Abstract
Biotin is an essential micronutrient that acts as a co-factor for biotin-dependent metabolic enzymes. In bacteria, the supply of biotin can be achieved by de novo synthesis or import from exogenous sources. Certain bacteria are able to obtain biotin through both mechanisms while others can only fulfill their biotin requirement through de novo synthesis. Inability to fulfill their cellular demand for biotin can have detrimental consequences on cell viability and virulence. Therefore understanding the transcriptional mechanisms that regulate biotin biosynthesis and transport will extend our knowledge about bacterial survival and metabolic adaptation during pathogenesis when the supply of biotin is limited. The most extensively characterized protein that regulates biotin synthesis and uptake is BirA. In certain bacteria, such as Escherichia coli and Staphylococcus aureus, BirA is a bi-functional protein that serves as a transcriptional repressor to regulate biotin biosynthesis genes, as well as acting as a ligase to catalyze the biotinylation of biotin-dependent enzymes. Recent studies have identified two other proteins that also regulate biotin synthesis and transport, namely BioQ and BioR. This review summarizes the different transcriptional repressors and their mechanism of action. Moreover, the ability to regulate the expression of target genes through the activity of a vitamin, such as biotin, may have biotechnological applications in synthetic biology.
Abbreviations: BCCP, biotin carboxyl carrier protein; BirA, biotin retention protein A; BPL, biotin protein ligase; EcBirA, Escherichia coli BirA; SaBirA, Staphylococcus aureus BirA
1. Introduction
Biotin (vitamin H or B7) is an important micronutrient that functions as a cofactor for biotin-dependent enzymes.1 These include the biotin-dependent carboxylases, decarboxylases and transcarboxylases, all of which are found in the microbial world. In the prototypical bacteria Escherichia coli, there is a single biotin-dependent enzyme, namely acetyl CoA carboxylase, that catalyzes the first committed step in the fatty acid biosynthesis pathway.2, 3 Other examples of biotin-dependent enzymes commonly found in prokaryotes include pyruvate carboxylase responsible for replenishing the TCA cycle with oxaloacetate,4 and propionyl CoA carboxylase required for the metabolism of certain amino acids and fatty acids.5 Micro-organisms, plants and some fungi are able to synthesize biotin de novo as well as importing it from their environment through the action of a biotin transport system. In contrast, humans and other mammals are biotin auxotrophs and rely solely on uptake from external sources, such as intestinal microflora or the diet.6 This genetic difference in biotin metabolism between humans and microbes provides potential drug targets for new antibiotic discovery (reviewed7). The biotin synthesis pathway is well characterized in E. coli and Bacillus subtilis and has recently been reviewed.8 In many bacteria the genes that encode the biotin biosynthetic enzymes are often clustered into an operon known as the bio operon.9 Briefly, the synthetic pathway commences with L-alanine and S-adenosyl-L- methionine being introduced into pimeloyl-ACP by the activities of 7-keto-8-aminopelargonic acid synthase (encoded by bioF) and 7,8-diaminopelargonic acid synthase (encoded by bioA), respectively, to generate 7,8-diaminopelargonic acid. Dethiobiotin synthetase (encoded by bioD) and biotin synthase (encoded by bioB) then catalyze the closure of the ureido and thiophane heterocycles, respectively, liberating biotin.
The de novo synthesis of biotin is metabolically costly, requiring 20 equivalents of ATP for each molecule of biotin and the activities of at least 4 metabolic enzymes.10 Therefore, transcriptional regulation of the biotin biosynthetic enzymes needs to be tightly controlled. In the model bacteria E. coli, the balance of biotin demand versus supply is maintained through the action of the biotin retention protein A (BirA); a bi-functional protein that is not only a transcriptional repressor but also serves as the biotin ligase that catalyzes the attachment of biotin onto the biotin-dependent carboxylases. In other microorganisms, such as Corynebacterium glutamicum and Agrobacterium tumefaciens, there is no BirA homolog to regulate biotin synthesis and transport. Instead, alternative DNA-binding proteins perform this function, namely BioQ and BioR respectively. The mechanisms by which BirA, BioQ and BioR regulate biotin biosynthesis and transport will be discussed in this review.
2. BirA is a bi-functional protein
BirA serves as both a transcriptional repressor and the enzyme responsible for protein biotinylation (outlined in Fig. 1). As both biotin ligase and transcriptional repressor activities are intimately linked, we provide an overview of both functions as background for the reader to understand the sophistication of this elegant system. Protein biotinylation is achieved through a conserved, two-step reaction mechanism that is catalyzed by biotin protein ligase (BPL) in all organisms. In the first partial reaction biotin and ATP are required to form biotinyl-5′-AMP that serves as both the reaction intermediate for protein biotinylation and corepressor for transcriptional regulation. The BirA: biotinyl-5′-AMP (holo) enzyme can then adopt one of two different fates. When the cellular demand for biotin is low holo BirA can dimerize and bind DNA where it functions as the transcriptional repressor of the biotin biosynthesis operon, thereby inhibiting the synthesis of more biotin. In contrast, in the presence of substrate requiring biotinylation the holo BirA functions as a biotin ligase. Here BPL recognizes and binds to a biotin carboxyl carrier protein (BCCP) present in the receiving enzyme that contains the lysine residue targeted for biotinylation.11 Protein biotinylation is an example of a post-translational modification that is performed with exquisite specificity. For example, the E. coli biotin ligase (BirA) modifies just one of the >4000 different proteins in the bacterial cell.12 Moreover, the biotin cofactor is covalently attached onto the side chain of one single, specific target lysine residue present in the active site of biotin-dependent enzymes. BPLs from a wide variety of species are able to modify BCCP from unrelated organisms,13, 14, 15 highlighting how highly conserved both the catalytic mechanism and the protein:protein interactions between enzyme and substrate have remained throughout evolution. The possible mechanisms through which BirA can switch between its two functions are described later in this review.
Fig. 1.
