Abstract
Thailanstatin A (TST-A) is a potent antiproliferative natural product discovered by our group from Burkholderia thailandensis MSMB43 through a genome-guided approach. The limited supply of TST-A, due to its low titer in bacterial fermentation, modest stability and very low recovery rate during purification, has hindered the investigations of TST-A as an anticancer drug candidate. Here we report the significant yield improvement of TST-A and its direct precursor, thailanstatin D (TST-D), through metabolic engineering of the thailanstatin biosynthetic pathway in MSMB43. Deletion of tstP, which encodes a dioxygenase involved in converting TST-A to downstream products including FR901464 (FR), resulted in 58% increase of the TST-A titer to 144.7 ± 2.3 mg/L and 132% increase of the TST-D titer to 14.6 ± 0.5 mg/L in the fermentation broth, respectively. Deletion of tstR, which encodes a cytochrome P450 involved in converting TST-D to TST-A, resulted in more than 7-fold increase of the TST-D titer to 53.2 ± 12.1 mg/L in the fermentation broth. An execution of 90 L pilot-scale fed-batch fermentation of the tstP deletion mutant in a 120-L fermentor led to the preparation of 714 mg of TST-A with greater than 98.5% purity. The half-life of TST-D in a phosphate buffer was found to be at least 202 h, significantly longer than that of TST-A or FR, suggesting superior stability. However, the IC50 values of TST-D against representative human cancer cell lines were determined to be greater than those of TST-A, indicating weaker antiproliferative activity. This work enabled us to prepare sufficient quantities of TST-A and TST-D for our ongoing translational research.
Keywords: Fermentation, Metabolic engineering, Natural product, Production, Thailanstatin
1. Introduction
Cancer has the second highest mortality rate only next to cardiovascular diseases and is usually associated with specific genetic and epigenetic changes in cellular processes.1, 2, 3 Pre-mRNA splicing inhibitors, including FR901464 (FR; Fig. 1),4, 5, 6 spliceostatins,7, 8 thailanstatins,9 pladienolides,10 FD-895,11 and herboxidiene (GEX1),12, 13, 14 are a promising class of bacterial natural products or their derivatives for cancer drug discovery and development. FR demonstrated not only potent antiproliferative activity against an array of human cancer cell lines with IC50 values in the low nanomolar (nM) range but also the ability to prolong the life of tumor-bearing mice.5 However, due to its instability as well as unacceptable levels of toxicity, FR has been abandoned from a drug development program.15 Mechanistically FR and spliceostatin A (SSA) inhibit pre-mRNA splicing by binding to SF3b, a subcomplex of the U2 small nuclear ribonucleoprotein in the spliceosome.16
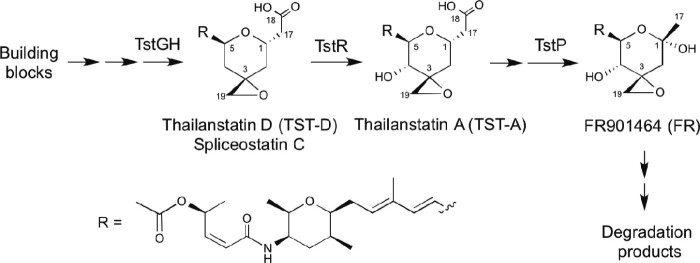
Fig. 1.
Structures and biosynthetic relationship of TST-D, TST-A and FR901464.
