ABSTRACT
Chromatin entry sites (CES) are 100- to 1,500-bp elements that recruit male-specific lethal (MSL) complexes to the X chromosome to upregulate expression of X-linked genes in male flies. CES contain one or more ∼20-bp GA-rich sequences called MSL recognition elements (MREs) that are critical for dosage compensation. Recent studies indicate that CES also correspond to boundaries of X-chromosomal topologically associated domains (TADs). Here, we show that an ∼1,000-kDa complex called the late boundary complex (LBC), which is required for the functioning of the Bithorax complex boundary Fab-7, interacts specifically with a special class of CES that contain multiple MREs. Mutations in the MRE sequences of three of these CES that disrupt function in vivo abrogate interactions with the LBC. Moreover, reducing the levels of two LBC components compromises MSL recruitment. Finally, we show that several of the CES that are physically linked to each other in vivo are LBC interactors.
KEYWORDS: insulators, boundaries, TADs, dosage compensation, Drosophila, male-specific lethal complexes, chromatin entry sites, MSL recognition elements, CLAMP, GAGA factor
INTRODUCTION
The three-dimensional organization of eukaryotic chromosomes plays a central role in diverse processes ranging from gene regulation to recombination and repair (1–3). Chromatin conformation capture (CCC) experiments have shown that a key organizing principle is the subdivision of chromosomes into a series of looped domains called topologically associated domains (TADs) (4, 5).
Though the scale of the loops is smaller, a similar subdivision of chromosomes into looped domains is present in Drosophila, and GA-rich cis elements are present at the bases of most loops (6). In both mammals and Drosophila, the endpoints of the loops are delimited by special architectural elements called boundaries or insulators. Boundaries in flies generate loops by pairing with each other. For example, pairing interactions between the two boundaries, homie and nhomie, that flank the even-skipped (eve) gene generate an ∼16-kb loop that encompasses eve and all of its regulatory elements (7). In addition to determining chromosome architecture, pairing interactions between boundaries have regulatory consequences. When interposed between enhancers/silencers and a target gene, boundary factors block regulatory interactions (8) or insulate regions of the genome against chromosomal position effects (9).
In addition to insulation, pairing interactions between boundaries can also facilitate long-distance gene regulation. In flies, this phenomenon was first observed for the su(Hw) and Mcp boundaries carried by transgene reporters inserted at distant sites (10–12). Pairing has subsequently been documented for many other boundaries in transgene boundary bypass experiments and in experiments in which boundaries, reporters, and enhancers are placed at distant locations using site-specific insertion techniques (7, 13, 14). For example, pairing interactions between the eve homie and nhomie boundaries can bring enhancers and reporters together over distances ranging from 140 kb to 2 Mb.
Another context in Drosophila in which communication over large chromosomal distances is thought to be critically important is dosage compensation, where a coordinated domain of active gene expression is established across the entire X chromosome (15–17). In Drosophila, expression of X-linked genes in males is upregulated to equalize dosage with the two X chromosomes in females. Dosage compensation depends upon a male-specific lethal (MSL) dosage compensation complex that contains five proteins (MSL1, MSL2, MSL3, MOF, and MLE) and one of two noncoding RNAs, roX1 or roX2 (RNA on the X) (16, 18). MSL complexes associate with actively transcribed genes on the male X chromosome, acetylating histone H4 lysine 16 (19) and increasing the amount of active RNA polymerase II over gene bodies (20).
At least two steps are involved in targeting the dosage compensation machinery to active X-linked genes. First, MSL complexes are recruited to the X chromosome by special 100- to 1,500-bp cis-acting elements called chromatin entry sites (CES) or high-affinity sites (HAS) (21–23). Next, MSL complexes spread from CES to the bodies of actively transcribed genes (24, 25). Most CES contain one or more smaller cis elements called MSL recognition elements (MREs) (23). MREs are ∼21-bp GA-rich sequences that include sequences that are similar to the recognition motif for the well-studied Trithorax-like GAGA factor (GAF), GAGAG (26). Two of the loci most enriched for the MSL complex are the CES within the roX1 and roX2 genes (21, 27, 28), both of which contain multiple MRE sequences.
Recent experiments reported by Ramírez et al. (29) have shown that many of the TAD boundaries on the X chromosome correspond to CES. As boundary elements, CES could provide hubs for the recruitment of MSL complexes and promote their bidirectional spread from the base of each loop toward active genes located within the loop. Consistent with an underlying structural role in organizing the topology of the X chromosome, the three-dimensional organization of the X chromosome is the same in male and female cells, as is the colocalization of CES with loop boundaries. Moreover, depleting MSL2 or MSL3 in male cells does not induce major alterations in the looping pattern. The only clear difference in the topological organization of male and female X chromosomes is in the frequency of long-distance interactions between roX1 and roX2 and sites elsewhere on the X chromosome, which is consistent with their male-specific transcriptional activity.
Other observations are consistent with the idea that CES are preexisting scaffolds for MSL recruitment and likely have properties in common with architectural elements elsewhere in the genome. Thus far, two zinc finger DNA binding proteins have been implicated in dosage compensation and/or CES function. One of the zinc finger proteins is chromatin-linked adaptor for MSL proteins (CLAMP). CLAMP is present at all CES, where it directly binds to MRE elements and is required for MSL complex recruitment (30–32). The other zinc finger protein is GAF. Like CLAMP, GAF also recognizes GA-rich motifs, including MREs. However, the connection between GAF and CES is so far only indirect. trans-Heterozygous combinations of strong and weak mutations in the GAF gene, Trithorax-like (Trl), have enhanced lethal effects in males compared to in females (33). Moreover, the male-specific lethal effects of Trl are related to the dosage compensation machinery, because they can be enhanced when the male flies are also heterozygous for mutations in one of the MSL complex-encoding genes. While these findings link CLAMP, and perhaps also GAF, to CES, both proteins have functions beyond their involvement in dosage compensation. GAF and CLAMP are essential proteins in both males and females (34) and recognize several thousand GA-rich sites on the X chromosome and autosomes (31).
Of potential relevance to the link between CES and TAD boundaries are the architectural functions of the GAF protein. GAF was first implicated as a boundary factor in transgene assays (35, 36). More recently, GAF was found to be a component of a very large (>780-kDa) protein complex, called the late boundary complex (LBC), that is critical for the functioning of a Drosophila Bithorax complex (BX-C) boundary, Fab-7 (37, 38). Though the identities of all the different components of the LBC are not yet known, antibody “supershift” electrophoretic mobility shift assays (EMSAs) indicate that it contains three proteins, GAF, Mod(mdg4), and E(y)2. While GAF and Mod(mdg4) are found associated with the Fab-7 boundary element using chromatin immunoprecipitation (ChIP) experiments, they are not the only proteins that localize to this boundary. In fact, one of the other DNA binding proteins found at Fab-7 in vivo is CLAMP.
Here, we show that CES share properties with previously characterized autosomal chromatin boundaries. We demonstrate that the Fab-7 boundary factor LBC binds specifically to several X-linked CES that contain multiple MREs, including roX1, roX2, and CES 5C2. Furthermore, LBC binding to these CES is abrogated by mutations in the MREs that disrupt CES activity in vivo. Connecting the architectural functions of CES to the LBC, we show that the LBC binds to CES that are known to interact with each other in vivo. We present biochemical evidence that CLAMP is a component of the LBC and use RNA interference (RNAi) depletion experiments to show that CLAMP and GAF bind interdependently in vivo. Moreover, we show that CLAMP and GAF are both important for the recruitment of MSL complexes to roX1, roX2, and CES 5C2. Overall, we provide a novel link between boundary-forming protein complexes and dosage compensation.
RESULTS
CLAMP binds to the Fab-7 boundary in vivo and in vitro.