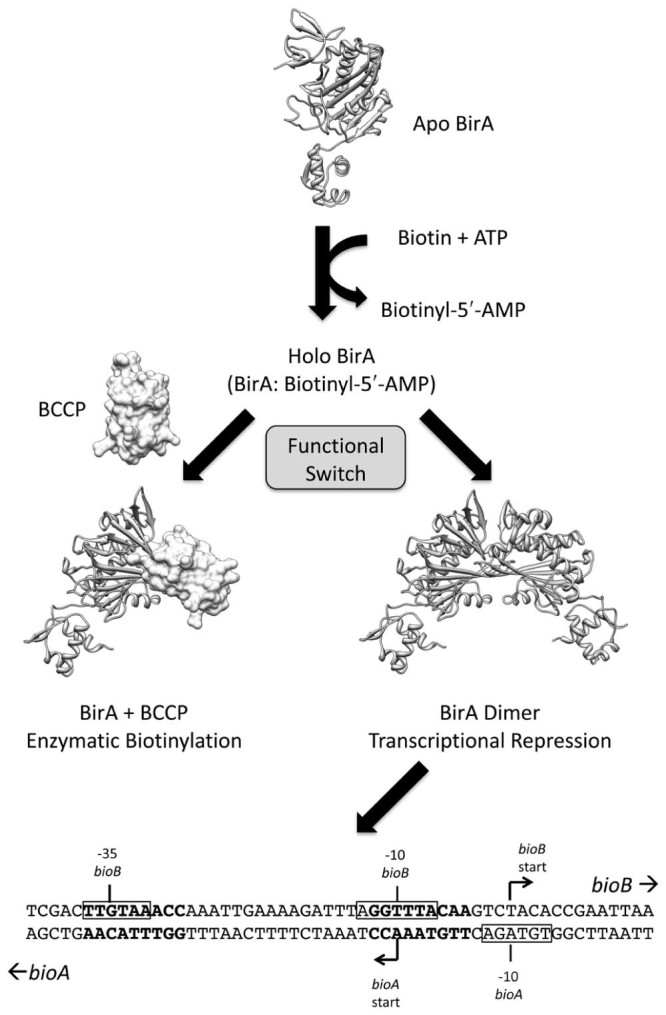
Bifunctional BirA from Escherichia coli. The schematic shows the two alternative functions for the protein. The bioO sequence from the biotin biosynthetic operon is shown below, with the BirA binding sites in bold text and the −10 and −35 sequences boxed.
All BPLs contain a conserved 2-domain catalytic core responsible for biotinyl-5′-AMP synthesis and protein biotinylation.16 The greatest divergence between the BPLs is in their N-terminal regions (see Fig. 2A). Class I BPLs are composed only of the conserved catalytic module that is required for protein biotinylation. Hence, these are mono functional enzymes. X-ray crystal structures of Class I BPLs have been reported for Mycobacterium tuberculosis19 and Pyrococcus horikoshii.21 In contrast, the Class II BPLs are truly bi-functional having both biotin ligase and transcriptional repressor activities due to an N-terminal DNA binding domain. BirA from E. coli is the most extensively studied representative of a Class II BPL, having been the subject of structural, genetic and biophysical studies (reviewed22, 23). Several recent reports of the homolog from S. aureus (SaBirA) have appeared in the literature and provide some interesting points of difference to the prototypical EcBirA that will be elaborated further below. Genetic knockout studies performed on both E. coli and S. aureus, among other bacteria, have demonstrated that BirA is an essential gene product.19, 24, 25, 26 Therefore, biotin ligases serve as promising targets for new antibacterials.27, 28, 29 Interestingly, a recent study reported that Francisella novicida expresses both a Class I and Class II BPL.30 In this bacterium the Class I enzyme functions as the biotin ligase whereas the Class II homolog is a transcriptional regulator essential for virulence in mouse infection models. However, this is exceptional as most bacteria only possess one BPL equivalent. The Class III BPLs, such as those found in mammals and certain eukaryotes, contain an extended N-terminal region making them at least twice the size of the Class II BPLs (Fig. 2A). This extension bears no sequence or functional similarity to the Class II DNA binding domain. Recent mutagenic, genetic and biophysical studies have demonstrated that the N-terminal extension contains a ‘proof reading’ activity to ensure that only the appropriate enzymes are selected for protein biotinylation.17, 18
Fig. 2.
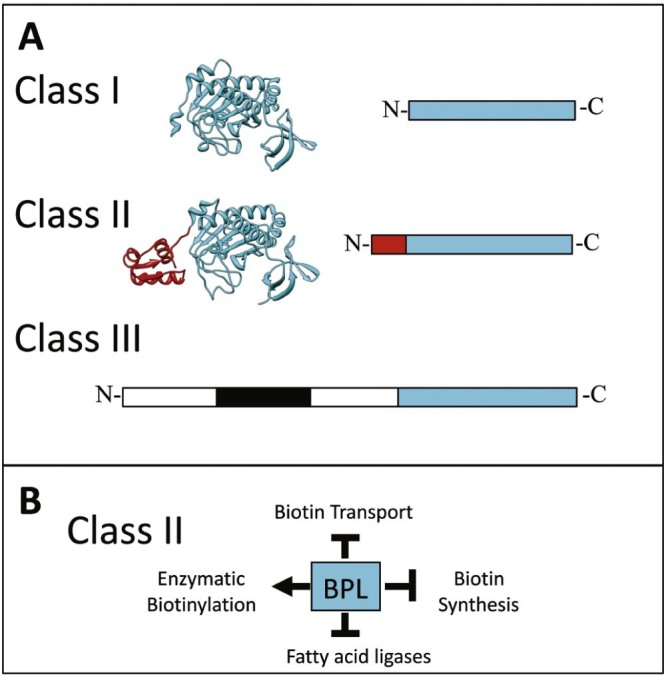
Biotin Protein Ligase. (A) The relative sizes of the three structural classes of BPLs are shown. The conserved catalytic region is depicted in blue, the DNA binding domain of Class II enzymes in red and the proof reading domain in human BPL is boxed black.17, 18 The structures of BPLs from M. tuberculosis [PDB 3RUX19] and E. coli [PDB 2EWN20] are highlighted. (B) Schematic overview showing the single protein model of protein biotinylation and transcriptional regulation in Class II BPLs.
3. Structural biology delineates the bifunctional activities of class II BPLs
The X-ray crystal structures of bifunctional Class II BPLs from E. coli and S. aureus reveal that the proteins are composed of three discrete domains. The N-terminal domain is a winged helix-turn-helix motif required for DNA binding. The central and C-terminal domains form the catalytic core of the enzyme that share high sequence homology with BPLs across all three kingdoms of life. The central domain of BirA is composed of α-helices and β-strands whereas the C-terminal domain is composed of antiparallel β-sheets.31 Although the function of the C-terminal domain has not been elucidated, it is believed that this region of the enzyme contributes to the binding of the BCCP substrate.32 The catalytic site of the enzyme is located in the central domain, where biotinyl-5′-AMP synthesis and biotinyl-transfer occur.33 Structural biology has provided further insights into the BPL catalyzed reaction. The adenylation of biotin proceeds in a sequential manner whereby biotin binds first to the enzyme followed by ATP, and its subsequent hydrolysis produces biotinyl-5′-AMP.20 Comparisons of the crystal structures of EcBirA in its unliganded (ie apo) and holo forms show that important conformational changes accompany ligand binding. Here the biotin-binding loop (amino acids 110–128) undergoes a disordered-to-ordered transition that closes over the biotin-binding pocket. This conformational change positions the side chain of a key tryptophan residue (Trp123) in the active site necessary for π–π stacking interactions with the adenylate moiety of ATP.20 Amino acids within this loop are also required for protein dimerization and, consequently, DNA binding.