FR was discovered by Nakajima et al. from Pseudomonas sp. No. 2663 (recently re-classified as Burkholderia sp. FERM BP-342117) through cell-based screenings;4 thailanstatin A (TST-A; Fig. 1) was discovered by us from Burkholderia thailandensis MSMB43 through genome mining.9 TST-A biosynthesis in MSMB43 and FR biosynthesis in FERM BP-3421 appear to use the same biosynthetic logic,9, 17, 18 raising the possibility that these two strains are either identical or very closely related. Three oxygenase activities, including a flavin-dependent monooxygenase (FMO) domain encoded by fr9H/tstGH, a cytochrome P450 encoded by fr9R/tstR and a Fe(II)/α-ketoglutarate-dependent dioxygenase encoded by fr9P/tstP, were proposed in the formation of a heavily decorated tetrahydropyran ring (Fig. 1). Specifically, the Fr9P/TstP dioxygenase was shown to convert a C1 acetic acid group in TST-A into a hemiketal group in FR through oxidative decarboxylation. As such, TST-A is a precursor of FR. The carboxylic acid moiety in TST-A is beneficial for drug development for the following reasons. First, this carboxylic acid moiety enables TST-A to be more stable than the hemiketal-containing FR.9 Second, when the carboxylic acid group of TST-A is modified to an ester linkage, which is more lipophilic and enhances the cell membrane permeability of the derived compound, the cytotoxicity of the compound is significantly increased.8
Our investigation of TST-A as an anticancer drug candidate encountered a shortage of compound supply, primarily due to its low titer in bacterial fermentation, modest stability and a very low recovery rate during purification.9 Here we report the significant yield improvement of TST-A through metabolic engineering of the thailanstatin biosynthetic pathway in MSMB43 and pilot scale fermentation. During the course of this research, a direct TST-A precursor was isolated and named as thailanstatin D in 2013 (TST-D; Fig. 1), which turned out to be identical to the recently reported spliceostatin C (SSC).17 The antiproliferative activity and stability of TST-D were assessed and reported as well.
2. Materials and methods
2.1. Strain, plasmid and media
All strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) agar or medium was used with appropriate concentration of antibiotics for routine cultivation of B. thailandensis MSMB43 strains or Escherichia coli strains. A 2S4G medium17 was used for bacterial fermentation in flasks; a slightly modified 2S4G medium composed of 40 g/L glycerol, 12.5 g/L HySoy soypeptone, 2 g/L (NH4)2SO4, 0.1 g/L MgSO4⋅7H2O and 2 g/L CaCO3 (pH 7.0) was used for fed-batch bacterial fermentation in a fermentor. The concentrated feed medium contained 400 g/L glycerol, 20 g/L (NH4)2SO4 and 1 g/L MgSO4⋅7H2O (pH 7.0).
Table 1.
Strains and plasmids used in this study.
| Strains and plasmids | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | General Escherichia coli host strain for DNA cloning | Lab stock |
| E. coli S17-1 | E. coli donor strain for interspecies conjugation | Lab stock |
| BthWT | Burkholderia thailandensis MSMB43 wild-type strain | CDC |
| BthΔtstP::FRT | B. thailandensis ΔtstP::FRT intermediate insertion mutant | This study |
| BthΔtstP | B. thailandensis ΔtstP final marker-free deletion mutant | This study |
| BthΔtstR::FRT | B. thailandensis ΔtstR::FRT intermediate insertion mutant | This study |
| BthΔtstR | B. thailandensis ΔtstR final marker-free deletion mutant | This study |
| Plasmids | ||
| pBS854-Tp | Tpr; donor of a trimethoprim-resistance cassette | 19 |
| pEX18Tc | TcroriT+sacB+; gene replacement vector with MCS | 20 |
| pBMTL3-Flp2 | Flp endonuclease expression vector | 21 |
| pEX18Tc-ΔtstP::Tp | TcrTproriT+sacB+; tstP gene replacement construct | This study |
| pEX18Tc-ΔtstR::Tp | TcrTproriT+sacB+; tstR gene replacement construct | This study |
CDC, US Centers for Disease Control and Prevention; Tpr, trimethoprim resistant; Tcr, tetracycline resistant.
2.2. Construction of gene deletion mutants of B. thailandensis MSMB43
A multiplex PCR method20 was used to create gene deletion mutant strains of MSMB43 (Fig. S1), as we routinely did in previous works.19, 22 PCR primers used for mutagenesis and for verification of mutant genotypes are listed in Table S1.
2.3. Bacterial fermentation, extraction and chromatographic purification of TST-A and TST-D
Bacterial fermentation, extraction, isolation and purification were performed according to schemes and associated description given in the Supplementary material (Figs. S2 and S3). TST-D was identified to be identical to spliceostatin C (SSC)17 with extensive UV, IR, HR-MS and NMR analyses (Figs. S4–S15).