The Fab-7 boundary spans a DNA segment that includes three chromatin-specific nuclease-hypersensitive sequences, HS*, HS1, and HS2 (Fig. 1B) (39–41). While all of these sequences contribute to boundary activity, replacement experiments indicated that HS1 alone is sufficient for full function in an otherwise wild-type background (37). Molecular and biochemical experiments have shown that one of the factors responsible for HS1 boundary activity is the LBC (37, 38). The LBC binds to the distal half of HS1, dHS1. This 240-bp sequence has 4 GAGAG motifs: GAGA3, GAGA4, GAGA5, and GAGA6 (Fig. 1B). Probes spanning all four GAGAG motifs, pairs of GAGAG motifs (GAGA3 and -4 and GAGA5 and -6), or single GAGAG motifs (GAGA3, GAGA4, and GAGA5 but not GAGA6) are shifted by the LBC (37). Deletion and mutational analysis of the GAGA3 and GAGA4 probes showed that the minimal length required for LBC binding is 65 bp and that the GAGA motifs are essential. Consistent with these in vitro findings, mutations in the GAGA3 and GAGA4 motifs disrupt Fab-7 boundary function in vivo (42). Not surprisingly, given the importance of the GAGA motifs, EMSA shifts generated by the LBC were found to be supershifted by GAF antibodies. Antibodies against Mod(mdg4) and E(y)2 also supershifted the LBC shift; however, antibodies against other known boundary proteins, such as Beaf, Zw5, and dCTCF, had no effect. For this reason, we sought other potential LBC-associated proteins.
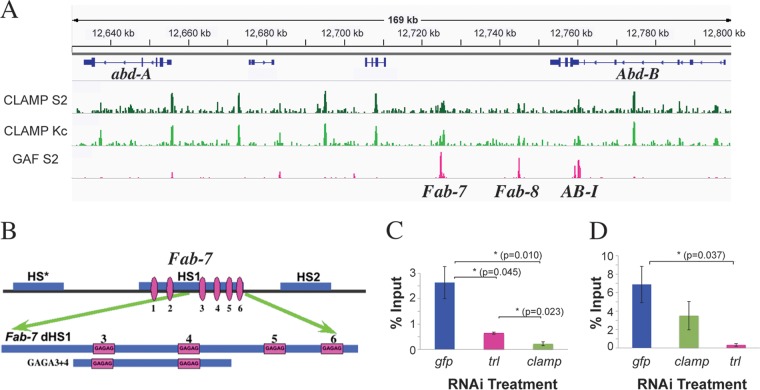
FIG 1.
CLAMP colocalizes with several boundaries in BX-C. (A) ChIP localization profiles for CLAMP in S2 and Kc cell lines (green) (31) and GAF (pink) (54) in a 160-kb region, including the Fab-7, Fab-8, and AB-I boundaries. (B) (Top) Chromatin organization of the Fab-7 boundary (1.2 kb) showing nuclease-hypersensitive regions. The distal 240 bp of Fab-7 HS1 (dHS1) contains 4 GAGAG motifs (pink). (Bottom) The 130-bp GAGA3+4 probe used for EMSA. (C and D) ChIP, followed by qPCR following clamp or trl knockdowns. The error bars represent standard errors obtained from 4 (gfp) or 3 (trl and clamp) independent experiments. P values from t tests are displayed for significance. *, P < 0.05, or **, P < 0.01 for differences between sample means. Comparisons between specific treatments are indicated by horizontal lines. For not statistically significant differences, P values are not displayed.
One plausible candidate is CLAMP. Like GAF, CLAMP recognizes GA-rich motifs, and ChIP sequencing (ChIP-seq) experiments revealed that many of the sequences bound by GAF and CLAMP on the X chromosome and autosomes closely overlap (31). In fact, one of the sites occupied by both CLAMP and GAF is Fab-7 (Fig. 1A; see Fig. S1 in the supplemental material). Aside from Fab-7, CLAMP localizes to other sequences within the Bithorax complex locus. Some of the CLAMP-bound loci are shared with GAF, while others are unique. Included among the common sites are two boundaries, Fab-8 and AB-I, from the Abd-B region of the complex, which both have LBC recognition sequences (37). Like Fab-7, Fab-8 and AB-I include GAGA motifs.
To determine if the presence of both CLAMP and GAF in the LBC has functional relevance, we asked whether CLAMP and GAF associate with the Fab-7 boundary interdependently. For this purpose, we used RNAi to reduce levels of either clamp or trl in tissue culture cells and then monitored their occupancy at Fab-7 in vivo by ChIP-quantitative PCR (qPCR). trl RNAi reduces CLAMP occupancy at Fab-7 (Fig. 1C), and conversely, although to a lesser extent, clamp RNAi reduces GAF occupancy at Fab-7 (Fig. 1D). These data suggest that CLAMP and GAF associations with Fab-7 are interdependent.
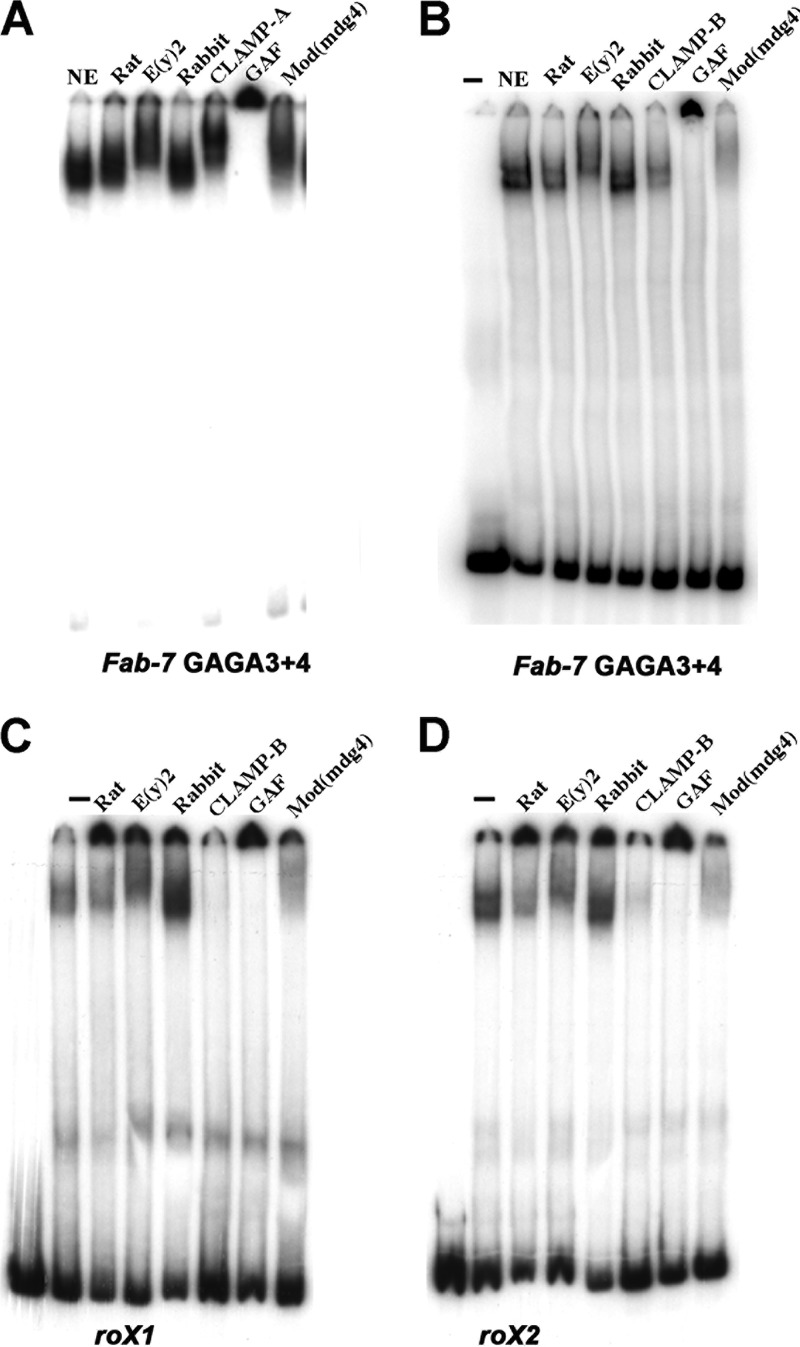
One plausible explanation for this relationship is that CLAMP is a component of the LBC. To explore this possibility, we used antibody supershift EMSAs with late-stage 6- to 22-h embryo nuclear extracts. Figure 2A and B show EMSA supershift experiments with the Fab-7 probe, GAGA3+4, spanning GAGA motifs 3 and 4, and two different CLAMP antibodies. As positive controls, we included antibodies against the other known LBC components [E(y)2, GAF, and Mod(mdg4)] in these EMSAs. The CLAMP antibody used for Fig. 2A (CLAMP-A) supershifts the GAGA3 and -4 LBC shift, while the CLAMP antibody shown in Fig. 2B (CLAMP-B) inhibits LBC binding to the probe. For CLAMP-B, we found that the extent of inhibition varies depending upon the ratio of antibody to extract and the length of time that the antibody is preincubated with the extract (not shown). For the Mod(mdg4) antibody, the inclusion of a preincubation step alters the relative balance between supershifting and inhibition (Fig. 2A and B). The LBC shift generated by the Fab-7 GAGA5+6 probe is also supershifted by CLAMP-A antibodies (see Fig. S2 in the supplemental material). In summary, CLAMP binds to Fab-7 both in vivo and in vitro.
