Abstract
Multi-drug resistance of pathogenic microorganisms is becoming a serious threat, particularly to immunocompromised populations. The high mortality of systematic fungal infections necessitates novel antifungal drugs and therapies. Unfortunately, with traditional drug discovery approaches, only echinocandins was approved by FDA as a new class of antifungals in the past two decades. Drug efflux is one of the major contributors to multi-drug resistance, the modulator of drug efflux pumps is considered as one of the keys to conquer multi-drug resistance. In this study, we combined structure-based virtual screening and whole-cell based mechanism study, identified a natural product, beauvericin (BEA) as a drug efflux pump modulator, which can reverse the multi-drug resistant phenotype of Candida albicans by specifically blocking the ATP-binding cassette (ABC) transporters; meantime, BEA alone has fungicidal activity in vitro by elevating intracellular calcium and reactive oxygen species (ROS). It was further demonstrated by histopathological study that BEA synergizes with a sub-therapeutic dose of ketoconazole (KTC) and could cure the murine model of disseminated candidiasis. Toxicity evaluation of BEA, including acute toxicity test, Ames test, and hERG (human ether-à-go-go-related gene) test promised that BEA can be harnessed for treatment of candidiasis, especially the candidiasis caused by ABC overexpressed multi-drug resistant C. albicans.
Keywords: Candida albicans, ABC transporter, Beauvericin, Virtual screening, Multi-drug resistance, Synergy
1. Introduction
The opportunistic fungal pathogen Candida albicans is one of the major causes of systemic severe infections with a high mortality. It represents an emerging global health threat, especially among the immunocompromised populations.1, 2 Unfortunately, only a limited number of effective classes of antifungals are available in the clinic:3 (i) azoles, such as fluconazole, which inhibit Erg11 and blockergosterol biosynthesis; (ii) polyenes, such as amphotericin B, which bind to ergosterol, forming pores in the cell membrane; (iii) echinocandins, such as caspofungin, which inhibit β-(1,3)-d-glucan synthase and disrupt cell-wall integrity; (iv) pyrimidine analogs, such as flucytosine, which inhibit DNA and RNA synthesis, and (v) griseofulvin, which deteriorate spindle and cytoplasmic microtubules, to influence bud formation. Unwanted side-effects remain a major problem in clinical use of these antifungal drugs. With increased prophylactic use of antifungal drugs in the expanding susceptible populations, including those immunocompromised patients, multi-drug resistance among fungal pathogens has significantly increased.3 There are various mechanisms that contribute to the multi-drug resistance of C. albicans.4, 5, 6 Among them, two are particularly predominant: (i) reducing intracellular drug accumulation by overexpression of drug efflux pumps: Cdr1 and Cdr2 belong to ABC transporters; and Mdr1 belongs to the major facilitator super family (MFS);7 (ii) use of alternative pathways or altered target enzymes when the primary one is blocked, e.g. acquiring mutations in ERG11, which encodes the azole target enzyme 14α-demethylase.8 Multi-drug resistant pathogens have significantly increased the frequency of treatment failures. Traditional strategy to deal with multi-drug resistance is increasing the drug dosage or finding another alternative drug. Unfortunately, the speed of resistance generation is much faster than that of new drug discovery, thus generating a vicious circle for more severe drug resistance. Some new antifungal agents were discovered during the past few decades, however, after consideration of toxicity and other side-effects, only one new class of antifungal drugs, echinocandins, is approved by FDA.9 We so urgently need new antifungals and antifungal therapies, as well as the means to find them. It is believed that modulating the activity of drug efflux pumps, together with synergistic drug combination can be one of the keys to control multi-drug resistant fungal pathogens. Inhibiting drug efflux pumps can sensitize multi-drug resistance; while aiming more than one target can slow down or even prevent the emergence of drug-resistance. Co-crystallization study showed that P-glycoprotein (P-gp), one kind of ABC transporters, the main contributor to cancer multi-drug resistance, can be inhibited by some chemicals.10 Traditional drug discovery demands massive screening, which is extremely time, labor, and cost consuming. Structure-based drug discovery is emerging, because it can deliver handleable candidates for experimental evaluation in a faster, low cost and highly efficient manner.11 As the structures of Cdr1 and Cdr2 are not elucidated yet, we established homology models of Cdr1 and Cdr2 for structure-based virtual screening of the customized sub-library of ZINC15.12 Interestingly, one of the high score ABC inhibitor candidates is BEA, which was reported previously to have strong synergy with some azole compounds, like miconazole against both drug susceptible and resistant C. albicans in vitro,13 and KTC against diverse fungal pathogens both in vitro and in vivo,14 the synergistic antifungal effect was not caused by their pharmacokinetic interactions.15 However, its synergism and the underlying mechanism of killing were unknown. In this study, combining virtual screening, docking and experimental evaluation, we revealed the synergistic mechanism of BEA, additionally, we also found that BEA alone has fungicidal activity. Together with in vivo testing and toxicity evaluation, BEA showed promising features as a new antifungal agent.
2. Materials and methods
2.1. Experimental subjects, chemicals, primers, and culture media
Cancer cell lines (A549, H1299, H157, H460, H358, H522, H1650, H1792, H226, Calu-1), C. parapsilosis (ATCC22019), and C. albicans SC5314 were maintained in our laboratory. Clinical isolates of C. albicans, C. krusei, and C. tropicalis were provided by a local hospital. All strains were maintained in 25% (v/v) glycerol at −80 °C. Specific pathogen-free ICR mice (white, 18–22 g, half females), were obtained from B & K Universal Group Limited, Shanghai, China.
RPMI 1640 (Invitrogen) was used according to the manufacturer’s protocol. YEPD liquid medium consisted of: yeast extract 1% (w/v), peptone 2% (w/v), dextrose 2% (w/v), 2% (w/v) agar will be added for plates.
The complete list of microbes used in this study is provided in Table S1. All primers used in this study are listed in Table S2. All chemicals were used in this study according to the manufacturer’s directions.
2.2. Virtual screening
The input files of receptor were prepared by AutoDock/Vina plugin of PyMOL,16 the sdf formatted ligands were downloaded from ZINC15 3D Tranches,12 under the parameters of “React. : Standard”, “Purch. : Wait OK”, “pH: Ref Mid”, and “Charge: −2 −1 0 +1 +2”. All tranches were imported into PyRx17 by Open Babel, then the molecules were processed by energy minimization and then all of them were converted to AutoDock Ligands. The virtual screening was carried out by Vina Wizard, the grid boxes were manually adjusted to center_x = 21.55, center_y = 65.64, center_z = 13.71, size_x = 62.13, size_y = 50.11, size_z = 52.25 for Cdr1; while center_x = 21.06, center_y = 65.66, center_z = 14.23, size_x = 62.51, size_y = 50.60, size_z = 52.23 for Cdr2. The exhaustiveness for both Cdr1 and Cdr2 is 8 (up to 9 poses).
2.3. Antifungal susceptibility and synergistic antifungal testing
Antifungal and synergistic antifungal tests were carried out as described previously,14, 18 using a broth microdilution protocol modified from the Clinical and Laboratory Standards Institute M-27A319 methods. All minimal inhibitory concentration (MIC) tests were undertaken in triplicate. MICs were determined as the concentration of drugs that inhibited microbial growth by 90% relative to the corresponding drug-free growth control.
