Abstract
Microbial-derived natural products are important in both the pharmaceutical industry and academic research. As the metabolic potential of original producer especially Streptomyces is often limited by slow growth rate, complicated cultivation profile, and unfeasible genetic manipulation, so exploring a Streptomyces as a super industrial chassis is valuable and urgent. Streptomyces sp. FR-008 is a fast-growing microorganism and can also produce a considerable amount of macrolide candicidin via modular polyketide synthase. In this study, we evaluated Streptomyces sp. FR-008 as a potential industrial-production chassis. First, PacBio sequencing and transcriptome analyses indicated that the Streptomyces sp. FR-008 genome size is 7.26 Mb, which represents one of the smallest of currently sequenced Streptomyces genomes. In addition, we simplified the conjugation procedure without heat-shock and pre-germination treatments but with high conjugation efficiency, suggesting it is inherently capable of accepting heterologous DNA. In addition, a series of promoters selected from literatures was assessed based on GusA activity in Streptomyces sp. FR-008. Compared with the common used promoter ermE*-p, the strength of these promoters comprise a library with a constitutive range of 60–860%, thus providing the useful regulatory elements for future genetic engineering purpose. In order to minimum the genome, we also target deleted three endogenous polyketide synthase (PKS) gene clusters to generate a mutant LQ3. LQ3 is thus an “updated” version of Streptomyces sp. FR-008, producing fewer secondary metabolites profiles than Streptomyces sp. FR-008. We believe this work could facilitate further development of Streptomyces sp. FR-008 for use in biotechnological applications.
1. Introduction
Microbial-derived natural products exhibit a diverse array of unique structures and biological activities, such as antibacterial, antifungal, antitumor, and immunosuppressant activities [1]. However, genetically engineering microbial strains to overproduce desired products was often limited by recalcitrance to genetic manipulation, poor growth characteristics, as well as limited understanding of a given strain's cultivation profile. Heterologous expression has been used as a strategy to overcome these limitations by providing a more amenable, well-characterized chassis for the production of natural products [2]. This method has also been proven a powerful tool for mining the metabolic potential of difficult-to-culture microorganisms and expressing cryptic gene clusters, particularly in the postgenomic age [3], [4].
One of the most crucial steps in the design of a heterologous expression system is the choice of a suitable host, which is usually determined heuristically. Escherichia coli and Saccharomyces cerevisiae are often used as general hosts, but heterologous expression of complex natural products in these host organisms often results in low titer and limits the types of chemicals [5]. These limitations are generally considered to be due to differences in the morphology, physiology, metabolism, and regulatory mechanisms between the above-mentioned hosts and the native producer, which is often an actinomycetes species [6]. Due to their similarity to most native strains, Streptomyces is “chassis” for heterologous expressing these compounds. To our knowledge, four Streptomyces species are commonly used, including Streptomyces lividans, Streptomyces coelicolor, Streptomyces avermitilis, and Streptomyces albus J1074. Streptomyces coelicolor is considered as the model strain and was used in development of the earliest genetic recombinant techniques for heterologous gene transfer [7], [8]. Streptomyces lividans, which is a plasmid-containing derivative of S. coelicolor that shares considerable genome sequence similarity, is often used for exogenous production of natural products. Derivatives of S. lividans and S. coelicolor in which the native biosynthesis pathways have been knocked out include S. coelicolor CH999 and S. lividans K4-114 [9], [10]. And S. coelicolor M1152 & M1154 with impressing manipulation on ribosome is successfully applied for improvement of natural product titers [11]. Streptomyces avermitilis, for which the genome sequence has been determined, is capable of producing macrolactone insecticides known as avermectins [12], [13]. Several S. avermitilis derivatives have been constructed in which a 1.4-Mb region of the 9.02-Mb linear chromosome has been deleted [14], and these strains have been evaluated based on the expression of 20 gene clusters [15]. Streptomyces albus J1074 is another commonly used Streptomyces host. Recent sequencing of the complete genome of S. albus revealed that at 6.84 Mb, it is the smallest Streptomyces genome determined thus far [16].
Although there are many reports of successful heterologous expression in the above-mentioned hosts, production sometimes fails when different hosts are employed [17], and it is difficult to predict what percentage of experiments will fail. There is, therefore, an urgent need for a standardized platform involving an efficient and reliable host organism to maximize the chances of successful heterologous expression. An optimal host should not only exhibit excellent growth characteristics and be readily amenable to genetic manipulation, but its genotype should also be consistent with the synthesis of molecules of the type and complexity of the product of interest. In addition, unfavorable or unnecessary elements of the genome should be removed when developing an organism as a microbial chassis [18].
In this study, Streptomyces sp. FR-008 was selected as a potential host organism for use as a chassis for heterologous expression. Streptomyces sp. FR-008 originated from the random protoplast fusion of two Streptomyces strains derived from Streptomyces hygroscopicus var. yingchengensis n. var. [19]. The mutant Streptomyces sp. FR-008 reportedly exhibits greater production of antibacterial factors than its parent strains and has been investigated extensively with respect to biosynthesis of the macrolide antibiotic candicidin. The candicidin biosynthesis gene cluster is 130-kb in size, and synthesis of candicidin involves a type I polyketide [20], suggesting that this organism can accommodate large biosynthetic machinery. More importantly, Streptomyces sp. FR-008 can produce 0.4 mg/L of candicidin via 3-days shake-flask fermentation in yeast extract-malt extract (YEME) medium [21], a medium normally used for growing Streptomyces rather than for fermentation. In addition, Streptomyces sp. FR-008 grows rapidly, which is a valuable characteristic for a chassis organism.