Surprisingly, the N-terminal helix-turn-helix domain of EcBirA is required for both catalytic function and DNA binding. Removal of the first 64 amino acids (Δ1–64) resulted in a truncated enzyme that had reduced affinity for biotin and biotinyl-5′-AMP.34 Hence, long-range interactions through the protein are believed to help stabilize the conformational changes associated with ligand binding. This observation was supported by recent studies by Chakravartty and Cronan that showed an E. coli ‘delta wing’ (Δ48–61) mutant strain resulted in the accumulation of ADP due to the hydrolysis of ATP.35 These cells exhibited slow growth under low biotin conditions. When the DNA binding domain from an unrelated protein, OmpR, was fused onto the N-terminus of EcBirA the chimeric protein restored growth of the E. coli delta wing strain in minimal media containing low biotin concentrations. The accumulation of ADP was also no longer observed. The authors propose that the wing in the helix-turn-helix structure is needed to stabilize the biotin-binding loop.35 Interestingly, in a recent study on the BirA from B. subtilis, deletion of the N-terminal region did not compromise enzyme activity in vitro36 suggesting that the role of the N-terminus in assisting catalysis is not conserved among all Class II BPLs. Indeed the recombinant expression of truncated B. subtilis BirA was able to complement a strain of E. coli expressing the N-terminally deleted EcBirA (Δ1–64).36
The N-terminal DNA-binding domains of both E. coli and S. aureus BirAs recognize specific palindromic sequences present in the operator site, bioO, upstream of the biotin biosynthesis operon. Bioinformatics analysis predicted that SaBirA is also responsible for regulating expression of the biotin transporter bioY and fatty acid biosynthetic enzymes yhfT and yhfS in S. aureus (summarized in Fig. 2B).9 The difference in target gene regulation between E. coli and S. aureus suggests that SaBirA is solely responsible for maintaining biotin levels within the bacteria by regulating expression of both biotin synthesis and transport proteins, as well as contributing to fatty acid synthesis through the transcriptional regulation of yhfT and yhfS and activation of acetyl CoA carboxylase. This is in contrast to E. coli where the BirA recognition sequence is only present in the promoter of the biotin biosynthesis operon. Consequently, we propose that BirA regulated gene expression is potentially more responsive to environmental stimuli in S. aureus than the bacterial model E. coli.
4. BirA dimerization is intimately linked to DNA binding
Homodimerization of EcBirA is a prerequisite for DNA binding. The binding of BirA to bioO is a co-operative event involving two BPL subunits and two bioO operator half-sites (Fig. 1).37, 38 The more stable the homodimer, the greater the affinity for DNA.39, 40 Mutation of amino acids that reside in the dimer interface of BirA results in loss of DNA binding activity.41 Sedimentation equilibrium studies have revealed the dimerization constant (KD) of apo BirA is greater than 1 mM39 and, thus, apo BirA is not likely to dimerize at the physiological concentrations present inside the bacterial cell which have been estimated at <10 molecules per cell.42 Similarly, biotin-bound BirA exhibits weak dimerization with a KD of 0.9 mM.39 In contrast, biotinyl-5′-AMP enhances dimerization free energy by −4.0 kcal/mol yielding a KD of 1 – 10 µM,39, 43, 44 suggesting that the co-repressor acts as an allosteric activator to dimer assembly and DNA binding.33
The crystal structures of both EcBirA and SaBirA show the dimers assemble in a side-by-side anti-parallel arrangement such that the two N-terminal HTH motifs are aligned for DNA binding.45, 46 An X-ray structure of any Class II BPL in complex with DNA has not yet been reported. However, molecular modeling studies propose that the N-terminal domain from one subunit of EcBirA binds to the major groove of the double helix while the other subunit binds to the minor groove.47 Mutation of amino acids Ser-32, Arg-33 and Ala-34 in the DNA-binding α-helices abolishes DNA binding and results in loss of repression activity.48 In E. coli, two face-to-face promoters drive expression of the bio operon. The recognition sequence for EcBirA (bioO) is an inverted repeat that is located in between the two promoters, at the −35 and −10 sites of the operator sequence (Fig. 1). Circular permutation analysis suggests that the double stranded DNA might be bent when in complex with EcBirA.47 On the other hand, small angle X-ray scattering analysis performed on the SaBirA:SabioO complex proposed that the DNA does not bend for this species.46 Hence, the footprint observed on the DNA is likely smaller for SaBirA than EcBirA.
5. Co-repressor induces BPL dimerization
Upon binding of the biotinyl-5′-AMP co-repressor, five loops located within the central domain of EcBirA undergo a disorder-to-order transition, with three of these loops (amino acids 110–128, 140–146 and 193–199) located in the dimerization interface.41 When dimerization occurs, an extended intermolecular β-sheet is formed involving residues 189–195 in the central domain.49 Following the structural changes induced by biotin, the biotin-binding loop encases the co-repressor and is stabilized through a network of hydrophobic interactions50 as well as direct hydrogen bonding interactions involving R118.49 Binding of the co-repressor leads to the ordering of the ATP-binding loop (residues 212–223) and better packing of the biotin-binding loop, which supports bonding interactions that stabilizes the dimer.20 Therefore, the biotin-binding loop must fold before dimerization.49 Direct interactions between the two monomers involve amino acids found in the loops, namely R118, R119 and D197.49
A recent study involving random mutagenesis to generate super-repressor mutants in E. coli identified an amino acid substitution with stronger DNA-binding to bioO than wildtype, namely G154 to aspartate.40 Interestingly, this amino acid is neither located in the helix-turn-helix motif nor the dimerization interface. This suggests that other residues within the central domain but located outside of the dimer interface can participate in long-range interactions that stabilize the dimer, resulting in tighter binding to DNA. No structural data for this mutant has yet been reported to fully understand these long-range bonding interactions. Likewise, a recent study focused on G142 that is present in the dimer interface but that does not directly contribute to dimerization.51 Substitution of G142 with alanine altered the structure of the 140–146 loop, and this in turn prevented the 193–199 loop from undergoing the disordered-to-ordered transition through long-range interactions. Together these studies highlight the importance of long-range allosteric interactions on dimerization and DNA binding.
In SaBirA, ligand binding induces similar conformational changes in the loops that are located at the dimerization interface. Like EcBirA, the biotin-binding loop in this interface (residues 118–129) undergoes a disorder-to-order transition to facilitate the interaction between the two-dimer subunits.46 The dimerization interfaces of biotinyl-5′-AMP bound EcBirA and SaBirA are illustrated in Fig. 3. The subunits are connected by an analogous intermolecular β-sheet interaction as observed for EcBirA, but the dimer is stabilized by additional intersubunit contacts.46 Of particular note is F123 that forms a hydrophobic interaction with the side chain of D200 from the opposing monomer (Fig. 3A).52 In EcBirA the homologous amino acid is R119 that forms a hydrogen bond with D197 on the partner subunit (Fig. 3B). Interestingly, substitution of R119 with an aromatic amino acid (R119W) has been shown to strongly disrupt homodimerization.44 Analytical ultra centrifugation studies on SaBirA revealed that replacing the F123 with either glycine or arginine abolished dimerization even in the presence of biotin, highlighting a role for this aromatic residue in homodimer assembly.52
Fig. 3.