2.4. Titer determination by LC–MS analysis
The titers of TST-A and TST-D in fermentation broth were quantified with an Agilent 6400 Series Triple Quadrupole LC–MS system equipped with an Agilent Eclipse Plus C18 column (φ 4.6 × 100 mm, 3.5 µm) and a UV detector. Briefly, each 0.5 mL of fermentation broth was sampled at various time points and was extracted twice with equal volume of ethyl acetate. Two extracts were combined, dried in a refrigerated CentriVap centrifugal vacuum concentrator (Labconco) and subsequently re-suspended in 0.5 mL of acetonitrile and filtered through a 0.22 µm filter. Two microliters of such acetonitrile solution was injected into the LC–MS system. The LC solvents included buffer A (water with 0.1% formic acid, FA) and buffer B (acetonitrile with 0.1% FA). The column was eluted with a linear gradient from 15% to 55% buffer B in 35 min, monitored at 235 nm and with a flow rate of 0.5 mL/min. MS signals were collected in positive mode under the following conditions: N2 gas temperature, 325 °C; gas flow, 10 L/min; nebulizer pressure, 20 psi; sheath gas temperature, 400 °C; sheath N2 gas flow, 12 L/min; capillary voltage, 4000 V; nozzle voltage, 500 V. TST-A and TST-D were eluted at 24 min and 31 min, respectively. The extracted ion species [M + H]+ for TST-A and TST-D were 536 m/z and 520 m/z, respectively. For the generation of standard curves, TST-A and TST-D were separately dissolved in phosphate buffer saline (PBS, pH 7.4) at a concentration series of 0.02, 0.01, 0.005, 0.0025, 0.00125, 0.000625 and 0.0003125 mg/mL. Then 0.5 mL of each sample of TST-A or TST-D was extracted twice with equal volume of ethyl acetate. The extracts were combined, tried under vacuum and re-suspended in 0.5 mL of acetonitrile, and 2 µL of which was applied into the LC–MS system. A standard curve was created by plotting the compound concentrations against the extracted ion peak areas and showed linearity within the range of concentrations tested (Figs. S16 and S17). The concentration of a sample was calculated using the equation generated from the standard curve.
For simple identification of TST-A and TST-D, the analysis was performed under the same LC–MS condition but with a different elution procedure: 0–9 min, from 15% to 100% buffer B; 9–12 min, in 100% buffer B; 12–13 min, from 100% to 15% buffer B; 13–18 min, in 15% buffer B. TST-A and TST-D were eluted at 8.2 and 9.2 min, respectively.
2.5. Stability and cytotoxicity assays
The stability and cytotoxicity of TST-D were assayed according to our previously described methods9 to determine the IC50 value and half-life (t1/2), using FR901464 as a reference.
3. Results
3.1. Targeted gene deletion
Marker exchange via double crossover of a 717-bp internal region of tstP in MSMB43 with the FRT-Tp-FRT cassette from pEX18Tc-ΔtstP::Tp resulted in an intermediate mutant strain BthΔtstP::FRT; subsequent marker removal by a site-specific Flp endonuclease overexpressed from pBMTL3-Flp2 resulted in the final marker-free BthΔtstP mutant strain. Similarly, a 933-bp internal region of tstR was deleted to generate BthΔtstR::FRT and BthΔtstR mutant strains (Table 1). Those genetic events were all verified by PCR diagnosis (Fig. S1).
3.2. Significant improvement of the production of TST-A and TST-D
Quantitative LC–MS analysis of samples from a 4-day flask fermentation showed that the titers of TST-A and TST-D produced by BthΔtstP strain increased 58% to 144.7 ± 2.3 mg/L and 132.0% to 14.6 ± 0.5 mg/L, respectively, and the titer of TST-D produced by BthΔtstR strain increased more than 7-fold to 53.2 ± 12.1 mg/L, all compared to the titers produced by BthWT strain. Meanwhile, deletion of tstP abolished the production of FR and deletion of tstR abolished the production of both TST-A and FR (Fig. 2).
Fig. 2.
