaR1 also indirectly regulates the expression of AveR.58 However, the exact mechanism of how this autoregulator signaling system influences avermectin production has yet to be determined. We also identified four TetR genes (A, B, C, and D) that directly regulate the transcription of aveR and indicate another GBL signaling molecule in this process (data unpublished).
Other regulators target genes that may be involved in avermectin precursor metabolism. The negative role of the TetR transcriptional regulatory gene SAV7471 in avermectin production was found by deletion, complementation, and overexpression experiments. SAV7471 directly represses the transcription of SAV7472-SAV7473, which has a positive effect on avermectin production. SAV7473 encodes a flavoprotein that is possibly involved in pantothenate and coenzyme A (CoA) metabolism. SAV7471 negatively regulates CoA biosynthesis, which may provide the precursors for avermectin biosynthesis.59 Deletion of the TetR transcriptional regulatory gene SAV151 results in 2-fold higher avermectin production than the wild type strain. SAV151 directly regulates the transcription of itself and the adjacent transcriptional unit SAV152-SAV153-SAV154. SAV152 encodes a putative dehydrogenase, and SAV154 encodes a putative hydrolase. These two genes may provide energy or precursors to promote avermectin production.60 However, the function of the target genes that may be involved in precursor or energy supply for avermectin biosynthesis need further clarification.
Some regulators affect both ave transcription and avermectin precursor metabolism. The TetR family transcriptional regulators SAV576 and SAV577 both have negative effects on avermectin production. The double deletion of SAV576-SAV577 produces an additional enhancing effect on avermectin yield. These two regulators indirectly downregulate the transcription of ave genes and reciprocally repress each other's transcription. They both directly repress the transcription of the adjacent gene SAV575 by competitively binding the same region, and SAV576 represses the transcription of its own gene. SAV575 encodes a cytochrome P450/NADPH-ferrihemoprotein reductase, which may provide precursors and enhance avermectin production.37, 61 The deletion of aveI results in at least 10-fold more avermectin B1a than the wild type strain and increases the level of the aveR transcript.62 The aveI gene also negatively regulates the genes involved in precursor biosynthesis for avermectin based on global comparative transcriptomic analysis between the aveI deletion mutant and the wild-type.63 The response regulator PhoP in the two-component PhoR-PhoP system negatively affects avermectin biosynthesis in S. avermitilis by directly regulating the transcription of aveR. PhoP also regulates nitrogen metabolism and some key genes involved in morphological differentiation and antibiotic production.64
Additional regulators affect avermectin production via unknown mechanisms. Up-stream of the aveR gene are two genes, aveR1 and aveR2, that negatively influence avermectin production. Disruption of these two genes increases avermectin levels more than 3-fold. However, their relationship with aveR and the mechanism of regulation are still unknown.65 Three regulatory genes (SAV213, SAV3818, and SAV4023) have stimulatory effects on avermectin production via an unknown mechanism.66 Bacterial eukaryotic-type serine-threonine protein kinases (STPKs), AfsK of S. coelicolor A3 (2) and S. griseus, activate the AfsR orthologs, and their coding genes are located near the afsR gene.67 SAV3816, which localizes near one afsR homolog (SAV3804), is an afsK ortholog (afsK-av). Avermectin production is abolished in an afsK-av deletion mutant and restored with complementation of the intact afsK-av or the 900-nt catalytic domain region.68 Further, tandem phosphorylation on Thr-165 and Thr-168 in afsK-av is responsible for the response to SAM accumulation to modulate avermectin production.
AfsR2 of S. lividans, also known as a target gene called AfsS in S. coelicolor, was introduced into S. avermitilis and increases avermectin production.69 Further experiments show that S. lividans AfsR2 targets several genes, such as glyceraldehyde-3-phosphate dehydrogenase, polyribo-nucleotide and superoxide dismutase, indicating that AfsR2 may be a pleiotropic regulator that controls differential expressions of various kinds of genes in Streptomyces species.70 However, a better understanding of the regulatory mechanism of the afs-gene family in avermectin production would provide new strategies for yield improvement.
Engineering drug efflux pumps
Drug efflux pumps are very important for self-protection to overcome the toxic effects of natural products and to reduce feedback inhibition to increase production.71 The avtAB operon, which is located upstream of the avermectin biosynthetic gene cluster, encodes the ABC transporter AvtAB, which is also an avermectin exporter. The inactivation of avtAB has no effect on avermectin production. However, avermectin production is increased both in the wild type and in industrial strains by increasing the concentration of avtAB mRNA. The ratio of intracellular to extra cellular accumulation of avermectin B1a drops from 6:1 to 4.5:1, and the overall productivity of avermectin B1a is improved by ~50%, from 3.3 to 4.8 g/L by increasing the transcriptional level of AvtAB.72 However, there is no effect on oligomycin levels. Regardless, whether this transporter can affect the production of other compounds requires further research.