To evaluate the potential of Streptomyces sp. FR-008 as a host for heterologous expression, the genome was first thoroughly characterized via PacBio sequencing and transcriptome analysis. In addition, a simplified conjugation method was developed and promoter element usage was assessed. Finally, all three endogenous polyketide biosynthesis gene clusters, comprising 150-kb of the chromosome, including the candicidin biosynthesis gene cluster, were knocked out in the absence of resistance markers to generate a relatively clean host free of polyketide competition. Based on these experiments, we were able to engineer Streptomyces sp. FR-008 to serve as a chassis with a simplified, less complex genotypic background. The organism can be further engineered via gene transfer and promoter element selection as a platform for heterologous expression of desired products.
2. Materials and methods
2.1. Microorganisms, plasmids, and culture conditions
Details regarding the bacterial strains and plasmids used in this study are provided in Table S1. The primers used in the study are listed in Table S2. Streptomyces sp. FR-008 was obtained from the collection of Professor Zixin Deng [20]. E. coli ET12567, harboring pUZ8002, was used as a vector donor for intergeneric conjugation [22]. All E. coli strains were cultivated in LB medium, whereas soy flour mannitol (SFM) solid medium (2% agar, 2% mannitol, 2% soybean powder [pH 7.2]) and 3% tryptone soy broth (TSB) (pH 7.2) liquid medium were used to cultivate Streptomyces strains and mutants. Fermentation was performed in liquid YEME medium (0.3% yeast extract, 0.5% Bacto-peptone, 0.3% malt extract, 1% glucose, 10.3% sucrose [pH 7.2]). E. coli strains were grown at 37 °C, and Streptomyces strains were cultured at 30 °C.
2.2. Complete genome sequencing and assembly
The PacBio sequencing platform was utilized for determining the genome sequencing of Streptomyces sp. FR-008 [23]. First, gDNA was fragmented to an average size of 10 kb and sequenced using PacBio RSII. Nine single-molecule real-time sequencing (SMRT) cells were used for each 3-h sequencing reaction. The PacBio consensus calling feature was used for correction of PacBio long reads, and the reads were assembled according to the Hierarchical Genome Assembly Process (HGAP) method [24]. An interpolated Markov model was used for whole-genome gene predictions. Protein-coding sequences (CDSs) were scanned using GLIMMER 3.0 [25], and their functions were predicted based on BLASTP homology searching under the following criteria: alignment of E-values, <1e−5; identity, >35%. Each gene was cataloged according to biological function in the cluster of orthologous groups (COG) database. Additionally, genes involved in secondary metabolite production were identified using antiSMASH 3.0 online software [26]. Data regarding the complete genome of Streptomyces sp. FR-008 were submitted to GenBank under accession numbers CP009802–CP009804.
2.3. Biomass measurement
Cell density was determined by measuring dry cell weight (dcw). A 2-mL culture was collected in a pre-weighed Eppendorf tube and immediately centrifuged at 10,000 × g for 20 min at 4 °C. The residual cells were washed twice with distilled water and then the supernatant was discarded. The pellet was dried to a constant weight at 105 °C, and the mass gain was measured.
2.4. Transcriptome sequencing
Cell pellets were rapidly harvested by centrifugation and flash frozen in liquid nitrogen. Total RNA was extracted using a bacteria kit (SBS Genetech Inc., Beijing, China). SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) and a TruSeq RNA sample prep kit v2 (Illumina) were used for the first- and second-strand cDNA synthesis. Library purification was performed using AMPure XP beads (Beckman), and quality was verified using an Agilent 2100 Bioanalyzer (Agilent Technologies, High Sensitivity DNA kit) and a Qubit® 2.0 fluorometer (Invitrogen, dsDNA HS Assay kit). Sequencing cluster generation and sequencing were performed on a Genome Analyzer IIx (Illumina) at Bohao Biotechnology Company (Shanghai, China). A single-end, 50-bp sequencing strategy was employed, and more than 89 million reads for each sample were sequenced using a Hiseq 2500 system (Illumina). Reads were aligned to the genome of Streptomyces sp. FR-008 using SOAPaligner/soap2 (version 2.21) with only unique matches, and the transcript level of each sample was quantified as reads per kilobase per million mapped reads (RPKM) [27]. Further comparisons between samples were normalized by taking the log2 of the RPKM value. The RNA-Seq data were submitted to the NCBI under accession number SRX1022665.
2.5. Conjugation
To optimize the conjugation procedure, the shuttle vector pIB139 [28] was transferred to E. coli ET12567/pUZ8002 by electroporation. A single colony was picked, cultured overnight, and then inoculated at a 1:10 dilution in fresh LB medium and cultured until the OD600 reached 0.8. These cells served as the donors. Spores harvested from an SFM plate were subjected to streamlined heat-shock (50 °C, 10 min) and pre-germination (37 °C, 2 h) steps to prepare them for use as recipient cells. The recipient and donor cells were counted using a dilution plating method and then mixed at various ratios and inoculated onto an SFM plate supplemented with 20 mM MgCl2. The plate was incubated at 30 °C for 11 h and then overlaid with 1 mL of sterile water containing apramycin and nalidixic acid. Approximately 30 h later, white exconjugants were detected. Several exconjugants were picked into TSB liquid medium with apramycin and then examined by PCR to confirm the presence of apramycin-resistance genes (using the primer pairs Apr-checkF and Apr-checkR).