FIG 2.
The LBC binds to Fab-7 GAGA3 and -4, roX1, and roX2. Probe Fab-7 GAGA3+4 (A and B), roX1 (C), or roX2 (D) was incubated with 6- to 22-h nuclear extracts (NE) with or without (−) either a control preimmune rat or rabbit serum, a rat polyclonal E(y)2 antibody, two different rabbit polyclonal CLAMP antibodies (A and B), rabbit polyclonal GAF antibody, or a rabbit polyclonal Mod(mdg4) antibody. (A and B) The nuclear extract, antibody, and labeled probe were added together, and the reaction mixture was then incubated for 30 min prior to gel electrophoresis. (C and D) The nuclear extract was first preincubated with the indicated antibody for 30 min; labeled probe was then added, and the reaction mixture was incubated for an additional 30 min prior to gel electrophoresis.
CLAMP is an integral LBC component.
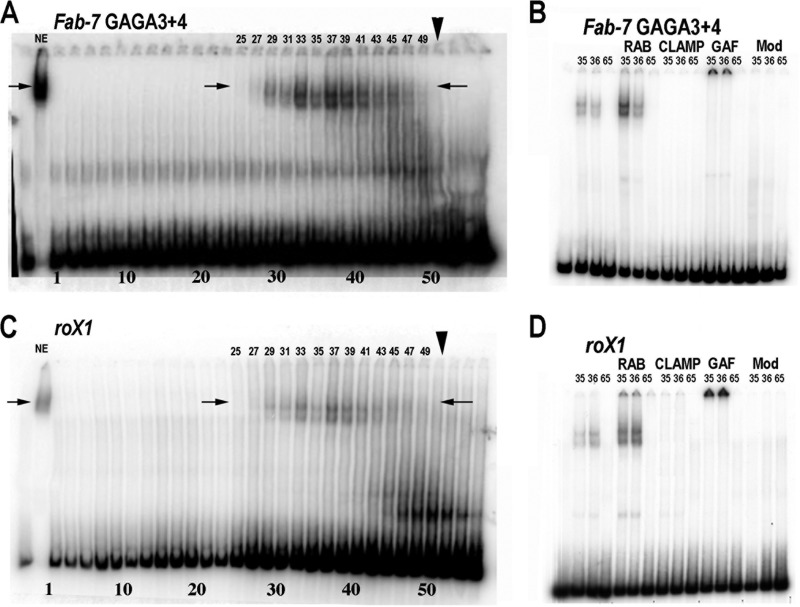
Our previous gel filtration experiments showed that the LBC exists as a very large preassembled complex in nuclear extracts (37). However, we did not determine whether the proteins found associated with the LBC shift in nuclear extracts by antibody supershift experiments cofractionate with the complex on gel filtration columns. In fact, since the pattern and yield of LBC shifts can differ from one probe to the next (Fig. 2B and 3), it is possible that some factors are not stably associated with the LBC. To further define the relationship between CLAMP and the other components of the LBC, we size fractionated nuclear extracts on a Superose 6 gel filtration column and assayed odd-numbered fractions for DNA binding activity using the GAGA3+4 probe. Figure 3A shows EMSAs of early fractions from the gel filtration column. The peak fraction for the 670-kDa marker is 51 (arrowhead), while the peak fractions for the LBC shift are between 33 and 41. This would be consistent with a molecular mass of approximately 1,000 kDa, which is larger than our previous estimates for the size of the LBC. The mobility of the two shifts generated by the size-fractionated LBC (Fig. 3A, arrows) is slightly more rapid than the LBC shift in nuclear extracts. This finding indicates that the LBC GAGA3 and -4 shift detected in nuclear extracts is likely generated by a core complex of integral LBC proteins plus proteins that are not stably associated with the complex and/or associate with the complex only when the LBC binds DNA.
FIG 3.
The LBC is a >1,000-kDa complex that contains CLAMP, GAF, and Mod(mdg4). Nuclear extracts were fractionated on a Superose 6 column and then used for EMSAs with Fab-7 GAGA3+4 (A and B) or roX1 (C and D) probe. (A and C) Ten microliters of odd-numbered column fractions (1 to 57) were incubated with GAGA3+4 (A) or roX1 (C). The peak fraction for the 670-kDa marker used as a standard is indicated by the arrowhead. The arrows indicate center positions of LBC shift in nuclear extracts. (B and D) EMSAs using 10 μl of the indicated column fractions preincubated with no antibody, rabbit serum (RAB), CLAMP-B, GAF, or Mod(mdg4) antibodies for 30 min. The labeled probe was then added, and the incubation continued for 30 min prior to gel electrophoresis.
To determine whether CLAMP, GAF, and Mod(mdg4) are components of the core LBC complex, two fractions, 35 and 36, which contain the LBC, and a control fraction, 65, distant from the LBC peak, were used for supershift experiments (Fig. 3B). Like the experiment shown in Fig. 2B, we preincubated the antibody with the fractions prior to the addition of labeled probe. As was found previously, the addition of serum alone to the peak fractions enhanced LBC binding. For the GAF antibody, we observed a supershift like that seen in nuclear extracts. For CLAMP and Mod(mdg4), the antibodies completely blocked LBC binding. The inhibition of binding seen with the gel filtration fractions likely reflects differences in the relative amounts of CLAMP and Mod(mdg4) in extracts compared to that in the column fractions containing LBC activity. Consistent with the inhibition of binding, CLAMP was present in column fractions containing LBC binding activity (see Fig. S3A in the supplemental material).
The LBC binds to the roX1 and roX2 CES.