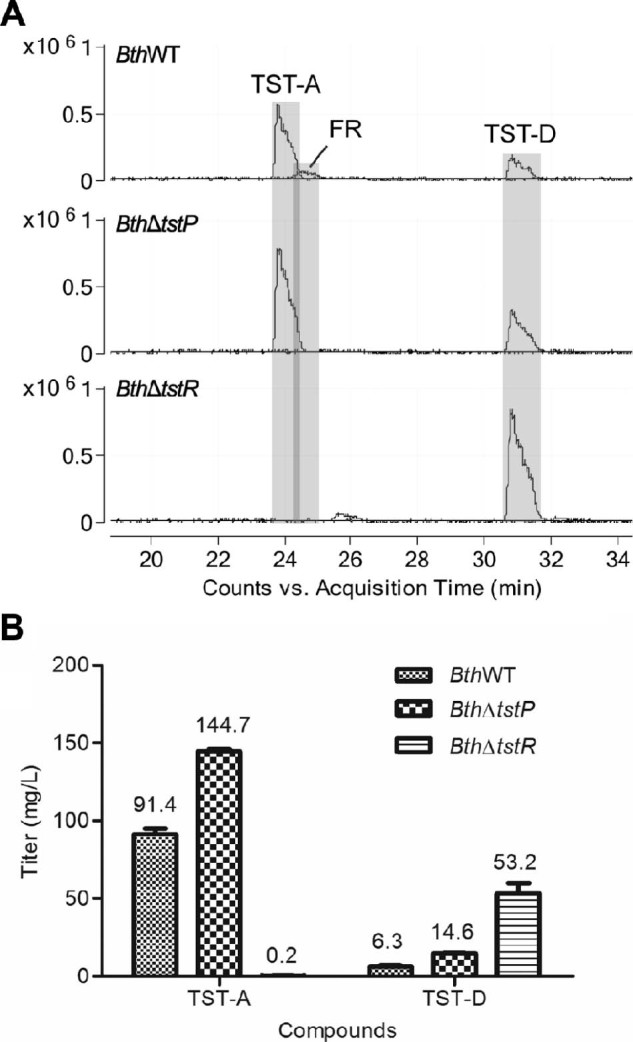
Detection (A) and quantification of the titers (B) of TST-A, TST-D and FR in the fermentation broths of BthWT, BthΔtstP and BthΔtstR strains with LC–MS.
Time-course monitoring of the production of TST-A and TST-D by BthΔtstP strain during pilot scale fed-batch fermentation demonstrated that both compounds reached their highest titers (181.9 mg/L for TST-A and 19.3 mg/L for TST-D) at 96 h, after which the titers declined (Fig. 3).
Fig. 3.
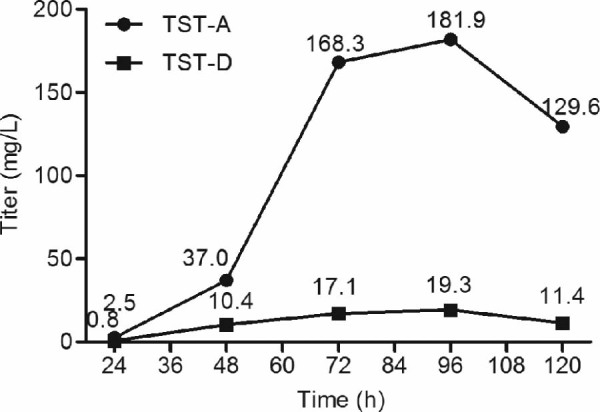
Time-course monitoring of the production titers of TST-A and TST-D during pilot scale fed-batch fermentation.