Protoplast fusion
Protoplast fusion is a method commonly used for natural product yield improvement. Chen et al.73 use the high avermectin producer 76-05 obtained through a continuous strain improvement program and the genetically engineered strain 73-12 that produces only B components and no oligomycin as the parental strains for intraspecific protoplast fusion. They created two genetically stable recombinant strains, F23 and F29, with both parental merits. The avermectin production of F23 and F29 is increased by 2.66- and 3.50-fold compared to parental strain 73-12, and reaches 84.20 and 103.45% of the parental strain 76-05. Further, F29 is very tolerant of fermentation conditions, such as temperature and aeration, which makes it a promising strain for industrial applications.73
Synthetic biology methods
Unlike metabolic engineering, which focuses on the rational design of natural regulation or metabolic networks, synthetic biology aims to design and build new biological systems from standard interchangeable parts for specific functions.74 With developments in molecular biology technology and systems biology, synthetic biology has been successfully applied to improving the yield of natural products. For instance, Artemisinin, a sesquiterpene lactone endoperoxide extracted from Artemisia annua L, is a highly effective anti-malaria drug that is in short supply.75 Paddon et al. report the engineering of Saccharomyces cerevisiae to produce high titers (up to 25 g/L) of artemisinic acid using a redesigned biosynthetic pathway.76 And the characterization of fumitremorgin B endoperoxidase (FtmOx1) may help to unravel the novel mechanism of endoperoxide formation reaction for the conversion of artemisinic acid to artemisinin.77 Similarly, taxol (paclitaxel) is a potent anticancer drug with cost-efficient production that was first isolated from the Taxus brevifolia Pacific yew tree. Ajikumar et al. report an Escherichia coli strain to increase taxadiene, the first committed Taxol intermediate, by 15,000-fold by using a multivariate modular approach to metabolic-pathway engineering.78
The application of synthetic biology to natural product yield improvement involves two main steps: (1) engineering cells used as specialized chassis; and (2) improving standard parts for optimization of the biosynthetic pathway.79 There are some Streptomyces chassis that can be used for avermectin production improvement, such as S. coelicolor80 and S. avermitilis.81, 82 Avermectin production was detected by using a new recombination cloning method to clone the 81-kb avermectin biosynthetic gene cluster into the linear plasmid of the model organism S. coelicolor.83 This confirms that the avermectin metabolite pathway is available in the new host S. coelicolor.
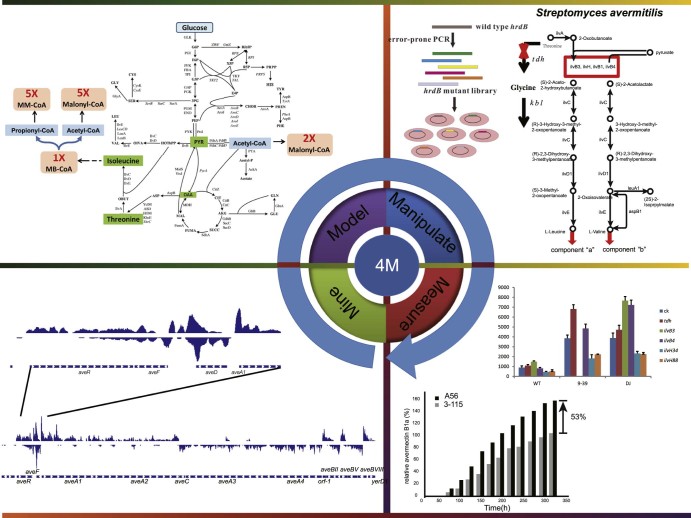
Synthetic biology research consists of iterative cycles of experimentation and computation characterized as the 4Ms Strategy: Mine, Model, Manipulation, and Measurement. This method was first applied in systems biology research at MIT.84 Systems biology research in natural biological systems will form the foundation for synthetic biology to redesign new biological systems.85 Synthetic biology will aid systems biology to understand and control the biological systems. Here, we introduce the 4Ms Strategy of synthetic biology and review avermectin yield improvement (Fig. 4).
Fig. 4.
The 4Ms Strategy used for avermectin improvement.
Mine
Mining is a way to identify the underlying relationships among a large number of datasets, such as genome, transcriptome, and proteome data. Chou and co-workers identify a synthase for a new sesquiterpene called Avermitilol with genome mining.86 Ikeda reviewed the biosynthetic gene cluster for secondary metabolites in S. avermitilis and provides the information needed for development of genome-minimized hosts.87 Proteomic analysis shows that fatty acid metabolism and the TCA cycle are repressed during avermectin biosynthesis.36, 37, 38 These data also revealed the association between hyphal morphology and avermectin production. This indicates that avermectin production is globally regulated and responds to environmental stresses. After transcription comparison between wild-type and avermectin overproducing strains, the global regulator σHrdB36 and several TetR family transcriptional regulators, such as SAV576,32 SAV577, and SAV15137, 61 were characterized. Other pieces of the genome were mined as well, such as the ABC transporter malEFG-α,39 AvtAB,72 the signaling molecule Avenolide,56 and additional regulators like σ25 52 and SAV7471.59
Model
The underlying relationships that come from mining can form hypotheses, which are reflected in predicted models. There are different types of models according to the type of questions that one seeks to answer.88 One type of models involves gene transcription regulation by regulators. The aveR promoter is predicted to be bound by the global regulator σHrdB.36 The interactions between two regulators SAV576 and SAV577 were modeled and considered to regulate the expression of each other and co-regulate avermectin production.37, 61 The other two regulators, SAV15160 and SAV7471,59 are assumed to regulate the adjacent genes that may be involved in the precursor's synthesis of avermectin. Luo et al. model the σ25-SmrAB (the down-stream genes) signal transduction system and the σ25 regulation of avermectin and oligomycin.52 Our group has also devoted efforts to construct a computational model of the primary metabolic variation toward avermectin biosynthesis (data unpublished).