2.6. Gene knockout
For targeted gene knockout, pJTU1278 was used on the basis of homologous recombination [29]. First, the two flanking arms were designed using Primer Premier5, with an additional digestion site included for cloning (primer pairs LA1-F/LA1-R and RA1-F/RA1-R for constructing LQ1; primer pairs LA2-F/LA2-R and RA1-F/RA1-R for constructing LQ2; and primer pairs LA3-F/LA3-R and RA1-F/RA1-R for constructing LQ3). The pJTU1278-derived construct was generated through cloning, with the apramycin-resistance marker included between the flanking arms. This construct was transformed into E. coli ET12567/pUZ8002 for first-round conjugation. Thiostrepton-resistant exconjugants were selected as single-crossover mutants. After sporulation on SFM medium without antibiotic selection, double-crossover mutants were selected based on thiostrepton-sensitive and apramycin-resistant and then verified by PCR analysis with flanking primers (primer pairs Check-F1 and Check-R1 for verifying LQ1, Check-F2 and Check-R2 for verifying LQ2, and Check-F3 and Check-R3 for verifying LQ3). In the next round of conjugation, the same flanking arms were ligated into pJTU1278. The thiostrepton-resistant exconjugants were selected as single-crossover mutants. After nonselective growth, apramycin- and thiostrepton-sensitive mutants were selected as targeted knockout mutants without the use of resistance markers. The genotype of the mutants was verified by PCR analysis with the flanking primers used in the first-round conjugation.
2.7. Test of promoter strength based on GusA activity
A total of 20 promoters were selected for evaluation based on previous reports and synthesized by Genewiz Corp. (Suzhou, China) [30], [31], [32] (Table S3). The promoters were then cloned into pSET152 [33] upstream of the gusA reporter gene by simple cloning. The resulting constructs (pLH1–pLH20) were transformed into Streptomyces sp. FR-008 using the simplified conjugation method developed in this study. Positive exconjugants were picked into TSB liquid medium containing apramycin, and introduction of gusA was verified by PCR amplification with primer pairs Gus-F and Gus-R. The selective strength of the promoter harbored by each transformant was assessed based on the enzymatic activity of the reporter protein GusA, according to a previous report [34].
3. Results
3.1. Complete Streptomyces sp. FR-008 genome sequence
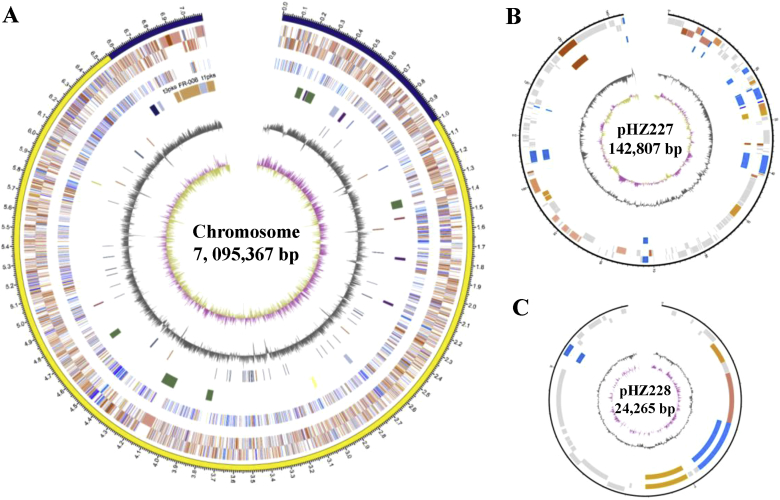
In order to obtain the detailed genetic information, the complete genome sequence of Streptomyces sp. FR-008 was determined using PacBio single-molecule real-time sequencing. Using the updated PacBio RSII, nine SMRT cells yielded 1.40 Gb of raw data (N50 length 7604 bp) in a 3-h sequencing reaction. Genome assembly performed directly from the PacBio data yielded three contigs (Fig. 1; GenBank accession numbers CP009802–CP009804). In addition to the linear chromosome, Streptomyces sp. FR-008 harbors two linear plasmids: pHZ227 and pHZ228. With a total length of 7,258,032 bp, the Streptomyces sp. FR-008 genome is one of the smallest Streptomyces genomes thus far reported.
Fig. 1.
