Abstract
Microbial natural products are a crucial source of bioactive molecules and unique chemical scaffolds. Despite their importance, rediscovery of known natural products from established productive microbes has led to declining interest, even while emergent genomic data suggest that the majority of microbial natural products remain to be discovered. Now, new sources of microbial natural products must be defined in order to provide chemical scaffolds for the next generation of small molecules for therapeutic, agricultural, and industrial purposes. In this work, we use specialized bioinformatic programs, genetic knockouts, and comparative metabolomics to define the genus Legionella as a new source of novel natural products. We show that Legionella spp. hold a diverse collection of biosynthetic gene clusters for the production of polyketide and nonribosomal peptide natural products. To confirm this bioinformatic survey, we create targeted mutants of L. pneumophila and use comparative metabolomics to identify a novel polyketide surfactant. Using spectroscopic techniques, we show that this polyketide possesses a new chemical scaffold, and firmly demonstrate that this unexplored genus is a source for novel natural products.
Keywords: Natural products, Legionella, Polyketides, PRISM
Abbreviations: LC, liquid chromatography; MS, mass spectrometry; NRPS, nonribosomal peptide synthetase; PKS, polyketide synthase; PCA, principal component analysis
1. Introduction
Microbial natural products have been the most important source of chemical scaffolds and drugs for the last century.1 Culturing random collections of bacteria and fungi isolated from the environment led to the identification of prolific natural product producing genera that then received extensive focus, including Streptomyces,2 Bacillus,3 Myxobacteria,4 and Cyanobacteria.5 However, this tight focus, along with over-reliance on classical bioactivity-guided isolation approaches, has led to diminishing returns from natural products drug discovery efforts,6 resulting in the closure of most industrial natural products programs. In spite of this trend, untargeted sequencing of bacterial genomes has revealed that biosynthetic potential is much more widespread than was previously suspected,7 and that the majority of microbial natural products are still awaiting discovery.8, 9 In addition, it is now known that only a small fraction of bacteria can be readily cultured in laboratory conditions,10 and that many talented natural products producers were likely missed in initial screening efforts. In light of this information, new genome-guided discovery efforts are a promising way to reveal valuable chemical scaffolds from previously uncharted bacteria.
Extensive studies on the biosynthesis of bacterial natural products have led to informatic strategies for their discovery based on extant genomic information.11, 12, 13, 14 Two of the most diverse and abundant classes of microbial natural products, polyketides and nonribosomal peptides, are produced by multi-enzyme assembly lines15 – referred to as polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs) respectively – that can be readily detected in bacterial genomes. Genome sequencing has revealed a number of interesting bacterial families that could be candidates for second generation genome-guided natural products discovery efforts. Recent examples from Clostridia,16 Eleftheria,17 and Entotheonella18 have demonstrated that exotic bacteria that had been challenging to culture can be a valuable resource of newmolecules that were not discovered in previous random screening efforts.
In 2004, researchers described a unique polyketide fluorophore from a species of Legionella,19 a member of the diverse Gamma Proteobacteria that had not previously been investigated for natural products. In its native environment, Legionella reproduces by infecting environmental amebas, replicating inside a specialized vacuole following phagocytosis.20 Legionella is also able to infect some mammalian phagocytes, causing a pneumonia known as Legionnaire's disease in immunocompromised individuals. Legionella was first isolated following an outbreak in 1977, and was found to require highly specialized media for growth;21, 22 relatively little remained known about the biology of this organism until the late 1990s. In part because of its relevance as a potential pathogen, a number of Legionella genomes are now sequenced, demonstrating a great deal of intra-genus differentiation, as well as a number of polyketide and nonribosomal peptide biosynthetic gene clusters. Given its niche culture requirements, relatively late discovery, and genetic diversity, Legionella is a strong candidate for the identification of new chemical scaffolds using genome-guided discovery efforts. In this work, we use bioinformatic tools to chart the diversity of natural product biosynthetic gene clusters in Legionella, and use targeted mutagenesis and comparative metabolomics to reveal a novel natural product chemical scaffold from this under-explored bacterium.
2. Material and methods
2.1. General experimental procedures
One-dimensional (1H,13C) and two-dimensional (1H-1H and 13C-1H HMBC, HMQC, and COSY) NMR spectra were recorded on a Bruker AVIII 700 MHz NMR spectrometer in deuterated dimethyl sulfoxide (Cambridge Isotope Laboratories). High-resolution MS spectra were collected on a Thermo LTQ OrbiTrap XL mass spectrometer (ThermoFisher Scientific, USA) with an electrospray ionization source (ESI). LCMS data were collected using a Bruker AmazonX ion trap mass spectrometer coupled with a Dionex UltiMate 3000 HPLC system, using a Luna C18 column (250 mm × 4.6 mm, Phenomenex) for analytical separations, running acetonitrile and ddH2O with 0.1% formic acid as the mobile phase.
