ABSTRACT
DNA repair pathways are aberrant in cancer, enabling tumor cells to survive standard therapies—chemotherapy and radiotherapy. Our group previously reported that, upon irradiation, the membrane-bound tyrosine kinase receptor TIE2 translocates into the nucleus and phosphorylates histone H4 at Tyr51, recruiting ABL1 to the DNA repair complexes that participate in the nonhomologous end-joining pathway. However, no specific molecular mechanisms of TIE2 endocytosis have been reported. Here, we show that irradiation or ligand-induced TIE2 trafficking is dependent on caveolin-1, the main component of caveolae. Subcellular fractionation and confocal microscopy demonstrated TIE2/caveolin-1 complexes in the nucleus, and using inhibitor or small interfering RNAs (siRNAs) against caveolin-1 or Tie2 inhibited their trafficking. TIE2 was found in caveolae and directly phosphorylated caveolin-1 at Tyr14 in vitro and in vivo. This modification regulated the generation of TIE2/caveolin-1 complexes and was essential for TIE2/caveolin-1 nuclear translocation. Our data further demonstrate that the combination of TIE2 and caveolin-1 inhibitors resulted in significant radiosensitization of malignant glioma cells, which will guide the development of combinatorial treatment with radiotherapy for patients with glioblastoma.
KEYWORDS: TIE2, caveolin-1, nuclear translocation, radioresistance, brain tumor, nuclear transport, radiosensitivity
INTRODUCTION
TIE2 is a membrane-bound receptor tyrosine kinase (RTK) that is expressed in endothelial cells and hematological systems, and it was unexpectedly found in several cancers, including malignant gliomas (1). The function of TIE2 in endothelial cells has been reported to be related to kinase signaling transduction regulating cell survival and vessel integrity and permeability (1, 2). Specifically in cancer, TIE2 activity is associated with multicompartment cross talk, tumor invasion, and multidrug resistance (1, 3–5). Recently, we described cell membrane-to-nucleus trafficking of TIE2 upon irradiation (IR) that involves secretion of Ang1, its agonist ligand. In the nucleus, TIE2 phosphorylates histone H4 at Tyr51, recruiting the ABL1 proto-oncogene to the DNA repair protein complex, resulting in a radioresistant phenotype (3).
The mechanisms of RTK trafficking are of major significance, as they regulate signaling, receptor organization, and degradation (6, 7). Nuclear RTKs function as transcription factors (8), and interestingly, recent reports have indicated a relationship between nuclear RTKs and epigenetic modifications (3, 9). Several studies have shown that RTK internalization is dependent on ligand binding and that, upon stimulation, RTKs translocate from the membrane to cytoplasmic vesicles (7, 10, 11). The internalization of RTKs might result in a positive or negative effect in receptor signaling, degradation, and recycling (12). Clathrin-coated and caveola pits are the most studied specialized domains in the plasma membrane, related to endocytosis and modulation of cell signaling, the clathrin-dependent endocytosis being a main internalization pathway for ligand-occupied RTKs, such as epidermal growth factor receptor (EGFR), RET, MET, insulin receptor, and NRTK1 (6, 13). The main integral protein of caveolae, caveolin-1 (14, 15), has been reported to be associated with several RTKS, such as Vascular endothelial growth factor receptor 2 (VEGFR2), EGFR, PDGR, EphR, and insulin receptor (13, 16, 17), although the role of this molecular association is generally related to enhancing or negatively regulating receptor signaling (13). Interestingly, endocytosis and trafficking seem to be bidirectionally linked, and several mechanisms by which signaling regulates endocytosis have been reported (7), including Src kinase activity downstream of RTK being involved in phosphorylating the clathrin heavy chain (7) or caveolin-1 (15). Studies of TIE2 trafficking have been limited to its internalization and degradation pathways. Thus, activation of TIE2 by Ang1 results in colocalization of the receptor with the clathrin heavy chain at the endothelial cell membranes, resulting in internalization and degradation (18, 19) and suggesting a mechanism that attenuates receptor activity and signaling (7). However, no specific molecular regulatory mechanisms of TIE2 endocytosis have been reported.
Our research group previously described Ang1-mediated cell membrane-to-nucleus TIE2 trafficking following IR, a pathway that is essential in conferring radioresistance to cancer and endothelial cells (3). Here, we identified the mechanism that underlies the TIE2 nuclear translocation observed upon IR and ligand upregulation. TIE2 is actively involved in regulating its cell membrane-to-nucleus movement by associating with caveolae and phosphorylating caveolin-1 at the Tyr14 residue, which regulates the generation of TIE2/caveolin-1 complexes. Of further interest, the combined inhibition of TIE2 and caveolin-1 resulted in the radiosensitization of glioma cells, suggesting that the tightly regulated membrane trafficking of TIE2 might be a useful target for developing radiosensitizers or neutralizing the aberrant DNA repair mechanisms involved in the resistance of brain tumors to therapy.
RESULTS
Caveolin-1 is expressed in GB cells.
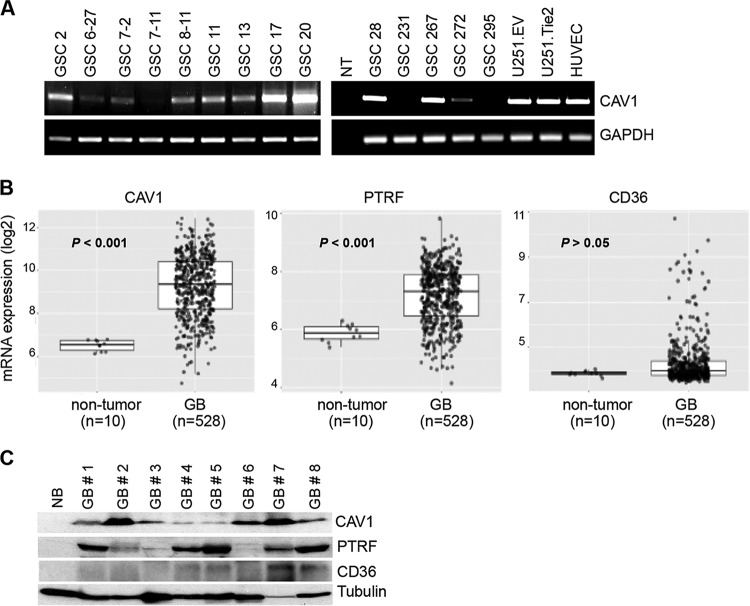
To elucidate the mechanisms underlying TIE2 trafficking, we first explored the involvement of caveolin-1, an essential component of caveolae (14, 15). We previously defined TIE2 as a tyrosine kinase receptor that is expressed in malignant gliomas and glioma stem cells (GSCs) (4). In this study, we first screened caveolin-1 expression levels in a panel of GSCs and observed that the caveolin-1 transcript was present in 78.6% of the GSCs analyzed (11 of 14) (Fig. 1A). An analysis of levels of caveolin-1 and other main components of caveolae in surgical specimens showed that caveolin-1 was overexpressed in human glioblastoma (GB) specimens (n = 528) compared to nontumoral tissues (n = 10; P < 0.001; data from the TCGA research network) (Fig. 1B). An analysis of other two proteins targeted to lipid rafts revealed that PTRF (cavin1) (15) but not CD36 (platelet glycoprotein 4) expression levels follow a similar expression pattern (Fig. 1B). Similar data were obtained by analyzing caveolin-1, PTRF, and CD36 protein levels by Western blot analysis in a panel of surgical human GBs. Caveolin-1 and PTRF were expressed at higher levels than in normal brain in most of the material examined, and CD36 was detected in only 4 of them, at low levels of expression (Fig. 1C). These data demonstrate the overexpression of two essential proteins in the functional caveolae in malignant gliomas.
FIG 1.
Caveola-related proteins are overexpressed in glioblastomas. (A) Analysis of the expression of caveolin-1 mRNA in a panel of GSCs, U251 glioma cells, and human umbilical vein endothelial cells was performed by reverse transcription-PCR (RT-PCR). In brief, cDNA was generated and subjected to RT-PCR, as described previously (3). GAPDH mRNA levels of expression were used as a control. The primers used for this project are listed in Table S2 in the supplemental material. NT, no template. (B) mRNA levels of caveolin-1, PTRF, and CD36 in normal tissue and glioblastomas, analyzed using the publicly available TCGA_GBM data set with the HG-U133A platform (TCGA Research Network, http://gliovis.bioinfo.cnio.es). P values were calculated using two-tailed Student's t test. (C) Protein levels of caveolin-1, PTRF, and CD36 analyzed by Western blotting of normal brain tissue (NB) and surgical human glioblastoma (GB). Tubulin expression was used as a loading control.