2.4. Rhodamine 6G efflux assay
Fungal strains were cultivated in YEPD liquid medium at 200 rpm (30 °C) for 16 hours. The harvested cells were washed twice with ice-cold glucose-free PBS. Cells were suspended in ice-cold glucose-free phosphate buffered saline (PBS) and incubated at 200 rpm (30 °C) for 4 hours under starvation conditions to reduce ABC pump activity. Cells were then washed twice and diluted to 108 cells/ml in ice-cold glucose-free PBS, as determined with a hemocytometer. BEA at a concentration of 16 µg/ml (about 2 × MIC) was added when necessary, while PBS was added as the negative control. All samples were incubated for another 2 hours at 200 rpm (30 °C). Then, 10 µM (final concentration) rhodamine 6G was added, and cells were incubated for further 1.5 hours at 200 rpm (30 °C). The external rhodamine 6G was then removed by washing with glucose-free PBS and glucose was added to the samples (to a final concentration of 3 mM) to reactivate the ABC efflux pumps, with PBS as the negative control. Cell samples (1 ml) were taken at designated time points, centrifuged and 100 µl of each supernatant was transferred into black 96-well microtiter plate with clear bottoms (Greiner, Germany). Rhodamine 6G fluorescence was measured with a Multilabel Plate Reader (Perkin Elmer, USA) at 510 nm excitation/535 nm emission wavelengths. Experiments were carried out at least in triplicate.
2.5. Intracellular calcium and ROS measurement
C. albicans SC5314 cells were cultivated in YEPD liquid medium at 200 rpm (30 °C) for about 16 hours. The harvested cells were washed twice with ice-cold PBS and diluted to 5 × 107 cells/ml with ice-cold PBS, as determined with a hemocytometer and confirmed by cfu counting. For calcium measurement, designated concentrations of each drug were added to 1 ml samples, and co-incubated at 200 rpm (30 °C) for 4 hours to get dose-dependent data. For the time-dependent assay, 64 µg/ml of each drug was added to 1 ml samples, and incubation was stopped at specific time points. For ROS detection, after co-incubation with each drug at 200 rpm (30 °C) for 4 hours, DCFH-DA (2′,7′-dichlorofluorescein diacetate) was added to a final concentration of 10 µM and cells were incubated for another 30 min at 200 rpm (30 °C). The cells were then washed to remove external drugs, resuspended in Ca NW working solution (according to DiscoverX HitHunter™ Calcium No WashPLUS Assay Kit protocol) and incubated at 37 °C, 200 rpm for 2 hours. Then 100 µl of each sample was transferred into black 96-well microtiter plate with clear bottoms (Greiner, Germany). The plate was equilibrated at room temperature for 6 hours, but this step was not necessary for ROS detection. The fluorescence was measured with a Multilabel Plate Reader (Perkin Elmer, USA) at 485 nm excitation/510 nm emission wavelengths for calcium measurement, and 485 nm excitation/525 nm emission wavelengths for ROS detection. Experiments were carried out at least in triplicate.
2.6. Mouse model and ethics statement
Animal related experiments were carried out in Guangdong Laboratory Animal Monitoring Institute, which has earned AAALAC accreditation (001469).
Specific-pathogen-free female ICR [Crl: CD-1] mice (female, white, 20–22 g) were used in this study. Experiments were carried out as described previously.14, 20 Animal use protocols were reviewed and approved by IACUC of Guangdong Laboratory Animal Monitoring Institute in accordance with the Guide for the Care and Use of Laboratory Animals. Animals were bred in negative pressure isolation cages in an animal negative pressure facility with an approval of and oversight by the Local Provincial Institutional Environmental Health and Safety Office,20 the whole project was carried out under the license IACUC2012006.
The animals, housed in cages of five mice per group and fed with standard rodent chow ad libitum, were allowed to acclimate for 1 week before active experimentation carried out. Before infection, mice were rendered neutropenic by i.p. injection of cyclophosphamide daily for 3 consecutive days at a dosage of 100 mg/kg body weight. Mice were then infected with 0.1 ml of 5 × 105 cfu/ml of C. albicans or C. parapsilosis in warmed saline (35 °C) by the lateral tail vein. Test compound(s) either alone or in combination (0.5 mg/kg BEA + KTC, and 50 mg/kg KTC) were administered orally by gavage 6 hours post infection and once daily thereafter for 5 days. A control group received 0.1 ml of saline by the same route as the placebo regimens. Animals were observed thrice daily for signs of drug-related morbidity or morality. Mice that became immobile or otherwise showed signs of severe illness were terminated and recorded as dying on the following day.
2.7. Necropsy and histopathology
Necropsy and histopathological analysis were carried out as described previously,20 briefly speaking, immediately after euthanasia in the 19th day of infection, the brain, heart, lungs, liver, spleen, and right kidney were immersed in buffered 10% formalin. After paraffin embedding and sectioning, standard 5 µm sections were cut and stained with hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS).
2.8. Quantification of differentially expressed genes by quantitative real time PCR
The primers used are listed in Table S2. RNA isolation, cDNA synthesis, and PCR amplification were carried out as manufacturer’s directions. Independent quantitative real time PCRs were performed in triplicate using the ABIPrism7300 Real-Time PCR System (Applied Biosystems, USA). The gene expression level relative to the calibrator was expressed as 2−ΔΔCT.
2.9. Acute toxicity assay
Forty specific pathogen-free ICR mice (white, 18–22 g, half females), housed in cages of five per group and fed with standard rodent chow ad libitum, were allowed to acclimatize for 1 week. Experiments were carried out in negative pressure stainless steel isolators at 24 ± 2 °C on mice that had been starved for the previous 12 hours. BEA was dissolved in 0.5% sodium carboxymethyl cellulose at the concentration of 0.05 g/ml. Oral administration of 0.4 ml/10g.w BEA (equivalent the dose of 2 g/kg) was performed by gavage. For 14 days, the mice were weighed thrice daily and observed for signs of drug-related morbidity or mortality. After the observation period, animals were killed by CO2 exposure followed by cervical dislocation.
2.10. Ames test and hERG test
The Ames test and the hERG test were done by Shanghai InnoStar Bio-Tech Co., Ltd.
2.11. BEA-resistant mutants screening
To select BEA-resistant mutants, we used a rapid selection regime in which SC5314 cells were plated onto YEPD containing a high concentration of BEA (512 µl/ml). Only colonies of the largest size (>1.6 mm2) that had acquired robust, reproducible resistance with MIC increasing more than 100 fold were chosen.