Manipulation
In experimental manipulation, it is very important to test a computational model before it is adopted for practical application. Zhuo et al. use in vitro transcription assays to verify that the transcription of aveR is specifically recognized and activated by σHrdB.36 Moreover, a library was constructed by using error-prone PCR for random mutagenesis of the hrdB gene to identify the effect of σHrdB on aveR and avermectin biosynthesis. Gene disruption, complementation, and overexpression are used to identify the effects on avermectin production. Real-time RT-PCR, chromatin immunoprecipitation (ChIP) assays, and electrophoretic mobility shift assays (EMSAs) are used to determine the regulatory relationship between two genes. DNase I footprinting is used to confirm the binding sequence of the regulators.37, 61 Ikeda's group has constructed genome-minimized Streptomyces hosts by using two complementary strategies, including general homologous recombination and site-specific recombination (Cre-loxP) in S. avermitilis. They also test this chassis by heterologous expression of biosynthetic gene clusters for secondary metabolites.81, 82 Additionally, the advent of new recombinant DNA technologies, such as Gibson Assembly,89 Red/ET,90 and TAR (transformation-assisted recombination),91 have facilitated synthetic biology applications, including the reconstruction of the biosynthetic pathway of natural products.
Measurement
After genetic manipulation, the final step in the 4Ms Strategy is the measurement of the change in phenotype. A56, an avermectin yield-improved strain, was obtained through high-throughput screening,36 and the yield of avermectin B1a in A56 is increased by 53% relative to the parental strain in a 180-m3 fermentor. Avermectin production has also been evaluated after the disruption, complementation, or overexpression of the regulator and ABC transporter.37, 72 The measurement of exogenous secondary metabolites in genome-minimized S. avermitilis hosts confirms engineered S. avermitilis as a viable chassis for natural and unnatural metabolite biosynthesis.81, 82 A new quantitative method based on flow cytometry and a superfolder green fluorescent protein (sfGFP) at single-cell resolution in Streptomyces will also facilitate the functional optimization of biosynthetic gene clusters in Streptomyces.92
These measurement data obtained from experimental manipulation were subsequently mined to build more optimal models. After iterative cycles of the 4Ms Strategy, the models will be improved to verify the hypotheses and predict the outcome. Indeed, the signaling molecule, transcriptional factor, and ABC transporter that are relevant to avermectin biosynthesis were all identified using this strategy. These parts could be assembled into devices that perform simple and defined functions, such as activation, regulation, and transportation. Further, these devices can form a simple system, i.e., avermectin synthesis. With cell growth, the signaling molecule accumulates and activates quorum sensing. This activates the positive regulators and efflux pump of avermectin. It also represses the negative regulators and inhibits the biosynthesis of other second metabolites. Ultimately, a high yield of avermectin is acquired.
Conclusions and future perspectives
The application of avermectin as an anthelmintic agent and the new discovery of antibacterial activity will increase the requirement for this compound. Here, we reviewed various methods that are widely used to improve avermectin fermentation production and the great progress that has been made in this field. The traditional fermentation and random mutagenesis methods are crucial for industrial fermentation and have a long history of success for natural product improvement. However, they are labor intensive and unsuitable for rational engineering. In contrast, metabolic engineering tunes the metabolic and regulatory networks in a rational way to improve production, while providing a better understanding of this natural biological system. With the development of genetic manipulation technologies and the understanding of the natural biology of the system, synthetic biology became available. Synthetic biology redesigns or constructs a more efficient biological system from new biological parts for specific functions. To synthesize a bioactive product, the whole system will be more predictable and controllable when using a specialized chassis and improved biosynthetic pathway. Further, it may optimize the industrial fermentation in such a way as to utilize fewer nutrient supplies and shorten the fermentation time. Synthetic biology has created new opportunities for particular strains that are recalcitrant to genetic manipulation and/or contain cryptic biosynthetic gene clusters. We anticipate that synthetic biology will be considered a promising strategy for the improvement of avermectin yield and other natural products in the future.