3.3. Recovery, isolation and purification of TST-A and TST-D
Totally, 236 g of crude extract containing an estimated 11.7 g of TST-A and 1.0 g of TST-D was obtained from about 90 L of fed-batch fermentation broth of BthΔtstP. A quantitative comparison between the compound titers in the fermentation broth harvested at 120 h (titers 129.6 mg/L for TST-A and 11.4 mg/L for TST-D) and after one round of 2% HP-20 resin absorption in the spent broth indicated that at least 96% of TST-A and 98% of TST-D were recovered by HP-20 absorption (Fig. S18). After one round of low pressure silica gel column separation, two rounds of flash chromatography separation and one round of preparative HPLC, 714 mg of TST-A with greater than 98.5% purity was obtained (Figs. S19–S22), reflecting an overall 6% recovery rate for TST-A from the fermentation broth. By using a similar purification protocol, 61.5 mg of TST-D was purified from 54 L of BthΔtstR fermentation in flasks, reflection a very low recovery rate of 2% (Figs. S23 and S24), mainly due to the interference of FK228, a known compound co-produced by the MSMB43 strains23 and having a similar retention time with that of TST-D.
3.4. TST-D is less cytotoxic but more stable than FR or TST-A
In vitro assay showed that TST-D possesses potent antiproliferative activities with IC50 values in the single nM range, yet it is a weaker compound compared to FR or TST-A (Table 2; Fig. S25). Interestingly, TST-D demonstrated a half-life in phosphate buffer (pH 7.4) at 37 °C greater than 202 h, significantly longer than that of FR (~7.8 h) (Fig. 4) or TST-A (~78 h).9
Table 2.
A comparison of antiproliferative activities of TST-D, FR and TST-A.
| Compound | IC50* (nM) | |||
|---|---|---|---|---|
| DU-145 | NCI-H232A | MDA-MB-231 | SKOV-3 | |
| Thailanstatin D | 6.35 ± 1.10 | 7.56 ± 0.57 | 9.93 ± 0.99 | 7.43 ± 0.99 |
| FR901464 | 0.68 ± 0.10 | 0.61 ± 0.07 | 0.84 ± 0.07 | 0.83 ± 0.09 |
| Thailanstatin A** | 1.11 ± 0.02 | 2.26 ± 0.17 | 2.58 ± 0.11 | 2.69 ± 0.37 |
IC50, half-maximal growth inhibition concentration provided as the average of triplicate well results with standard deviation.
Historical data from our previous publication.9
Fig. 4.
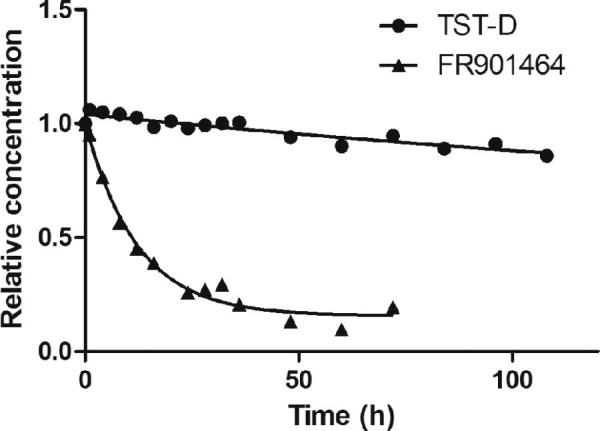
Stability of TST-D and FR in a phosphate buffer.
4. Discussion
Pre-mRNA splicing inhibitors have emerged as a promising class of anticancer drug candidates.24 However, the development of FR, a prototypic compound of splicing inhibitors, has suffered a setback due to its instability as well as toxicity.15 TST-A, a natural analog of FR discovered by our group, exhibited comparable activity as FR but much greater stability than FR.9 Therefore, we have been engaging TST-A in translational research as a cancer drug candidate. This current work was designed to solve the supply issue of TST-A.