TIE2 nuclear translocation is caveolin-1 dependent.
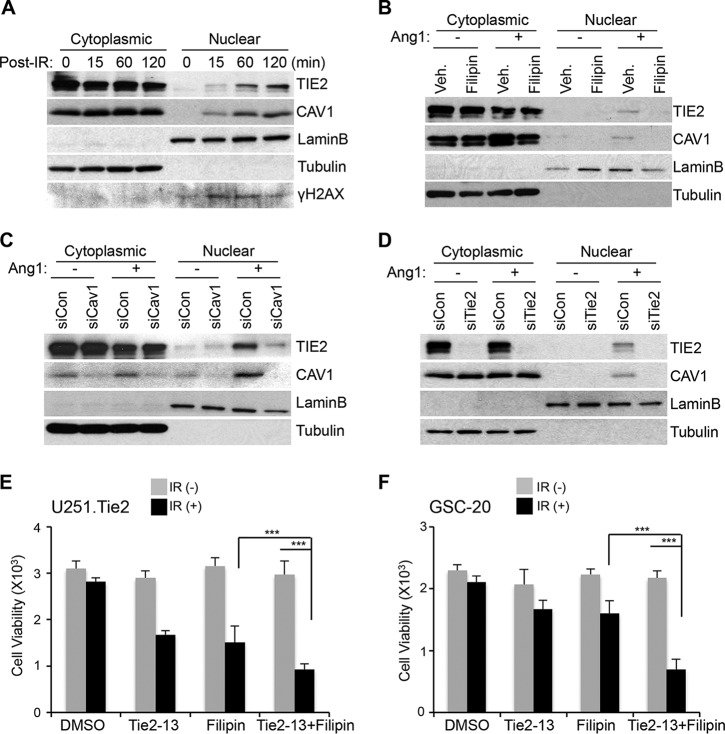
To analyze the putative relationship between TIE2 nuclear translocation and caveolin-1, we first examined the trafficking of caveolin-1 after IR. We found caveolin-1 expression in the nuclear fraction of IR-treated cells, and as expected (3), we observed the presence of TIE2 in the nucleus after IR exposure (Fig. 2A; see also Fig. S1A in the supplemental material). As reported, Ang1 ligand exposure also resulted in TIE2 nuclear translocation (Fig. 2B; see also Fig. S1B in the supplemental material). To test the role of caveolin-1 in TIE2 nuclear translocation, we inhibited caveolin-1 expression using pharmacological and biological strategies. First, we treated the cells with a chemical inhibitor of caveolin-1, filipin, known to bind cholesterol in the plasma membrane and impair caveola invagination and internalization (20), and we observed that both TIE2 and caveolin-1 nuclear translocations were inhibited when cells were treated with filipin (Fig. 2B; see also Fig. S1B in the supplemental material). These results were corroborated using two different small interfering RNAs (siRNAs) against caveolin-1 (Fig. 2C; see also Fig. S2A and S3A in the supplemental material). Of note, the detection of caveolin-1 in the nuclear fraction after Ang1 exposure was jeopardized by using two different siRNAs against Tie2 (Fig. 2D; see also Fig. S2B and S3B in the supplemental material). Collectively, these data demonstrate that nuclear translocation of TIE2, as observed upon IR or Ang1 exposure, is caveolin-1 dependent; they also suggest a mechanistic link between the two molecules.
FIG 2.
TIE2 nuclear translocation is caveolin-1 dependent. (A) U251.Tie2 cells were IR treated (137Cs; 10 Gy) for the indicated time periods, and nuclear and cytoplasmic protein extraction was performed by subcellular fractionation, as previously described (3). Proteins were analyzed by Western blotting, as reported previously (3), to assess TIE2 and caveolin-1 (CAV1) expression levels. LaminB and tubulin expression levels were used as nuclear and cytoplasmic protein loading controls, respectively. γH2AX expression levels were used as an indicator of DNA damage after IR. (B) U251.Tie2 cells were serum starved for 24 h in serum-free medium and then incubated in fresh serum-free medium containing the caveolin-1 inhibitor filipin (5 μg/ml; Sigma-Aldrich); 60 min later, Ang1 (400 ng/ml; R&D Systems) was added for 30 min. Proteins were extracted after nuclear and cytoplasmic fractionation, as previously reported (3), and levels of TIE2 and caveolin-1 expression were assessed by Western blotting. DMSO was used as the vehicle (Veh.). LaminB and tubulin expression levels were used as nuclear and cytoplasmic protein loading controls, respectively. (C) siRNA against caveolin-1 was transfected into U251.Tie2 cells, and 48 h later, cytoplasmic and nuclear TIE2 and caveolin-1 were analyzed by Western blotting after Ang1 exposure (400 ng/ml; 30 min). Bovine serum albumin (0.1% in phosphate-buffered saline) was used as the vehicle/control in Ang1(−)-treated samples. siRNAs were transfected into cells using the INTERFERin transfection reagent (PolyPlus Transfection) at concentrations of 10 nM; after 48 h, subcellular fractions of nuclear and cytoplasmic proteins were analyzed by Western blotting. Nontargeted siRNA (siCon) was used as the control. (D) Western blot analysis with cytoplasmic and nuclear extract from U251.Tie2 cells after transfection of siRNA against Tie2 (10 nM) and Ang1 stimulus, as explained for panel C. The antibodies and working dilutions are listed in Table S3. The siRNA sequences used are listed in Table S1. (E) U251.Tie2 cells were plated in a 96-well plate at subconfluent density and treated with Tie2-13 (5 μM) and filipin (2 μg/ml) (36), alone or in combination; 24 h later, they were irradiated (15 Gy) and kept in an incubator for an additional 48 h at 37°C. Cell viability was assessed using the CellTitre-Blue cell viability assay (Promega) according to the manufacturer's instructions. Data represent means ± standard deviations (SD) from three independent experiments. P values were calculated using Welch's modified t test; ***, P ≤ 0.001. (F) GSC-20 cells were treated as described for panel B with Tie2-13 (5 μM) and filipin (1 μg/ml), alone or in combination for 1 h, and then irradiated (20 Gy) and kept in an incubator for an additional 72 h at 37°C. Data represent means ± SD from three independent experiments. P values were calculated using Welch's modified t test; ***, P ≤ 0.001.
The results of our previous study clearly show that nuclear, not cytoplasmic or cell membrane-bound, TIE2 is related to radioresistance by regulating the nonhomologous end-joining (NHEJ) DNA repair pathway (3) and that decreasing TIE2 expression levels or jeopardizing the nuclear entry of RTK elicits radiosensitivity to previously radioresistant cultures (3). Since the nuclear translocation of TIE2 is caveolin-1 dependent, we investigated the role of caveolin-1 in TIE2-mediated radioresistance. To this end, we pharmacologically inhibited TIE2 and caveolin-1 using a TIE2 inhibitor, Tie2-13, or the caveolin-1 inhibitor, filipin. First, we determined the effect of Tie2-13 inhibition on TIE2 expression and nuclear translocation upon Ang1; after subcellular fractionation, we observed that Tie2-13 decreased the levels of nuclear TIE2 in a dose-dependent manner (see Fig. S4A in the supplemental material). We treated TIE2-expressing U251.Tie2 glioma cells with Tie2-13 and filipin, alone or in combination, and compared their sensitivity to IR by analyzing cell viability. TIE2-expressing cells exhibited radioresistance (3), and the inhibition of TIE2 or caveolin-1 resulted in radiosensitization of these cultures. Of interest, U251.Tie2 cells were significantly more sensitive to IR when treated with both inhibitors (Fig. 2E; see also Fig. S4B and C in the supplemental material). These results were corroborated by analyzing the radioresistance of GSC20, a GSC line that expresses endogenous TIE2 (3) (Fig. 2F). These results suggest that TIE2 or caveolin-1 inhibitors counter the aberrant DNA repair pathways that are present in gliomas and place caveolin-1 as a putative mediator of TIE2-mediated radioresistance.
TIE2 associates with caveolin-1 in caveolae.