2.2. Strains and culture conditions
All experiments and genetic manipulations were conducted using the genetically amenable Legionella pneumophila Philadelphia-1 variant LP02. Mutants were constructed in LP02 with or without a pBH6119 plasmid enabling GFP-expression through an upstream icmR promoter, maintained through complementation of LP02's natural thymidine auxotrophy.23 L. pneumophila was grown at 37 °C for all liquid cultures in either BYE (10 g/L ACES, 10 g/L yeast extract, 1 g/L monosodium α-ketoglutarate, 0.4 g/L L-cysteine, 0.25 g/L ferric pyrophosphate, 0.1 g/L thymidine, pH = 6.9) or chemically defined Legionella media (350 mg/L L-arginine, 510 mg/L L-aspartic acid, 400 mg/L L-cysteine, 600 mg/L L-glutamic acid, 150 mg/L L-histidine, 470 mg/L L-isoleucine, 640 mg/L L-leucine, 650 mg/L L-lysine, 200 mg/L L-methionine, 350 mg/L L-phenylalanine, 115 mg/L L-proline, 650 mg/L L-serine, 330 mg/L L-threonine, 100 mg/L L-tryptophan, 400 mg/L L-tyrosine, 480 mg/L L-valine, 315 mg/L ammonium chloride, 50 mg/L sodium chloride, 20 mg/L calcium chloride, 1.18 g/L potassium phosphate monobasic, 70 mg/L magnesium sulfate, 250 mg/L ferric pyrophosphate, 100 mg/L thymidine, 10 g/L ACES). When visualizing sliding motility, plates were incubated at 30 °C for roughly three weeks. All media were supplemented with 0.1 g/L thymidine to support the auxotrophy of LP02.
2.3. Comparative metabolomic analysis
To generate samples for LCMS analysis wild type, Δlpg1939, Δlpg2186, and Δlpg2228 LP02 strains were grown in 50 mL of chemically defined Legionella media at 37 °C for one week. Following this, cultures were harvested by centrifugation, pellets were extracted with methanol, and supernatants were extracted with 20 g/L HP20 resin. Extracts were pooled and were subsequently dried by rotary evaporation and resuspended in methanol (2 mL). Samples were processed by LCMS with a 25 cm Luna C18 column (250 mm × 4.6 mm), using water and acetonitrile with 0.1% formic acid as the mobile phase. Acetonitrile was held at 2% for the first 2 min, then steadily ramped to 100% by 45 min, held until 53 min, then reset to 2% and held until 60 min, at a flow rate of 1.2 mL/min. Principal component analysis of Legionella extracts was carried out using Bruker Daltonics Profile Analysis with the following parameters: Rt range: 3–58 min; mass range: m/z 200–1200; rectangular bucketing: 10 sec (Δm/z of 2); normalized by using the sum of bucket values in the analysis. Chromatogram subtractions were performed using Bruker Daltonics MetaboliteDetect software using the eXpose mode to reveal differences in excess of 5-fold, with Δm/z of 0.5 and Δt of 0.5 min.
2.4. Isolation and purification of legionellol A
Wild type LP02 colonies from BCYE plates were inoculated into BYE cultures (5 mL) in sterile 50 mL Falcon tubes and grown for two days at 250 rpm and 37 °C. These cultures were used to inoculate sterile 2.8 L Fernbach flasks containing BYE (1.5 L). Cultures were grown at 37 °C with shaking at 200 rpm for roughly one week or until two days after peak melanin production. Following growth, cells were pelleted by centrifugation at 6000 rpm for 30 min. Supernatants were mixed with 20 g/L washed HP20 resin (Diaion) for 2 h at room temperature. Resins were harvested using Buchner funnel vacuum filtration, and washed with 10% methanol to remove highly polar melanins. Resin was eluted with excess 100% methanol which was then dried by rotary vacuum. Extracts were resuspended in methanol and separated by LCMS using a Luna C18 column (250 mm × 10 mm) with HPLC grade water and acetonitrile with 0.1% formic acid as the mobile phase. To purify legionellol, acetonitrile began at 5% for the first 2 min, then ramped to 30% by 5 min and held until 27 min, followed by a shallow ramp to 45% by 40 min, followed by a wash of 100% from 42 to 52 min. Flow was maintained at 6 mL/min, and legionellol A eluted at 33 min.
2.5. Incorporation of 13C ornithine
To assess the origins of the modified amino acid present in the legionellol metabolites, cultures of L. pneumophila in chemically defined media were grown for five days in the presence of 13C ornithine (2 mM; Cambridge Isotope Laboratories) or 12C ornithine (2 mM). Supernatants of cultures containing 12C and 13C ornithine were extracted with HP-20 resin and analyzed by LCMS.
2.6. Reconstitution of sliding motility
To assess whether various legionellol species were responsible for sliding motility, legionellol A or acylated legionellol (10 µg) was dissolved in methanol (10 µL) and added as a drop to a 0.5% agar plate of BCYE media. After the drops of legionellol, acyl legionellol, or methanol alone had dried, an overnight culture of LP02 Δlpg2228 was dropped (10 µL) over top of it and subsequently dried. Plates were incubated at 28 °C for one week before imaging.