3. Results
3.1. Homology modeling of C. albicans ABC transporters and virtual screening for inhibitors
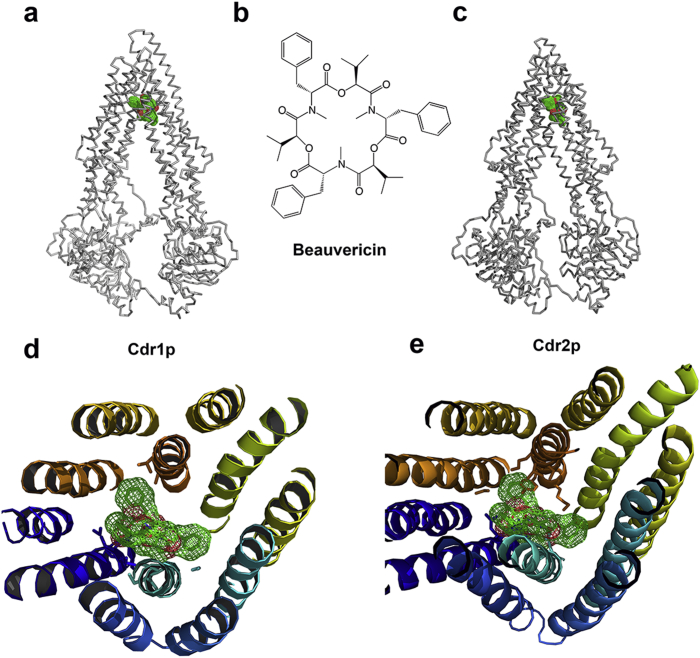
The homology models of the C. albicans ABC transporters, Cdr1 and Cdr2 were built using Alignment Mode algorithm of SwissModel server,21, 22, 23 taking a known mouse P-glycoprotein crystal structure (PDB code: 3G60)10 as the template, which shares 29% sequence identity and 50% similarity to Cdr1 and 31% sequence identity and 51% similarity to Cdr2, respectively. As co-crystallization showed QZ59-RRR (cyclic-tris-(R)-valineselenazole, Mol. wt. 687.42 Daltons) and QZ59SSS (cyclic-tris-(S)-valineselenazole, Mol. wt. 687.42 Daltons) can be poly-specifically bond to the P-gp internal cavity to block its efflux function.10 Based on the molecular weight of QZ59, we decided to screen the 3D ZINC15 database12 with >500 Daltons virtually. Meantime, QZ59-RRR and QZ59-SSS were added into our customized database manually as positive control. The virtual screening was carried out using the Vina Wizard of PyRx.17 No surprise that QZ59-RRR docked nicely to both Cdr1 and Cdr2 in a reasonable cavity position with the binding affinity of −9.0 kcal/mol and −8.3 kcal/mol, respectively; while the binding affinity for QZ59-SSS is −8.3 kcal/mol and −8.5 kcal/mol, respectively. In total, 912 compounds with more than 500 Daltons that have available 3D information from ZINC15 database12 were used for virtual screening. Each compound was screened for up to 9 poses for each receptor, the binding affinity of those around 16200 poses varied from −13.1 kcal/mol to −5.1 kcal/mol for Cdr1, and from −13.8 kcal/mol to −5.2 kcal/mol for Cdr2 (Table S3). By manually checking the reasonable docking poses with the UCSF Chimera,24 we interestingly found that 8 BEA compounds have even better binding affinities to the internal cavity of both Cdr1 and Cdr2 than QZ59 compounds do (Table 1). BEA was previously reported to have highly effective synergistic antifungal activity against diverse fungal pathogens, but its precise molecular mechanism of synergy is yet unclear.14
Table 1.
Binding affinity of BEA compounds with Cdr1 and Cdr2.
| ZINC Id | Chemical name | Binding affinity kcal/mol |
rmsd/ub |
rmsd/lb |
|||
|---|---|---|---|---|---|---|---|
| Cdr1 | Cdr2 | Cdr1 | Cdr2 | Cdr1 | Cdr2 | ||
| ZINC85643633 | Beauvericin | −9.8 | −8.4 | 0.000 | 3.036 | 0.000 | 5.568 |
| ZINC95540658 | Beauvericin A | −10.3 | −10.1 | 0.000 | 0.000 | 0.000 | 0.000 |
| ZINC95607714 | Beauvericin H1 | −9.1 | −9.5 | 5.899 | 0.000 | 9.572 | 0.000 |
| ZINC95542396 | Beauvericin H2 | −9.6 | −10.7 | 16.142 | 0.000 | 20.455 | 0.000 |
| ZINC95613104 | Beauvericin H3 | −10.5 | −10.4 | 0.000 | 0.000 | 0.000 | 0.000 |
| ZINC95607711 | Beauvericin G1 | −11.0 | −8.8 | 0.000 | 21.689 | 0.000 | 25.254 |
| ZINC95613105 | Beauvericin G2 | −10.3 | −9.6 | 0.000 | 0.000 | 0.000 | 0.000 |
| ZINC28974061 | Beauvericin G3 | −10.3 | −9.0 | 0.000 | 10.572 | 0.000 | 14.905 |
| QZ59-RRR | −9.0 | −8.3 | 21.093 | 17.523 | 23.909 | 20.773 | |
| QZ59-SSS | −8.8 | −8.5 | 14.237 | 24.328 | 19.356 | 28.751 | |
The virtual screening results here indicated that BEA can block the internal cavity of ABC transporter, and serve as potential inhibitors. We used PyMOL25 together with UCSF Chimera24 to display the docking models of BEA with Cdr1 and Cdr2, respectively (Fig. 1).
Fig. 1.
Docking models of BEA with Cdr1 and Cdr2.
3.2. BEA indeed blocks the efflux function of C. albicans ABC transporters
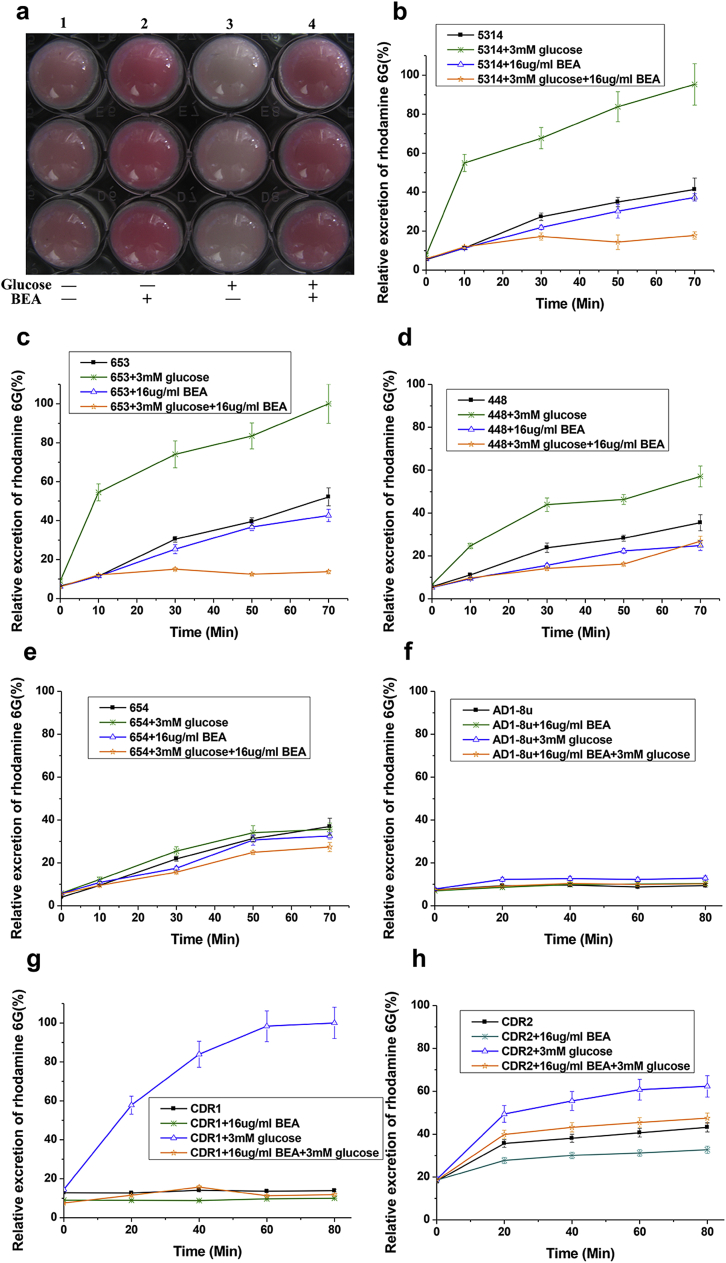
In order to validate the virtual screening results, we used rhodamine 6G, a specific substrate of Cdr1 and Cdr2, as an indicator to study the efflux function of C. albicans ABC transporters. All testing C. albicans strains were starved for 4 hours in glucose-free PBS, and then 3 mM (final concentration) glucose was added to reactivate the ABC transporters when specified. When SC5314 was treated with 16 µg/ml of BEA, twice the MIC, the efflux of rhodamine 6G by ABC transporters was totally inhibited, even after 3 mM glucose was added (Fig. 2a, 2b). To confirm this observation, C. albicans strains with the following genes were knocked-out: CDR1 (strain 448), CDR2 (strain 653) and both CDR1 and CDR2 (strain 654)26, 27 were subjected to the assay. Strain 653 behaved just like SC5314 did, while the efflux of rhodamine 6G in strain 448 was also sharply inhibited, but not as much as in strain 653, because the efflux function of Cdr1 is stronger than that of Cdr2 as reported in Ref. 28. Strain 654 did not show the inhibition since CDR1 and CDR2 were both knocked out (Fig. 2c–e).