Schematic representation of the Streptomyces sp. FR-008 chromosome and the two linear plasmids. (A) The chromosome atlas. From the outside in, circles 1 and 2: predicted genes (reverse and forward strands, respectively) colored according to cluster of orthologous groups (COG) function categories; circle 3: essential genes (cell division and chromosome partitioning, replication, transcription, translation, amino acid/nucleotide transport and metabolism, color coding as for circles 1 and 2); circle 4: biosynthetic gene clusters (three PKS gene clusters are labeled in orange); circle 5: tRNA and rRNA (deep blue and brown, respectively); circle 6: GC content; circle 7: GC skew ([G – C/G + C], khaki indicates values > 0, purple values < 0). (B) and (C) Atlases of linear plasmids pHZ227 and pHZ228, respectively. Circles 1 and 2: predicted genes (reverse and forward strands, respectively) colored according to COG function categories; circle 3: essential genes (cell division and chromosome partitioning, replication, transcription, translation, amino acid/nucleotide transport and metabolism, color coding as for circles 1 and 2); circle 4: GC content; circle 5: GC skew (G − C/G + C).
Basic information regarding the Streptomyces sp. FR-008 genome is shown in Table 1. The linear chromosome is comprised of 6920 predicted CDSs, with an average G + C content of 73.41%. The replication origin, oriC, is located near the middle of the chromosome. In comparison to other sequenced Streptomyces chromosomes, the center essential “core region” of the Streptomyces sp. FR-008 chromosome was predicted to be relatively small, at 5.56 Mb. The two linear plasmids, pHZ227 and pHZ228, contain 180 and 27 CDSs, respectively. It should be noted that the chromosome was found to lack the conserved dnd genes responsible for DNA phosphothiolation. In addition, along with the CRISPR-Cas bacterial immune system, a set of genes involved in arsenic resistance was identified on pHZ227 [35]. These components may protect the cells against the toxic effects of arsenic, as well as damage from exogenesis sources, including phages and other invasive genetic elements.
Table 1.
General features of the complete genome sequence of Streptomyces sp. FR-008.
| Value | Chromosome | pHZ227 | pHZ228 |
|---|---|---|---|
| Topology | Linear | Linear | Linear |
| Length (bp) | 7, 090, 956 | 142, 804 | 24, 272 |
| GC content (%) | 73.41% | 69.34% | 72.87% |
| CDS (no.) | 6920 | 178 | 29 |
| Coding (%) | 76.10% | 37.78% | 25.91% |
| rRNA operon (no.) | 7 | ||
| tRNA (no.) | 65 (43 species) |
For a more in-depth analysis of the Streptomyces sp. FR-008 genome, COG function category and antiSMASH analyses were carried out [26] (Fig. S1). The highest proportion of genes was found to be that involved in transcription (11%), and genes associated with 12 diverse regulatory families were identified, providing evidence of an inherent multi-level transcription regulatory network in Streptomyces sp. FR-008. The global transcript gamma-butyrolactone system was not identified in the Streptomyces sp. FR-008 genome, however. The second richest COG category included genes involved in carbohydrate (9%) and amino acid (8%) transport and metabolism. In addition, nearly 6% of CDSs were found to be associated with transport systems, primarily the ATP-binding cassette (ABC) and major facilitator superfamily transporters. Moreover, a combined antiSMASH analysis and NCBI homology search revealed 23 putative secondary metabolic gene clusters on the Streptomyces sp. FR-008 chromosome, which is consistent with the high biosynthesis potential of most Streptomyces (Table S4). Eleven of the 21 putative secondary metabolite gene clusters were found to be involved in non-ribosomal peptide synthetases/polykeide (NRPS/PKS) biosynthesis, two of which was defined as encoding type I PKS and the other as encoding type III PKS. Compared with other annotated Streptomyces genomes, that of Streptomyces sp. FR-008 encodes a relatively small PKS gene pool, which may explain the strain's efficient production of the macrolide candicidin with minimal homologous competition.
3.2. Transcriptional profiling during different growth phases
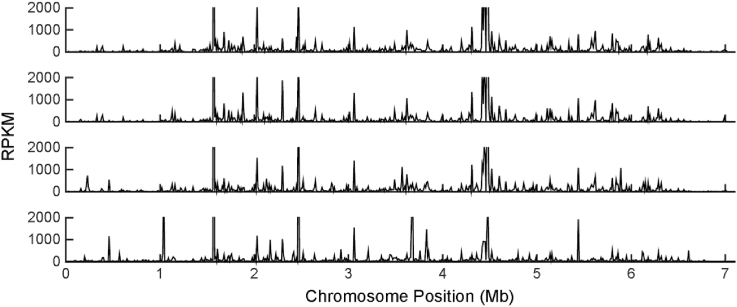
Genome annotation analyses were augmented by comprehensive transcriptional profiling using RNA-Seq analysis of Streptomyces sp. FR-008 over time. Cells were grown in liquid TSB medium and harvested at four time points: 14, 18, 24, and 36 h, representing the lag, exponential, transitional, and stationary growth phases, respectively (Fig. S2). As reported, the chromosome site of heterologous gene integration could affect gene expression as a consequence to influence targeted production [36]. To locate the chromosomal region exhibiting high expression, we conducted a global transcriptome analysis (Fig. 2). The majority of genes that maintained a high expression level at all four time points examined was found to be involved in translation and energy production and was located on different regions of the chromosome. Besides, individual genes keep at highly expression levels under all four time points can be selected as candidate of strong constitutive promoters as reported in S. albus J1074 case [32]. Here, we selected top 1% of the most highly expressed genes as conclude in Table S5. And the promoter sequence upstream of these 1% cutoff genes can be predictable by bioinformatics tools [37], in order to generate a panel of strong constitutive promoters.
Fig. 2.