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgements
The authors would like to thank Changming Zhao, Chunbo Lou, Yong Tao, Feng Xie, Xiaopeng Jia, and Tao Zhang for helpful discussions. This work was supported in part by the National Program on Key Basic Research Project (973 program, 2013CB734000 and 2012CB725203), the National Natural Science Foundation of China (31170095, 31000057, and 31100075), the Ministry of Science and Technology of the People's Republic of China (2011ZX11102-011-11 and 2013ZX10005004-005), and China Ocean Mineral Resources R & D Association (DY125-15-T-07). L.Z. is a recipient of the National Distinguished Young Scholar Program in China.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Miller T.W., Chaiet L., Cole D.J., Cole L.J., Flor J.E., Goegelman R.T. Avermectins, new family of potent anthelmintic agents: isolation and chromatographic properties. Antimicrob Agents Chemother. 1979;15:368–371. doi: 10.1128/aac.15.3.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen B. A new golden age of natural products drug discovery. Cell. 2015;163:1297–1300. doi: 10.1016/j.cell.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thuan N.H., Pandey R.P., Sohng J.K. Recent advances in biochemistry and biotechnological synthesis of avermectins and their derivatives. Appl Microbiol Biotechnol. 2014;98:7747–7759. doi: 10.1007/s00253-014-5926-x. [DOI] [PubMed] [Google Scholar]
- 4.Egerton J., Ostlind D., Blair L., Eary C., Suhayda D., Cifelli S. Avermectins, new family of potent anthelmintic agents: efficacy of the B1a component. Antimicrob Agents Chemother. 1979;15:372–378. doi: 10.1128/aac.15.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim L.E., Vilchèze C., Ng C., Jacobs W.R., Ramón-García S., Thompson C.J. Anthelmintic avermectins kill M. tuberculosis, including multidrug resistant clinical strains. Antimicrob Agents Chemother. 2013;57:1040–1046. doi: 10.1128/AAC.01696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H., Ren B., Dai H., Dai S., Zhang Y., Liu Y. Reversal of meticillin resistance in Staphylococcus aureus by the anthelmintic avermectin. Int J Antimicrob Agents. 2014;44:274–276. doi: 10.1016/j.ijantimicag.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Liu X., Bolla K., Ashforth E.J., Zhuo Y., Gao H., Huang P. Systematics-guided bioprospecting for bioactive microbial natural products. Antonie Van Leeuwenhoek. 2012;101:55–66. doi: 10.1007/s10482-011-9671-1. [DOI] [PubMed] [Google Scholar]
- 8.Burg R.W., Miller B.M., Baker E.E., Birnbaum J., Currie S.A., Hartman R. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother. 1979;15:361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siddique S., Syed Q., Adnan A., Nadeem M., Irfan M., Ashraf Qureshi F. Production of avermectin B1b from Streptomyces avermitilis 41445 by batch submerged fermentation. Jundishapur J Microbiol. 2013;6 [Google Scholar]
- 10.Xu Z., Cen P. Enhanced production of avermectin B1a by medium optimization and glucose feeding with Streptomyces avermilitis. Bioprocess Eng. 1999;20:67–71. [Google Scholar]
- 11.Gao H., Liu M., Liu J., Dai H., Zhou X., Liu X. Medium optimization for the production of avermectin B1a by Streptomyces avermitilis 14-12A using response surface methodology. Bioresour Technol. 2009;100:4012–4016. doi: 10.1016/j.biortech.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Xu Z., Cen P. Stimulation of avermectin B1a biosynthesis in Streptomyces avermilitis by feeding glucose and propionate. Biotechnol Lett. 1999;21:91–95. [Google Scholar]
- 13.Ikeda H., Kotaki H., Tanaka H., Omura S. Involvement of glucose catabolism in avermectin production by Streptomyces avermitilis. Antimicrob Agents Chemother. 1988;32:282–284. doi: 10.1128/aac.32.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulman M., Acton S., Valentino D.L., Arison B.H. Purification and identification of dTDP-oleandrose, the precursor of the oleandrose units of the avermectins. J Biol Chem. 1990;265:16965–16970. [PubMed] [Google Scholar]
- 15.Wohlert S.-E., Lomovskaya N., Kulowski K., Fonstein L., Occi J.L., Gewain K.M. Insights about the biosynthesis of the avermectin deoxysugar L-oleandrose through heterologous expression of Streptomyces avermitilis deoxysugar genes in Streptomyces lividans. Chem Biol. 2001;8:681–700. doi: 10.1016/s1074-5521(01)00043-6. [DOI] [PubMed] [Google Scholar]