Fig. 2.
BEA inhibits rhodamine 6G efflux in C. albicans CDR null mutants and in S. cerevisiae overexpressed CDRs. Error bars indicate standard deviation.
To further investigate the effect of BEA on individual drug efflux pumps, Saccharomyces cerevisiae strains independently overexpressing C. albicans CDR1 (strain CDR1), C. albicans CDR2 (strain CDR2), and C. albicans MDR1 (strain MDR1)29, 30 were used to carry out the rhodamine 6G efflux assay. We found that BEA couldsignificantly inhibit rhodamine 6G efflux in strain CDR1 and strain CDR2 (Fig. 2g, 2h). No difference of rhodamine 6G efflux was observed in strain AD1-8u- and strain MDR1, and both served as negative controls (rhodamine 6G is not a substrate of C. albicans Mdr1), with or without glucose (3 mM) (Fig. 2f, and Fig. S1f). Additionally, no inhibition of Nile Red efflux in strain MDR1 by BEA was observed, which is a substrate of C. albicans Mdr1.31
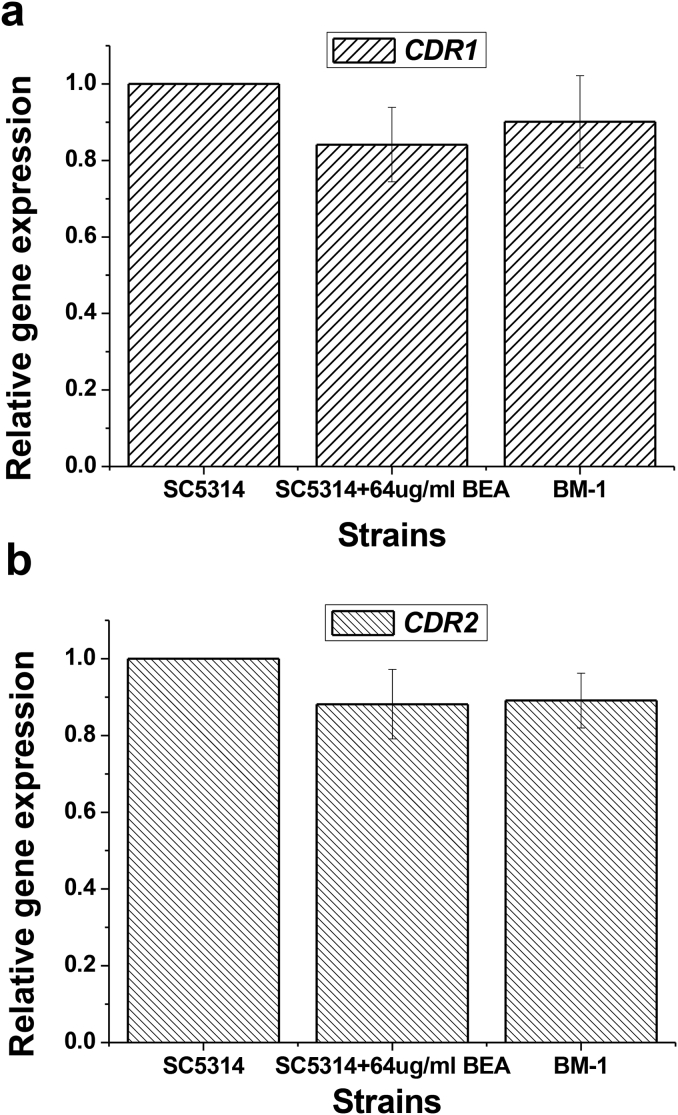
Furthermore, we also observed that when BEA was added to the strains actively effluxing rhodamine 6G, the inhibition effect happened immediately (within 10 min) (Fig. S1). This result indicated that BEA acted on ABC transporters directly, rather than indirectly (through inhibition of gene transcription or/and translation). To confirm this observation, we first used quantitative real time PCR to verify the mRNA levels of CDR1 and CDR2 in SC5314 before and after BEA-treatment. No significant changes in gene transcription were observed. Moreover, we observed almost the same transcription level of ABC transporter genes in a laboratory acquired BEA-resistant strain BM-1 as that in its parental strain SC5314 (Fig. 3).
Fig. 3.
ABC genes expression level with and without BEA treatment.
The results of rhodamine 6G efflux assay indicated that BEA can indeed specifically act on protein and inhibit the efflux function of C. albicans ABC transporters, but not MFS transporters. It is the mechanism of synergy we are looking for.