2.7. Insertional inactivation of genes in Legionella
Targeted insertional inactivation of biosynthetic genes in LP02 was performed as previously described,24 and will be summarized below. All primers and plasmids used in this process are described in Table S3. If not stated explicitly, genetic manipulations and molecular biology techniques followed those from Cold Spring Harbor Protocols, available at http://www.molecularcloning.com/.
In short, two neighboring 500 bp–1000 bp fragments of target genes were cloned into pBlueScript KSII or a modified pBlueScript KSII bearing a chloramphenicol resistance cassette, swapped with the ampicillin resistance cassette through two flanking BspHI sites. Plasmids were digested with Xba1 and Sac1, and gene fragments were digested with Xba1 and Kpn1, or with Sph1 and Sac1. Chloramphenicol or kanamycin resistance cassettes were amplified with flanking Kpn1 and Sph1 cut sites from pRE112 or pET-28b respectively. The digested plasmid, two digested gene fragments, and resistance cassette were ligated simultaneously and transformed into chemically competent DH5α (Invitrogen). All plasmids were confirmed by sequencing from either end of the homology arms and resistance cassettes. To transform into Legionella, fresh colonies of LP02 were re-streaked as a dime-sized patch on fresh BCYE and 10 µL of 100 ng/µL knockout vector solution was added on top, followed by incubation at 30 °C for 48 h. Following incubation, colonies were re-streaked onto BCYE with kanamycin or chloramphenicol to select for transformants. Genomic integration was confirmed through PCR with primers for the resistance cassette and a region of the genome just outside the homology arm. PCR with primers for amplifying the original gene fragment was used to verify the exclusive presence of the insertionally inactivated allele.
2.8. High resolution mass spectrometry
Legionellol A was dissolved in a mixture of HPLC grade methanol and water with 0.1% formic acid providing a final concentration of roughly 10 µg/mL. The sample was infused directly into a Thermo LTQ OrbiTrap XL mass spectrometer running Xcaliber 2.07 and TunePlus 2.4 SP1 at a flow rate of 3 µL/min and ionized using an electrospray ionization source. The mass spectrometer was operated in positive mode with a maximum resolution of 100,000. The high resolution mass for legionellol A was an average of 29 scans.
| Compound | Molecular formula | Calculated m/z | Observed m/z | Δppm |
|---|---|---|---|---|
| Legionellol A [M+H]+ | C23H43N2O7 | 459.30650 | 459.30665 | 0.327 ppm |
2.9. PRISM analysis of Legionella genomes
Legionella genomes were downloaded from NCBI and loaded into PRISM12 (http://www.magarveylab.ca/prism) using standard settings. Of the assembled genomes used in this analysis, 24 were draft genomes and 10 were fully assembled. Polyketide and nonribosomal peptide gene clusters were stored, annotated, and confirmed by secondary analysis using the BLAST function of Integrated Microbial Genomes (IMG; http://img.jgi.doe.gov/), which also checked for fragmentation resulting from incomplete genome assemblies. A phylogenetic tree of Legionella spp. was generated with 16S rRNA sequences collected from Legionella genomes, using the Geneious tree builder program with Tamura-Nei as the genetic distance model and neighbor-joining as the tree build method.
3. Results
3.1. Bioinformatic assessment of biosynthetic potential in Legionella
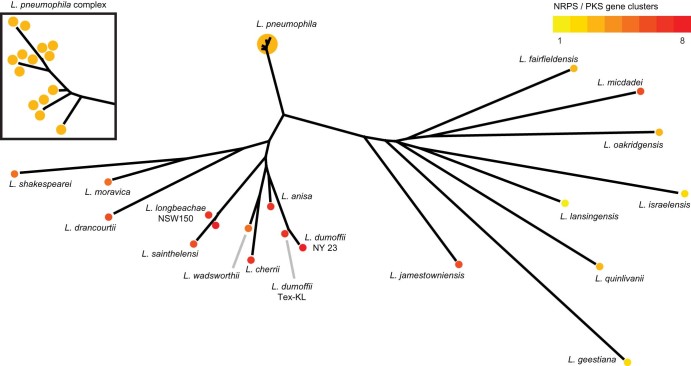
To assess the diversity of polyketide and nonribosomal peptide gene clusters present in Legionella, we profiled all sequenced genomes available for this genus through our software for PRediction Informatics for Secondary Metabolomes (PRISM;12 Fig. S1). This in-house web application is able to efficiently identify polyketide and nonribosomal peptide gene clusters from bacterial genomes, compare them to known biosynthetic gene clusters, and predict the structures of their small molecule products. We used PRISM to analyze 34 sequenced Legionella genomes – including 15 L. pneumophila strains – revealing a diverse array of NRPS, PKS, and hybrid biosynthetic gene clusters (Fig. 1). From our sample of 34 genomes, we identified 141 biosynthetic gene clusters related to nonribosomal peptides (46), polyketides (7), or hybrid (88) systems, with an average of 4 biosynthetic gene clusters per genome. In addition to the raw number of hybrid assembly systems found in Legionella genomes, these clusters were also the most diverse, with 18 distinct hybrid biosynthetic gene clusters from these 34 genomes, compared to 13 distinct nonribosomal peptide gene clusters and only 3 purely polyketide gene clusters. In sharp contrast to the massive canonical assembly line architectures,15 biosynthetic assembly lines in Legionella are highly fragmented and small, with many comprised of individual domains or lone multi-domain modules. This was