- 16.Schulman M.D., Valentino D., Nallin M., Kaplan L. Avermectin B2 O-methyltransferase activity in “Streptomyces avermitilis” mutants that produce increased amounts of the avermectins. Antimicrob Agents Chemother. 1986;29:620–624. doi: 10.1128/aac.29.4.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulman M.D., Valentino D., Streicher S., Ruby C. “Streptomyces avermitilis” mutants defective in methylation of avermectins. Antimicrob Agents Chemother. 1987;31:744–747. doi: 10.1128/aac.31.5.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda H., Wang L.-R., Ohta T., Inokoshi J., Ōmura S. Cloning of the gene encoding avermectin B 5-O-methyltransferase in avermectin-producing Streptomyces avermitilis. Gene. 1998;206:175–180. doi: 10.1016/s0378-1119(97)00581-7. [DOI] [PubMed] [Google Scholar]
- 19.Yin P., Wang Y.-H., Zhang S.-L., Chu J., Zhuang Y.-P., Chen N. Effect of mycelial morphology on bioreactor performance and avermectin production of Streptomyces avermitilis in submerged cultivations. J Chin Inst Chem Eng. 2008;39:609–615. [Google Scholar]
- 20.Liang J.-G., Chu X.-H., Xiong Z.-Q., Chu J., Wang Y.-H., Zhuang Y.-P. Oxygen uptake rate regulation during cell growth phase for improving avermectin B1a batch fermentation on a pilot scale (2 m3) World J Microbiol Biotechnol. 2011;27:2639–2644. [Google Scholar]
- 21.Liang J.-G., Chu X.-H., Chu J., Wang Y.-H., Zhuang Y.-P., Zhang S.-L. Oxygen uptake rate (OUR) control strategy for improving avermectin B1a production during fed-batch fermentation on industrial scale (150 m3) Afr J Biotechnol. 2010;9:7186–7191. [Google Scholar]
- 22.Chen J., Zhang S., Chu J., Zhuang Y., Luo J., Bai H. Ethanol evolution rate: a new parameter to determine the feeding rate for the production of avermectins by Streptomyces avermitilis. Biotechnol Lett. 2004;26:109–113. doi: 10.1023/b:bile.0000012887.12874.fb. [DOI] [PubMed] [Google Scholar]
- 23.Wang L.Y., Huang Z.L., Li G., Zhao H.X., Xing X.H., Sun W.T. Novel mutation breeding method for Streptomyces avermitilis using an atmospheric pressure glow discharge plasma. J Appl Microbiol. 2010;108:851–858. doi: 10.1111/j.1365-2672.2009.04483.x. [DOI] [PubMed] [Google Scholar]
- 24.Gao H., Liu M., Zhuo Y., Zhou X., Liu J., Chen D. Assessing the potential of an induced-mutation strategy for avermectin overproducers. Appl Environ Microbiol. 2010;76:4583–4586. doi: 10.1128/AEM.01682-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu M., Gao H., Shang P., Zhou X., Ashforth E., Zhuo Y. Magnetic field is the dominant factor to induce the response of Streptomyces avermitilis in altered gravity simulated by diamagnetic levitation. PLoS ONE. 2011;6:e24697. doi: 10.1371/journal.pone.0024697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aikawa M., Lopes-Shikida S., Lemos M., Pradella J., Padilla G. Screening of spontaneous and induced mutants in Streptomyces avermitilis enhances avermectin production. Appl Microbiol Biotechnol. 1999;52:558–562. [Google Scholar]
- 27.Gao H., Liu M., Zhou X., Liu J., Zhuo Y., Gou Z. Identification of avermectin-high-producing strains by high-throughput screening methods. Appl Microbiol Biotechnol. 2010;85:1219–1225. doi: 10.1007/s00253-009-2345-5. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda H., Nonomiya T., Usami M., Ohta T., Ōmura S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc Natl Acad Sci USA. 1999;96:9509–9514. doi: 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeda H., Ishikawa J., Hanamoto A., Shinose M., Kikuchi H., Shiba T. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 30.Sun P., Zhao Q., Yu F., Zhang H., Wu Z., Wang Y. Spiroketal formation and modification in avermectin biosynthesis involves a dual activity of AveC. J Am Chem Soc. 2013;135:1540–1548. doi: 10.1021/ja311339u. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda H., Omura S. Avermectin biosynthesis. Chem Rev. 1997;97:2591–2610. doi: 10.1021/cr960023p. [DOI] [PubMed] [Google Scholar]
- 32.Yoon Y., Kim E.-S., Hwang Y.-S., Choi C.-Y. Avermectin: biochemical and molecular basis of its biosynthesis and regulation. Appl Microbiol Biotechnol. 2004;63:626–634. doi: 10.1007/s00253-003-1491-4. [DOI] [PubMed] [Google Scholar]
- 33.Stutzman-Engwall K., Conlon S., Fedechko R., Kaczmarek F., McArthur H., Krebber A. Engineering the aveC gene to enhance the ratio of doramectin to its CHC-B2 analogue produced in Streptomyces avermitilis. Biotechnol Bioeng. 2003;82:359–369. doi: 10.1002/bit.10578. [DOI] [PubMed] [Google Scholar]