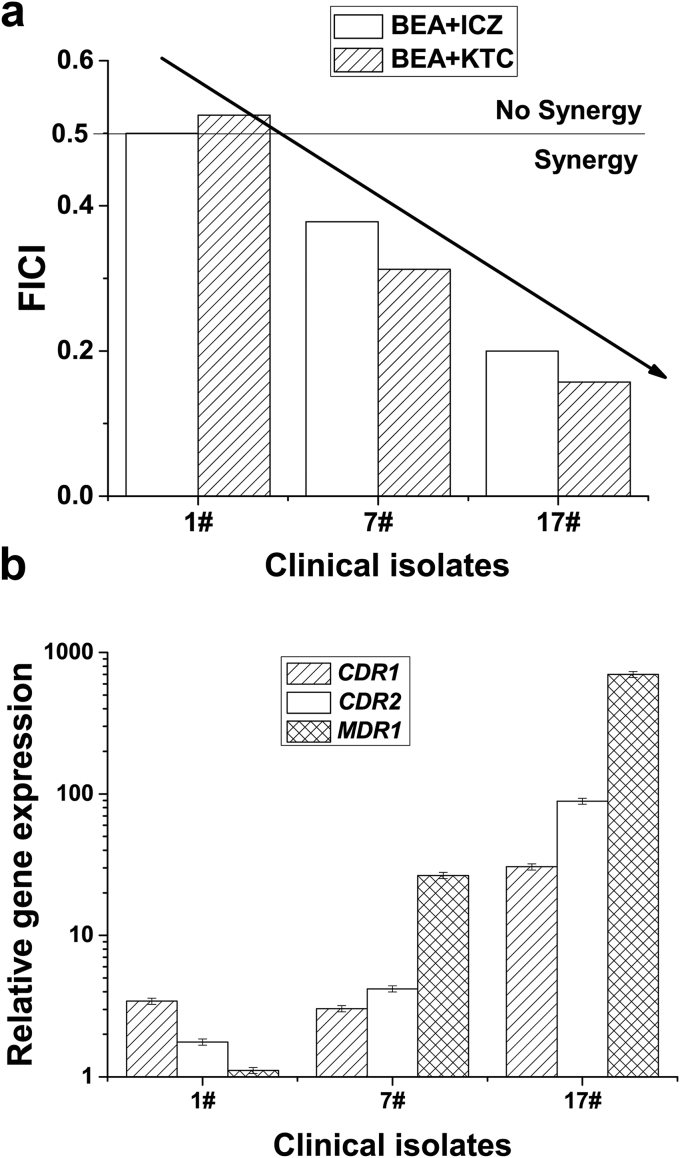
3.3. BEA has better synergistic antifungal effect with azoles against multi-drug resistant C. albicans
We hypothesized that as an efflux inhibitor, BEA might have better synergistic antifungal effect on those drug efflux pumps overexpressed multi-drug resistant strains. We tested the combination of BEA with KTC, and BEA with itraconazole (ICZ) against diverse Candida isolates, both drug sensitive and resistant (Table 2, Fig. 4). As expected, we found that BEA showed synergy with both azoles in a very low dosage on these yeast pathogens. Interestingly, the antifungal effect of BEA and azoles was indeed better in those drug efflux pumps overexpression strains than in drug sensitive strains, according to the fractional inhibitory concentration index (FICI)32 (Fig. 4).
Table 2.
Broad spectrum synergistic antifungal activity of BEA.
| Strain | MIC (µg/ml)b |
FICIc | MIC (µg/ml)b |
FICIc | |||
|---|---|---|---|---|---|---|---|
| BEA[a] | KTC[a] | KTC[c] | ICZ[a] | ICZ[c] | |||
| Candida albicans SC5314 | 8 | 0.008 | 0.002 | 0.5 | 0.032 | 0.004 | 0.375 |
| Candida tropicalisa | 16 | 1.6–3.2 | <0.064 | <0.145 | 0.032 | 0.008 | 0.5 |
| Candida kruseia | >32 | 0.8–1.6 | 0.025 | <0.28 | 0.5 | 0.064 | 0.378 |
| Candida parapsilosis ATCC14054 | 16 | 0.025 | <0.0064 | <0.5 | 0.25 | 0.004 | 0.266 |
Clinical isolates;
[a], alone, [c], combination with 1/4MIC of BEA;
FICI = (MICdrug A in combination/MICdrug A alone) + (MICdrug B in combination/MICdrug B alone), (FICI > 4: Antagonism; FICI < 0.5: Synergy; 0.5 < FICI < 4: Additive).
Fig. 4.
BEA synergism correlates with efflux pump expression. Quantitative real time PCR analysis of gene transcription was performed in triplicate. Mean values from three independent experiments are shown. Error bars indicate standard deviation.
3.4. BEA elevates intracellular calcium concentrations and triggers cell death in C. albicans
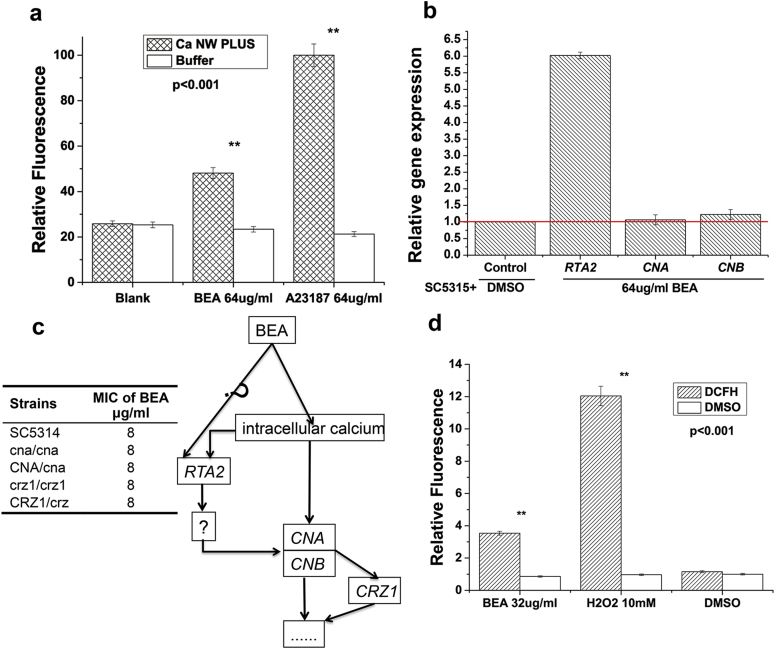
BEA alone showed some antifungal activity as well (Table 2). However, the inhibition of C. albicans ABC transporters is unlikely to be responsible for this antifungal activity, because these genes are not essential for the survival of C. albicans. Thus, there must be another target of BEA. Chemically, BEA is a cyclic hexadepsipeptide compound. Its ion-complexing capability might allow BEA to transport alkaline earth metal ions and alkaline metal ions across cell membranes. Previous studies had demonstrated that BEA is a potent calcium ionophore, can trigger apoptosis in lung cancer cells and human acute lymphoblastic leukemia.33, 34 We want to see if there is similar effect in C. albicans. We compared the intracellular calcium concentrations of C. albicans SC5314 after treatment with BEA and A23187 (a known calcium ionophore), independently. Results showed that BEA could significantly (P < 0.001) increase the intracellular calcium concentration (Fig. 5a) in a clear dose- and time-dependent manner, but the different patterns between BEA and A23187 (Fig. S2) suggested that BEA and A23187 might have different modes of action. In our previous work, RTA2 was found up-regulated by increased intracellular calcium in a calcineurin-dependent manner.35, 36 After 4 hours of BEA treatment (64 µg/ml), RTA2 transcription level indeed increased about 5-fold (P < 0.001) (Fig. 5b), which confirmed our observation that BEA can elevate the concentration of intracellular calcium. While the transcription level of calcineurin pathway-related genes CNA, CNB, and CRZ1 did not change significantly after BEA treatment, while the sensitivities of BEA to the calcineurin pathway mutants did not change either (Fig. 5c), indicating that BEA might target the upstream of calcineurin pathway (Fig. 5c).
Fig. 5.
BEA elevates intracellular calcium and triggers the apoptosis pathway of C. albicans. The experiment was performed in triplicate. Error bars indicate standard deviation.
Intracellular calcium, a secondary messenger, is a multi-functional molecule, and under some conditions, it could trigger programmed cell death.33, 34 It has been demonstrated that reactive oxygen species (ROS) is one of the typical hallmarks of early apoptosis in C. albicans.37, 38 Thus, we studied the effect of BEA on ROS generation. Not surprising, intracellular ROS was significantly induced (P < 0.001) by BEA; hydrogen peroxide was used as a positive control (Fig. 5d).