Global view of gene expression profiles through physical location of chromosome. The X-axis is indicated the site on chromosome, and the Y-axis is indicated the gene expression value of RPKM based on RNA-Seq data.
The expression profiles of 23 predicted gene clusters were also examined (Fig. S3). The results showed that the expression of five NRPS-related gene clusters began during different growth phases. The expression of candicidin biosynthesis genes began before the lag phase and continued until the stationary phase, consistent with the short fermentation time observed in a previous experiment [21]. Expression of transcripts for other secondary metabolite genes was weak or absent in cells grown under these conditions in TSB medium. In addition, based on hypergeometric distribution and COG functional enrichment, the up- or down-regulated genes were assigned to 21 functional groups (Fig. S4). Genes involved in metabolism of amino acids, nucleotides, and inorganic ions were all up-regulated before the exponential phase and then down-regulated in the stationary phase. In contrast, genes involved in carbohydrate metabolism were up-regulated in the stationary phase, suggesting that there is a high demand for carbohydrate products during the later growth stages compared with earlier stages, when primary metabolism is sufficient. Genes involved in transcription were markedly up-regulated in the stationary phase, indicating that specific transcription regulators play important roles in regulating metabolism during the late stages of growth.
3.3. Genetic manipulation
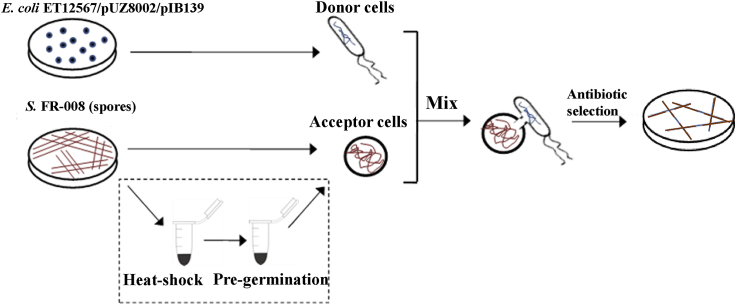
Various practical challenges have limited the genetic manipulation of actinomyces. Based on the high conjugation efficiency of Streptomyces sp. FR-008 observed in our past work, in the present study, we sought to optimize its conjugation procedure, focusing on the heat-shock and pre-germination steps of recipient. First, the optimal ratio of donor to recipient cells was assessed. The results of these analyses showed that an equal ratio provided the highest conjugation efficiency (Table S6). Thus, subsequent optimizations were carried out at an equal donor to recipient ratio. We then examined the effect of omitting the heat-shock treatment of recipient spores. Surprisingly, omitting the heat-shock step had no significant effect on efficiency. Similar optimization analyses of the pre-germination steps were performed, and the results showed that omitting the pre-germination steps also had no appreciable effect on conjugation efficiency. The conjugation efficiency afforded by this simplified procedure involving only the mixing of donors and acceptors, without the heat-shock and pre-germination steps (Fig. 3), was approximately 20% higher (Table 2). Notably, due to the rapid growth rate of Streptomyces sp. FR-008, the time required for the appearance of exconjugants or resistance selection was reduced to approximately 30 h, far less than the 3–5 days required for most Streptomyces strains.
Fig. 3.
Schematic diagram of workflow for comparing the simplified and general conjugation procedures. Refer to the text in the corresponding sections for detailed descriptions of each step. The procedures in the dashed box included the heat-shock and pre-germination steps, which were omitted in the simplified conjugation procedure developed in this study.
Table 2.
Conjugation efficiency at different recipient treatments omitted with heat-shock or pre-germinationa.
| Donor | Conjugation efficiency with different recipient treatments ( × 10−6) |
|||
|---|---|---|---|---|
| Heat-shock (+) pre-germination (+) | Heat-shock (+) pre-germination (−) | Heat-shock (−) pre-germination (+) | Heat-shock (−) pre-germination (−) | |
| E. coli ET12567 (pUZ8002)/pIB139 | 1.64 ± 0.19 | 1.56 ± 0.18 | 1.68 ± 0.16 | 2.07 ± 0.17 |
All data are mean values of three independent experiments, and the error bars indicate the standard deviation.
3.4. Assessment of promoter strength in the host
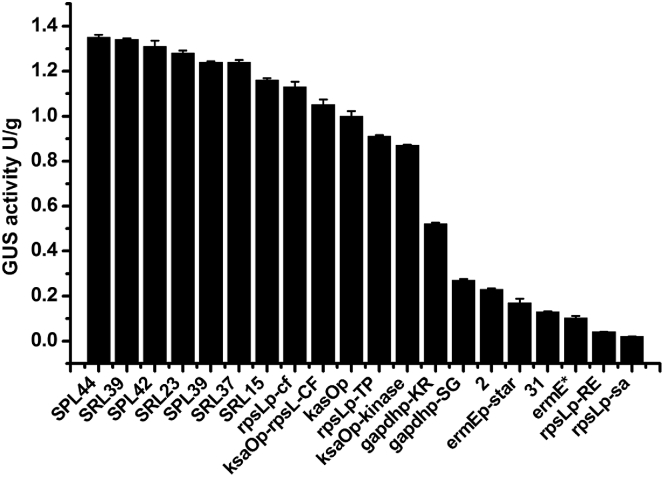
In engineering a heterologous expression pathway, a panel of well-characterized strong promoters is typically used to fine-tune expression of the desired genes. Previously, the promoter ermEp was the only available option for Streptomyces sp. FR-008 [21], [38]. Recently, however, synthetic promoters stronger than ermEp have been used to overproduce various natural products [30], [39]. To provide more promoter options for Streptomyces sp. FR-008, we chose 20 strong promoters as candidates and evaluated them with respect to regulation of GusA activity in Streptomyces sp. FR-008. The selected promoters played different regulatory roles, included promoters derived from temporally expressed kasOp for a SARP family regulator in S. coelicolor [30], [31], and constitutive promoters identified through transcriptome analyses in S. albus J1074 [32] (Table S3). These promoters exhibited reproducible and homogeneous activity ranging from 60 to 860% of the activity of ermE*-p (Fig. 4).