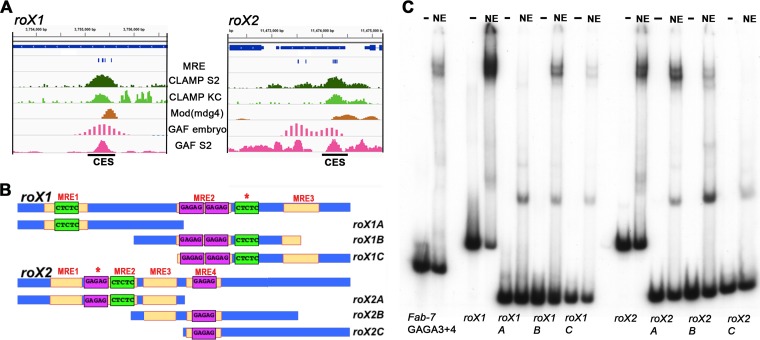
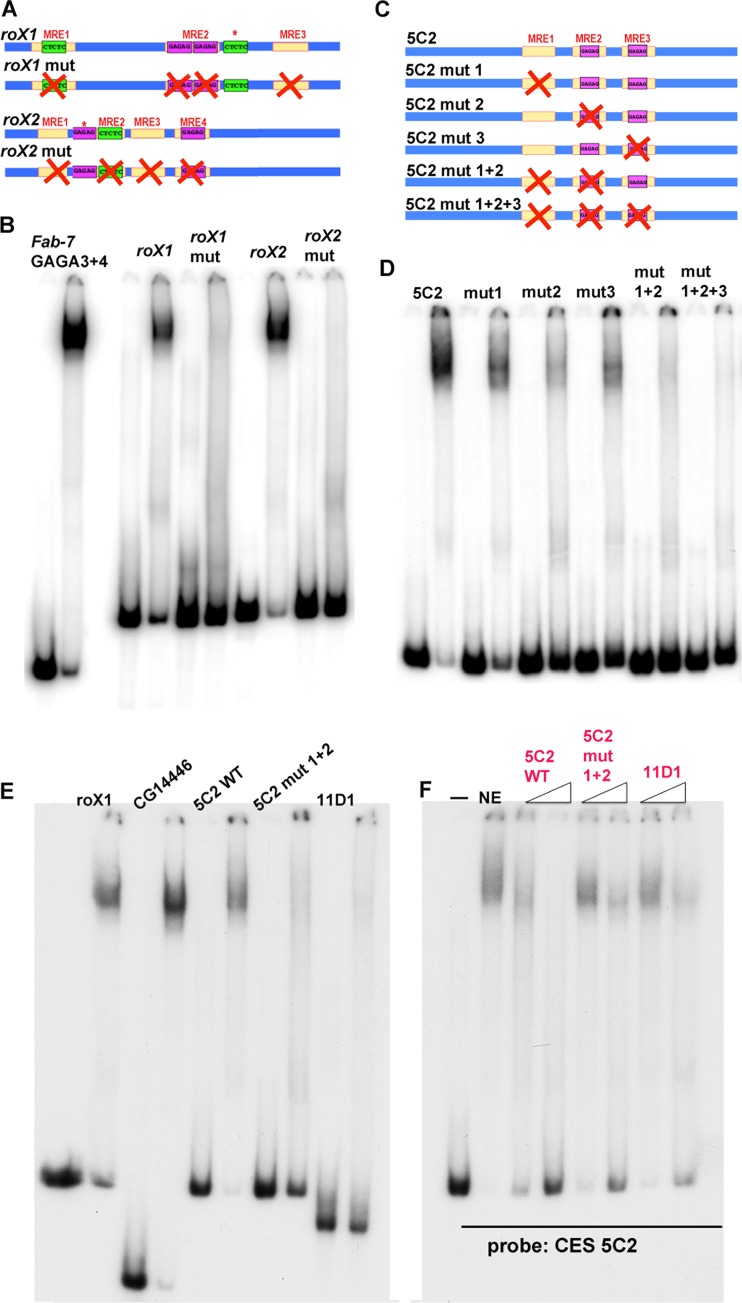
The in vivo localization of CLAMP to the Fab-7 boundary and its presence within the LBC raise the possibility that CES on the X chromosome utilizes the same boundary complex. To test this hypothesis, we first focused on two canonical CES, roX1 and roX2. As shown in Fig. 4B, the roX1 CES has three MREs, while the roX2 CES has four. The central roX1 MRE has two GAGAG motifs, while the two flanking MREs each have a single GAGAG motif. For roX2, two of the four MREs have a single GAGAG motif. Both CES have an additional GAGAG motif (Fig. 4B, asterisk) that is not classified as an MRE because the flanking sequences do not match the 21-bp MRE consensus sequence.
FIG 4.
The LBC can bind to roX1 and roX2 CES. (A) ChIP profiles at the rox1 and rox2 CES. Tracks displayed are for CLAMP ChIP-seq in S2 and Kc cells (31), GAF ChIP-seq in S2 cells (54), and Mod(Mdg4) and GAF embryo ChIP with microarray technology (ChIP chip) (modENCODE). (B) Schematic of the sequence organization of the CES roX1 and roX2 (200 bp) and the probes used for EMSAs. MRE motifs, yellow; GAF binding sites in forward orientation, purple; GAF binding sites in reverse orientation, green. The labeling of the GA-rich elements represents the exact sequences that are present within each element. Asterisks denote a GA-rich sequence that does not match the position weight matrix for the MRE. (C) EMSAs of full-length and truncated roX1 and roX2 probes. Probes: Fab-7 GAGA3+4, roX1, roX2, and roX1/roX2 subfragments incubated with (NE) or without (−) late embryo nuclear extracts. Note that the input of labeled roX1C was not equivalent to that of the other roX1 probes. For this reason, the yield of the LBC roX1C shift appears reduced relative to the other probes.
We first examined the ChIP profiles of CLAMP, GAF, and Mod(mdg4) in the regions around the roX1 and roX2 genes. Within the resolution of available genome-wide data sets, GAF and Mod(mdg4) colocalize with CLAMP at the CES located within the roX1 and roX2 genes in vivo (Fig. 4A). Since all three of the known DNA binding components of the LBC [CLAMP, GAF, and Mod(mdg4)] associate with the roX1 and roX2 CES in vivo, we next tested for LBC binding in vitro. We generated 200-bp probes spanning the core roX1 and roX2 CES DNA sequences. We also subdivided each CES into three smaller, ∼100-bp overlapping probes (Fig. 4B). Figure 4C shows EMSAs with these CES probes and, as a positive control, the 134-bp Fab-7 GAGA3+4 probe. For the two full-length roX1 and roX2 CES probes, we observed two major, slowly migrating bands whose mobilities closely matched those seen for the GAGA3 and -4 LBC shift. For the smaller subfragments, two of the roX1 and two of the roX2 probes generate shifts that comigrate with the GAGA3 and -4 LBC shifts. The relative yield and precise pattern of shifted bands differed with these smaller probes. For roX1, the highest yields were for probe B, which has MRE2 plus part of MRE3, and probe C, which has MRE2 plus MRE3. For rox2, the highest binding efficiency was seen for probe A, which has three MREs. In contrast, the LBC bound poorly to probes with only a single MRE (roX1-A and roX2-C). Similar variations in the relative yield and pattern of bands generated by the LBC were also noted when smaller Fab-7 probes were used (37, 43). In addition to the LBC shift, the smaller probes gave a collection of faster-migrating shifts.
We used several different experimental approaches to determine whether the LBC binds to roX1 and roX2. First, if the complex binding to the roX1 and roX2 CES is the same as the complex that binds to Fab-7 GAGA3+4, then binding should be sensitive to cross-competition. Figure S4A in the supplemental material shows that LBC binding to the 200-bp roX1 probe is competed not only by itself but also by excess cold roX2 and Fab-7 GAGA3+4 DNA. Similar results were observed when the labeled probes were roX2 or Fab-7 GAGA3+4 (see Fig. S4B and C in the supplemental material); LBC binding to these two probes was competed by excess cold roX1, roX2, and Fab-7 GAGA3+4. Second, if the factor binding to the roX1 and roX2 probes in nuclear extracts corresponds to the LBC, then it should contain the four known protein components: GAF, Mod(mdg4), CLAMP, and E(y)2. Figure 2C and D shows that antibodies against these four LBC proteins either supershifted the roX1 and roX2 LBC shifts or inhibited LBC binding to DNA (Fig. 2C and D). In each case, the effects were similar to those observed with the Fab-7 probe GAGA3+4 (Fig. 2A and B). Third, we incubated the 200-bp roX1 probes with fractions from the Superose 6 gel filtration column. Peak fractions for the roX1 LBC shift (Fig. 3C, lanes 33 to 41) corresponded to the peak fractions for the Fab-7 GAGA3 and -4 LBC shift (Fig. 3A). Finally, as was observed for Fab-7 GAGA3 and -4, supershift experiments using gel filtration fractions containing LBC activity showed that CLAMP, GAF, and Mod(mdg4) are core components of the complex that generates both the roX1 (Fig. 3D) and the roX2 (see Fig. S3B in the supplemental material) LBC shifts. Taken together, these findings demonstrate that the LBC binds to two key CES, roX1 and roX2.
LBC binding to CES is MRE dependent.