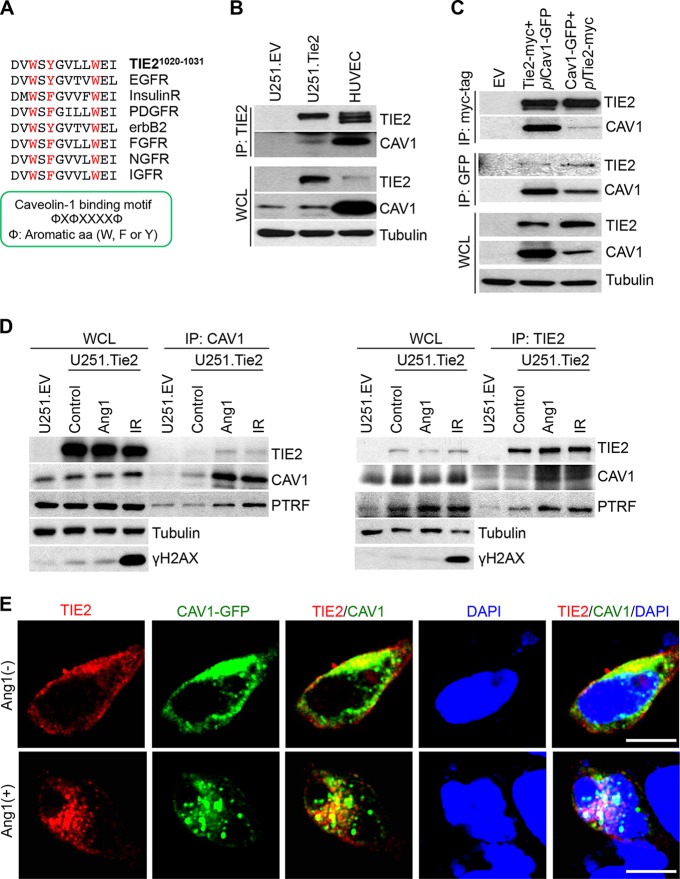
To further analyze the relationship between TIE2 and caveolin-1 during the nuclear translocation of TIE2, we examined a possible association between TIE2 and caveolin-1. It has been reported that the caveolin-1 scaffolding domain interacts with proteins such as G-protein, Src-like kinases, and several receptor kinases by specifically binding the aromatic-rich caveolin-1 binding motif (ΦXΦXXXXΦ, where Φ represents aromatic amino acids [W, F, or Y]) (21). We found that TIE2 protein contains a caveolin-1 binding motif located in amino acids (aa) 1020 to 1031, DVWSYGVLLWEI (Fig. 3A). To explore a TIE2/caveolin-1 interaction, we coimmunoprecipitated the TIE2 complexes that are present in Ang1-treated U251.TIE2 glioma cells and HUVEC cells, which express endogenous TIE2 (3), and we detected caveolin-1 in TIE2 complexes in both cell lines (Fig. 3B). To further confirm the interaction between TIE2 and caveolin-1, we generated stable transfected HEK293 cells that expressed Tie2-myc (3) or caveolin-1–GFP (where GFP is green fluorescent protein) and determined the association between TIE2 and caveolin-1 in each of the two cell lines after the transient transfection of Cav1-GFP plasmid (plCav1-GFP) or Tie2-myc plasmid (plTie2-myc), respectively. Immunoprecipitation of the tracking proteins (myc and GFP) revealed the presence of TIE2/caveolin-1 complexes (Fig. 3C).
FIG 3.
TIE2 is associated with caveolin-1 in caveolae. (A) Description of a putative binding motif (21) of TIE2 to caveolin-1. Note that the motif is common to other RTKs. (B) Coimmunoprecipitation was performed as previously described (3), with anti-TIE2 antibody, in U251.EV, U251.Tie2, and HUVEC cultures in Ang1-containing medium. The presence of caveolin-1 in TIE2 complexes was analyzed by Western blotting. WCL, whole-cell lysates. (C) HEK293.Tie2-myc and HEK293.Cav1-GFP cells that constitutively expressed TIE2 and caveolin-1, respectively, were transfected with plCav1-GFP and plTie2-myc plasmids using the X-tremeGENE HP DNA transfection reagent (Roche Applied Science) according to the manufacturer's protocol. Forty-eight hours later, Ang1 (400 ng/ml; 30 min) was added and immunoprecipitation was performed with anti-myc tag or anti-GFP antibodies, followed by a Western blot analysis of TIE2/caveolin-1 complexes. (D) U251.Tie2-myc cells were treated with Ang1 (400 ng/ml; 30 min) or IR (10 Gy; 60 min), and immunoprecipitation was performed with anti-caveolin-1 or anti-TIE2 antibodies. Levels of TIE2, caveolin-1, and PTRF were analyzed by Western blotting. U251.EV cells were used as a control. γH2AX expression levels were used as an indicator of DNA damage after IR. (E) HEK293.Tie2-myc cells were transfected with plCav1-GFP plasmid as explained for panel C, and 48 h later, cells were transferred to chamber slides (Lab-Tek) with 60% to 70% confluence. One day after being plated, cells were serum starved for 4 h and treated with Ang1 (400 ng/ml) for 30 min. Cell fixation and immunofluorescence studies were performed as previously described (3), with anti-myc tag antibody (red fluorescence). Green fluorescence from the GFP signal was used as a reporter for caveolin-1 expression. DAPI was used for nuclear staining. Merged images are provided for visualization of TIE2/caveolin-1 nuclear complexes. Images were captured using a confocal microscope (Olympus FluoView FV1000). Bar, 10 μm. A list of antibodies and working dilutions is provided in Table S3.
Since caveolin-1 is associated with PTRF in the functional caveola structures, while noncaveolar caveolin-1 is free from PTRF complexes (22), we investigated whether the association between TIE2 and caveolin-1 occurs in caveola structures. To this end, we coimmunoprecipitated cell lysates from cultures treated with Ang1 or IR with both anti-TIE2 and anti-caveolin-1 antibodies, and we detected the presence of PTRF in both TIE2 and caveolin-1 complexes, suggesting that these proteins interact in caveola structures (Fig. 3D). To further visualize the TIE2–caveolin-1 interaction, we performed immunofluorescence of HEK293 cells expressing TIE2 and caveolin-1, which showed that after Ang1 stimulus, both TIE2 and caveolin-1 colocalized in the nucleus (Fig. 3E). These data suggest that TIE2 and caveolin-1 associate in the caveolae and further confirm that upon exposure to Ang1 ligand, both proteins translocate to the nucleus.
TIE2 phosphorylates caveolin-1 at Tyr14 in vitro and in vivo.