particularly pronounced for nonribosomal peptide gene clusters, which nearly always possessed only a single monomer-activating adenylation domain (11 of 13). The majority of Legionella polyketide synthases, either in pure polyketide systems or in hybrid systems, were fully or partially trans-AT25 (14 of 21), where the monomer-loading acyl transferase enzyme is sourced from outside the multi-domain assembly line. Although trans-AT polyketide synthases are becoming less rare with continued sequencing, Legionella has an unusual twist, in that nearly all of the trans-AT systems do not have an associated acyltransferase in the biosynthetic gene cluster, which is exceptionally rare.26 In addition to the unusual organization, size, and monomer-loading strategies of these biosynthetic clusters, they also possess a large number of non-canonical termination mechanisms. In canonical multi-domain assembly line polyketide or nonribosomal peptide biosynthesis the small molecule chain is released from the enzyme assembly line via thioester hydrolysis through a C-terminal thioesterase domain.15 Although thioesterases are still the most abundant chain release enzyme (12 of 33 complete clusters), most Legionella biosynthetic gene clusters do not possess thioesterases, and instead appear to rely on NADH-dependent reductases27 (5 of 33) or condensation domains (9 of 33), the latter of which are exceptionally rare in described bacterial polyketide and nonribosomal peptide biosynthetic gene clusters.28, 29, 30 Most notably, many gene clusters did not possess analogs of any of these established chain release enzymes, suggesting alternative means of terminating thio-template natural product biosynthesis.
Fig. 1.
PRISM analysis reveals that Legionella is a diverse genus with conserved biosynthetic potential. PRISM was used to identify polyketide and nonribosomal peptide gene clusters in 34 sequenced Legionella genomes, which was confirmed by manual inspection. PKS and NRPS gene clusters were sorted as a heat map and overlaid onto a phylogenetic tree constructed using 16S rRNA sequences to highlight the conserved biosynthetic potential of Legionella.
During this bioinformatic analysis, we did not uncover any biosynthetic gene clusters that were substantially similar to established or sequenced gene clusters from other bacteria, including from well-studied genera such as Streptomyces or relatively closely related natural product producers, such as Pseudomonas. Despite the unusual architecture of most of these biosynthetic gene clusters – which often lack conventional monomer-loading or chain-release enzymes – many of these assemblages can be found in different species and appear to be well-conserved. Legionella is known to have a patchwork genome,31 which rapidly sheds and acquires genes in accordance with survival needs,32 suggesting that these conserved unconventional biosynthetic gene clusters are likely still functional. Moreover, transcriptomics studies in L. pneumophila have demonstrated that these biosynthetic gene clusters are transcriptionally active in a variety of culture conditions.33, 34 To assess whether any of these unusual gene clusters produced new natural product scaffolds, we chose to pursue a genomic and metabolomic strategy with the genetically-tractable L. pneumophila strain LP02.
3.2. Genetic and metabolic profiling of L. pneumophila uncovers a novel natural product
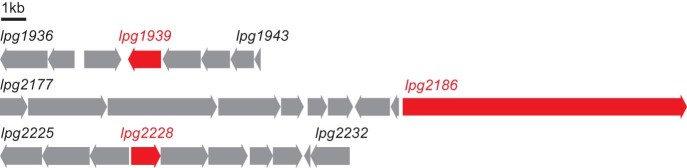
We constructed a series of genetic knockouts in L. pneumophila strain LP02, targeting ketosynthases in each of the 3 hybrid polyketide-nonribosomal peptide gene clusters visible in the genome (Fig. 2, Table 1, Tables S1 and S2). Each of these gene clusters is well-conserved in L. pneumophila, represented in nearly all of the 15 complete or partially-sequenced strain genomes. However, each cluster has a highly unusual architecture, as two (represented by Δlpg1939 and Δlpg2228) are composed exclusively of individual biosynthetic domains and lack chain terminating enzymes, while the third (represented by Δlpg2186) encodes a minimal NRPS and trans-AT PKS without an apparent trans-acting acyltransferase present in the genome. Following insertional activation, we observed that deletion of a ketosynthase (Δlpg2228) in one of these minimal gene clusters caused a loss of sliding motility, indicating the production of a surfactant, consistent with a previous microbiological study35 (Fig. 3A).
Fig. 2.
Hybrid polyketide-nonribosomal peptide gene clusters of Legionella pneumophila targeted for mutagenesis. PRISM identified three hybrid PKS-NRPS gene clusters in the genome of L. pneumophila LP02, roughly defined as lpg1936-lpg1943, lpg2177-lpg2186, and lpg2225-lpg2232. Ketosynthases in each gene cluster were insertionally inactivated by insertion of a kanamycin resistance gene, disrupting expression of lpg1939, lpg2186, and lpg2228 (red).
Table 1.