3.5. In vivo synergistic antifungal effect of BEA
To extend the in vitro observations, we evaluated the potential synergistic activity of BEA with a low dosage of KTC in a cyclophosphamide pre-treated immunocompromised murine model.14 Placebo-treated mice, infected with either C. albicans SC5314 or C. parapsilosis ATCC 14054, had demonstrated significant hypertrophy, bleeding, and fibrinoid necrosis in kidney, heart, spleen, lung, liver and brain tissues, yeast cells, and/or pseudohyphal forms, were visible (Fig. S3).
High dosage (50 mg/kg body weight) of KTC alone increased mean survival time (from 8.2 ± 0.4 d to 19.4 ± 2.0 d) of the treated mice, and caused a change in the relative number of histological foci of infection, but did not reduce the fungal burden in tissues comparing to the placebo-treated mice. Histopathologically, abscesses of renal body, and necrosis of glomerulus membrane, appeared with an absence of the renal capsule. Three types of foci were noticed: (1) abundant yeast cells and mycelia occupied the parenchyma which had been dissolved; (2) inflammatory cells infiltrated with fragmentation of necrotic cells; and (3) the boundary between the fungus and the host contained inflammatory, necrotic, and intumescent cells. Similar phenomena were observed in other organs. In this murine model, even the high dosage of KTC alone had limited therapeutic efficacy because KTC is fungistatic and the mice were pre-treated with immunosuppressant, cyclophosphamide.
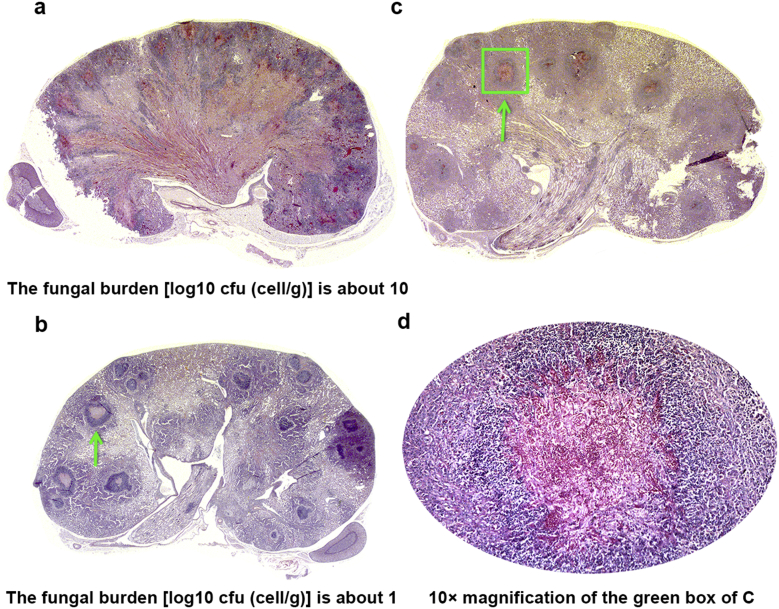
As one of the target organs in experimental candidiasis, kidney contains the greatest number of pathogens and has the most severe lesions, including diffuse foci of infection, acute inflammation, and necrosis. The cortical and medullar regions displayed acute diffuse glomerulonephritis; nephrons and proximal tubes were destroyed or even dissolved. Interestingly, kidneys from mice treated with 0.5 mg/kg BEA and KTC showed significant reduction in tissue damage and inflammatory cell infiltration as compared to those of placebo-treated mice control. Occasionally, abscesses of the renal body and granulomas of the glomerulus membrane were observed as well as limited necrosis and exfoliation of cells. Few fungal cells were observed and they were associated with only a limited number of inflammatory cells (mostly neutrophils, lymphocytes, monocytes, and fibroblasts). The periphery of these foci of infection contained numerous keratinocytes and fibrotic scars that formed peculiar immunological rings (Fig. 6). Even though there was limited tissue damage evident by microscopic evaluation. 0.5 mg/kg BEA and KTC had a significant therapeutic effect in the infected mice. By day 12 of post-infection, calculated by [log10 cfu (cell/g)], fungal burden of kidney was reduced by about 85%.14
Fig. 6.
Therapeutic effect of BEA synergizes with a low dosage of KTC on the infected mouse kidney.
3.6. Toxicity evaluation of BEA
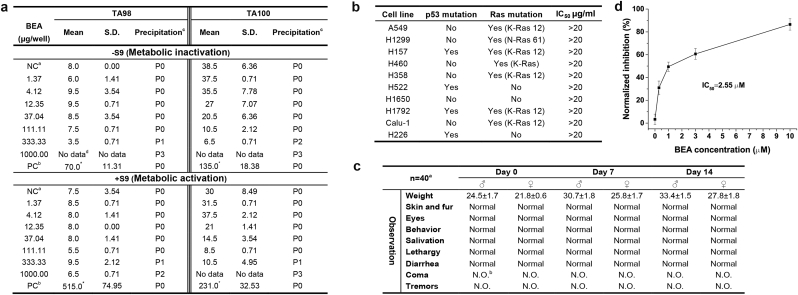
Thus, BEA was proofed to be a high potential synergistic antifungal agent, and only a HepG2 toxicity evaluation14 is not sufficient to address that BEA is safe to use in clinic. An Ames test (Salmonella typhimurium TA98 and TA100)39, 40 was adopted to evaluate the mutagenic potential of BEA. No compound related bacterial reverse mutagenicity was observed in both metabolic activation (+S9) and inactivation conditions (−S9) (Fig. 7a). Besides Ames test, ten lung cancer lines with different backgrounds were used to test the cytotoxicity of BEA, all IC50 are more than 20 µg/ml, indicating BEA only had limited cytotoxicity (Fig. 7b), but highly selective for Candida spp. Furthermore, we used a healthy murine model to test the acute toxicity of BEA. All testing mice survived without a toxic shock syndrome and observable signs of drug-related morbidity or mortality in a 2-week-observation period after a single-dose of BEA (2 g/kg body weight) gavage. Their body weight even increased (Fig. 7c). The effective therapeutic concentration of BEA is 0.5 mg/kg body weight.14 Therefore, the therapeutic index is very high. More and more structurally and functionally unrelated drugs have been found that can block the hERG potassium channel, prolong cardiac action potentials and lead to long QT syndrome, causing cardiac disease. So, the hERG test now is widely used as a predictor of cardiac risk in vivo41, 42 for drug discovery. The hERG test of BEA showed that 0.3 µM and higher concentration of BEA could inhibit the hERG potassium channel in a dose dependent manner; the IC50 of BEA is about 2.55 µM (Fig. 7d), which is considered controllable cardiac risk. All the toxicity tests indicated that BEA has good pharmacological potential with minimal toxicity.
Fig. 7.
Toxicity evaluation of BEA.