- 34.Stutzman-Engwall K., Conlon S., Fedechko R., McArthur H., Pekrun K., Chen Y. Semi-synthetic DNA shuffling of aveC leads to improved industrial scale production of doramectin by Streptomyces avermitilis. Metab Eng. 2005;7:27–37. doi: 10.1016/j.ymben.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Yin P., Wang Y.-H., Zhang S.-L., Chu J., Zhuang Y.-P., Wang M.-L. Isolation of soluble proteins from an industrial strain Streptomyces avermitilis in complex culture medium for two-dimensional gel electrophoresis. J Microbiol Methods. 2008;73:105–110. doi: 10.1016/j.mimet.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 36.Zhuo Y., Zhang W., Chen D., Gao H., Tao J., Liu M. Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis. Proc Natl Acad Sci USA. 2010;107:11250–11254. doi: 10.1073/pnas.1006085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo J., Zhang X., Luo S., He F., Chen Z., Wen Y. A novel TetR family transcriptional regulator, SAV576, negatively controls avermectin biosynthesis in Streptomyces avermitilis. PLoS ONE. 2013;8:e71330. doi: 10.1371/journal.pone.0071330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin P., Li Y.-Y., Zhou J., Wang Y.-H., Zhang S.-L., Ye B.-C. Direct proteomic mapping of Streptomyces avermitilis wild and industrial strain and insights into avermectin production. J Proteomics. 2013;79:1–12. doi: 10.1016/j.jprot.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 39.Li M., Chen Z., Zhang X., Song Y., Wen Y., Li J. Enhancement of avermectin and ivermectin production by overexpression of the maltose ATP-binding cassette transporter in Streptomyces avermitilis. Bioresour Technol. 2010;101:9228–9235. doi: 10.1016/j.biortech.2010.06.132. [DOI] [PubMed] [Google Scholar]
- 40.Skinner D.D., Morgenstern M.R., Fedechko R.W., Denoya C.D. Cloning and sequencing of a cluster of genes encoding branched-chain alpha-keto acid dehydrogenase from Streptomyces avermitilis and the production of a functional E1 [alpha beta] component in Escherichia coli. J Bacteriol. 1995;177:183–190. doi: 10.1128/jb.177.1.183-190.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denoya C.D., Fedechko R.W., Hafner E.W., McArthur H.A., Morgenstern M.R., Skinner D.D. A second branched-chain alpha-keto acid dehydrogenase gene cluster (bkdFGH) from Streptomyces avermitilis: its relationship to avermectin biosynthesis and the construction of a bkdF mutant suitable for the production of novel antiparasitic avermectins. J Bacteriol. 1995;177:3504–3511. doi: 10.1128/jb.177.12.3504-3511.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutton C.J., Gibson S.P., Goudie A.C., Holdom K.S., Pacey M.S., Ruddock J.C. Novel avermectins produced by mutational biosynthesis. J Antibiot. 1991;44:357–365. doi: 10.7164/antibiotics.44.357. [DOI] [PubMed] [Google Scholar]
- 43.Wang F., Wang Y., Ji J., Zhou Z., Yu J., Zhu H. Structural and functional analysis of the loading acyltransferase from avermectin modular polyketide synthase. ACS Chem Biol. 2015;10(4):1017–1025. doi: 10.1021/cb500873k. [DOI] [PubMed] [Google Scholar]
- 44.Yoon G.-S., Ko K.-H., Kang H.-W., Suh J.-W., Kim Y.-S., Ryu Y.-W. Characterization of S-adenosylmethionine synthetase from Streptomyces avermitilis NRRL8165 and its effect on antibiotic production. Enzyme Microb Technol. 2006;39:466–473. [Google Scholar]
- 45.Zhao X., Wang Q., Guo W., Cai Y., Wang C., Wang S. Overexpression of metK shows different effects on avermectin production in various Streptomyces avermitilis strains. World J Microbiol Biotechnol. 2013;29:1869–1875. doi: 10.1007/s11274-013-1350-0. [DOI] [PubMed] [Google Scholar]
- 46.Chiang P., Gordon R., Tal J., Zeng G., Doctor B., Pardhasaradhi K. S-Adenosylmethionine and methylation. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- 47.Kim D.-J., Huh J.-H., Yang Y.-Y., Kang C.-M., Lee I.-H., Hyun C.-G. Accumulation of S-adenosyl-L-methionine enhances production of actinorhodin but inhibits sporulation in Streptomyces lividans TK23. J Bacteriol. 2003;185:592–600. doi: 10.1128/JB.185.2.592-600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikeda H., Takada Y., Pang C.-H., Tanaka H., Omura S. Transposon mutagenesis by Tn4560 and applications with avermectin-producing Streptomyces avermitilis. J Bacteriol. 1993;175:2077–2082. doi: 10.1128/jb.175.7.2077-2082.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitani S., Ikeda H., Sakamoto T., Noguchi S., Nihira T. Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl Microbiol Biotechnol. 2009;82:1089–1096. doi: 10.1007/s00253-008-1850-2. [DOI] [PubMed] [Google Scholar]
- 50.Guo J., Zhao J., Li L., Chen Z., Wen Y., Li J. The pathway-specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively affects oligomycin biosynthesis. Mol Genet Genomics. 2010;283:123–133. doi: 10.1007/s00438-009-0502-2. [DOI] [PubMed] [Google Scholar]
- 51.Jiang L., Liu Y., Wang P., Wen Y., Song Y., Chen Z. Inactivation of the extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis. Biotechnol Lett. 2011;33:1955–1961. doi: 10.1007/s10529-011-0673-x. [DOI] [PubMed] [Google Scholar]