Genes of a Legionella pneumophila hybrid biosynthetic gene cluster (lpg2225-2234).
| Gene name | Locus tag | Predicted function | Strand | Amino acids |
|---|---|---|---|---|
| lol1 | lpg2225 | GH3-family auxin responsive protein | − | 509 |
| lol2 | lpg2226 | Isovaleryl CoA dehydrogenase | − | 563 |
| lol3 | lpg2227 | Propionyl-CoA carboxylase | − | 479 |
| lol4 | lpg2228 | 3-oxoacyl-(acyl carrier protein) synthase III | + | 353 |
| lol5 | lpg2229 | Acyl CoA synthetase | + | 581 |
| lol6 | lpg2230 | Acyl CoA ligase | + | 464 |
| lol7 | lpg2231 | 3-oxoacyl reductase | + | 250 |
| lol8 | lpg2232 | 3-oxoacyl-(acyl carrier protein) synthase III | + | 336 |
| lol9 | lpg2233 | Acyl carrier protein | − | 75 |
| lol10 | lpg2234 | Major facilitator superfamily efflux pump | − | 455 |
Fig. 3.
Mutations in a hybrid polyketide gene cluster result in motility and metabolomic alterations. (A) Extended growth of Legionella pneumophila LP02 on 0.5% agar plates results in pronounced sliding motility, which is absent in the Δlpg2228 strain. (B) Comparative metabolomic analysis of wild type and Δlpg2228 cultures with LCMS highlights a series of molecules which are absent in Δlpg2228.
To identify polyketides or nonribosomal peptides associated with the identified gene clusters, we used a comparative metabolomic strategy. All L. pneumophila LP02 wild type and knockout strains were grown in rich or chemically defined media until several days past maturity, before cell pellets were collected by centrifugation and extracted with methanol, and supernatants were extracted with absorbent HP20 resin. Cell pellet and supernatant extracts were pooled and processed for small molecule contents using liquid chromatography paired with mass spectrometry (LCMS). Data files and chromatograms were analyzed to identify distinguishing molecular features between varying genetic backgrounds using Bruker MetaboliteDetect and ProfileAnalysis software, which enabled comprehensive chromatographic analysis and principal component analysis (PCA) respectively. The results of these analyses indicated no metabolomic differences between Δlpg2186, Δlpg1939, and wild type LP02. However, the Δlpg2228 mutant was shown to have significant metabolomic deviations from both wild type and the other mutants (Fig. 3B, Fig. S2). This strain had previously beenobserved to be deficient in sliding motility (Fig. 3A), and appears to lack a series of metabolites which would correspond to the absent surfactant and biosynthetic intermediates. This series was present in all other biosynthetic mutants, which likewise showed no deviation from the wild type sliding colony phenotype, indicating that the unusual and minimal gene cluster associated with these metabolites likely does not rely on polyketide-derived precursors supplied in trans, at least not from the identified gene clusters. Although these surfactants ionize quite well, ionization intensity during mass spectrometry is not necessarily correlated with high abundance, and we found during initial isolation attempts that these molecules were present in extremely low quantities.
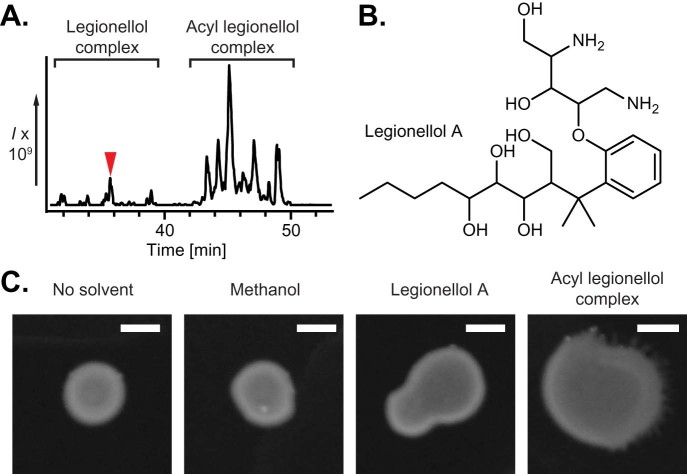
Close analysis of our metabolites detected by comparative metabolomics and PCA indicated two complexes of natural products (Fig. 4A), likely corresponding to a series of core metabolites and acylated variants as indicated by an increased retention time and mass, but highly similar MS/MS fragmentation (Fig. S3). Although each unacylated compound was generally present as a single predominant peak for each mass, each acylated molecule was present as three to five individual peaks, indicating considerable heterogeneity. In hopes of obtaining a pure compound for NMR structure elucidation, we chose to isolate the most abundant core scaffold metabolite (459.2 [M+H]+) through serial rounds of LCMS purification from 100 L of wild type LP02 culture. As a further demonstration of the remarkably low abundance of this molecule, preparative scale isolation yielded <500 µg of this metabolite. The structure of this pure compound was then elucidated by a combination of 1D and 2D NMR experiments and high resolution mass spectrometry, which provided a definitive molecular formula of C23H42N2O7 (0.327 Δppm; see Material and Methods). 1H, 1H-correlation spectroscopy (COSY) identified a short chain acyl unit, a number of secondary alcohols, one primary alcohol, and a system of four aromatic protons. Importantly, this also revealed that a prominent 133 m/z [M+H]+ fragment observed during MS/MS was a 2,5-diaminopentane-1,3,4-triol. 13C-amino acid feeding experiments demonstrated that this hydroxylated diaminopentane was derived from ornithine (Fig. S4). 1H, 13C-heteronuclear multiple-bond correlation spectroscopy (HMBC) revealed a gem-di-methyl group between the phenol and primary alcohol, and was used to link pieces identified with COSY, leading to a highly unusual molecule that we named legionellol A (Fig. 4B; Supplementary Note – Structure characterization). Reanalysis of the legionellol series of metabolites identified through PCA indicated at least 10 distinct chemical entities, including smaller, more hydrophilic legionellol variants which appear to have shorter acyl tails or lack the gem-di-methyl. In addition, MS/MS revealed that a minor series of legionellol variants likely possess an ornithine to arginine substitution in the modified amino acid portion of the molecule, and likewise possess a similar series of structural variations. Importantly, acylated forms of legionellol could also be observed, and MS/MS indicates that these are modified with a hydroxy-fatty acid, primarily C12 and C14 (Fig. S3). To determine which molecular species was responsible for sliding motility, a 10 µg sample of an acyl legionellol or legionellol A was dried onto a 0.5% agar plate (as well as methanol alone), and the Δlpg2228 strain was deposited on top. The results of the corresponding outgrowth indicate that sliding motility was restored more effectively by supplementation with the acylated legionellol, rather than the scaffold alone, indicating that the variably acylated legionellol complex is likely the active surfactant (Fig. 4C).