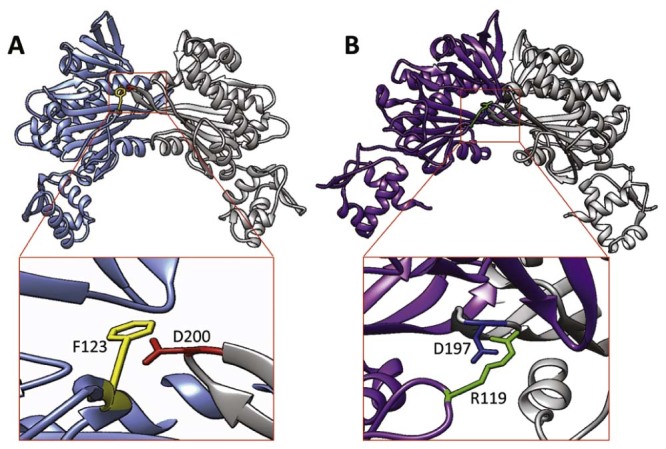
Intersubunit contacts of BirA. The structures of dimeric holo BirA from (A) S. aureus [PDB 3RIR46] and (B) E. coli [PDB 2EWN20] are shown, with one subunit colored while the other subunit is in gray ribbon. Key amino acids in the dimerization interface are highlighted.
Recent studies on SaBirA dimerization indicate that the non-liganded form of the enzyme is also able to dimerize at low concentrations with a KD of 29 ± 1.8 µM. This is a sharp contrast to apo EcBirA, which only dimerizes at millimolar concentrations.39, 43, 44 Apo-SaBirA dimer was also shown to bind DNA in an electrophoretic mobility shift assay with KD = 649 ± 43 nM, which is only 6-fold weaker than the binding of the holo-enzyme (KD = 108 ± 6.0 nM).52 Hence, allosteric regulation of the BirA switch in S. aureus may be more complex than originally thought. These data highlight key differences between E. coli and S. aureus BirAs that may have important physiological consequences that impact the bacteria's ability to sense their surroundings and adapt to the niche microenvironments they inhabit.
6. Switching between enzymatic and repressor functions of BPL
Both the biotin ligase and transcriptional repressor activities of BPL are critical for cell metabolism and survival. Therefore, controlling the switch between these two mutually exclusive functions is likely important for virulence. This raises a key question; how does the enzyme switch between enzymatic and DNA binding modes? One hypothesis proposed by Weaver and co-workers53 is based on the observation that EcBirA utilizes the same β sheet for both homodimerization and the interaction with the substrate BCCP (Fig. 4). A co-complex of the enzyme with BCCP has not yet been crystallized with a Class II BPL, but has been achieved with the Class I enzyme from P. horikoshii.21 The model proposes that when there is an excess of non-biotinylated substrate, holo-BirA will preferentially bind BCCP, thereby preventing BirA homodimerization. Alternatively, when the concentration of BCCP is low, holo-BirA will accumulate and homodimerize, leading to DNA binding and subsequent repression of transcription. Therefore, in this model, the regulatory switch between the enzymatic and transcriptional repressor functions of BirA is governed by competing protein:protein interactions and the intracellular concentration of non-biotinylated BCCP.46, 53 Recent DNase I footprint studies performed in the presence of BCCP provide support for this model.55
Fig. 4.
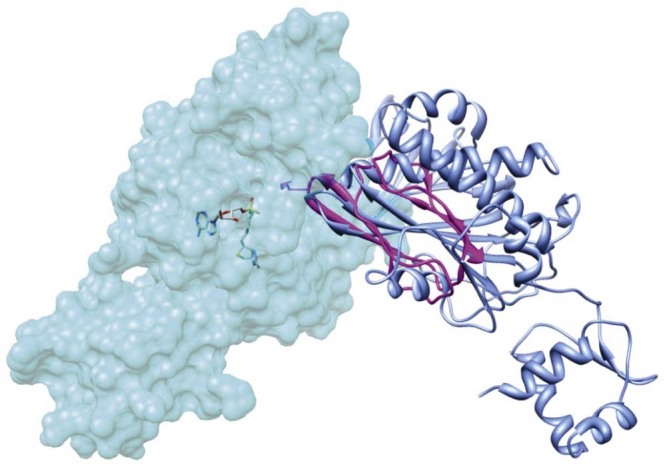
Competing protein:protein interactions. The structure of holo E. coli BirA is shown with one subunit in space filled mode and the other in blue ribbon [PDB 2EWN20]. The BCCP substrate bound to BirA (pink ribbon) has been modeled using the BPL:BCCP complex from P. horikoshii [PDB 2EJG32] with UCSF Chimera software.54
In contrast, Solbiati and Cronan56 proposed an alternative mechanism whereby biotinyl-5′-AMP is the key regulator of alternative protein:protein interactions. These authors argue that in order to compete with homodimerization, the BirA:BCCP complex must be strong and long lived in the cell. However, the enzyme-substrate interaction is believed to be transient as there has been no evidence to support a stable BirA:BCCP complex. Furthermore, these authors showed that a small 14 amino acid synthetic peptide was effective at de-repressing a EcBirA regulated reporter construct in vivo.56 Given the small size of this biotin-accepting substrate, extensive protein–protein interactions were not required to effectively disrupt EcBirA binding to DNA. The authors propose that the regulatory switch between the two functions is, therefore, the removal of the biotinyl-5′-AMP co-repressor from the active site of EcBirA rather than the competing protein-protein interactions.
A study conducted by Pendini and colleagues suggests that the level of the apo-BCCP is likely to be the key switch between the two mutually exclusive functions, at least in S. aureus.46 Small-angle X-ray scattering data showed that in the absence of BCCP, SaBPL formed a homodimer that was receptive to binding DNA. However, when apo-BCCP was included in the same reaction mixture, DNA binding activity was disrupted. These authors highlight that X-ray crystal structures demonstrated that upon the removal of biotin from the enzyme's catalytic site, the dimerization interface is destabilized by the conformational changes in the biotin-binding loop. Presumably this mechanism allows for the release of BCCP following biotinylation.