- 52.Luo S., Sun D., Zhu J., Chen Z., Wen Y., Li J. An extracytoplasmic function sigma factor, σ25, differentially regulates avermectin and oligomycin biosynthesis in Streptomyces avermitilis. Appl Microbiol Biotechnol. 2014:1–16. doi: 10.1007/s00253-014-5759-7. [DOI] [PubMed] [Google Scholar]
- 53.Yu Q., Bai L., Zhou X., Deng Z. Inactivation of the positive LuxR-type oligomycin biosynthesis regulators OlmRI and OlmRII increases avermectin production in Streptomyces avermitilis. Chin Sci Bull. 2012;57:869–876. [Google Scholar]
- 54.Omura S., Ikeda H., Ishikawa J., Hanamoto A., Takahashi C., Shinose M. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L., Guo J., Wen Y., Chen Z., Song Y., Li J. Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J Ind Microbiol Biotechnol. 2010;37:673–679. doi: 10.1007/s10295-010-0710-0. [DOI] [PubMed] [Google Scholar]
- 56.Kitani S., Miyamoto K.T., Takamatsu S., Herawati E., Iguchi H., Nishitomi K. Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc Natl Acad Sci USA. 2011;108:16410–16415. doi: 10.1073/pnas.1113908108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyamoto K.T., Kitani S., Komatsu M., Ikeda H., Nihira T. The autoregulator receptor homologue AvaR3 plays a regulatory role in antibiotic production, mycelial aggregation and colony development of Streptomyces avermitilis. Microbiology. 2011;157:2266–2275. doi: 10.1099/mic.0.048371-0. [DOI] [PubMed] [Google Scholar]
- 58.Wang J.-B., Zhang F., Pu J.-Y., Zhao J., Zhao Q.-F., Tang G.-L. Characterization of AvaR1, an autoregulator receptor that negatively controls avermectins production in a high avermectin-producing strain. Biotechnol Lett. 2014;36:813–819. doi: 10.1007/s10529-013-1416-y. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y., Yan T., Jiang L., Wen Y., Song Y., Chen Z. Characterization of SAV7471, a TetR-family transcriptional regulator involved in the regulation of coenzyme A metabolism in Streptomyces avermitilis. J Bacteriol. 2013;195:4365–4372. doi: 10.1128/JB.00716-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He F., Liu W., Sun D., Luo S., Chen Z., Wen Y. Engineering of the TetR family transcriptional regulator SAV151 and its target genes increases avermectin production in Streptomyces avermitilis. Appl Microbiol Biotechnol. 2014;98:399–409. doi: 10.1007/s00253-013-5348-1. [DOI] [PubMed] [Google Scholar]
- 61.Guo J., Zhang X., Chen Z., Wen Y., Li J. Two adjacent and similar TetR family transcriptional regulator genes, SAV577 and SAV576, co-regulate avermectin production in Streptomyces avermitilis. PLoS ONE. 2014;9:e99224. doi: 10.1371/journal.pone.0099224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen L., Lu Y., Chen J., Zhang W., Shu D., Qin Z. Characterization of a negative regulator AveI for avermectin biosynthesis in Streptomyces avermitilis NRRL8165. Appl Microbiol Biotechnol. 2008;80:277–286. doi: 10.1007/s00253-008-1545-8. [DOI] [PubMed] [Google Scholar]
- 63.Chen L., Chen J., Jiang Y., Zhang W., Jiang W., Lu Y. Transcriptomics analyses reveal global roles of the regulator AveI in Streptomyces avermitilis. FEMS Microbiol Lett. 2009;298:199–207. doi: 10.1111/j.1574-6968.2009.01721.x. [DOI] [PubMed] [Google Scholar]
- 64.Yang R., Liu X., Wen Y., Song Y., Chen Z., Li J. The PhoP transcription factor negatively regulates avermectin biosynthesis in Streptomyces avermitilis. Appl Microbiol Biotechnol. 2015:1–11. doi: 10.1007/s00253-015-6921-6. [DOI] [PubMed] [Google Scholar]
- 65.Stutzman-Engwall K.J., Price B.S. 2001. Streptomyces avermitilis regulatory genes for increased avermectin production. Google Patents. [Google Scholar]
- 66.Duong C., Lee H.-N., Choi S.-S., Lee S.Y., Kim E.-S. Functional expression of SAV3818, a putative TetR-family transcriptional regulatory gene from Streptomyces avermitilis, stimulates antibiotic production in Streptomyces species. J Microbiol Biotechnol. 2009;19:136–139. doi: 10.4014/jmb.0806.387. [DOI] [PubMed] [Google Scholar]
- 67.Umeyama T., Lee P.-C., Ueda K., Horinouchi S. An AfsK/AfsR system involved in the response of aerial mycelium formation to glucose in Streptomyces griseus. Microbiology. 1999;145:2281–2292. doi: 10.1099/00221287-145-9-2281. [DOI] [PubMed] [Google Scholar]
- 68.Rajkarnikar A., Kwon H.-J., Ryu Y.-W., Suh J.-W. Catalytic domain of AfsKav modulates both secondary metabolism and morphologic differentiation in Streptomyces avermitilis ATCC 31272. Curr Microbiol. 2006;53:204–208. doi: 10.1007/s00284-006-0062-1. [DOI] [PubMed] [Google Scholar]