Fig. 4.
An unusual PKS gene cluster in Legionella pneumophila encodes for legionellol, a novel surfactant scaffold. (A) Metabolites absent from Δlpg2228 appear to separate as two complexes by LCMS, comprised of a smaller hydrophilic series of metabolites, and a larger hydrophobic series of metabolites. The most abundant of these hydrophilic scaffold molecules – legionellol A – is indicated with an arrow. (B) Structure of the novel L. pneumophila surfactant legionellol A as deduced by NMR and MS experiments. (C) To assess the impact of legionellol metabolites on sliding motility, 10 µL of Δlpg2228 overnight culture was added to a 0.5% agar BCYE plate alone, or over top of dried 10 µL drops of methanol, 1 µg/µL acyl legionellol (684 Da), or 1 µg/µL legionellol A. Plates were grown for one week at 30 °C, and demonstrate that acyl legionellol is able to recapitulate sliding motility. Scale bars are equal to 5 mm.
Legionellol is a novel natural product which arises from a minimal gene cluster composed of discrete domains often associated with polyketide biosynthesis (Fig. 2; Table 1). From this gene cluster, enzymes can be identified which appear responsible fatty acid adenylation and CoA-ligation, and for the tethering, condensation, reduction, and dehydration of polyketide extending units such as malonate. However, genes required for several biosynthetic processes – including chain release and generation of the modified amino acid – are notably absent, suggesting that the enzymes required for these processes are located elsewhere in the genome, or that these functions are performed by new classes of biosynthetic enzymes. To assess whether unusual enzymes may account for some absent biosynthetic processes, we investigated an unusual GH3 family protein that was present in one of the two operons in the legionellol gene cluster, and which we suspected may be involved with attaching the modified ornithine residue. GH3 family proteins are known from plants, where they act as indole-3-acetic acid amido-synthetases during auxin biosynthesis.36 More specifically, these enzymes use ATP to adenylate a free carboxylic acid before catalyzing the displacement of the adenosine monophosphate by a nucleophilic amine from a free amino acid. While this mechanism is common in plants it has yet to be implicated in the biosynthesis of bacterial secondary metabolites, despite occasionally occurring in PKS biosynthetic gene clusters.37, 38 To assess whether this conspicuous enzyme was responsible for coupling the modified ornithine to the legionellol polyketide precursor, we generated an insertionally inactivated mutant using the same approach we had demonstrated previously. Despite its presence in the legionellol biosynthetic operon, insertional inactivation of this unusual enzyme did not alter legionellol production, although it remains to be seen whether other unusual biosynthetic enzymes may play a role in the construction of this novel natural product.
4. Discussion
Natural products have provided chemical scaffolds and molecular innovation that has driven drug discovery and development efforts for the last century.1 Despite their inherent value, rediscovery of known natural products has led to declining discovery rates and a loss in industrial interest.6 By pursuing challenging bacteria that had not been previously studied as natural products producers, we reasoned that we could identify new, exotic chemistry arising from novel biosynthetic gene clusters. In this work, we used bioinformatics and advanced natural products chemistry techniques to investigate Legionella as a source of novel natural product scaffolds, yielding the new molecule legionellol A.