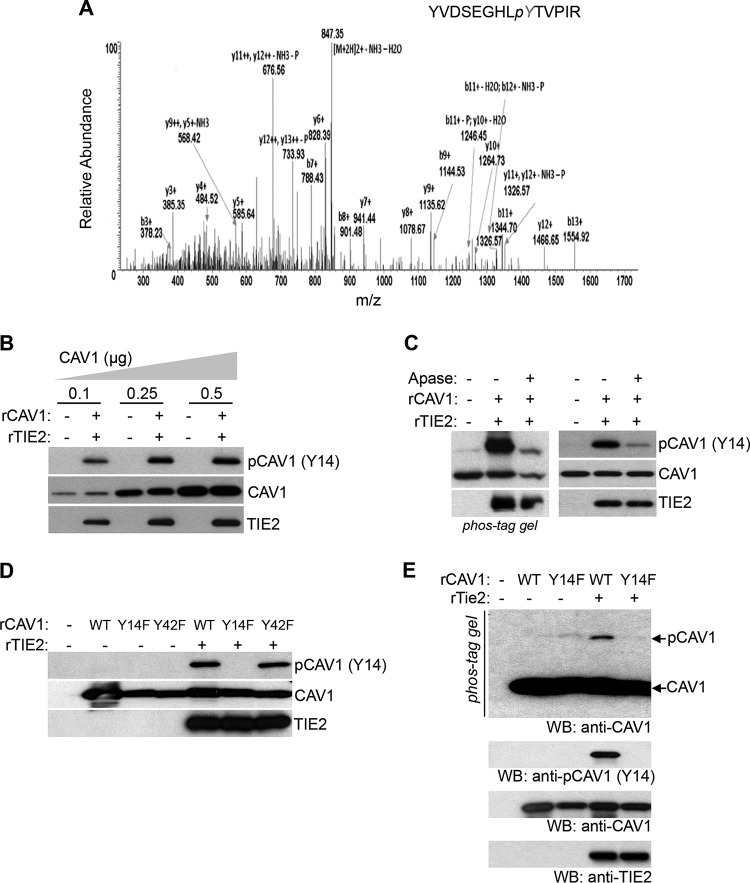
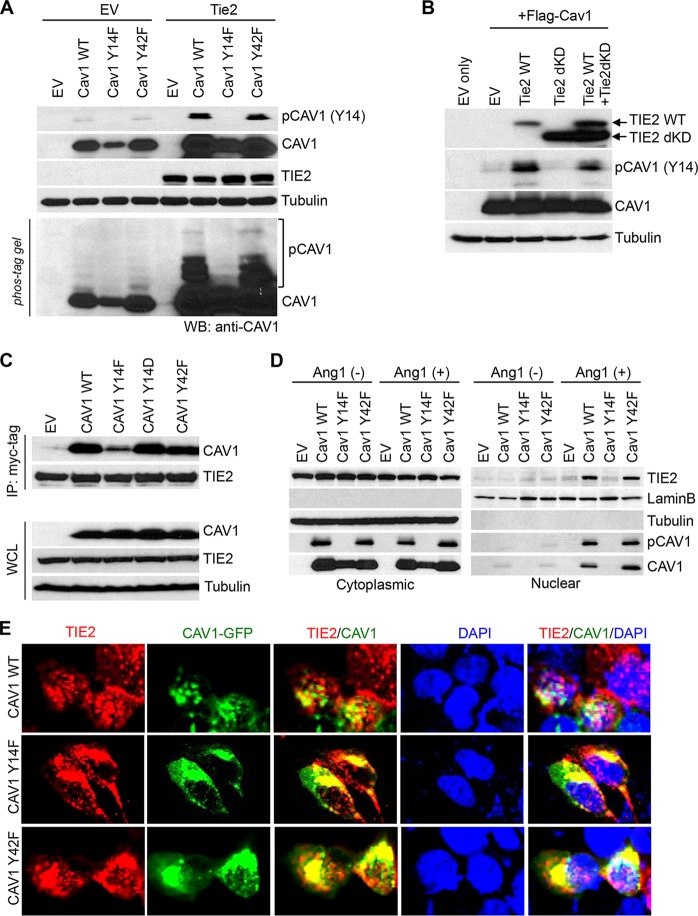
Caveolin-1 phosphorylation plays important roles in caveola formation, caveola-mediated mechanosensing, and endocytosis (14, 23). To determine whether TIE2 regulates caveolin-1 phosphorylation, we performed a mass spectrometry (MS) analysis using samples from an in vitro kinase assay. A total ion chromatogram of tryptic peptide from recombinant caveolin-1 showed a phosphorylated peptide, YVDSEGHLpyTVPIR (Fig. 4A; see also Fig. S5A to C in the supplemental material), identifying the phosphorylation of Tyr14 in caveolin-1 in the presence of active TIE2. The data also showed that additional Tyr residues of calveolin-1 were not phosphorylated by TIE2 (Fig. S5C in the supplemental meterial). To validate these data, we incubated recombinant TIE2 (rTIE2) protein with several doses of recombinant caveolin-1 (rCAV1) protein, followed by a Western blot analysis with a specific antibody against phospho-caveolin-1 (pY14). In this dose-dependent experiment, we found that TIE2 specifically phosphorylated caveolin-1 at the Tyr14 residue (Fig. 4B). Caveolin-1 phosphorylation by TIE2 was validated using a Phos-tag gel analysis that clearly separated the unphosphorylated from the phosphorylated caveolin-1 forms (3), with the latter identified as caveolin-1-pY14 using a standard Western blot analysis (Fig. 4C). The use of alkaline phosphatase treatment showed a decrease in this phosphorylation, confirming that TIE2 was responsible for the phosphorylation of caveolin-1 at the Tyr14 residue (Fig. 4C). To analyze the specificity of this regulation, we constructed two mutant forms of caveolin-1 (Y14F and Y42F) (see Fig. S6 in the supplemental material) and exposed them to rTIE2 in an in vitro kinase assay, followed by a Western blot analysis in standard gel or Phos-tag gel. The results showed that TIE2 specifically phosphorylated caveolin-1 at residue Tyr14 (Fig. 4D and E).
FIG 4.
TIE2 phosphorylates caveolin-1 at Tyr-14 residue in vitro. (A) Tyr14 phosphorylation of caveolin-1 by TIE2 was detected by mass spectrometry. In vitro kinase assays were conducted with purified GST-tagged TIE2 active protein (200 ng; no. TEK-7296H; Creative Biomart) and recombinant wild-type caveolin-1 (500 ng; no. H00000857-P01; Novus Biologicals), as previously reported (3). The resulting caveolin-1 band was subjected to nano-LC/MS/MS analysis, as previously reported (3), which detected a tryptic fragment at an m/z mass/charge ratio of 864.90, matched to the doubly charged phosphopeptide YVDSEGHLpyTVPIR, suggesting that Y9 was phosphorylated. Mascot ion score was 40, with an expectation value of 0.0014 MS/MS. (B) TIE2-mediated caveolin-1 phosphorylation was analyzed after in vitro kinase assays with different concentrations of recombinant caveolin-1. Immunoblots for p-caveolin-1 (pY14), total caveolin-1, and TIE2 are shown. (C) TIE2-mediated phosphorylation of caveolin-1 was analyzed by an in vitro kinase assay and caveolin-1 detection in a SuperSep Phos-tag (Wako Pure Chemicals Industries, Ltd.) gel, as previously reported (3), and by standard Western blot analysis using a specific antibody against p-caveolin-1 at the Y14 site. Alkaline phosphatase (Apase; 2 U; Roche) was used for 1 h at 37°C. (D, E) In vitro kinase assays of TIE2 and recombinant wild-type and mutant caveolin-1 proteins, followed by standard Western blot (WB) analysis to analyze the caveolin-1, pY14 caveolin-1, and TIE2 levels of expression (D) or immunoblot using a SuperSep Phos-tag gel to detect unphosphorylated and phosphorylated forms of caveolin-1 (E). Lower panels, standard immunoblotting with indicated antibodies.
We also validated caveolin-1 phosphorylation at Tyr14 in vivo by transfecting wild-type and Y14F and Y42F mutant caveolin-1 into HEK293 and HEK293.Tie2 cells and determining caveolin-1 pY14 phosphorylation levels (Fig. 5A). Further proving the role of TIE2 in phosphorylating caveolin-1 in vivo, we genetically impaired the corresponding catalytic kinase domain of TIE2 protein. We transfected both wild-type Tie2 and Tie2 harboring a deleted kinase domain (Tie2dKD) together with caveolin-1-expressing plasmid into HEK293 cells. We found that wild-type TIE2 phosphorylated caveolin-1 at Tyr14 but the death kinase mutant did not. Of note, we detected caveolin-1 phosphorylation in rescue experiments by transfecting wild-type Tie2 (Fig. 5B). Our results showed that upon ligand stimulus, TIE2 directly phosphorylates caveolin-1 at the Tyr14 residue.
FIG 5.
TIE2 phosphorylation of caveolin-1 at Tyr14 is essential for TIE2/caveolin-1 nuclear translocation. (A) In vivo detection of Tyr14 phosphorylation in caveolin-1 by TIE2 was analyzed using cell lysates from HEK293 and HEK293.Tie2-myc cells transfected with wild-type or mutant Y14F or Y42F caveolin-1 plasmids and exposed to Ang1 (400 ng/ml; 30 min). Immunoblots for the presence of caveolin-1, phospho-caveolin-1, and TIE2 levels are shown. (B) TIE2 catalytic function is essential for caveolin-1-Tyr14 phosphorylation, as shown by performing kinase assays followed by transfecting HEK293 cells with wild-type Tie2 or a mutant harboring a deleted kinase domain (TIE2dKD), together with a transfection of a plasmid expressing caveolin-1. Cells were exposed to Ang1 (400 ng/ml) for 30 min. Immunoblots for the presence of caveolin-1, phospho-caveolin-1, TIE2, and tubulin levels are shown. (C) HEK293.Tie2-myc cells were transfected with Flag-Cav1 wild type (WT), Flag-Cav1 Y14F, Flag-Cav1 Y14D, and Flag-Cav1 Y42F, and 36 h later cells were exposed to Ang1 (400 ng/ml; 30 min); coimmunoprecipitation with TIE2 antibody was performed. Caveolin-1 and TIE2 protein were detected with anti-Flag tag and anti-myc tag antibody, respectively. (D) HeLa cells were transfected with Tie2 expression plasmids; 16 h later, they were cotransfected with wild-type or mutant Flag-tagged caveolin-1 plasmids. Forty-eight hours later, cultures were exposed to Ang1 (400 ng/ml; 30 min) and cytoplasmic and nuclear proteins, obtained by subcellular fractionation, were subjected to Western blot analysis using the indicated antibodies. Bovine serum albumin (0.1% in phosphate-buffered saline) was used as a vehicle/control in Ang1(−)-treated samples. LaminB and tubulin expression levels were used as nuclear and cytoplasmic protein loading controls, respectively. EV, empty vector. (E) HEK293.Tie2-myc cells were transfected with Flag-Cav1 WT, Flag-Cav1 Y14F, or Flag-Cav1 Y42F, and 36 h later cells were serum starved for 4 h before treatment with Ang1 (400 ng/ml; 30 min). Cell fixation and immunofluorescence studies were performed as previously described (3), using anti-myc tag antibody (red fluorescence). Green fluorescence from the GFP signal was used as a reporter for caveolin-1 expression. DAPI was used for nuclear staining. Merged images are provided for visualization of TIE2/caveolin-1 colocalization. Images were captured using a confocal microscope (Olympus FluoView FV1000). The antibodies and working dilutions are listed in Table S3.