During the course of this research, TST-D was discovered as the direct precursor of TST-A and was shown to have even greater stability than TST-A. We also observed several physicochemical factors that affect the production and/or purification of TST-A and/or TST-D, and learned valuable lessons. First, in the presence of high concentrations of sodium chloride in a fermentation medium (such as M8 medium used previously9), significant portion of the epoxide-containing TST-A is spontaneously converted to the chlorohydrin-containing TST-B at ambient temperature, confirming an independent assessment.8 We learned to omit sodium chloride in the fermentation medium (i.e. 2S4G medium17) and from reagents or solvents used the downstream purification steps for TST-A. Second, TST-A appeared unstable in the crude extract if let unprocessed at room temperature for a period of days. Empirically we found that TST-A can be stabilized in the crude extract by mixing with silica gel immediately after having been concentrated with a rotary evaporator. Third, formic acid (0.1%, v/v) in chromatographic solvents is necessary for effective separation of TST-A in ODS columns. Fourth, as noted above, MSMB43 strains also produce high levels of FK228,23 and TST-D and FK228 showed similar retention times and overlapping peaks in ODS chromatography, which hampered the separation of TST-D from FK228 and results in a very low recovery rate of TST-D. Fortunately, FK228 can be crystallized from the semi-purified FPLC fractions. We learned to insert a crystallization step to effectively remove the majority of FK228 from the semi-purified TST-D fractions prior to ODS column HPLC separation (Fig. S3). Lastly, final concentrating of large volume of preparative HPLC fractions containing purified TST-A or TST-D in acetonitrile–water should be achieved through a short period of rotary evaporation at 35 °C to remove acetonitrile followed by lyophilization to remove water completely.
Considering that both TST-A and TST-D contain a stabilizing C17-carboxylic acid group, that a C4-hydroxyl group is the only difference between TST-A and TST-D, and that a C3-epoxide group is known to be critical for bioactivity9 and is present in all three compounds FR, TST-A and TST-D (Fig. 1), the observed results suggest a bipolar contribution of the C4-hydroxyl group to the structure–activity relationship of those compounds. On the one hand, this C4-hydroxyl group appeared to synergize with the C3-epoxide group in FR and TST-A for their superb antiproliferative activities; on the other hand, this C4-hydroxyl group is also a labile factor for FR and TST-A.
In conclusion, we provided a successful scale-up production method for the preparation of adequate quantity of TST-A and TST-D for translational studies, employing engineered strains of B. thailandensis MSMB43 and a pilot-scale fed-batch fermentation system. We also observed that TST-D is more stable than TST-A in a physiologically relevant phosphate buffer but is less potent than TST-A against several cancer cell lines tested.
Acknowledgments
This work was supported in part by a grant from the US National Institute of Health/National Cancer Institute (R01 CA152212) and a pilot grant from the US National Institute of Health/National Center for Advancing Translational Sciences through the University of Texas Southwestern Medical Center (award number 5UL1TR001105).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at doi:10.1016/j.synbio.2016.02.002.
Appendix. Supplementary material
The following is the supplementary data to this article:
Figs. S1–S25 and Table S1.
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Dawson M.A., Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima H., Sato B., Fujita T., Takase S., Terano H., Okuhara M. New antitumor substances, FR901463, FR901464 and FR901465. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J Antibiot. 1996;49:1196–1203. doi: 10.7164/antibiotics.49.1196. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima H., Hori Y., Terano H., Okuhara M., Manda T., Matsumoto S. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J Antibiot. 1996;49:1204–1211. doi: 10.7164/antibiotics.49.1204. [DOI] [PubMed] [Google Scholar]
- 6.Nakajima H., Takase S., Terano H., Tanaka H. New antitumor substances, FR901463, FR901464 and FR901465. III. Structures of FR901463, FR901464 and FR901465. J Antibiot. 1997;50:96–99. doi: 10.7164/antibiotics.50.96. [DOI] [PubMed] [Google Scholar]