Legionella is a diverse genus, and relatively limited genome sequencing has revealed species with significant biosynthetic potential. In this work, we chose to use the genetically-tractable L. pneumophila strain LP02 to pursue a knockout and comparative metabolomics approach to discover new molecules. However, the L. pneumophila complex is not the most impressive candidate for natural product discovery, as species related to L. dumoffii and L. longbeachae typically possess at least twice the number of biosynthetic gene clusters present in L. pneumophila. Lone representative genomes of more exotic Legionella spp. such as L. micdadei and L. shakespearei also indicate that more extensive sequencing will reveal substantial numbers of new biosynthetic gene clusters from these unexplored organisms, as both species possess large and unique hybrid assembly lines. Using PRISM, we can now rapidly profile emergent genomes from this promising genus, identify biosynthetic gene clusters automatically, and prioritize candidates for discovery in the hopes of continuing to reveal interesting new natural products.
Although the rules for polyketide and nonribosomal peptide biosynthesis are well defined, exotic organisms often deviate from established systems.16, 18 Legioliulin, the only example of a polyketide (or nonribosomal peptide) known from Legionella prior to this study, highlights several biosynthetic oddities that frequently occur in Legionella biosynthetic gene clusters, including a propensity for trans-AT PKS logic that does not include an associated acyl transferase.26 This situation is more bizarre in L. pneumophila, which has three hybrid polyketide and nonribosomal peptide gene clusters, but only has a single malonyl-acyltransferase (lpg1394), which is associated with the fatty acid synthase operon, suggesting that there may be an alternative monomer loading strategy to create polyketide metabolites such as legionellol. Legionella also possesses a number of unusual chain termination strategies, including the use of C-terminal condensation domains to facilitate trans-esterification and thioester cleavage, which had only been reported previously in a handful of bacterial natural products, including the Myxobacterial natural product crocacin,30 FK520,28 and the C-1027 enediyne.29 The frequency of these unusual biosynthetic domains, combined with the unorganized lone-domain structure of Legionella's biosynthetic gene clusters, limits the predictability of their polyketide and nonribosomal peptide products. This is well illustrated by legionellol, which is a remarkably complex natural product that arises from a seemingly simple biosynthetic gene cluster. Given its unusual structure and minimal biosynthetic gene cluster, the precise biosynthetic pathway of legionellol is still unclear, but the origins of some units can be speculated. Generation of the modified ornithine residue would require hydroxylation of the ornithine β- and γ-carbons, followed by activation of the carboxylic acid and iterative reduction to the observed alcohol. How the putative polyketide portion of legionellol is constructed is much less clear, and must include a number of exotic monomers and transformations. Construction of the polyketide likely begins with activation of a short-chain fatty acid of varying length, giving rise to the observed range of legionellol variants. Chain elongation with glycerate-derived hydroxymalonate followed by ketoreduction could then result in the hydroxylated acyl chain. Beyond this however, biosynthetic processes involved in constructing legionellol or its monomers become difficult to predict, such as the origins of the unusual primary alcohol and neighboring gem-dimethyl. Hopefully, further study of this promising genus may reveal innate biosynthetic logic involved in creating both legionellol and other unique natural products, facilitating prediction and discovery of new chemical scaffolds.
Acknowledgements
This work was supported by generous gifts from McMaster University and the Canadian Institute of Health Research (CIHR). C. Johnston is funded through a CIHR Doctoral Research Award. N. Magarvey is funded by a CIHR New Investigator Award. We wish to thank Dr. Cyril Guyard for providing L. pneumophila LP02, as well as the GFP-expression plasmid pBH6119.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at doi:10.1016/j.synbio.2015.12.001.
Appendix. Supplementary material
The following is the supplementary data to this article:
Supplementary information.
References
- 1.Newman D.J., Cragg G.M. Natural products as a source of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hopwood D.A. Oxford University Press; NY, USA: 2007. Streptomyces in nature and medicine: the antibiotic markers. [Google Scholar]
- 3.Cochrane S.A., Vederas J.C. Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med Res Rev. 2014:1–28. doi: 10.1002/med.21321. [DOI] [PubMed] [Google Scholar]
- 4.Weissman K.J., Müller R. A brief tour of myxobacterial secondary metabolism. Bioorg Med Chem. 2009;17:2121–2136. doi: 10.1016/j.bmc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 5.Nunnery J.K., Meyers E., Gerwick W.H. Biologically active secondary metabolites from marine cyanobacteria. Curr Opin Biotechnol. 2010;21:787–793. doi: 10.1016/j.copbio.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koehn F.E., Carter G.T. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 7.Donadio S., Monciardini P., Sosio M. Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep. 2007;21:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- 8.Nett M., Ikeda H., Moore B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skinnider M.A., Johnston C.W., Zvanych R., Magarvey N.A. Automated identification of depsipeptide natural products by an informatic search algorithm. Chembiochem. 2015;16:223–227. doi: 10.1002/cbic.201402434. [DOI] [PubMed] [Google Scholar]