To further study the functional importance of caveolin-1-Y14 phosphorylation in TIE2 trafficking, we first examined the binding status of caveolin-1 with TIE2, by transfecting several mutant forms of caveolin-1 (phosphodefective Y14F, phosphomimicking Y14D, and a phosphodefective Y42F for specificity) followed by coimmunoprecipitation with TIE2 antibody and Western blot analysis. The results showed that phosphorylation at Tyr14 is, at least partially, required for the generation of TIE2/caveolin-1 complexes (Fig. 5C). We then determined the role of TIE2-mediated caveolin-1-Y14 phosphorylation in nuclear TIE2 translocation by analyzing the expression of TIE2 and caveolin-1 in nuclear and cytoplasmic fractions of cells exposed to Ang1. We found that TIE2 nuclear translocation was challenged when cells expressed the Y14F mutant caveolin-1 (Fig. 5D). In addition, the lack of Y14 phosphorylation jeopardized the nuclear translocation of caveolin-1 and the TIE2/caveolin-1 complexes (Fig. 5D and E). These results show that TIE2 phosphorylates caveolin-1 at Tyr14, both in vitro and in vivo, and that this modification is essential for the formation of TIE2/caveolin-1 complexes and the nuclear translocation of TIE2.
DISCUSSION
In previous studies, we uncovered an important role of the RTK TIE2 in cancer radioresistance, i.e., its participation in the epigenetic regulation and DNA repair mechanisms (3). Here we extend the scope of our investigations to include detailed molecular-level characterization of the cellular trafficking of TIE2. Our results showed that Ang1 stimulus or ionizing radiation led to the localization of TIE2 in caveola structures and to the trafficking of TIE2/caveolin-1 complexes into the nucleus. TIE2 phosphorylates caveolin-1 at Tyr-14, and this modification is essential for the complex trafficking into the nucleus.
Clathrin-dependent endocytosis is considered the main internalization pathway for ligand-occupied RTKs (6, 13, 24). In fact, previous reports showed that TIE2 colocalizes with the clathrin heavy chain, resulting in internalization and degradation (18, 19). This mechanism is in accordance with the prevailing consensus that endocytosis is related to signaling attenuation (7, 12). Our data, however, showed that upon IR stress or Ang1 stimulus, TIE2 internalized into the nucleus after associating with and phosphorylating caveolin-1 (Fig. 6). Caveolin-1 phosphorylation at Tyr-14 is reported to be associated with endocytosis and trafficking of caveolae by changing the spatial conformation of caveolae (25). In this study, TIE2 phosphorylated caveolin-1 at Tyr-14, facilitating the formation of TIE2/caveolin-1 complexes and their nuclear translocation. Further studies will be necessary to understand which factors govern the TIE2 sorting to the clathrin or caveola internalization pathways (Fig. 6).
FIG 6.
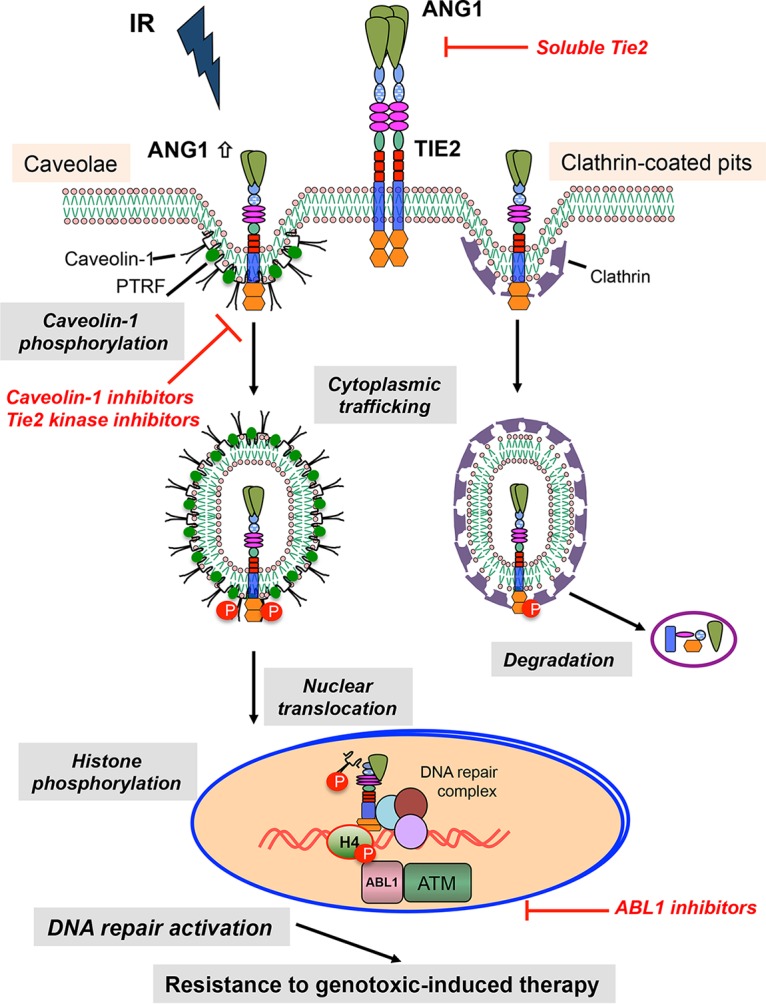
Schematic model for proposed TIE2 internalization pathways. Here we depict the model for TIE2 cellular trafficking. A previous internalization model of TIE2 upon Ang1 stimulus described the localization of TIE2 to clathrin-coated pits and degradation. Following ionizing radiation and subsequent Ang1 upregulation, TIE2 localizes to caveola structures and phosphorylates caveolin-1 at Tyr 14, regulating their nuclear trafficking. In the nucleus, TIE2 binds chromatin and DNA repair complexes, directly phosphorylating histone H4 at Tyr51, which is read by the ABL1 proto-oncogene. This pathway is involved in enhancing a nonhomologous end-joining mechanism resulting in radioresistance of cancer cells. The specific signals involved in the sorting of TIE2 toward clathrin-coated pits or caveola structures are not known. Shown are possible avenues for inhibition of the described process.
While RTK trafficking has been associated in some instances to caveolin-1, the pathway described here differs in several aspects from those previously reported. Under oxidative stress, EGFR is colocalized with caveolin-1 and is sorted to a perinuclear compartment, escaping from degradation and remaining active (16). Interestingly, ligand-induced phosphorylation of EGFR has been reported to induce dissociation of EGFR from caveolae as a mechanism of autoregulation of EGF signaling (26). VEGF ligand stimulus of endothelial cells induces caveolin-1 nuclear translocation, together with VEGFR2 and endothelial NO synthase (eNOS), downmodulating VEGFR2 activity (17, 27). In addition, although caveolin-1 was found to be phosphorylated at Tyr 14 in VEGFR and EGFR signaling pathways, it seems to be, in both cases, Src kinase mediated (17, 28), differing from the pathway described here, in which TIE2 directly phosphorylates caveolin-1, regulating its own cellular trafficking.
The role of caveolin-1 Tyr14 in binding protein partners has been described previously. Thus, Src-dependent caveolin-1 Tyr14 phosphorylation is involved in the generation of eNOS/caveolin-1, which promoted eNOS inhibition as part of a feedback regulation (29). Further describing the mechanism underlying TIE2 cellular trafficking, our results showed that TIE2-mediated caveolin-1 Tyr14 phosphorylation is regulating the generation of TIE2/caveolin-1 complexes, which in turn will regulate TIE2/caveolin-1 nuclear translocation.