7. Regulation of biotin biosynthesis and biotin transport in organisms with class I BPL
In organisms containing Class I BPLs and no BirA homolog, such as α-proteobacteria, the transcriptional regulation of biotin biosynthesis has to be fulfilled by other proteins. A comparative genome study revealed co-localization of biotin biosynthetic genes with a recognition sequence for a GntR-type transcription factor called BioR in many α-proteobacterial genomes.57 Additionally, a protein belonging to the TetR family of transcription factors, BioQ, has recently been identified as the key player in the regulation of biotin biosynthesis in Mycobacterium smegmatis58 and biotin transport in Corynebacterium glutamicum.59 The target genes regulated by BioQ and BioR are summarized in Fig. 5A. As these two proteins have only been identified recently, they have not yet been as extensively studied as BirA. For both BioR and BioQ, the repressor function of both proteins appears to be independent of biotin, and no ligands have yet been identified for these transcription factors.58 A two-protein model involving BPL together with either BioQ or BioR has been proposed as a possible mechanism for biotin sensing and regulation in bacteria containing Class I BPLs (Fig. 5B).58 This model assumes that there is cross talk between the biotin sensor (BPL) and the transcriptional regulator (BioQ/R). However, the molecular details supporting this model still require experimental validation.
Fig. 5.
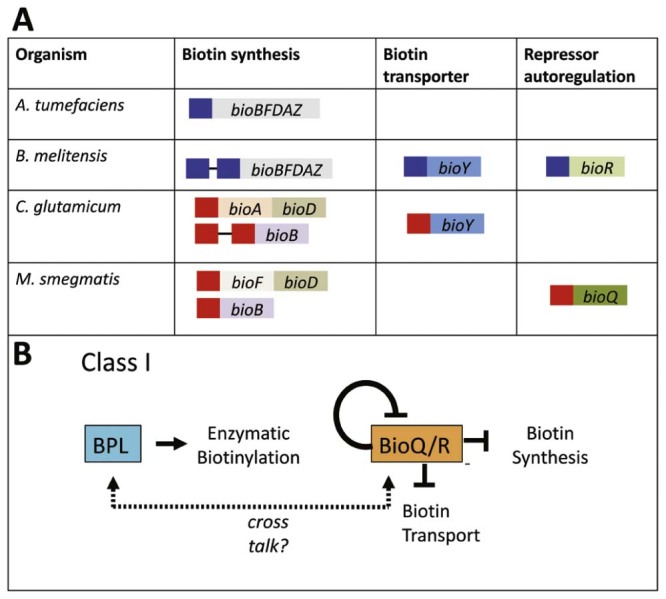
Summary of BioR and BioQ transcriptional regulation. (A) The genes and metabolic pathways regulated by BioR (blue boxes) and BioQ (red boxes) are shown. Each box represents an individual operator. (B) Schematic overview showing the two-protein model of protein biotinylation and transcriptional regulation in Class I BPLs.
8. BioR mediated gene expression
Bioinformatic analysis of genomic sequences from α-proteobacteria suggested that biotin synthesis in these bacteria is regulated by BioR.57 It was also observed that the DNA recognition sequence for BioR was found upstream of the bioY gene biotin transporter in several other α-proteobacteria such as M. loti, B. melitensis, Silicibacter sp. TM1040 and S. pomeroyi. The BioR recognition sequence also co-localized with bioR genes in certain organisms such as Mesorhizobium loti, Brucella melitensis, Bradyrizhobium japonicum, Silicibacter pmeroyi and Rhodobacder sphaeroides, suggesting auto-regulation – a feature not observed with BirA/BPL. However this auto-regulation is not completely conserved as the BioR recognition sequence was not present upstream of the bioR gene in Agrobacterium tumefaciens. Moreover, in A. tumefaciens, BioR does not control the expression of the BioY biotin transporter protein. Fig. 6 outlines the localization of BioR recognition sequence in different α-proteobacteria.
Fig. 6.
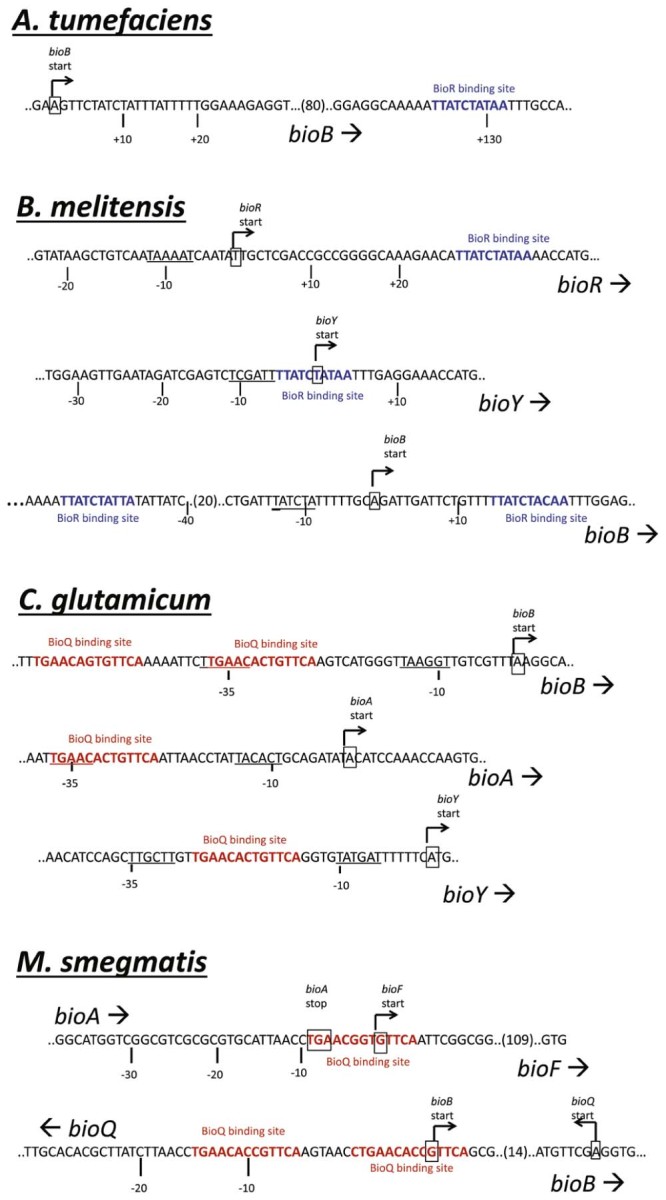
BioR and BioQ. Sequences of the binding sites for BioR and BioQ are shown. Transcription start sites are boxed, and −35 and −10 sequences are underlined. Sequences of the binding sites for BioR (blue) and BioQ (red) are colored.