Previous studies have shown that the MRE sequences in the roX1 and roX2 CES are required to recruit the MSL complex to ectopic autosomal sites. If the LBC plays an important role in the functioning of these two CES, then mutations in the MREs that disrupt MSL recruitment in vivo should reduce or eliminate the LBC shift in nuclear extracts. Figure 5B shows that this prediction is correct: MRE mutations, which abrogate CES activity in vivo, eliminate LBC binding in vitro. To confirm the effects of the MRE mutations, we used wild-type or mutant roX1 and roX2 CES DNAs as competitors for LBC binding to the Fab-7 GAGA3+4 probe. Figure S4D in the supplemental material shows that unlabeled wild-type roX1 and roX2 competed for LBC binding to GAGA3+4, whereas the corresponding mutant DNAs failed to compete effectively.
FIG 5.
LBC binding is MRE dependent. (A) Schematic drawing of the wild-type (WT) (top) and mutated (bottom) versions of the 200-bp roX1/roX2 sequences used for EMSAs. Asterisks denote a GA-rich sequence that does not match the position weight matrix for the MRE. (B) Mutations in the roX1 and roX2 MREs abrogate LBC binding. (C) Schematic of the wild-type and mutant versions of the 5C2 probes. (D) Single and combination mutations in three 5C2 MREs differentially affect LBC binding. (E) Comparison of LBC binding to CES 11D1 with other LBC substrates as indicated. (F) Excess unlabeled wild-type 5C2, 5C2 mut1+2, and CES 11D1 DNAs were used to compete for LBC binding to labeled CES 5C2. Fold excess of cold competitor added (left to right): CES 5C2, 12.5× and 100×; CES 5C2 mut1+2, 25× and 200×; CES 11D1, 25× and 200×.
Other than roX1 and roX2, the most thoroughly characterized multi-MRE CES is CES 5C2 (23). As indicated in Fig. 5C, CES 5C2 contains three 21-bp MREs. Two of these MREs (MRE2 and -3) have a GAGAG motif, while the third, MRE1, has a GA-rich sequence but no GAGAG motif. Like roX1 and roX2, a shift that comigrates with the Fab-7 GAGA3 and -4 LBC shift was observed with a probe spanning the three MREs in CES 5C2 (Fig. 5D). We used competition and supershift experiments to demonstrate that the CES 5C2 shift is generated by the LBC. Figure S5A in the supplemental material shows that excess unlabeled 5C2 DNA competes for LBC binding to labeled GAGA3+4 and roX1 probes. Figure S6A in the supplemental material shows that, as was observed for roX1, roX2, and Fab-7 GAGA3 and -4, the 5C2 shift is generated by a complex that contains CLAMP, GAF, and Mod(mdg4).
Previous studies have shown that mutations in one or a combination of the 5C2 MREs abrogate CES function in vivo to different extents (23). Two different assays were used to measure CES function. One was a luciferase assay in S2 tissue culture cells, while the other was a ChIP assay of MSL2 binding to the 5C2 sequence inserted into an autosomal locus in fly larvae. Mutations in a single 5C2 MRE weakened CES function in one or both of these assays but did not completely eliminate activity. In contrast, the combination mutants (mut1+2 and mut1+2+3) essentially eliminated both the stimulatory activity of CES 5C2 on transcription and Msl2 recruitment.
The effects of different MRE mutations on the binding of the LBC to the CES 5C2 probe in nuclear extracts parallel those observed in the two in vivo assays (Fig. 5D). LBC binding was reduced in the three single mutations, 5C2 mut1, 5C2 mut2, and 5C2 mut3, while binding was largely or completely lost in the double (5C2 mut1+2) or triple (5C2 mut1+2+3) mutant, respectively. Consistent with the elimination of LBC binding, excess unlabeled 5C mut1+2+3 DNA failed to compete for LBC binding to either the roX1 or Fab-7 GAGA3+4 probe (see Fig. S5A in the supplemental material). It is interesting that though MRE1 does not contain a precise GAGAG motif, mutation of the GA-rich core weakens LBC binding, just like mutations in MRE2 and -3.
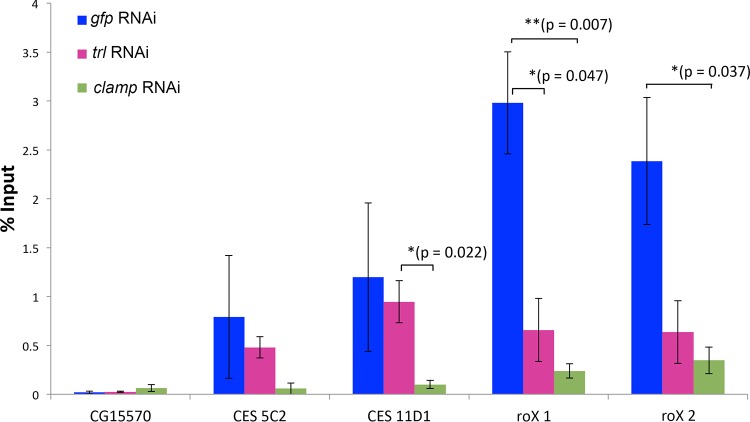
Knockdowns of CLAMP and GAF reduce MSL3 binding to the roX1, roX2, and 5C2 CES.
The experiments described above indicated that mutations in roX1, roX2, and 5C2, which weaken or disrupt CES function in vivo, compromise or eliminate LBC binding in vitro. To further test the connection between LBC and the recruitment of the dosage compensation machinery to these three CES, we used RNAi to reduce levels of clamp and trl in male S2 tissue culture cells and measured MSL3 occupancy in vivo. As previously documented (31), we found that MSL3 association with roX1, roX2, and CES 5C2 is CLAMP dependent (Fig. 6). Unlike the roX loci, it is important to note that the changes in occupancy at CES 5C2 are not statistically significant due to high variability within the control sample specifically at this locus. This is consistent with lower occupancy of CLAMP and GAF at CES 5C2 than at the other two CES. As would be expected if MSL recruitment depends upon the LBC, MSL association with the roX1 and roX2 CES is also reduced when GAF levels are reduced.
FIG 6.
GAF and CLAMP contribute to MSL complex recruitment. ChIP-qPCR was conducted using an antibody specific for the MSL3 protein after RNAi treatment targeting trl and clamp (or gfp as a control). CG15570 was used as a negative-control target. The CES 5C2, 11D1, roX1, and rox2 were tested for a change in MSL3 binding. P values from t tests are displayed for significance: *, P < 0.05, or **, P < 0.01 for differences between sample means. Horizontal bars indicate treatment comparisons for displayed P values. P values are not displayed for those >0.05. The error bars indicate standard errors of the mean.
CES 11D1, an example of a CES with only a single MRE.
The three CES we have tested for LBC binding thus far (roX1, roX2, and 5C2) fall into the subclass that has multiple MREs; however, most of the CES identified by ChIP experiments have only a single MRE sequence (23). Since the 5C2 mutant 5C2 mut1+2, gave only a barely detectable LBC shift, we wondered whether CES with a single MRE are bound by the LBC. The only CES with a single MRE that has been studied in detail is CES 11D1. It differs from 5C2, and also roX1 and roX2, in that a small, 150-bp fragment spanning the 11D1 MRE has only minimal CES function. Instead, a larger, 500-bp element is needed for full CES activity, and in this context, the MRE is required (23). Based on these in vivo results and the EMSAs described above, a plausible expectation is that the 150-bp 11D1 fragment would be a poor substrate for the LBC. This is the case. In the experiment shown in Fig. 5E, we compared the shift generated by the 11D1 probe with that observed for three multi-MRE probes. Two are the 200-bp roX1 and 5C2 sequences discussed above, while the third is a CES at CG14446 (see below). The CES CG14446 is only 100 bp and was included in the experiment to control for the possible effects of probe length. In comparison to the three positive controls, the 11D1 probe generates only a very weakly labeled LBC shift. In fact, the yield of the LBC-shifted DNA is not much different from that observed for the MRE 5C2 mutant, 5C2 mut1+2, which retains only a single MRE. To confirm these findings, we used excess cold 5C2, 5C2 mut1+2, and 11D1 DNAs to compete for LBC binding to labeled 5C2 (Fig. 5F). Although a 200-fold excess of unlabeled 11D1 DNA reduced LBC binding to 5C2, a significant fraction (∼50%) of the 5C2 probe was still shifted by the LBC. A similar result was observed when the 5C2 mut1+2 DNA was used as the cold competitor. In comparison to these two single-MRE DNAs, a 12.5-fold excess of the wild-type 5C2 DNA was sufficient to give an equivalent, if not a slightly greater, degree of competition. Therefore, CES with a single MRE, like 11D1 and 5C2 mut1+2, bind to the LBC much more poorly than CES that have multiple MRE sequences.