- 7.Liu X., Biswas S., Tang G.L., Cheng Y.-Q. Isolation and characterization of spliceostatin B, a new analogue of FR901464, from Pseudomonas sp. No. 2663. J Antibiot. 2013;66:555–558. doi: 10.1038/ja.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He H., Ratnayake A.S., Janso J.E., He M., Yang H.Y., Loganzo F. Cytotoxic spliceostatins from Burkholderia sp. and their semisynthetic analogues. J Nat Prod. 2014;77:1864–1870. doi: 10.1021/np500342m. [DOI] [PubMed] [Google Scholar]
- 9.Liu X., Biswas S., Berg M.G., Antapli C.M., Xie F., Wang Q. Genomics-guided discovery of thailanstatins A, B, and C As pre-mRNA splicing inhibitors and antiproliferative agents from Burkholderia thailandensis MSMB43. J Nat Prod. 2013;76:685–693. doi: 10.1021/np300913h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizui Y., Sakai T., Iwata M., Uenaka T., Okamoto K., Shimizu H. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. III. In vitro and in vivo antitumor activities. J Antibiot. 2004;57:188–196. doi: 10.7164/antibiotics.57.188. [DOI] [PubMed] [Google Scholar]
- 11.Seki-Asano M., Okazaki T., Yamagishi M., Sakai N., Takayama Y., Hanada K. Isolation and characterization of a new 12-membered macrolide FD-895. J Antibiot. 1994;47:1395–1401. doi: 10.7164/antibiotics.47.1395. [DOI] [PubMed] [Google Scholar]
- 12.Miller-Wideman M., Makkar N., Tran M., Isaac B., Biest N., Stonard R. Herboxidiene, a new herbicidal substance from Streptomyces chromofuscus A7847. Taxonomy, fermentation, isolation, physico-chemical and biological properties. J Antibiot. 1992;45:914–921. doi: 10.7164/antibiotics.45.914. [DOI] [PubMed] [Google Scholar]
- 13.Sakai Y., Yoshida T., Ochiai K., Uosaki Y., Saitoh Y., Tanaka F. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. I. Taxonomy, production, isolation, physicochemical properties and biological activities. J Antibiot. 2002;55:855–862. doi: 10.7164/antibiotics.55.855. [DOI] [PubMed] [Google Scholar]
- 14.Sakai Y., Tsujita T., Akiyama T., Yoshida T., Mizukami T., Akinaga S. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. II. The effects on cell cycle progression and gene expression. J Antibiot. 2002;55:863–872. doi: 10.7164/antibiotics.55.863. [DOI] [PubMed] [Google Scholar]
- 15.Kotake Y., Kaida D., Mizui Y., Yoshida M. [Discovery of splicing inhibitors and its impact on drug development] Tanpakushitsu Kakusan Koso. 2008;53:28–35. [PubMed] [Google Scholar]
- 16.Kaida D., Motoyoshi H., Tashiro E., Nojima T., Hagiwara M., Ishigami K. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 17.Eustaquio A.S., Janso J.E., Ratnayake A.S., O'Donnell C.J., Koehn F.E. Spliceostatin hemiketal biosynthesis in Burkholderia spp. is catalyzed by an iron/alpha-ketoglutarate-dependent dioxygenase. Proc Natl Acad Sci U S A. 2014;111:E3376–85. doi: 10.1073/pnas.1408300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F., He H.Y., Tang M.C., Tang Y.M., Zhou Q., Tang G.L. Cloning and elucidation of the FR901464 gene cluster revealing a complex acyltransferase-less polyketide synthase using glycerate as starter units. J Am Chem Soc. 2011;133:2452–2462. doi: 10.1021/ja105649g. [DOI] [PubMed] [Google Scholar]
- 19.Wang C., Henkes L.M., Doughty L.B., He M., Wang D., Meyer-Almes F.J. Thailandepsins: bacterial products with potent histone deacetylase inhibitory activities and broad-spectrum antiproliferative activities. J Nat Prod. 2011;74:2031–2038. doi: 10.1021/np200324x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi K.H., Schweizer H.P. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 2005;5:30. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C., Wesener S.R., Zhang H., Cheng Y.-Q. An FAD-dependent pyridine nucleotide-disulfide oxidoreductase is involved in disulfide bond formation in FK228 anticancer depsipeptide. Chem Biol. 2009;16:585–593. doi: 10.1016/j.chembiol.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Potharla V.Y., Wesener S.R., Cheng Y.-Q. New insights into the genetic organization of the FK228 biosynthetic gene cluster in Chromobacterium violaceum no. 968. Appl Environ Microbiol. 2011;77:1508–1511. doi: 10.1128/AEM.01512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X.Y., Wang C., Cheng Y.-Q. FK228 from Burkholderia thailandensis MSMB43. Acta Cryst Sect E Struct Rep Online. 2012;68:o2757–8. doi: 10.1107/S160053681203601X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonnal S., Vigevani L., Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1–S25 and Table S1.