Feng and co-workers further investigated the role of BioR in regulating biotin synthesis by conducting a series of electromobility gel shift assays.10 In addition to binding the recognition sequence in its own genome, BioR from A. tumefaciens was able to bind DNA probes with sequences derived from B. japonicum, R. sphaeroides and B. melitensis. In addition, the B. melitensis BioR was able to repress expression of A. tumefaciens bioB in vivo.10 These data support the hypothesis that the BioR:operator interaction is well conserved in α-proteobacteria. It also revealed that the expression of the biotin operon (bioBFDAZ) in wildtype A. tumefaciens was 10–15 fold lower relative to a BioR knockout when the bacteria were grown in high (1 µM) biotin media. This observation further validates the role of BioR as a transcriptional repressor.10
In B. melitensis, the BioR recognition sequence is located upstream of the genes encoding both BioR and the BioY biotin transporter (Fig. 6). Two BioR sites are also present in the bioBFDAZ operon, indicating a complex regulatory network of biotin metabolism involving BioR.10 Electromobility gel shift assays also confirmed that B. melitensis BioR was able to bind to a DNA probe containing the BioY promoter sequence, providing the first evidence that BioR can mediate expression of the biotin transporter in these bacteria.10 In contrast, A. tumefaciens BioR does not regulate biotin transport and only binds weakly to the recognition site present within the coding region of bioB, in vivo.10 As a consequence, the amount of biotin produced by A. tumefaciens is greater than their minimum growth requirement, which presumably is beneficial for survival in their environment.10
A search for a ligand and co-repressor of BioR concluded that biotin is not the natural ligand.10 Complete removal of biotin from the protein preparations had no effect upon DNA-binding in vitro. Conversely, the hypothesis that biotin can serve as a dissociation factor that disrupts DNA-BioR complex was also tested. Again, the addition or removal of biotin had no effect on DNA-binding activity. Various biotin metabolites were also tested and none showed evidence of being a ligand for BioR. Hence, further studies are required to define the links between biotin sensing by BPL and downstream control by BioR in the two protein model of biotin-regulated gene expression.
9. BioQ mediated gene expression
In Corynebacterium glutamicum, the biotin synthesis pathway is incomplete, thereby rendering the bacteria biotin auxotrophic. However, the expression of the bio genes is proposed to be controlled by a transcription factor belonging to the TetR protein family, namely BioQ. Although the TetR family of regulators has been well characterized, none have previously been shown to regulate biotin synthesis.60 Bioinformatic analysis of the C. glutamicum genome revealed co-localization of the bioQ coding region with the biotin biosynthetic genes (Fig. 6).59 The presence of the BioQ recognition sequence in the promoter of the bioQ gene suggests auto-regulation of the transcription factor (Fig. 6). In addition, the same regulatory sequence is also found upstream of the bioY biotin transporter, suggesting BioQ regulates its expression. This is crucial for biotin auxotrophic C. glutamicum that relies on supply of the micronutrient from the external environment. In addition, the study showed that increasing the biotin concentration in the growth media only had modest repression on the transcription of the biotin operon in this organism.59
In contrast to C. glutamicum, BioQ is believed to regulate expression of the transporter genes in Mycobacterium smegmatis. The recognition sites are also localized upstream of the bioF and bioQ/B genes suggesting regulation of biotin biosynthesis and auto-regulation by BioQ in this species (Fig. 6).58 Tang et al. also showed that BioQ binding sites are also found in other mycobacterium species such as M. abcessus, M. gilvum, M.JLS, M. massiliense, M. rhodesiae and M. vanbaalenii, but not in the clinically important human pathogen M. tuberculosis.58 The same study also confirmed the role of BioQ as a functional repressor. Increased mRNA levels of bioF, bioB and bioD were measured in a ΔbioQ mutant strain of M. smegmatis compared to the wildtype parent. LacZ-based reporter assays using the bioFD promoter also showed expression was highly increased in ΔbioQ strain whereas no LacZ activity was observed in the wildtype strain.58 In addition, increasing levels of biotin in the growth media resulted in decreasing expression of biotin biosynthesis genes bioF, bioD and bioB for the wildtype bacteria whereas there was no significant change in the ΔbioQ strain. These findings underline the biotin sensing ability of BioQ.
10. Potential biotechnological applications
Synthetic biology facilitates us to better understand life through the dissection then reconstruction of complex biological systems.61 At the heart of this endeavor are engineered genetic circuits that allow us to dissect the interplay between genes, proteins, cells and systems. Synthetic biology is currently used to deliver valuable bio-products and therapeutic molecules, such as fine chemicals, peptides, proteins and antibodies. For example, most monoclonal antibodies are produced recombinantly using genetically engineered Chinese hamster ovary cells as bio-factories.62 Many of the reagents that have been developed by industry are becoming valuable tools in academia. Underpinning synthetic biology are well-characterized transcriptional regulators required to engineer the genetic circuits and tightly control bio-production. The TetR inducible expression system is an example that is widely employed due to its high specificity toward its recognition system and the high affinity to tetracycline, a well characterized antibiotic.63 There is now a need fill our discovery toolbox with a greater variety of well-characterized transcriptional regulators with utility in systems biology. In this review, the mechanisms of BirA, BioQ and BioR in regulating their target genes are discussed. The high specificity of BirA to its target operator, as well its ability to regulate gene expression in response to external biotin, provides an attractive approach to developing novel ligand-regulated gene expression systems for use in bacteria, yeast plants and animal systems. To date this approach has not yet been exploited. Biotin provides a highly attractive ligand to regulate transcription due to its low cost, solubility in aqueous solutions, low toxicity to many cell types and has no regulatory issues. While the well-studied Class II BirA enzyme/repressor from E. coli provides one useful example for generating a controllable genetic switch, the emergence of other biotin responsive transcription factors extends our repertoire of potential systems. Other Class II BPLs may be more responsive to environmental biotin levels, such as that from S. aureus. The BioR and BioQ proteins provide alternative repressors with distinctive mechanisms of action to BirA/BPL. These may potentially be advantageous when generating new genetic circuits that are highly responsive to external stimuli, such as the addition of a vitamin. Further research on biotin regulated transcriptional factors promise to replenish our toolkit with greater variety of new agents for systems biology.