Given the low affinity of the LBC for CES 11D1, we wondered whether CLAMP and GAF are important for recruiting MSL complexes to this sequence in vivo. Figure 6 shows that CES 11D1 differs from the multi-MRE CES in that knocking down CLAMP compromises MSL accumulation, while knocking down GAF does not. A plausible interpretation of this finding is that the LBC is not critical for the CES function of CES11D1.
The LBC binds to topologically linked CES.
The CCC experiments performed by Ramírez et al. (29) showed that paired CES often define the endpoints of chromatin looped domains on the X chromosomes of both males and females and thus are likely to have architectural functions equivalent those of classical boundary elements or insulators. Although the basis for pairing specificity is not understood, pairing in transgene assays been shown to be mediated by protein-protein self-interactions. For example, boundary bypass experiments indicated that boundary elements consisting of multimerized dCTCF sites pair with multimerized dCTCF sites but not do not pair with multimerized sites for Su(Hw) (44). Thus, a plausible expectation was that the LBC would bind to both partners in at least a subset of the paired CES elements on the X chromosome.
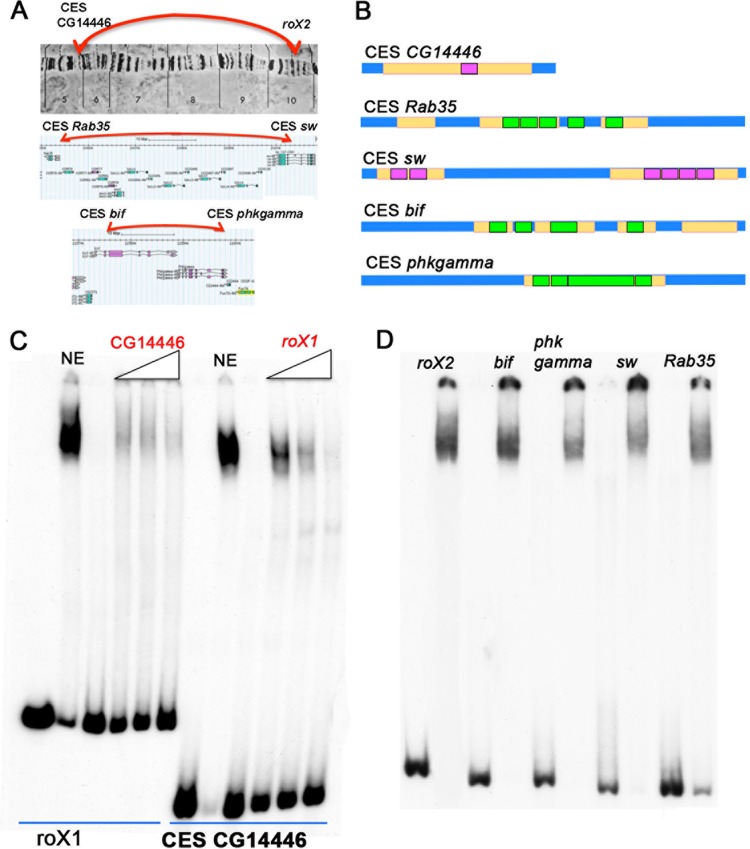
To test this hypothesis, we selected three of the interacting CES pairs Ramírez et al. identified. These CES pairs were chosen because ChIP experiments indicated that they are bound by both CLAMP and GAF in vivo and because they have multiple GA-rich MRE sequences. The first interacting pair is roX2 and the CES associated with CG14446. These two CES are quite distant from each other, approximately 5 Mb apart. The 100-bp CG14446 CES has a large, 72-bp region that contains multiple overlapping MREs but only one GAGAG motif (Fig. 7). The second CES pair corresponds to the CES associated with rab35a and sw. These two CES are much closer, approximately 50 kb apart. The 200-bp CES rab35a has three MREs that have a total of 6 GAGAG motifs (CTCTC). The CES sw has two long GA-rich MREs (36 bp and 68 bp) and a total of six GAGAG motifs. The third pair are the CES associated with bif and phkgamma. They are separated from each other by about 15 kb. CES bif has four MREs, three of which have a GAGAG motif (CTCTC). CES phkgamma has a single, long MRE-containing locus containing five GAGAG motifs (CTCTC) (Fig. 7).
FIG 7.
The LBC binds to topologically linked CES. (A) Schematic of long-range interactions between three pairs of CES in a polytene X chromosome (CG14446↔roX2) (top) and in Flybase GBrowser (rab35↔sw; bif↔phkgamma) (bottom). (B) Schematic of the 5 CES probes that were used for EMSAs. MRE motifs, yellow; GAF motifs in forward orientation, purple; GAF motifs in reverse orientation, green. (C) Cross-competition experiment with increasing amounts of cold CES CG14446 or roX1 probes showing that the same complex binds to the different CES. Cold competitor was added in 5×, 10×, and 50× excess. (D) LBC binds to physically interacting CES.
Figure 7 shows that these physically linked CES generate an LBC-like shift when incubated with 6- to 22-h nuclear extracts. We used several experimental approaches to confirm that the observed shifts correspond to the LBC. In the case of the CES that pairs with roX2, CG14446, antibody supershift experiments indicated that, as expected, the shift is generated by a complex that contains the same proteins as the LBC: GAF, Mod(mdg4), E(y)2, and CLAMP (see Fig. S6B in the supplemental material). Precisely the same result was obtained for the rab35a CES, which physically pairs with the sw CES (see Fig. S6C in the supplemental material). The identification of the LBC is further confirmed by assays of fractions from Superose 6 gel filtration columns. Figure S7 in the supplemental material shows that the elution profile of the LBC shift for the CG14446 CES probe is the same as that observed for the Fab-7 and roX1 probes. We also used cross-competition experiments to demonstrate that the LBC complex binds to the paired CES probes (see Fig. S6B in the supplemental material).
DISCUSSION
Boundary elements (or insulators) function to subdivide chromosomes in multicellular eukaryotes into a series of topologically independent looped domains. The importance of these architectural elements in the regulation of gene expression was first established in experiments dating back nearly 30 years (8, 9). These experiments showed that boundaries have blocking activity that can restrict the action of enhancers and silencers to the loops in which they reside. Subsequent work revealed yet another function of these architectural elements, namely, promoting long-distance regulatory interactions (13, 45). One context in which long-distance regulation is especially important is X chromosome dosage compensation. In male flies, MSL complexes recruited to the X chromosome by CES spread to nearby genes and upregulate their transcription. Recent experiments have drawn a connection between the recruitment and spreading of the MSL complexes from CES and an architectural function in which the CES define boundaries of looped domains (TADs) along the X chromosome in both sexes (29). However, it is not clear from these experiments whether CES share other properties with classical fly boundary elements. Additionally, the factors responsible for the architectural activities of the CES were not identified. Here, we show that a canonical Bithorax complex boundary factor, the LBC, is deployed by a subset of CES for their regulatory and architectural functions.
LBC and boundaries in BX-C.
A combination of genetic and biochemical experiments have implicated the LBC in the functioning of three BX-C boundaries: Fab-7, Fab-8, and AB-I (37, 42–44). The LBC has several unusual properties: (i) the relatively long, ∼65-bp DNA minimal binding length, (ii) the diversity of the sequences recognition element, and (iii) the size and composition of the LBC.