Caveolin-1 expression seems to be dependent on tumor type, and while it has been found to be overexpressed in breast, prostate, and lung cancer, it is downregulated in colon and ovarian carcinomas (15). A study using nonquantitative immunohistochemistry detected caveolin-1 expression in a cohort of 20 glioblastomas (30). Our results show for the first time the quantitative expression of caveolin-1 and PTRF in a large set of glioblastomas and the expression of caveolin-1 in brain tumor stem cells. Furthermore, our data suggest a positive relationship between gliomas and two essential proteins in the functional caveolae, caveolin-1 and PTRF.
Reports describing the localization of caveolin-1 in the nuclear compartment are scarce, limited to the report of exogenously expressed caveolin-1 in SKOV3 and IGtC3 ovarian carcinoma cells (31) and upon VEGF ligand stimulus of endothelial cells (17, 27). Moreover, there is a lack of description of the mechanisms underlying such translocation into the nucleus. Thus, our report uncovers a new protein-trafficking regulation involving both the RTK TIE2 and the major player in functional caveolae, caveolin-1.
Our results might complement reports that show that caveolin-1 may play a role in the radioresistant phenotype of various tumors, such as nonsmall cell lung carcinoma and prostate, pancreas, and breast cancers (32). It has been reported that caveolin-1 expression increases after IR in several cancer cell lines and that its expression confers resistance to DNA damage by modulating homologous recombination and NHEJ systems (33). We propose that this role of caveolin-1 might be related to its effects on RTK trafficking to the nucleus. However, further studies are required to determine whether caveolin-1 has any additional role in the nucleus after cotranslocating with TIE2, such as transcription regulation (31). Of interest, caveolin-1 is a substrate for ABL1, and we have previously reported the role of ABL1 in reading the TIE2-mediated modification of histone 4 at Tyr51 (3); however, a link between ABL1 and nuclear caveolin-1 has not yet been described.
In summary, after ligand Ang1 binding or irradiation, the tyrosine kinase receptor TIE2 routes to the caveola vesicles and phosphorylates caveolin-1 at Tyr14, which facilitates the translocation of both membrane proteins to the nucleus. We previously showed that nuclear TIE2 phosphorylates histone H4Y51 and recruits DNA repair proteins to the DNA damage site (3). Because TIE2 has been found to be expressed in several cancers, including gliomas (1), our research provides evidence that targeting this molecular mechanism of translocation might be an effective strategy to prevent radioresistance of brain tumor and other malignant tumors.
MATERIALS AND METHODS
Cell culture conditions.
U251 human glioma cells were maintained in Dulbecco's modified Eagle medium (DMEM)–F-12, supplemented with 10% (vol/vol) fetal bovine serum and 1% penicillin and streptomycin. Human umbilical vein endothelial cells (HUVEC) were maintained in EGM-2 Bullet kit medium (Lonza). Glioma stem cell cultures were characterized as previously reported (34) and maintained in DMEM–F-12, supplemented with B27 (Life Technologies), epidermal growth factor (EGF), and basic fibroblast growth factor (20 ng/ml each; Sigma-Aldrich). HEK293 and HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum (vol/vol) and 1% penicillin-streptomycin. U251.EV, U251.Tie2, U251.Tie2-myc, HEK293.EV, and HEK293.Tie2-myc cell lines were maintained in the presence of 300 μg/ml G418 (Corning).
RNA interference.
siRNAs were transfected into cells using the INTERFERin transfection reagent (Polyplus transfection) at concentrations of 10 nM or otherwise indicated; after 48 h of transfection, the knockdown efficiency was evaluated by determining the protein levels in whole-cell lysates. The siRNA sequences used are listed in Table S1 in the supplemental material.
Reverse transcription-PCR analysis.
Total RNA was extracted with TRIzol (Invitrogen), and 2 μg of RNA was reverse transcribed to cDNA with the High Capacity RNA-to-cDNA kit (Applied Biosystems). For reverse transcription-PCR expression, cDNA was subjected to a PCR cycle analyzer with a Kapa PCR master mixture (Kapa Biosystems) using the following PCR conditions: 95°C for 2 min, 95°C for 15 s, 64°C for 15 s, and 72°C for 15 s for 32 cycles, with a final extension at 72°C for 10 min and holding at 4°C. Primer sequences are provided in Table S2 in the supplemental material.
Lysate preparation, immunoprecipitation, and Western blot analysis.
Cells were treated with ANG1 or radiation. After the indicated time periods at 37°C, cells were washed with phosphate-buffered saline (PBS), collected with a cell scraper, and lysed with IPH buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 5% glycerol, 0.2% NP-40, 1 mM Na3VO4 containing phosphatase, and protease inhibitor cocktails). After incubation on ice for 15 min, the lysates were subjected to sonication with 30× amplitude for 10 s for three cycles (total, 30 s) in a Qsonica sonicator (VWR). The supernatant was cleaned by centrifugation at 14,000 rpm for 10 min at 4°C to obtain total cell lysates. The lysate was subjected to immunoprecipitation with the specific antibody overnight at 4°C. The resulting mixture was incubated with protein A-agarose (for rabbit host) (Millipore) or protein G-agarose (for mouse host) (Millipore) for 30 min, washed, and separated by 4 to 20% SDS-PAGE. The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane, and Western blot analysis was conducted with the indicated antibodies; representative results of 2 or 3 independent experiments are shown. To isolate proteins from brain tumor tissue, 0.1 g to 0.2 g tissue was resuspended with 400 to 600 μl of IPH buffer containing 0.2% SDS and sonicated for 30 s. The supernatant was cleaned by centrifugation (14,000 rpm; 10 min; 4°C) to obtain total protein lysates. Forty micrograms of protein lysates was used for Western blot analysis. Normal human brain tissue lysate was purchased from Abcam (catalog number ab29466). Restore PLUS Western blot stripping buffer (Thermo Scientific) was used to reblot the membranes as recommended by the manufacturer. The antibodies used are listed in Table S3 in the supplemental material.
Subcellular fractionation.
Cells were washed with ice-cold PBS, harvested by scraping or trypsinization, and collected in a hypotonic lysis buffer (20 mM HEPES [pH 7.0], 10 mM KCl, 2 mM MgCl2, 0.5% NP-40, 1 mM Na3VO4, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and aprotinin [2 mg/ml]) for 10 min on ice. Cells were then homogenized by 20 strokes in a tight-fitting Dounce homogenizer (Kontes disposable pestle with microtubes; Fisher Scientific). After centrifugation at 4,500 rpm for 5 min to sediment the nuclei, the supernatant was transferred to a new tube and centrifuged at 14,000 rpm for 20 min. The resulting supernatant was collected as a cytoplasmic fraction and transferred to a prechilled tube. The nuclear pellet was washed three times with hypotonic lysis buffer, resuspended, and periodically vortexed in nuclear extraction buffer (20 mM HEPES [pH 7.9], 400 mM NaCl, 1 mM EDTA [pH 8.0], 1 mM EGTA [pH 7.0], phosphatase inhibitor cocktails 2 and 3 [Sigma-Aldrich], and a protease inhibitor cocktail [Sigma-Aldrich]) on ice for 30 min. After centrifugation at 14,000 rpm for 10 min at 4°C, the nuclear lysates were collected in prechilled tubes.
Cell viability and clonogenic assays.
Cells were plated at subconfluent density with TIE2-13 (TIE2 inhibitor, 1 μM in dimethyl sulfoxide [DMSO]) (35) and filipin (a caveolin-1 inhibitor, 1 to 2 μg/ml in DMSO) (36) in a 96-well plate, and 24 h later they were irradiated (15 Gy) and kept in an incubator for an additional 48 to 72 h at 37°C. A CellTitre-Blue cell viability assay (Promega) was used according to the manufacturer's instructions. When indicated, cells were stained with crystal violet solution (0.1% crystal violet and 20% methanol) for 15 min at room temperature (RT), and the results were imaged. To perform a clonogenic assay, 25 cells were plated per well in six-well plates, treated with the indicated inhibitors for 24 h, and then irradiated (2.5 Gy). Cells were incubated for 14 days, and the visible colonies were counted after crystal violet staining, as described above.
Plasmid construction, transfection, and primers.