Acknowledgements
This work was supported by the National Health and Medical Research Council of Australia (application APP1068885) and the Centre for Molecular Pathology, University of Adelaide. We are grateful to the Carthew and Wallace families for their financial support of this work.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Polyak S.W., Chapman-Smith A. Biotin. In: Lennarz W.J., Lane M.D., editors. Encyclopeaedia of biological chemistry. Academic Press; San Diego (CA): 2013. pp. 221–225. [Google Scholar]
- 2.Cronan J.E., Jr, Waldrop G.L. Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res. 2002;41:407–435. doi: 10.1016/s0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- 3.Polyak S.W., Abell A.D., Wilce M.C., Zhang L., Booker G.W. Structure, function and selective inhibition of bacterial acetyl-coa carboxylase. Appl Microbiol Biotechnol. 2012;93:983–992. doi: 10.1007/s00253-011-3796-z. [DOI] [PubMed] [Google Scholar]
- 4.Jitrapakdee S., St Maurice M., Rayment I., Cleland W.W., Wallace J.C., Attwood P.V. Structure, mechanism and regulation of pyruvate carboxylase. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deodato F., Boenzi S., Santorelli F.M., Dionisi-Vici C. Methylmalonic and propionic aciduria. Am J Med Genet C Semin Med Genet. 2006;142C:104–112. doi: 10.1002/ajmg.c.30090. [DOI] [PubMed] [Google Scholar]
- 6.Polyak S.W., Bailey L.M., Azhar A., Booker G.W. Biotin (Vitamin H or B7) In: Betancourt A.I., Gaitan H.F., editors. Micronutrients: sources, properties and health benefits. Nova Publishers; New York: 2012. pp. 65–93. [Google Scholar]
- 7.Salaemae W., Azhar A., Booker G.W., Polyak S.W. Biotin biosynthesis in Mycobacterium tuberculosis: physiology, biochemistry and molecular intervention. Protein Cell. 2011;2:691–695. doi: 10.1007/s13238-011-1100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S., Cronan J.E. Closing in on complete pathways of biotin biosynthesis. Mol Biosyst. 2011;7:1811–1821. doi: 10.1039/c1mb05022b. [DOI] [PubMed] [Google Scholar]
- 9.Rodionov D.A., Mironov A.A., Gelfand M.S. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Y., Zhang H., Cronan J.E. Profligate biotin synthesis in alpha-proteobacteria – a developing or degenerating regulatory system? Mol Microbiol. 2013;88:77–92. doi: 10.1111/mmi.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman-Smith A., Morris T.W., Wallace J.C., Cronan J.E., Jr Molecular recognition in a post-translational modification of exceptional specificity. Mutants of the biotinylated domain of acetyl-CoA carboxylase defective in recognition by biotin protein ligase. J Biol Chem. 1999;274:1449–1457. doi: 10.1074/jbc.274.3.1449. [DOI] [PubMed] [Google Scholar]
- 12.Chapman-Smith A., Cronan J.E., Jr The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem Sci. 1999;24:359–363. doi: 10.1016/s0968-0004(99)01438-3. [DOI] [PubMed] [Google Scholar]
- 13.Cronan J.E., Jr, Wallace J.C. The gene encoding the biotin-apoprotein ligase of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1995;130:221–229. doi: 10.1111/j.1574-6968.1995.tb07724.x. [DOI] [PubMed] [Google Scholar]
- 14.Leon-Del-Rio A., Leclerc D., Akerman B., Wakamatsu N., Gravel R.A. Isolation of a cDNA encoding human holocarboxylase synthetase by functional complementation of a biotin auxotroph of Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:4626–4630. doi: 10.1073/pnas.92.10.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polyak S.W., Chapman-Smith A., Mulhern T.D., Cronan J.E., Jr, Wallace J.C. Mutational analysis of protein substrate presentation in the post-translational attachment of biotin to biotin domains. J Biol Chem. 2001;276:3037–3045. doi: 10.1074/jbc.M003968200. [DOI] [PubMed] [Google Scholar]
- 16.Pendini N.R., Bailey L.M., Booker G.W., Wilce M.C., Wallace J.C., Polyak S.W. Microbial biotin protein ligases aid in understanding holocarboxylase synthetase deficiency. Biochim Biophys Acta. 2008;1784:973–982. doi: 10.1016/j.bbapap.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 17.Mayende L., Swift R.D., Bailey L.M., Soares da Costa T.P., Wallace J.C., Booker G.W. A novel molecular mechanism to explain biotin-unresponsive holocarboxylase synthetase deficiency. J Mol Med. 2012;90:81–88. doi: 10.1007/s00109-011-0811-x. [DOI] [PubMed] [Google Scholar]
- 18.Campeau E., Gravel R.A. Expression in Escherichia coli of N- and C-terminally deleted human holocarboxylase synthetase. Influence of the N-terminus on biotinylation and identification of a minimum functional protein. J Biol Chem. 2001;276:12310–12316. doi: 10.1074/jbc.M009717200. [DOI] [PubMed] [Google Scholar]
- 19.Duckworth B.P., Geders T.W., Tiwari D., Boshoff H.I., Sibbald P.A., Barry C.E., 3rd Bisubstrate adenylation inhibitors of biotin protein ligase from Mycobacterium tuberculosis. Chem Biol. 2011;18:1432–1441. doi: 10.1016/j.chembiol.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Wood Z., Weaver L.H., Brown P.H., Beckett D., Matthews B.W. Co-repressor induced order and biotin repressor dimerization: a case for divergent followed by convergent evolution. J Mol Biol. 2006;357:509–523. doi: 10.1016/j.jmb.2005.12.066. [DOI] [PubMed] [Google Scholar]
- 21.Bagautdinov B., Kuroishi C., Sugahara M., Kunishima N. Crystal structures of biotin protein ligase from Pyrococcus horikoshii OT3 and its complexes: structural basis of biotin activation. J Mol Biol. 2005;353:322–333. doi: 10.1016/j.jmb.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 22.Beckett D. Biotin sensing at the molecular level. J Nutr. 2009;139:167–170. doi: 10.3945/jn.108.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beckett D. Regulating transcription regulators via allostery and flexibility. Proc Natl Acad Sci U S A. 2009;106:22035–22036. doi: 10.1073/pnas.0912300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapman-Smith A., Turner D.L., Cronan J.E., Jr, Morris T.W., Wallace J.C. Expression, biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier protein of Escherichia coli acetyl-CoA carboxylase. Biochem J. 1994;302(Pt 3):881–887. doi: 10.1042/bj3020881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsyth R.A., Haselbeck R.J., Ohlsen K.L., Yamamoto R.T., Xu H., Trawick J.D. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 26.Payne D.J., Gwynn M.N., Holmes D.J., Pompliano D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 27.Paparella A.S., Soares da Costa T.P., Yap M.Y., Tieu W., Wilce M.C., Booker G.W. Structure guided design of biotin protein ligase inhibitors for antibiotic discovery. Curr Top Med Chem. 2014;14:4–20. doi: 10.2174/1568026613666131111103149. [DOI] [PubMed] [Google Scholar]
- 28.Soares da Costa T.P., Tieu W., Yap M.Y., Pendini N.R., Polyak S.W., Sejer Pedersen D. Selective inhibition of biotin protein ligase from Staphylococcus aureus. J Biol Chem. 2012;287:17823–17832. doi: 10.1074/jbc.M112.356576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tieu W., Soares da Costa T.P., Yap M.Y., Keeling K.L., Wilce M.C.J., Wallace J.C. Optimising in situ click chemistry: the screening and identification of biotin protein ligase inhibitors. Chem Sci. 2013;4:3533–3537. [Google Scholar]