4. Discussion
Historically, infectious diseases were a huge threat to human health before antibiotics were discovered, but now, they are returning with a powerful weapon, multi-drug resistance, which is a worldwide threat.43, 44 As eukaryotes, fungal pathogens share lots of features with human cells, which restrict the antifungals discovery. With prophylactic and excessive use of these limited antifungals, multi-drug resistance is rapidly emerging, fungal pathogens adopted intricate strategies to avoid the lethal effects of antifungals,4, 5 and the most well-known and studied strategy is overexpression of drug efflux pumps to increase drug excretion.7 Among the drug efflux pumps, ABC transporters are considered to be the major contributor to multi-drug resistance.28 Novel antifungal drugs and therapeutic strategies are urgently needed. But with traditional drug discovery approach, only one class of antifungals is approved by FDA during the past two decades.9 Although the development of modern high throughput screening methods makes a massive screening in a relatively short time possible, it still needs special equipment, and consumes valuable compounds. The whole process is considered as relatively time, labor and cost consuming, not to mention how low the chance is to finally screen out something. We definitely need new approaches for antifungals discovery. Structure based virtual screening provides us the possibility to virtually find the specific molecules for experimental validation in a fast and low cost way. In this study, we demonstrated that this strategy is doable. We virtually identified BEA as the potential inhibitor of C. albicans ABC transporters, which was further confirmed by real experiments. In fact, BEA can re-sensitize multi-drug resistance by blocking the drug efflux function of ABC transporters. This is the mechanism of synergistic antifungal activity of BEA. The more the drug efflux pumps, the better the synergistic antifungal activity of BEA. The synergistic antifungal activity of BEA and azoles was also evaluated in vivo.
After BEA + KTC treatment, in the kidney, one of the target organs of fungal infection, the amyloidosis and necrosis decreased, no broad hemorrhages were observed, and nephrons remained intact. Inflammatory cells were also noted, but to a lesser extent. Remarkably, rare fungal cells were besieged by the immunological rings formed by inflammatory cells, mostly neutrophils and lymphocytes. A few foci were infiltrated with monocytes and fibroblasts. The periphery of these foci was surrounded with keratinocytes and fibrotic cells forming peculiar immunological rings. We assume that the synergistic treatment kills most of the pathogens, and the remaining pathogens probably will be taken care of by the recovering immune system, because the effect of cyclophosphamide can only last about 7 days after administration. On the other hand, whether BEA can enhance the immune response or not is under study.
It is interesting to note that enniatin, an analog of BEA, was found to inhibit S. cerevisiae Pdr5, a homolog of Cdr1, in vitro.45 We have determined which domains of Cdr1 and Cdr2 that enniatin binds to inhibit the function of C. albicans ABC in vitro.46 We also found that when CDR4, a homolog of CDR1 in Neurospora crassa, was knocked out, the synergistic effect of BEA and a low dosage of KTC was lost. These results indicate that BEA is an inhibitor of fungal ABC transporters. The same effect was observed in some cancer cells with P-glycoprotein, a Cdr1 and Cdr2 homolog.47 With all the knowledge we got, we believe that BEA can act as a special inhibitor of ABC transporters to reverse multi-drug resistance. This finding could also inspire us a new and effective strategy for cancer therapy. BEA alone is fungicidal. Previous studies demonstrated BEA as a potent calcium ionophore in some mammalian cells.33, 34 In this study, BEA was also found to act as a calcium ionophore in C. albicans to elevate the intracellular calcium concentration. However, it is not clear whether the increased intracellular calcium was from the extracellular environment or from the intracellular calcium stores. The imbalance of intracellular calcium has been connected to apoptosis pathway. Mitochondria may act as buffers of the intracellular calcium concentration, and regulate vital processes, including apoptosis.48 In our study, we found that one of the early apoptosis markers, ROS, was sharply induced after the intracellular calcium concentration increased. ROS has been considered to be responsible for the killing effects of antibiotics in some cases.49, 50 However, the correlation between ROS and antibiotic-killing mechanisms has become controversial.51, 52 In our case, BEA had multi-antifungal targets. ROS was partially responsible for its fungicidal activity. Antibiotics had largely contributed to human health, but during the wide usage, drug resistance has weakened the antibiotic effects, and made infections that were once easily curable very dangerous again, so we need to put more efforts to control those drug resistant pathogens, as antibiotics discovery is entering into the resistance era, and to find synergistic antibiotics is one way out of the looming antibiotic-resistance crisis.53 Another advantage of drug combination is it can take advantage of drug resistances to reverse them.54 BEA in this study is such a vivid example. A summary of the synergistic antifungal mechanisms of BEA is shown in Fig. S4. Based on our work, we believe that BEA has a great potential to be a new antifungal agent.
Acknowledgments
This work was supported by the National Program on Key Basic Research Project (973program, 2013CB734000), in part by grants from the National Natural Science Foundation of China [31670052, 31430002, 31320103911, 31400090, 81302678 and 31125002], the Ministry of Science and Tech-nology of the People’s Republic of China [2011ZX09102-011-11, 2013ZX10005004-005], China Ocean Mineral Resources R & D Association (Grant No. DY125-15-T-07), and the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 312184. YT is partially supported by a grant from the Novo Nordisk Foundation. LZ is an awardee for the National Distinguished Young Scholar Program in China.
The authors are very grateful to Theodore C. White for providing drug-resistant clinical C. albicans isolates and Dominique Sanglard for providing CDR1 and CDR2 knockout strains. We also thank Elizabeth Ashforth for critical reading and helpful discussions.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at doi:10.1016/j.synbio.2016.10.001.
Contributor Information
Yaojun Tong, Email: yaojun.tong@gmail.com.
Lixin Zhang, Email: lzhang03@gmail.com.
Appendix. Supplementary material
The following are the supplementary data to this article:
Virtual screening results.
Tables S1 and S2 and Figs. S1–S4.
References
- 1.Brown G.D., Denning D.W., Levitz S.M. Tackling human fungal infections. Science. 2012;336:647. doi: 10.1126/science.1222236. [DOI] [PubMed] [Google Scholar]
- 2.Pfaller M.A., Diekema D.J. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20(1):133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui J., Ren B., Tong Y., Dai H., Zhang L. Synergistic combinations of antifungals and anti-virulence agents to fight against Candida albicans. Virulence. 2015;6:362–371. doi: 10.1080/21505594.2015.1039885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson J.B. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat Rev Microbiol. 2005;3(7):547–556. doi: 10.1038/nrmicro1179. [DOI] [PubMed] [Google Scholar]
- 5.Cowen L.E. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol. 2008;6(3):187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- 6.Prasad R., Kapoor K. Multidrug resistance in yeast Candida. Int Rev Cytol. 2005;242:215–248. doi: 10.1016/S0074-7696(04)42005-1. [DOI] [PubMed] [Google Scholar]
- 7.Prasad R., Gaur N.A., Gaur M., Komath S.S. Efflux pumps in drug resistance of Candida. Infect Disord Drug Targets. 2006;6(2):69–83. doi: 10.2174/187152606784112164. [DOI] [PubMed] [Google Scholar]
- 8.Sanglard D., Ischer F., Parkinson T., Falconer D., Bille J. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob Agents Chemother. 2003;47(8):2404–2412. doi: 10.1128/AAC.47.8.2404-2412.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L., Demain A.L. Humana Press; Totowa (NJ): 2005. Natural products: drug discovery and therapeutic medicine. [Google Scholar]