The pcDNA3-Tie2, which contains full-length human Tie2 cDNA, pcDNA3.1-Tie2-myc, and pcDNA3.1-Tie2dKD-myc plasmids, has been previously described (37, 38). pcDNA3.1-Flag, pcDNA3.1-Flag-Cav1WT, pcDNA3.1-Flag-Cav1Y14F, pcDNA3.1-Flag-Cav1Y14D, pcDNA3.1-Flag-Cav1Y42F, pET32a-EV, pET32a-Cav1WT, pET32a-Cav1Y14F, pET32a-Cav1Y42F, pEGFP-N1, pEGFP-Cav1WT, pEGFP-Cav1Y14F, and pEGFP-Cav1Y42F were constructed using the QuikChange II site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's recommended protocol. Cells were transfected using the X-tremeGENE HP DNA transfection reagent (Roche Applied Science) according to the manufacturer's protocol. The primers used are listed in Table S2.
Immunofluorescence microscopy.
Cells were seeded in chamber slides (Lab-Tek), and after the indicated treatments, they were washed with PBS, fixed with 4% paraformaldehyde, permeabilized for 30 min with 0.2% Triton X-100 in PBS, and blocked for 30 min in a blocking buffer (3% bovine serum albumin [BSA] and 2% horse serum) at RT. After incubation with the indicated primary antibodies in blocking buffer overnight at 4°C, the cells were incubated with secondary fluorescent antibodies for 60 min at RT. After a final washing, Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) was added. Images were captured using a confocal microscope (Olympus FluoView FV1000).
Mass spectrometry analysis.
In vitro kinase assays were performed with active TIE2 and recombinant CAV1 protein, after a gel-run caveolin-1 band was cut and sent for mass spectrometry analysis. Nano-liquid chromatography/tandem mass spectrometry (nano-LC-MS/MS) was performed on a Thermo Finnigan LTQ Orbitrap Velos (Thermo Scientific), coupled with an Eksigent NanoLC Ultra nano-HPLC (AB SCIEX). The sample was injected onto a nano-trap column (100-mm inner diameter by 1 cm, C18 PepMap100) with an autosampler and then passed down a C18 reversed-phase home-packed column (SB-C18, Zorbax, 5 mm; Agilent). The flow rate was 400 nl/min with a 60-min LC gradient, and the mobile phases were as follows: A, 5% acetonitrile (ACN) and 0.1% formic acid (FA); B, 100% ACN and 0.1% FA. Eluted peptides were sprayed through a charged emitter tip (PicoTip emitter, 10 ± 1 mm; New Objective) into the mass spectrometer. Parameters included the following: spray voltage at +2.0 kV, Fourier transform MS mode for MS acquisition of precursor ions (60,000 resolution); ion trap MS mode for subsequent MS/MS of top six precursors selected; and fragmentation accomplished via collision-induced dissociation. Proteome Discoverer version 1.2 (Thermo Scientific) was used for protein identification and modification analysis. Parameters included the following: selection of the enzyme as trypsin; maximum missed cleavages, 2; variable modifications included oxidation (M) and tyrosine phosphorylation; precursor ion tolerance was at 0.02 Da; and MS/MS fragment tolerance was at 0.6 Da. The significance of a peptide match was based on expectation values (P < 0.05).
In vitro kinase assay.
In vitro kinase assays were conducted with 200 ng of purified glutathione S-transferase (GST)-tagged TIE2 active protein (no. TEK-7296H; Creative Biomart) in kinase buffer consisting of 25 mM MOPS (morpholinepropanesulfonic acid) (pH 7.2), 12.5 mM β-glycerol phosphate, 20 mM MgCl2, 12.5 mM MnCl2, 2 mM EDTA (pH 8.0), 5 mM EGTA (pH 7.0), and 0.25 mM dithiothreitol (DTT) with the addition of 20 mM ATP. The substrate, recombinant caveolin-1 protein (catalog no. H00000857-P01; Novus Biologicals), was added to the reaction mixture, which was incubated at 37°C for 30 min. The kinase reaction was stopped by the addition of 4× protein sample buffer (Invitrogen), and the mixture was boiled for 5 min, followed by SDS-PAGE (4 to 20%) analyses. Phosphorylated caveolin-1 protein was detected by Western blotting.
Phos-tag gel analysis.
SuperSep Phos-tag (50 mM) 12.5%, 13-well gels (Wako Pure Chemicals Industries Ltd.) were used to detect phosphorylated proteins by SDS-PAGE using an analogous Phos-tag complex with two manganese (II) ions, Mn2+-Phos-tag. To analyze the phosphoproteins, an in vitro kinase assay was performed, and the samples were then loaded onto a Phos-tag gel; the proteins were transferred to PVDF membranes according to the manufacturer's protocol. Western blot analyses were conducted using specific antibodies. Alkaline phosphatase (Roche) was used to dephosphorylate the proteins after the in vitro kinase assay for 1 h at 37°C.
Purification of caveolin-1 recombinant proteins.
Full-length human CAV1 cDNA was cloned into the pET-32a(+) vector (no. 69015-3; Novagen) between BamHI and XhoI sites. Cav1 wild-type and two other mutant (Cav1Y14F and Cav1Y42F) plasmids were used as the template to generate protein in a cell-free protein system (AccuRapid cell-free protein expression kit, no. K7250; Bioneer) according to the manufacturer's protocol. Caveolin-1 protein expression was confirmed by colloidal blue staining (Invitrogen) in addition to Western blotting using antibodies against caveolin-1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ann M. Sutton (Department of Scientific Publications, MD Anderson, Houston, TX) for scientific editorial assistance and Xuejun Fan and Joy Gumin (Brain Tumor Program, MD Anderson, Houston, TX) for technical assistance. We acknowledge Federico Bussolino (University of Turin, Turin, Italy) for providing the pcDNA3-Tie2 plasmid and William A. Barton (Virginia Commonwealth University, Richmond, VA) for providing pcDNA3.1-Tie2-myc.
We declare that we have no conflict of interest.
Author contributions were as follows: M.B.H., R.S., J.F., and C.G.-M. developed the original hypothesis. M.B.H., R.S., J.F., C.G.-M., and J.L. designed the study, performed experiments, and analyzed data. X.L. carried out and interpreted the mass spectrometry experiments. K.R.H. carried out the statistical analysis. Y.R.-M., F.P.M., H.J., and M.-C.H. participated with the discussion of results. F.F.L. provided GSCs. The manuscript was written by M.B.H., R.S., J.F., and C.G.-M.
This work was supported by National Institutes of Health (NIH) grant R01 NS069964, grant P50 CA127001, and Cancer Center Support Grant P30CA016672 (Sequencing and Microarray Facility).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00142-17.