Fig. 4.
Strength of selected promoters in Streptomyces sp. FR-008. The strength of each promoter (black bars) was assessed based on GusA activity. All data are mean values of three independent experiments, and error bars indicate the standard deviation.
3.5. Streamlining the Streptomyces sp. FR-008 genome by knockout of PKS gene clusters
Candicidin is the main secondary metabolite of Streptomyces sp. FR-008 [20]. To generate a smaller and more streamlined genome encoding a minimum of extracellular metabolites, we deleted all of the PKS genes in Streptomyces sp. FR-008. Analyses of the complete genome identified three PKS gene clusters. The type I PKS cluster is responsible for candicidin biosynthesis. Although the compounds produced by another type I and type III PKS clusters are unknown, it is necessary to remove similar genes to generate a clean polyketide background. Because of the relatively restricted size of the “arm region” in Streptomyces sp. FR-008, it is difficult to predict whether deletion of a large segment of the genome would result in a significant change in the chromosome structure. Therefore, we proceeded cautiously, sequentially deleting the targeted PKS genes. As shown in Fig. S5, all three PKS gene clusters were sequentially deleted, producing strains LQ1, LQ2, and LQ3. The genes were deleted in the following order: candicidin biosynthesis gene cluster, SFR6781 (type III PKS), and SFR6902–SFR6904 (type I PKS). Complete deletion of these gene clusters and joining of the neighboring sequences were confirmed by PCR analysis (Fig. S5). The genome of host LQ3 was streamlined by the removal of nearly 156 kb of the genome sequence while avoiding the competing sinks of metabolic flux and enabling the characterization of transfer of exogenous gene clusters. It should also be noted that in construction of LQ2, in which the type III PKS was deleted, we obtained the targeted double-crossover mutant without first distinguishing the single-crossover mutant, illustrating that the host is amenable to genetic manipulation.
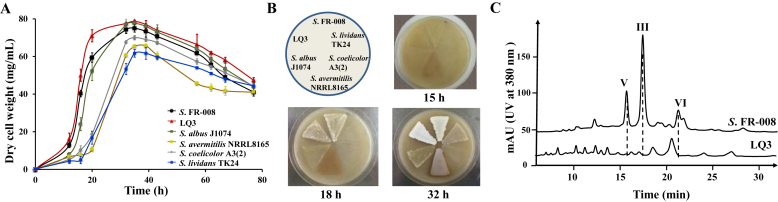
We examined the growth and sporulation characteristics of host LQ3 in comparison with the parental strain and four other widely used hosts. LQ3 exhibited a slightly higher growth rate than Streptomyces sp. FR-008 in TSB liquid medium, reaching a higher biomass yield in a shorter time (Fig. 5A). However, both strains exhibited similar sporulation rates on SFM agar plates (Fig. 5B). Under the conditions used here, Streptomyces sp. FR-008 and LQ3 exhibited the best growth profiles of all the strains examined. YEME medium, typically used for candicidin fermentation, was used to compare the production of secondary metabolites by Streptomyces sp. FR-008 and LQ3. High-performance liquid chromatography analysis of whole-broth ethyl acetate extracts confirmed the absence of major endogenous metabolites (Fig. 5C).
Fig. 5.
Comparison of growth phenotype and high-performance liquid chromatography profiles of Streptomyces sp. FR-008 and its targeted deletion mutant, LQ3. (A) Growth curves of Streptomyces sp. FR-008 and LQ3 along with four other general Streptomyces hosts grown in TSB liquid medium. (B) Sporulation phenotype of Streptomyces sp. FR-008 and LQ3 along with four other general Streptomyces hosts grown on SFM agar medium. (C) High-performance liquid chromatography traces of ethyl acetate extracts of Streptomyces sp. FR-008 and LQ3 cultured in YEME liquid medium for 4 days. Peaks labeled V, III, and VI are indicated as candicidin-V, –III, and –VI respectively, which are three main candicidin components in the extracts of the wild-type. mAU, milliabsorbance units.
To further evaluate differences between wild type Streptomyces sp. FR-008 and LQ3, RNA sequencing was employed to analyze their gene expression profiles in the stationary phase, when the majority of secondary metabolites are produced. Overall, 101 genes were either up- or down-regulated at least 2-fold in LQ3 (39 genes were up-regulated and 62 genes were down-regulated). Most of these genes (66/101) were found to encode hypothetical proteins. Genes encoding ABC transporter permease and CoA synthetase were slightly up-regulated, and genes encoding some transporter and transmembrane proteins were down-regulated, with weak expression in LQ3. No expression of PKS genes was detected, verifying the clean PKS background of LQ3. Thus, the targeted deletion of PKS to produce mutant LQ3 did not affect transcription from a global perspective. These data suggested that the genome of LQ3 is stable under the conditions examined in this study.