The gel filtration experiments reported here suggest that the LBC is significantly larger than our previous estimate of 670 kDa. These experiments also indicate that the complex detected after gel filtration differs from that observed in nuclear extracts in that it generates a more rapidly migrating shift. The change in mobility does not seem to be probe specific and was observed with all three of the DNAs we tested (Fab-7 GAGA3 and -4, roX1, and CES CG14446). This finding suggest that some proteins dissociate from the LBC during gel filtration or, alternatively, interact with the LBC only after it binds to DNA. Included in the group of proteins that remain in the LBC during gel filtration are the two DNA binding proteins that we detected previously in nuclear extracts, GAF and Mod(mdg4). In the studies reported here, we identified a third LBC-associated DNA binding protein, CLAMP. Like the other two proteins, CLAMP cofractionates with LBC DNA binding activity and thus appears to be an integral component of the LBC. Strengthening the connection between CLAMP and the LBC, ChIP experiments showed that CLAMP interacts in vivo with three BX-C boundaries (Fab-7, Fab-8, and AB-I) that are recognized by the LBC in nuclear extracts. Additionally, CLAMP and GAF binding to Fab-7 in tissue culture cells is interdependent, suggesting that they are part of the same protein complex that is destabilized in the absence of either component.
LBC and CES.
A key question posed by the discovery that CES are architectural elements that link distant X chromosome sequences to each other (29) is their relationship to chromatin boundaries. Our finding that CLAMP is an integral component of the BX-C boundary factor LBC provides a plausible link between CES and boundary elements. Previous experiments have shown that CLAMP binds to CES and that this association is critical for the recruitment of MSL complexes to the elements (31). However, these experiments did not illuminate how CLAMP interacts with CES. Here, we show that in nuclear extracts CLAMP binds to nine multi-MRE CES, not independently, but rather, as a component of the LBC chromatin boundary factor.
A number of lines of evidence argue that LBC binding to roX1, roX2, and 5C2 in vitro is relevant to their ability to recruit MSL complexes in vivo. The first is the requirement for both GAF and CLAMP in recruitment of the MSL complex to several key CES. With the important caveat that there could be indirect effects caused by gradually depleting GAF and CLAMP, knockdowns of both proteins diminish MSL recruitment to these CES in vivo. The second line of evidence is the close match between the sequence recognition properties of the LBC and the requirements for CES function in vivo. A feature common to most CES is the presence of one or more GA-rich MREs in a DNA segment of between ∼100 and 250 bp. The MRE sequences have been shown to be required for the CES functions of roX1, roX2, CES 5C2, and CES 11D1. Of these four CES, the relationship between CES function in vivo and MREs has been analyzed in the most detail for CES 5C2. Alekseyenko et al. (23) showed that mutations in individual MREs within a 150-bp CES 5C2 element reduced, but did not eliminate, CES activity, while CES activity was disrupted by mutations in two or all three of the MREs. Consistent with the in vivo effects on CES activity, we found that mutations in individual 5C2 MREs weakened LBC binding, whereas binding was disrupted when all three of the MREs were mutant. Importantly, the failure of LBC to bind to CES 5C2 mut1+2+3 in nuclear extracts also recapitulates the effects of the triple mutant on CLAMP occupancy in vivo (31).
In the cases of roX1 and roX2, we found that mutations in all of their MREs eliminated the LBC shift in nuclear extracts. Though we did not test the effects of mutations in individual MREs, experiments with smaller, 100-bp subfragments indicated that LBC binding to roX1 and roX2 probes requires the presence of at least two MREs. This conclusion was reinforced by our experiments with CES 11D1. This CES differs from roX1, roX2, and CES 5C2 in that it has only a single MRE and no GAGAG motifs. Unlike other CES with multiple MREs, the 11D1 element is a poor substrate for the LBC.
The observation that the LBC binds preferentially to CES that have multiple MREs, which span a relatively large 100- to 200-bp sequence, is also consistent with other in vivo experiments on these elements. For example, Gilfillan et al. (46) identified an ∼500-bp CES element, DBF-DHS. Within this element, they found that a 40-bp GA-rich sequence, DBF 12-L15, is essential for recruiting the MSL complex in flies. Though required for the functioning of the full-length CES, this 40-bp sequence is not sufficient on its own. On the other hand, when multimerized, the 40-bp element is able to recruit MSL complexes. In the case of roX2, Park et al. (47) found that a 134-bp fragment spanning the four roX2 MREs (MRE1 to -4) functions as a CES at ectopic sites on autosomes. As can be seen in Fig. 4C, the 100-bp roX2A probe, which contains the first three roX2 MREs, bound the LBC almost as well as the larger, 200-bp roX2 probe.
While we have not tested the architectural functions of CES in the studies presented here, previous work on Fab-7 and Fab-8 (37, 42, 48) demonstrated that the LBC sequences in both of these boundaries have classical insulator activity and can mediate long-distance pairing interactions. Consequently, a reasonable expectation is that the LBC will have similar architectural functions when associated with other sequences in the genome, including CES. Consistent with this notion, we have shown that three of the pairs of interacting CES identified as “TAD boundaries” in tissue culture cells by Ramírez et al. (29) are recognized by the LBC in nuclear extracts. These interacting pairs are CES roX2↔CES CG14446, CES rab35↔CES sw, and CES bif1↔CES phkgamma.
Boundaries versus CES.
Although our findings clearly implicate the LBC in CES function, many questions remain.
(i) Is LBC association a common feature of CES?
We tested only a small subset of sequences that have been classified as CES. With the exception of CES 11D1, all of them have multiple, closely clustered MREs, and the LBC binds to these elements with an affinity at least equivalent to that observed for Fab-7 probes. For three of the multi-MRE CES (roX1, roX2, and CES 5C2), there is in vivo evidence that they function as CES and can recruit MSL complexes to ectopic sites. The five remaining sequences were classified as CES on the basis of ChIP experiments and the presence of MREs. Consequently, we do not know whether they can function as CES in vivo. However, if they do, it seems likely that both MSL recruitment and LBC binding depend on the clustered MREs, as is the case for roX1, roX2, and CES 5C2.
While other multi-MRE CES should also have properties similar to those studied here, nearly two-thirds of the CES that have been identified in ChIP experiments have a single MRE (23). The only member of this group that we tested is CES 11D1, and its interaction with the LBC clearly differs from that of CES containing multiple MREs. Whereas all of the multi-MRE CES we tested bound strongly to the LBC, the LBC bound only weakly to a 150-bp probe containing the single CES 11D1 MRE. The limited affinity for the LBC could explain why CES 11D1 requires a 500-bp fragment for CES activity while smaller, 100- to 200-bp fragments spanning the clustered MREs are sufficient for the CES activity of roX1, roX2, and CES 5C2. One question is whether other single-MRE CES have properties similar to those of CES 11D1. Since probes derived from the Fab-7 dHS1 sequence are bound with relatively high affinity by the LBC yet do not have any MRE consensus motifs, it is possible that some of the single-MRE CES have other motifs that promote LBC association.
On the other hand, there are reasons to think that the distinct properties of CES 11D1 may be typical of many single-MRE CES. In ChIP experiments, Soruco et al. (31) found that CLAMP association with single-MRE CES is dependent on synergistic interactions with MSL. In contrast, CLAMP associates with multi-MRE sequences independently of the dosage compensation machinery. Furthermore, nucleosome occupancy of single-MRE CES differs between the two sexes; it is typically lower in male cells and higher in female cells (31). In contrast, occupancy rates of multi-MRE CES are similar in the two sexes, suggesting that these are constitutive binding sites. These differences, taken together with the limited affinity for the LBC and the persistence of MSL association after GAF RNAi treatment, support a model in which single- and multi-MRE CES play distinct roles in dosage compensation (31). In this model, the LBC would bind to multi-MRE CES, and they would provide a platform for recruiting MSL complexes. Chromatin modifications induced by MSL association with multi-MRE CES along the X chromosome would then open up the single-MRE CES, potentially permitting the binding of CLAMP and presumably yet other factors. The presence of these proteins would set up a positive-feedback loop by facilitating the recruitment of MSL complexes.
(ii) What distinguishes CES from autosomal boundaries like Fab-7?