REFERENCES
- 1.Martin V, Liu D, Fueyo J, Gomez-Manzano C. 2008. Tie2: a journey from normal angiogenesis to cancer and beyond. Histol Histopathol 23:773–780. doi: 10.14670/HH-23.773. [DOI] [PubMed] [Google Scholar]
- 2.Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. 2000. Vascular-specific growth factors and blood vessel formation. Nature 407:242–248. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 3.Hossain MB, Shifat R, Johnson DG, Bedford MT, Gabrusiewicz KR, Cortes-Santiago N, Luo X, Lu Z, Ezhilarasan R, Sulman EP, Jiang H, Li SS, Lang FF, Tyler J, Hung MC, Fueyo J, Gomez-Manzano C. 2016. TIE2-mediated tyrosine phosphorylation of H4 regulates DNA damage response by recruiting ABL1. Sci Adv 2:e1501290. doi: 10.1126/sciadv.1501290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee OH, Xu J, Fueyo J, Fuller GN, Aldape KD, Alonso MM, Piao Y, Liu TJ, Lang FF, Bekele BN, Gomez-Manzano C. 2006. Expression of the receptor tyrosine kinase Tie2 in neoplastic glial cells is associated with integrin beta1-dependent adhesion to the extracellular matrix. Mol Cancer Res 4:915–926. doi: 10.1158/1541-7786.MCR-06-0184. [DOI] [PubMed] [Google Scholar]
- 5.Liu D, Martin V, Fueyo J, Lee OH, Xu J, Cortes-Santiago N, Alonso MM, Aldape K, Colman H, Gomez-Manzano C. 2010. Tie2/TEK modulates the interaction of glioma and brain tumor stem cells with endothelial cells and promotes an invasive phenotype. Oncotarget 1:700–709. doi: 10.18632/oncotarget.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goh LK, Sorkin A. 2013. Endocytosis of receptor tyrosine kinases. Cold Spring Harb Perspect Biol 5:a017459. doi: 10.14670/HH-23.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorkin A, von Zastrow M. 2009. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol 10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen MK, Hung MC. 2015. Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases. FEBS J 282:3693–3721. doi: 10.1111/febs.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou RH, Wang YN, Hsieh YH, Li LY, Xia W, Chang WC, Chang LC, Cheng CC, Lai CC, Hsu JL, Chang WJ, Chiang SY, Lee HJ, Liao HW, Chuang PH, Chen HY, Wang HL, Kuo SC, Chen CH, Yu YL, Hung MC. 2014. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev Cell 30:224–237. doi: 10.1016/j.devcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conner SD, Schmid SL. 2003. Regulated portals of entry into the cell. Nature 422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 11.Doherty GJ, McMahon HT. 2009. Mechanisms of endocytosis. Annu Rev Biochem 78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 12.Tomas A, Futter CE, Eden ER. 2014. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol 24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delos Santos RC, Garay C, Antonescu CN. 2015. Charming neighborhoods on the cell surface: plasma membrane microdomains regulate receptor tyrosine kinase signaling. Cell Signal 27:1963–1976. doi: 10.1016/j.cellsig.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Parton RG, del Pozo MA. 2013. Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol 14:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Outschoorn UE, Sotgia F, Lisanti MP. 2015. Caveolae and signalling in cancer. Nat Rev Cancer 15:225–237. doi: 10.1038/nrc3915. [DOI] [PubMed] [Google Scholar]
- 16.Khan EM, Heidinger JM, Levy M, Lisanti MP, Ravid T, Goldkorn T. 2006. Epidermal growth factor receptor exposed to oxidative stress undergoes Src- and caveolin-1-dependent perinuclear trafficking. J Biol Chem 281:14486–14493. doi: 10.1074/jbc.M509332200. [DOI] [PubMed] [Google Scholar]
- 17.Labrecque L, Royal I, Surprenant DS, Patterson C, Gingras D, Beliveau R. 2003. Regulation of vascular endothelial growth factor receptor-2 activity by caveolin-1 and plasma membrane cholesterol. Mol Biol Cell 14:334–347. doi: 10.1091/mbc.E02-07-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogdanovic E, Coombs N, Dumont DJ. 2009. Oligomerized Tie2 localizes to clathrin-coated pits in response to angiopoietin-1. Histochem Cell Biol 132:225–237. doi: 10.1007/s00418-009-0603-3. [DOI] [PubMed] [Google Scholar]
- 19.Bogdanovic E, Nguyen VP, Dumont DJ. 2006. Activation of Tie2 by angiopoietin-1 and angiopoietin-2 results in their release and receptor internalization. J Cell Sci 119:3551–3560. doi: 10.1242/jcs.03077. [DOI] [PubMed] [Google Scholar]
- 20.Torgersen ML, Skretting G, van Deurs B, Sandvig K. 2001. Internalization of cholera toxin by different endocytic mechanisms. J Cell Sci 114:3737–3747. [DOI] [PubMed] [Google Scholar]
- 21.Couet J, Li S, Okamoto T, Ikezu T, Lisanti MP. 1997. Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J Biol Chem 272:6525–6533. [DOI] [PubMed] [Google Scholar]
- 22.Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, Walser P, Abankwa D, Oorschot VM, Martin S, Hancock JF, Parton RG. 2008. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132:113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fridolfsson HN, Roth DM, Insel PA, Patel HH. 2014. Regulation of intracellular signaling and function by caveolin. FASEB J 28:3823–3831. doi: 10.1096/fj.14-252320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HH, Wang YN, Hung MC. 2015. Non-canonical signaling mode of the epidermal growth factor receptor family. Am J Cancer Res 5:2944–2958. [PMC free article] [PubMed] [Google Scholar]
- 25.Zimnicka AM, Husain YS, Shajahan AN, Sverdlov M, Chaga O, Chen Z, Toth PT, Klomp J, Karginov AV, Tiruppathi C, Malik AB, Minshall RD. 2016. Src-dependent phosphorylation of caveolin-1 Tyr-14 promotes swelling and release of caveolae. Mol Biol Cell 27:2090–2106. doi: 10.1091/mbc.E15-11-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abulrob A, Giuseppin S, Andrade MF, McDermid A, Moreno M, Stanimirovic D. 2004. Interactions of EGFR and caveolin-1 in human glioblastoma cells: evidence that tyrosine phosphorylation regulates EGFR association with caveolae. Oncogene 23:6967–6979. doi: 10.1038/sj.onc.1207911. [DOI] [PubMed] [Google Scholar]
- 27.Feng Y, Venema VJ, Venema RC, Tsai N, Caldwell RB. 1999. VEGF induces nuclear translocation of Flk-1/KDR, endothelial nitric oxide synthase, and caveolin-1 in vascular endothelial cells. Biochem Biophys Res Commun 256:192–197. doi: 10.1006/bbrc.1998.9790. [DOI] [PubMed] [Google Scholar]
- 28.Dittmann K, Mayer C, Kehlbach R, Rodemann HP. 2008. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol Cancer 7:69. doi: 10.1186/1476-4598-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Bakhshi FR, Shajahan AN, Sharma T, Mao M, Trane A, Bernatchez P, van Nieuw Amerongen GP, Bonini MG, Skidgel RA, Malik AB, Minshall RD. 2012. Nitric oxide-dependent Src activation and resultant caveolin-1 phosphorylation promote eNOS/caveolin-1 binding and eNOS inhibition. Mol Biol Cell 23:1388–1398. doi: 10.1091/mbc.E11-09-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parat MO, Riggins GJ. 2012. Caveolin-1, caveolae, and glioblastoma. Neuro Oncol 14:679–688. doi: 10.1093/neuonc/nos079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanna E, Miotti S, Mazzi M, De Santis G, Canevari S, Tomassetti A. 2007. Binding of nuclear caveolin-1 to promoter elements of growth-associated genes in ovarian carcinoma cells. Exp Cell Res 313:1307–1317. doi: 10.1016/j.yexcr.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Mahmood J, Zaveri SR, Murti SC, Alexander AA, Connors CQ, Shukla HD, Vujaskovic Z. 2016. Caveolin-1: a novel prognostic biomarker of radioresistance in cancer. Int J Radiat Biol 92:747–753. doi: 10.1080/09553002.2016.1222096. [DOI] [PubMed] [Google Scholar]
- 33.Zhu H, Yue J, Pan Z, Wu H, Cheng Y, Lu H, Ren X, Yao M, Shen Z, Yang JM. 2010. Involvement of Caveolin-1 in repair of DNA damage through both homologous recombination and non-homologous end joining. PLoS One 5:e12055. doi: 10.1371/journal.pone.0012055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang H, Gomez-Manzano C, Aoki H, Alonso MM, Kondo S, McCormick F, Xu J, Kondo Y, Bekele BN, Colman H, Lang FF, Fueyo J. 2007. Examination of the therapeutic potential of Delta-24-RGD in brain tumor stem cells: role of autophagic cell death. J Natl Cancer Inst 99:1410–1414. doi: 10.1093/jnci/djm102. [DOI] [PubMed] [Google Scholar]
- 35.Gourzoulidou E, Carpintero M, Baumhof P, Giannis A, Waldmann H. 2005. Inhibition of angiogenesis-relevant receptor tyrosine kinases by sulindac analogues. Chembiochem 6:527–531. doi: 10.1002/cbic.200400192. [DOI] [PubMed] [Google Scholar]
- 36.Lu Z, Ghosh S, Wang Z, Hunter T. 2003. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell 4:499–515. doi: 10.1016/S1535-6108(03)00304-0. [DOI] [PubMed] [Google Scholar]
- 37.Audero E, Cascone I, Maniero F, Napione L, Arese M, Lanfrancone L, Bussolino F. 2004. Adaptor ShcA protein binds tyrosine kinase Tie2 receptor and regulates migration and sprouting but not survival of endothelial cells. J Biol Chem 279:13224–13233. doi: 10.1074/jbc.M307456200. [DOI] [PubMed] [Google Scholar]
- 38.Seegar TC, Eller B, Tzvetkova-Robev D, Kolev MV, Henderson SC, Nikolov DB, Barton WA. 2010. Tie1-Tie2 interactions mediate functional differences between angiopoietin ligands. Mol Cell 37:643–655. doi: 10.1016/j.molcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.