4. Discussion
The results of a previous study showed that several Streptomyces strains and corresponding deletion mutants were suitable for use as chassis for industrial biosynthesis [2], [5]. Due to the differences in mechanism and requirements for each type of biosynthesis, and the complex nature of metabolic regulation, a well-defined chassis for improved heterologous expression is desirable. Hence, in this study, we focused on engineering a chassis derived from Streptomyces sp. FR-008.
Through PacBio sequencing, the complete Streptomyces sp. FR-008 genome was assembled directly into three contigs, suggesting that this sequencing platform and assembly method are ideally suited for GC-rich species. It was surprising to find that the size of the Streptomyces sp. FR-008 genome was naturally minimized, with only three PKS gene clusters, which is consistent with the strain's relatively rapid, high-titer candicidin fermentation capability. Additionally, the smaller and thus relatively simple genome of Streptomyces sp. FR-008 is ideally structured for genome-scale modeling. The rapid growth potential of Streptomyces sp. FR-008, in combination with the optimized conjugation system we developed and the promoter elements we characterized as ideally suited for heterologous expression, suggest that this strain would be an useful chassis. The rapid growth rate and amenability to the transfer of exogenesis genes provides a platform for simpler and less time-consuming heterologous expression without a loss of efficiency.
To provide for a less complex genomic and transcriptomic background, we deleted the PKS gene clusters located in the 0.53-Mb right subtelomeric region of the Streptomyces sp. FR-008 chromosome. Considering that the genome architecture could be affected by both the linear composition and three-dimensional structure of the chromosome, LQ3 was generated by sequential deletion of the three PKS gene clusters. Strain LQ3 thus represents an “updated” version of Streptomyces sp. FR-008, with a more stable and streamlined genome. The minimization of secondary metabolite production in LQ3 provides for simpler and more effective separation and purification processes. Similar to S. coelicolor M1152 and its parental strain, S. coelicolor M14, LQ3 lacks three native biosynthetic gene clusters compared with Streptomyces sp. FR-008, thus providing for heterologous expression yields that are dozens of times higher than those of the parental strain [11]. Combined with the convenient genetic operation system and fine-tuning of transcriptional control elements, further ‘plug-and-play’ manipulation of this chassis could be carried out to increase its metabolic potential.
5. Conclusion
Expressing heterologous native drug biosynthesis genes in easily manipulated host cells can overcome the unfavorable characteristics of many native producing organisms, which are often difficult to culture and manipulate genetically. The use of more amenable, well-characterized surrogate host organisms to express cryptic gene clusters from un-culturable strains could facilitate the production of novel chemicals. The availability of a well-characterized, fast growing, and genetically pliable organism as a biosynthesis “chassis” would greatly enhance the success of heterologous expression efforts. We evaluated the potential of Streptomyces sp. FR-008 to serve as an emerging chassis. We conducted genome sequencing and transcriptome analyses, developed a simplified conjugation method, and identified strong promoters to produce an improved PKS gene knockout mutant suitable as a platform for the expression of exogenous genes for a variety of biotechnology applications.
Acknowledgments
This research was supported by grants from J1 Biotech Co. Ltd., the 973 Project (2011CBA00800, 2012CB721000) and the 863 Project (2012AA02A701) from the Ministry of Science and Technology of China, and from the Natural Science Foundation of Hubei Province (2015CFB415).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.synbio.2016.07.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Patridge E., Gareiss P., Kinch M.S., Hoyer D. An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discov Today. 2016;21:204–207. doi: 10.1016/j.drudis.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Ongley S.E., Bian X., Neilan B.A., Müller R. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat Prod Rep. 2013;30:1121–1138. doi: 10.1039/c3np70034h. [DOI] [PubMed] [Google Scholar]
- 3.Zarins-Tutt J.S., Barberi T.T., Gao H., Mearns-Spragg A., Zhang L., Newman D.J. Prospecting for new bacterial metabolites: a glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat Prod Rep. 2016;33:54–72. doi: 10.1039/c5np00111k. [DOI] [PubMed] [Google Scholar]
- 4.Milshteyn A., Schneider J.S., Brady S.F. Mining the metabiome: identifying novel natural products from microbial communities. Chem Biol. 2014;21:1211–1223. doi: 10.1016/j.chembiol.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M.M., Wang Y., Ang E.L., Zhao H. Engineering microbial hosts for production of bacterial natural products. Nat Prod Rep. 2016;33:963–987. doi: 10.1039/c6np00017g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berdy J. Bioactive microbial metabolites. J Antibiot (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 7.Hopwood D.A. Genetic contributions to understanding polyketide synthases. Chem Rev. 1997;97:2465–2498. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 8.Kieser T., Bibb M.J., Buttner M.J., Chater K.F., Hopwood D.A. John Innes Foundation; Norwich, United Kingdom: 2000. Practical Streptomyces genetics. [Google Scholar]