- 10.Rappé M.S., Giovannoni S.J. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 11.Johnston C.W., Skinnider M.A., Wyatt M.A., Li X., Ranieri M.R., Yang L. An automated Genomes-to-Natural Products platform (GNP) for the discovery of modular natural products. Nat Commun. 2015;6:8421. doi: 10.1038/ncomms9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skinnider M.A., Dejong C.A., Rees P.N., Johnston C.W., Li H., Webster A.L. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM) Nucleic Acids Res. 2015:1–18. doi: 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohimani H., Liu W.T., Kersten R.D., Moore B.S., Dorrestein P.C., Pevzner P.A. NRPquest: coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J Nat Prod. 2014;77:1902–1909. doi: 10.1021/np500370c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R. antiSMASH 3.0 – a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–43. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischbach M.A., Walsh C.T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 16.Lincke T., Behnken S., Ishida K., Roth M., Hertweck C. Closthioamide: an unprecedented polythioamide antibiotic from the strictly anaerobic bacterium Clostridium cellulolyticum. Angew Chem Int Ed Engl. 2010;49:2011–2013. doi: 10.1002/anie.200906114. [DOI] [PubMed] [Google Scholar]
- 17.Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson M.C., Mori T., Rückert C., Uria A.R., Helf M.J., Takada K. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 19.Amemura-Maekawa J., Hayakawa Y., Sugie H., Moribayashi A., Kura F., Chang B. Legioliulin, a new isocoumarin compound responsible for blue-white autofluorescence in Legionella (Fluoribacter) dumoffii under long-wavelength UV light. Biochem Biophys Res Commun. 2004;323:954–959. doi: 10.1016/j.bbrc.2004.08.180. [DOI] [PubMed] [Google Scholar]
- 20.Hubber A., Roy C.R. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 21.Feeley J.C., Gibson R.J., Gorman G.W., Langford N.C., Rasheed J.K., Mackel D.C. Charcoal- yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol. 1979;10:437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren W.J., Miller R.D. Growth of Legionnaires disease bacterium (Legionella pneumophila) in chemically defined medium. J Clin Microbiol. 1979;10:50–55. doi: 10.1128/jcm.10.1.50-55.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammer B.K., Swanson M.S. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol. 1999;33:721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- 24.Sexton J.A., Vogel J.P. Regulation of hypercompetence in Legionella pneumophila. J Bacteriol. 2004;186:3814–3825. doi: 10.1128/JB.186.12.3814-3825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- 26.Ahrendt T., Miltenberger M., Haneburger I., Kirchner F., Kronenwerth M., Brachmann A.O. Biosynthesis of the natural fluorophore legioliulin from legionella. Chembiochem. 2013;14:1415–1418. doi: 10.1002/cbic.201300373. [DOI] [PubMed] [Google Scholar]
- 27.Reimmann C., Patel H.M., Serino L., Barone M., Walsh C.T., Haas D. Essential PchG-dependent reduction in pyochelin biosynthesis of Pseudomonas aeruginosa. J Bacteriol. 2001;183:813–820. doi: 10.1128/JB.183.3.813-820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gatto G.J., Jr, McLoughlin S.M., Kelleher N.L., Walsh C.T. Elucidating the substrate specificity and condensation domain activity of FkbP, the FK520 pipecolate incorporating enzyme. Biochemistry. 2005;44:5993–6002. doi: 10.1021/bi050230w. [DOI] [PubMed] [Google Scholar]
- 29.Lin S., Van Lanen S.G., Shen B. A free-standing condensation enzyme catalyzing ester bond formation in C-1027 biosynthesis. Proc Natl Acad Sci U S A. 2009;106:4183–4188. doi: 10.1073/pnas.0808880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller S., Rachid S., Hoffmann T., Surup F., Volz C., Zaburannyi N. Biosynthesis of crocacin involves an unusual hydrolytic release domain showing similarity to condensation domains. Chem Biol. 2014;21:855–865. doi: 10.1016/j.chembiol.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Cazalet C., Rusniok C., Brüggemann H., Zidane N., Magnier A., Ma L. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 32.O'Connor T.J., Adepoju Y., Boyd D., Isberg R.R. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A. 2011;108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hindré T., Brüggermann H., Buchrieser C., Héchard Y. Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology. 2008;154:30–41. doi: 10.1099/mic.0.2007/008698-0. [DOI] [PubMed] [Google Scholar]
- 34.Dalebroux Z.D., Yagi B.F., Sahr T., Buchrieser C., Swanson M.S. Distinct roles of ppGpp and DskA in Legionella pneumophila differentiation. Mol Microbiol. 2010;76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart C.R., Burnside D.M., Cianciotto N.P. The surfactant of Legionella pneumophila is secreted in a TolC-dependent manner and is antagonistic toward other Legionella species. J Bacteriol. 2011;193:5971–5984. doi: 10.1128/JB.05405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Q., Westfall C.S., Hicks L.M., Wang S., Jez J.M. Modulating plant hormones by enzyme action: the GH3 family of acyl acid amido synthetases. J Biol Chem. 2010;285:29780–29786. doi: 10.4161/psb.5.12.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olano C., Wilkinson B., Sánchez C., Moss S.J., Sheridan R., Math V. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tü4055: cluster analysis and assignment of functions. Chem Biol. 2004;11:87–97. doi: 10.1016/j.chembiol.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 38.Kong R., Liu X., Su C., Ma C., Qiu R., Tang L. Elucidation of the biosynthetic gene cluster and the post-PKS modification mechanism for fostriecin in Streptomyces pulveraceus. Chem Biol. 2013;20:45–54. doi: 10.1016/j.chembiol.2012.10.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.