- 10.Aller S.G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lionta E., Spyrou G., Vassilatis D.K., Cournia Z. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr Top Med Chem. 2014;14:1923–1938. doi: 10.2174/1568026614666140929124445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sterling T., Irwin J.J. ZINC 15-ligand discovery for everyone. J Chem Inf Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda T., Arai M., Yamaguchi Y., Masuma R., Tomoda H., Omura S. New beauvericins, potentiators of antifungal miconazole activity, produced by Beauveria sp FKI-1366 – I. Taxonomy, fermentation, isolation and biological properties. J Antibiot. 2004;57(2):110–116. doi: 10.7164/antibiotics.57.110. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L.X., Yan K.Z., Zhang Y., Huang R., Bian J., Zheng C.S. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc Natl Acad Sci USA. 2007;104(11):4606–4611. doi: 10.1073/pnas.0609370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei L., Zhang L., Dai R. An inhibition study of beauvericin on human and rat cytochrome P450 enzymes and its pharmacokinetics in rats. J Enzyme Inhib Med Chem. 2009;24:753–762. doi: 10.1080/14756360802362041. [DOI] [PubMed] [Google Scholar]
- 16.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 18.Song F., Dai H., Tong Y., Ren B., Chen C., Sun N. Trichodermaketones A-D and 7-O-methylkoninginin D from the marine fungus Trichoderma koningii. J Nat Prod. 2010;73:806–810. doi: 10.1021/np900642p. [DOI] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute . 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; Approved standard. 3rd ed, CLSI document M27-A3. Wayne, PA: National Committee for Clinical Laboratory Standards. [Google Scholar]
- 20.Wu Y., Min F., Pan J., Wang J., Yuan W., Zhang Y. Systemic Candida parapsilosis infection model in immunosuppressed ICR mice and assessing the antifungal efficiency of fluconazole. Vet Med Int. 2015;2015 doi: 10.1155/2015/370641. 370641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 23.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 24.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 25.Schrodinger L.L.C. 2010. The PyMOL Molecular Graphics System. Version 1.5.0.4. [Google Scholar]
- 26.Sanglard D., Ischer F., Monod M., Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanglard D., Ischer F., Monod M., Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143(Pt 2):405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 28.Holmes A.R., Lin Y.H., Niimi K., Lamping E., Keniya M., Niimi M. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrob Agents Chemother. 2008;52:3851–3862. doi: 10.1128/AAC.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Decottignies A., Grant A.M., Nichols J.W., de Wet H., McIntosh D.B., Goffeau A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J Biol Chem. 1998;273:12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]
- 30.Lamping E., Monk B.C., Niimi K., Holmes A.R., Tsao S., Tanabe K. Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae. Eukaryot Cell. 2007;6:1150–1165. doi: 10.1128/EC.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivnitski-Steele I., Holmes A.R., Lamping E., Monk B.C., Cannon R.D., Sklar L.A. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal Biochem. 2009;394:87–91. doi: 10.1016/j.ab.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eliopoulos G.M.M.R. In: Antibiotics in laboratory medicine. Lorian V., editor. Williams & Wilkins; Baltimore (MD): 1991. pp. 432–492. [Google Scholar]
- 33.Chen B.F., Tsai M.C., Jow G.M. Induction of calcium influx from extracellular fluid by beauvericin in human leukemia cells. Biochem Biophys Res Commun. 2006;340:134–139. doi: 10.1016/j.bbrc.2005.11.166. [DOI] [PubMed] [Google Scholar]
- 34.Jow G.M., Chou C.J., Chen B.F., Tsai J.H. Beauvericin induces cytotoxic effects in human acute lymphoblastic leukemia cells through cytochrome c release, caspase 3 activation: the causative role of calcium. Cancer Lett. 2004;216:165–173. doi: 10.1016/j.canlet.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Jia X.M., Ma Z.P., Jia Y., Gao P.H., Zhang J.D., Wang Y. RTA2, a novel gene involved in azole resistance in Candida albicans. Biochem Biophys Res Commun. 2008;373:631–636. doi: 10.1016/j.bbrc.2008.06.093. [DOI] [PubMed] [Google Scholar]
- 36.Jia X.M., Wang Y., Jia Y., Gao P.H., Xu Y.G., Wang L. RTA2 is involved in calcineurin-mediated azole resistance and sphingoid long-chain base release in Candida albicans. Cell Mol Life Sci. 2009;66(1):122–134. doi: 10.1007/s00018-008-8409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai B.D., Cao Y.Y., Huang S., Xu Y.G., Gao P.H., Wang Y. Baicalein induces programmed cell death in Candida albicans. J Microbiol Biotechnol. 2009;19:803–809. [PubMed] [Google Scholar]
- 38.Sharma M., Manoharlal R., Puri N., Prasad R. Antifungal curcumin induces reactive oxygen species and triggers an early apoptosis but prevents hyphae development by targeting the global repressor TUP1 in Candida albicans. Biosci Rep. 2010;30:391–404. doi: 10.1042/BSR20090151. [DOI] [PubMed] [Google Scholar]
- 39.Ames B.N., McCann J., Yamasaki E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat Res. 1975;31:347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- 40.Flamand N., Meunier J., Meunier P., Agapakis-Causse C. Mini mutagenicity test: a miniaturized version of the Ames test used in a prescreening assay for point mutagenesis assessment. Toxicol In Vitro. 2001;15:105–114. doi: 10.1016/s0887-2333(01)00003-0. [DOI] [PubMed] [Google Scholar]
- 41.Bowlby M.R., Peri R., Zhang H., Dunlop J. hERG (KCNH2 or Kv11.1) K+ channels: screening for cardiac arrhythmia risk. Curr Drug Metab. 2008;9:965–970. doi: 10.2174/138920008786485083. [DOI] [PubMed] [Google Scholar]
- 42.Priest B.T., Bell I.M., Garcia M.L. Role of hERG potassium channel assays in drug development. Channels (Austin) 2008;2:87–93. doi: 10.4161/chan.2.2.6004. [DOI] [PubMed] [Google Scholar]
- 43.Cosgrove S.E., Carmeli Y. The impact of antimicrobial resistance on health and economic outcomes. Clin Infect Dis. 2003;36:1433–1437. doi: 10.1086/375081. [DOI] [PubMed] [Google Scholar]
- 44.Smith P.A., Romesberg F.E. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat Chem Biol. 2007;3(9):549–556. doi: 10.1038/nchembio.2007.27. [DOI] [PubMed] [Google Scholar]
- 45.Hiraga K., Yamamoto S., Fukuda H., Hamanaka N., Oda K. Enniatin has a new function as an inhibitor of Pdr5p, one of the ABC transporters in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2005;328:1119–1125. doi: 10.1016/j.bbrc.2005.01.075. [DOI] [PubMed] [Google Scholar]
- 46.Tanabe K., Lamping E., Nagi M., Okawada A., Holmes A.R., Miyazaki Y. Chimeras of Candida albicans Cdr1p and Cdr2p reveal features of pleiotropic drug resistance transporter structure and function. Mol Microbiol. 2011;82(2):416–433. doi: 10.1111/j.1365-2958.2011.07820.x. [DOI] [PubMed] [Google Scholar]
- 47.Sharom F.J., Lu P., Liu R., Yu X. Linear and cyclic peptides as substrates and modulators of P-glycoprotein: peptide binding and effects on drug transport and accumulation. Biochem J. 1998;333(Pt 3):621–630. doi: 10.1042/bj3330621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizzuto R., De Stafani D., Raffaello A., Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 49.Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 50.Dwyer D.J., Kohanski M.A., Collins J.J. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009;12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y., Imlay J.A. Cell death from antibiotics without the involvement of reactive oxygen species. Science. 2013;339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keren I., Wu Y., Inocencio J., Mulcahy L.R., Lewis K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science. 2013;339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 53.Brown E.D., Wright G.D. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 54.Baym M., Stone L.K., Kishony R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science. 2016;351 doi: 10.1126/science.aad3292. aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Virtual screening results.
Tables S1 and S2 and Figs. S1–S4.