- 69.Lee J.-Y., Hwang Y.-S., Kim S.-S., Kim E.-S., Choi C.-Y. Effect of a global regulatory gene,afsR2, from Streptomyces lividans on avermectin production in Streptomyces avermitilis. J Biosci Bioeng. 2000;89:606–608. doi: 10.1016/s1389-1723(00)80065-1. [DOI] [PubMed] [Google Scholar]
- 70.Kim C.-Y., Park H.-J., Kim E.-S. Proteomics-driven identification of putative AfsR2-target proteins stimulating antibiotic biosynthesis in Streptomyces lividans. Biotechnol Bioprocess Eng. 2005;10:248–253. [Google Scholar]
- 71.Méndez C., Salas J.A. The role of ABC transporters in antibiotic-producing organisms: drug secretion and resistance mechanisms. Res Microbiol. 2001;152:341–350. doi: 10.1016/s0923-2508(01)01205-0. [DOI] [PubMed] [Google Scholar]
- 72.Qiu J., Zhuo Y., Zhu D., Zhou X., Zhang L., Bai L. Overexpression of the ABC transporter AvtAB increases avermectin production in Streptomyces avermitilis. Appl Microbiol Biotechnol. 2011;92:337–345. doi: 10.1007/s00253-011-3439-4. [DOI] [PubMed] [Google Scholar]
- 73.Chen Z., Wen J., Song Y., Wen Y., Li J. Enhancement and selective production of avermectin B by recombinants of Streptomyces avermitilis via intraspecific protoplast fusion. Chin Sci Bull. 2007;52:616–622. [Google Scholar]
- 74.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 75.Ro D.-K., Paradise E.M., Ouellet M., Fisher K.J., Newman K.L., Ndungu J.M. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 76.Paddon C.J., Westfall P., Pitera D., Benjamin K., Fisher K., McPhee D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 77.Yan W., Song H., Song F., Guo Y., Wu C.-H., Her A.S. Endoperoxide formation by an α-ketoglutarate-dependent mononuclear non-haem iron enzyme. Nature. 2015;527:539–543. doi: 10.1038/nature15519. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Ajikumar P.K., Xiao W.-H., Tyo K.E., Wang Y., Simeon F., Leonard E. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhuo Y., Zhang T., Wang Q., Cruz-Morales P., Zhang B., Liu M. Synthetic biology of avermectin for production improvement and structure diversification. Biotechnol J. 2014;9:316–325. doi: 10.1002/biot.201200383. [DOI] [PubMed] [Google Scholar]
- 80.Gomez-Escribano J.P., Bibb M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Komatsu M., Uchiyama T., Ōmura S., Cane D.E., Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci USA. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Komatsu M., Komatsu K., Koiwai H., Yamada Y., Kozone I., Izumikawa M. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol. 2013;2:384–396. doi: 10.1021/sb3001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang R., Xia H., Xu Q., Dang F., Qin Z. Recombinational cloning of the antibiotic biosynthetic gene clusters in linear plasmid SCP1 of Streptomyces coelicolor A3 (2) FEMS Microbiol Lett. 2013;345:39–48. doi: 10.1111/1574-6968.12183. [DOI] [PubMed] [Google Scholar]
- 84.Tadmor B., Tidor B. Interdisciplinary research and education at the biology–engineering–computer science interface: a perspective. Drug Discov Today. 2005;10:1183–1189. doi: 10.1016/S1359-6446(05)03540-3. [DOI] [PubMed] [Google Scholar]
- 85.Barrett C.L., Kim T.Y., Kim H.U., Palsson B.Ø., Lee S.Y. Systems biology as a foundation for genome-scale synthetic biology. Curr Opin Biotechnol. 2006;17:488–492. doi: 10.1016/j.copbio.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 86.Chou W.K., Fanizza I., Uchiyama T., Komatsu M., Ikeda H., Cane D.E. Genome mining in Streptomyces avermitilis: cloning and characterization of SAV_76, the synthase for a new sesquiterpene, avermitilol. J Am Chem Soc. 2010;132:8850–8851. doi: 10.1021/ja103087w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ikeda H., Shin-ya K., Omura S. Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol. 2014;41:233–250. doi: 10.1007/s10295-013-1327-x. [DOI] [PubMed] [Google Scholar]
- 88.MacDonald J.T., Barnes C., Kitney R.I., Freemont P.S., Stan G.-B.V. Computational design approaches and tools for synthetic biology. Integr Biol (Camb) 2011;3:97–108. doi: 10.1039/c0ib00077a. [DOI] [PubMed] [Google Scholar]
- 89.Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 90.Bird A.W., Erler A., Fu J., Hériché J.-K., Maresca M., Zhang Y. High-efficiency counterselection recombineering for site-directed mutagenesis in bacterial artificial chromosomes. Nat Methods. 2012;9:103–109. doi: 10.1038/nmeth.1803. [DOI] [PubMed] [Google Scholar]
- 91.Gibson D.G., Benders G.A., Axelrod K.C., Zaveri J., Algire M.A., Moodie M. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bai C., Zhang Y., Zhao X., Hu Y., Xiang S., Miao J. Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proc Natl Acad Sci USA. 2015;112:12181–12186. doi: 10.1073/pnas.1511027112. [DOI] [PMC free article] [PubMed] [Google Scholar]