- 30.Feng Y., Chin C.Y., Chakravartty V., Gao R., Crispell E.K., Weiss D.S. The atypical occurrence of two biotin protein ligases in Francisella novicida is due to distinct roles in virulence and biotin metabolism. MBio. 2015;6 doi: 10.1128/mBio.00591-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson K.P., Shewchuk L.M., Brennan R.G., Otsuka A.J., Matthews B.W. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci U S A. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagautdinov B., Matsuura Y., Bagautdinova S., Kunishima N. Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate. J Biol Chem. 2008;283:14739–14750. doi: 10.1074/jbc.M709116200. [DOI] [PubMed] [Google Scholar]
- 33.Eisenstein E., Beckett D. Dimerization of the Escherichia coli biotin repressor: corepressor function in protein assembly. Biochemistry. 1999;38:13077–13084. doi: 10.1021/bi991241q. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y., Beckett D. Evidence for interdomain interaction in the Escherichia coli repressor of biotin biosynthesis from studies of an N-terminal domain deletion mutant. Biochemistry. 1996;35:1783–1792. doi: 10.1021/bi952269e. [DOI] [PubMed] [Google Scholar]
- 35.Chakravartty V., Cronan J.E. The wing of a winged helix-turn-helix transcription factor organizes the active site of BirA, a bifunctional repressor/ligase. J Biol Chem. 2013;288:36029–36039. doi: 10.1074/jbc.M113.525618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henke S.K., Cronan J.E. Successful conversion of the Bacillus subtilis BirA Group II biotin protein ligase into a Group I ligase. PLoS ONE. 2014;9:e96757. doi: 10.1371/journal.pone.0096757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbott J., Beckett D. Cooperative binding of the Escherichia coli repressor of biotin biosynthesis to the biotin operator sequence. Biochemistry. 1993;32:9649–9656. doi: 10.1021/bi00088a017. [DOI] [PubMed] [Google Scholar]
- 38.Streaker E.D., Beckett D. Coupling of site-specific DNA binding to protein dimerization in assembly of the biotin repressor-biotin operator complex. Biochemistry. 1998;37:3210–3219. doi: 10.1021/bi9715019. [DOI] [PubMed] [Google Scholar]
- 39.Streaker E.D., Gupta A., Beckett D. The biotin repressor: thermodynamic coupling of corepressor binding, protein assembly, and sequence-specific DNA binding. Biochemistry. 2002;41:14263–14271. doi: 10.1021/bi0203839. [DOI] [PubMed] [Google Scholar]
- 40.Chakravartty V., Cronan J.E. Altered regulation of Escherichia coli biotin biosynthesis in BirA superrepressor mutant strains. J Bacteriol. 2012;194:1113–1126. doi: 10.1128/JB.06549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon K., Streaker E.D., Ruparelia S., Beckett D. Multiple disordered loops function in corepressor-induced dimerization of the biotin repressor. J Mol Biol. 2000;304:821–833. doi: 10.1006/jmbi.2000.4249. [DOI] [PubMed] [Google Scholar]
- 42.Taniguchi Y., Choi P.J., Li G.W., Chen H., Babu M., Hearn J. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao H., Beckett D. Kinetic partitioning between alternative protein-protein interactions controls a transcriptional switch. J Mol Biol. 2008;380:223–236. doi: 10.1016/j.jmb.2008.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao H., Naganathan S., Beckett D. Thermodynamic and structural investigation of bispecificity in protein-protein interactions. J Mol Biol. 2009;389:336–348. doi: 10.1016/j.jmb.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon K., Beckett D. Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Sci. 2000;9:1530–1539. doi: 10.1110/ps.9.8.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pendini N.R., Yap M.Y., Polyak S.W., Cowieson N.P., Abell A., Booker G.W. Structural characterization of Staphylococcus aureus biotin protein ligase and interaction partners: an antibiotic target. Protein Sci. 2013;22:762–773. doi: 10.1002/pro.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Streaker E.D., Beckett D. A map of the biotin repressor-biotin operator interface: binding of a winged helix-turn-helix protein dimer to a forty base-pair site. J Mol Biol. 1998;278:787–800. doi: 10.1006/jmbi.1998.1733. [DOI] [PubMed] [Google Scholar]
- 48.Buoncristiani M.R., Howard P.K., Otsuka A.J. DNA-binding and enzymatic domains of the bifunctional biotin operon repressor (BirA) of Escherichia coli. Gene. 1986;44:255–261. doi: 10.1016/0378-1119(86)90189-7. [DOI] [PubMed] [Google Scholar]
- 49.Weaver L.H., Kwon K., Beckett D., Matthews B.W. Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator. Proc Natl Acad Sci U S A. 2001;98:6045–6050. doi: 10.1073/pnas.111128198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eginton C., Naganathan S., Beckett D. Sequence-function relationships in folding upon binding. Protein Sci. 2015;24:200–211. doi: 10.1002/pro.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eginton C., Cressman W.J., Bachas S., Wade H., Beckett D. Allosteric coupling via distant disorder-to-order transitions. J Mol Biol. 2015;427:1695–1704. doi: 10.1016/j.jmb.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 52.Soares da Costa T.P., Yap M.Y., Perugini M.A., Wallace J.C., Abell A.D., Wilce M.C. Dual roles of F123 in protein homodimerization and inhibitor binding to biotin protein ligase from Staphylococcus aureus. Mol Microbiol. 2014;91:110–120. doi: 10.1111/mmi.12446. [DOI] [PubMed] [Google Scholar]
- 53.Weaver L.H., Kwon K., Beckett D., Matthews B.W. Competing protein:protein interactions are proposed to control the biological switch of the E coli biotin repressor. Protein Sci. 2001;10:2618–2622. doi: 10.1110/ps.32701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Adikaram P.R., Beckett D. Protein:protein interactions in control of a transcriptional switch. J Mol Biol. 2013;425:4584–4594. doi: 10.1016/j.jmb.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solbiati J., Cronan J.E. The switch regulating transcription of the Escherichia coli biotin operon does not require extensive protein-protein interactions. Chem Biol. 2010;17:11–17. doi: 10.1016/j.chembiol.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodionov D.A., Gelfand M.S. Computational identification of BioR, a transcriptional regulator of biotin metabolism in Alphaproteobacteria, and of its binding signal. FEMS Microbiol Lett. 2006;255:102–107. doi: 10.1111/j.1574-6968.2005.00070.x. [DOI] [PubMed] [Google Scholar]
- 58.Tang Q., Li X., Zou T., Zhang H., Wang Y., Gao R. Mycobacterium smegmatis BioQ defines a new regulatory network for biotin metabolism. Mol Microbiol. 2014 doi: 10.1111/mmi.12817. [DOI] [PubMed] [Google Scholar]
- 59.Brune I., Gotker S., Schneider J., Rodionov D.A., Tauch A. Negative transcriptional control of biotin metabolism genes by the TetR-type regulator BioQ in biotin-auxotrophic Corynebacterium glutamicum ATCC 13032. J Biotechnol. 2012;159:225–234. doi: 10.1016/j.jbiotec.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 60.Ramos J.L., Martinez-Bueno M., Molina-Henares A.J., Teran W., Watanabe K., Zhang X. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobrin A., Saxena P., Fussenegger M. Synthetic biology: applying biological circuits beyond novel therapies. Integr Biol (Camb) 2016 doi: 10.1039/c5ib00263j. [DOI] [PubMed] [Google Scholar]
- 62.Brown A.J., James D.C. Precision control of recombinant gene transcription for CHO cell synthetic biology. Biotech Adv. 2015 doi: 10.1016/j.biotechadv.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 63.Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]