- 9.McDaniel R., Ebert-Khosla S., Hopwood D.A., Khosla C. Engineered biosynthesis of novel polyketides. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 10.Ziermann R., Betlach M.C. Recombinant polyketide synthesis in Streptomyces: engineering of improved host strains. Biotechniques. 1999;26:106–110. doi: 10.2144/99261st05. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Escribano J.P., Bibb M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omura S., Ikeda H., Ishikawa J., Hanamoto A., Takahashi C., Shinose M. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci U. S. A. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda H., Ishikawa J., Hanamoto A., Shinose M., Kikuchi H., Shiba T. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 14.Komatsu M., Uchiyama T., Omura S., Cane D.E., Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U. S. A. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komatsu M., Uchiyama T., Omura S., Cane D.E., Ikeda H. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol. 2013;2:384–396. doi: 10.1021/sb3001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaburannyi N., Rabyk M., Ostash B., Fedorenko V., Luzhetskyy A. Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genomics. 2014;15:97. doi: 10.1186/1471-2164-15-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Mulert U., Luzhetskyy A., Hofmann C., Mayer A., Bechthold A. Expression of the landomycin biosynthetic gene cluster in a PKS mutant of Streptomyces fradiae is dependent on the coexpression of a putative transcriptional activator gene. FEMS Microbiol Lett. 2004;230:91–97. doi: 10.1016/S0378-1097(03)00861-9. [DOI] [PubMed] [Google Scholar]
- 18.Tyo K.E., Kocharin K., Nielsen J. Toward design-based engineering of industrial microbes. Curr Opin Microbiol. 2010;13:255–262. doi: 10.1016/j.mib.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang R.F., Zhou Q. Studies on fusion breeding of protoplasts from antibiotic producing strain 5102. Chin J Biotechnol. 1987;3:130–136. [PubMed] [Google Scholar]
- 20.Chen S., Huang X., Zhou X., Bai L., He J., Jeong K.J. Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem Biol. 2003;10:1065–1076. doi: 10.1016/j.chembiol.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Wang T., Bai L., Zhu D., Lei X., Liu G., Deng Z. Enhancing macrolide production in Streptomyces by coexpressing three heterologous genes. Enzyme Microb Technol. 2012;50:5–9. doi: 10.1016/j.enzmictec.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Mazodier P., Petter R., Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J Bacteriol. 1989;171:3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koren S., Harhay G.P., Smith T.P., Bono J.L., Harhay D.M., McVey S.D. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14:R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chin C.S., Alexander D.H., Marks P., Klammer A.A., Drake J., Heiner C. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 25.Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson C.J., Hughes-Thomas Z.A., Martin C.J., Bohm I., Mironenko T., Deacon M. Increasing the efficiency of heterologous promoters in actinomycetes. J Mol Microbiol Biotechnol. 2002;4:417–426. [PubMed] [Google Scholar]
- 29.He Y., Wang Z., Bai L., Liang J., Zhou X., Deng Z. Two pHZ1358-derivative vectors for efficient gene knockout in streptomyces. J Microbiol Biotechnol. 2010;20:678–682. doi: 10.4014/jmb.0910.10031. [DOI] [PubMed] [Google Scholar]
- 30.Bai C., Zhang Y., Zhao X., Hu Y., Xiang S., Miao J. Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proc Natl Acad Sci U. S. A. 2015;112:12181–12186. doi: 10.1073/pnas.1511027112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W., Li X., Wang J., Xiang S., Feng X., Yang K. An engineered strong promoter for streptomycetes. Appl Environ Microbiol. 2013;79:4484–4492. doi: 10.1128/AEM.00985-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y., Zhang L., Barton K.W., Zhao H. Systematic identification of a panel of strong constitutive promoters from streptomyces albus. ACS Synth Biol. 2015;4:1001–1010. doi: 10.1021/acssynbio.5b00016. [DOI] [PubMed] [Google Scholar]
- 33.Bierman M., Logan R., O'Brien K., Seno E.T., Rao R.N., Schoner B.E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 34.Siegl T., Tokovenko B., Myronovskyi M., Luzhetskyy A. Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab Eng. 2013;19:98–106. doi: 10.1016/j.ymben.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Wang L., Chen S., Xiao X., Huang X., You D., Zhou X. arsRBOCT arsenic resistance system encoded by linear plasmid pHZ227 in Streptomyces sp. strain FR-008. Appl Environ Microbiol. 2006;72:3738–3742. doi: 10.1128/AEM.72.5.3738-3742.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryant J.A., Sellars L.E., Busby S.J., Lee D.J. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014;42:11383–11392. doi: 10.1093/nar/gku828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siwo G., Rider A., Tan A., Pinapati R., Emrich S., Chawla N. Prediction of fine-tuned promoter activity from DNA sequence. F1000Research. 2016;5:158. doi: 10.12688/f1000research.7485.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y., Meng Q., You D., Li J., Chen S., Ding D. Selective removal of aberrant extender units by a type II thioesterase for efficient FR-008/candicidin biosynthesis in Streptomyces sp. strain FR-008. Appl Environ Microbiol. 2008;74:7235–7242. doi: 10.1128/AEM.01012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo Y., Huang H., Liang J., Wang M., Lu L., Shao Z. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun. 2013;4:2894. doi: 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.