Though our experiments implicate the LBC in the functioning of multi-MRE CES, how it promotes the recruitment of MSL complexes to these CES but not to Fab-7 is unclear. One possibility is that the LBC variants that bind to CES are distinct from those that interact with canonical boundary elements. While this may seem farfetched, until we know more about the composition of the LBC, it cannot be excluded. The reason for this is the presence of Mod(mdg4) in the LBC. There are 31 predicted Mod(mdg4) isoforms (49). All share an N-terminal Mod(mdg4) BTB domain that mediates the assembly of Mod(mdg4) octomers (50), while they have different C-terminal domains. Of these 31 isoforms, 27 have unique FLYWCH DNA binding domains. If Mod(mdg4) is incorporated into the LBC as an octomer, there could be many different versions of the complex with potentially different DNA binding and protein-protein interaction properties.
However, a more likely alternative is that the LBC provides a platform for other factors that are actually responsible for recruiting/assembling MSL complexes. For example, Villa et al. (51) have shown that MSL2 has sequence- and shape-specific DNA binding activities and that MSL-2↔DNA interactions are responsible for mediating the recruitment of the MSL dosage compensation machinery to a specific class of CES called PionX sites when they are present at high local concentrations. According to their model, MSL2 interacts specifically with an extension of the MRE sequence that is located 5′ to the GA-rich core. Moreover, in contrast to the known requirements for CES function in vivo, the interaction between MSL2 and PionX sequences in vitro appears to be largely independent of the GA-rich motifs. This model would potentially fit our findings, which show that LBC binding to X chromosome CES requires GA-rich sequences. In principle, LBC↔GAn-MRE interactions could provide a mechanism for presenting the 5′ PionX sequences in a configuration suitable for MSL2 interaction with adjacent sequences. Clearly, further studies will be needed to assess whether this model is correct. Overall, we have provided new mechanistic insights into how sequences on the X chromosome function to establish chromatin boundaries that are associated with an active domain of coordinate gene regulation.
MATERIALS AND METHODS
Nuclear extracts.
Nuclear extracts from 6- to 22-h embryos were prepared as described previously (52) with small modifications. Zero- to 12-h embryos from Oregon R were collected from apple juice plates and aged for 10 h at room temperature. The extraction was completed with the final concentration of KCl at 360 mM.
Probes for EMSA.
Probes were obtained by PCR and purified on agarose–1× Tris-acetate-EDTA (TAE) gels, followed by phenol-chloroform extraction. The probe sequences, as well as primer sequences, used for PCR are listed in Table S1 in the supplemental material.
EMSA.
Electrophoretic mobility shift assays were performed using 32P-labeled DNA probes under conditions described previously (37). See the supplemental material for details.
Gel filtration.
Fractionation of the nuclear extracts derived from 6- to 22-h embryos was performed by size exclusion chromatography using a Superose 6 10/330 GL column (GE Healthcare). Molecular mass markers ranging from 1,350 to 670,000 Da (Bio-Rad) were used as gel filtration standards.
Western blotting.
Either 0.1 μl of late-stage nuclear extract (about 2 μg of total protein) or 2 μl of the column-fractionated material was run on 10% SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane, probed with anti-CLAMP B antibody (1:250; a new antibody generated using Abcam custom services), and developed with Amersham ECL Prime Western blotting detection reagent (GE Healthcare), following the manufacturer's protocol.
Generation of dsRNA for RNAi treatment.
Templates for double-stranded RNA (dsRNA) to target the CLAMP gene (clamp), the GAF gene (Trl), and the control green fluorescent protein gene (gfp) sequence were generated by PCR using the following template sequences. The clamp RNAi template, DRSC03718, was designed as described in detail previously (30; http://www.flyrnai.org). gfp dsRNA, designed as previously described (53), was used as a control. The GAF (Trl) dsRNA target sequence used was also designed previously (54). The clamp, trl, and gfp constructs were synthesized using the T7 MEGAscript kit (Ambion), including DNase I treatment for 15 min at 37°C following dsRNA synthesis. The dsRNA was then purified using the RNeasy kit (Qiagen). Sequences are reported in Table S2 in the supplemental material.
RNAi treatment of SL2 cells.
RNAi treatment of SL2 cells was adapted from previous studies (30, 31). Briefly, RNAi was set up in T150 flasks as follows: 135 μg dsRNA was diluted in 3 ml UltraPure water (Invitrogen) with 6 ml SL2 cells at a concentration of 8 × 105 cells/ml in Schneider's medium (Gibco) for 45 min at 25°C; then, 21 ml Schneider's medium with 10% heat-inactivated fetal bovine serum (FBS) was added, and the RNAi-treated cells were grown for 6 days at 25°C until they were collected for chromatin preparation.
Chromatin preparation.
Chromatin preparation was based on previous work (55), modified for T150 cell culture flasks, in which cross-linking and quenching were performed. Samples were then washed three times, pelleted, and resuspended in lysis buffer for sonication. See Materials and Methods in the supplemental material for specific protocol and buffer details.
IP.
Either 5 μl CLAMP antibody (SDIX) (30), 10 μl GAF antibody (a gift from John Lis), or 4 μl MSL3 serum (a gift from Mitzi Kuroda) was added to each 1-ml immunoprecipitation (IP) sample and rotated at 4°C overnight. At the same time, 50 μl protein G Dynabeads per sample was incubated in bead-blocking buffer (10 mM PBS, 0.1% Triton X-100, and 3% bovine serum albumin [BSA]) with rotation at 4°C overnight. The lysate and beads were then incubated together for 2 h at 4°C. Samples were then washed and eluted from the beads, and a DNA cleanup was performed. See Materials and Methods in the supplemental material for details of wash steps and buffers and DNA cleanup information.
Quantitative real-time PCR.
All the target primer sequences used were previously designed (23, 37). Briefly, the average relative quantitation (threshold cycle [CT] value) of two replicate reactions was determined for all samples. Platinum SYBR master mix with ROX (Invitrogen) was used. Three independent chromatin preparations were conducted for each experiment, and standard deviations are reported. Dilutions of input to 1% were conducted to ensure that samples were in the linear range for quantitative PCR. To assess the overall ChIP enrichment, CT values were normalized with respect to the 1% input values, and then the averages and standard deviations of the biological replicates were reported for each primer target.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anton Golovnin, Pavel Georgiev, Elissa Lei, Victor Corces, Carl Wu, Mitzi Kuroda, John Lis, and David Gilmour for the gift of antibodies.
This work was supported by a grant from the NIH (R01GM098461-1), an American Cancer Society Research Scholar grant (123682-RSG-13-040-01-DMC), and a Pew Biomedical Scholars program grant awarded to E.L. and to P.S. (GM043432). A.K. and P.S. also acknowledge support from grants to the Gene Biology Institute by the Russian Federation Ministry of Education and Science (RF 14.B25.31.0022) and the Russian Scientific Foundation (16-14-10346).
We have no conflicts of interest to disclose.
Author contributions were as follows: conceptualization, E.G.K., A.K., D.W., P.S., and E.L.; investigations, E.G.K., A.K., D.W., and T.A.; writing of the original draft, P.S. and E.L.; writing, review, and editing, E.G.K., A.K., P.S., and E.L.; funding acquisition, P.S. and E.L.; supervision, P.S. and E.L.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00253-17.
REFERENCES
- 1.Hnisz D, Weintraub AS, Day DS, Valton A, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, Reddy J, Borges-Rivera D, Lee TI, Jaenisch R, Porteus MH, Dekker J, Young RA. 2016. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. 2014. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. Bioessays 36:163–172. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ribeiro de Almeida C, Hendriks RW, Stadhouders R. 2015. Dynamic control of long-range genomic interactions at the immunoglobulin κ light-chain locus. Adv Immunol 128:183–271. doi: 10.1016/bs.ai.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Dixon JR, Gorkin DU, Ren B. 2016. Chromatin domains: the unit of chromosome organization. Mol Cell 62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker J, Heard E. 2015. Structural and functional diversity of topologically associating domains. FEBS Lett 589:2877–2884. doi: 10.1016/j.febslet.2015.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eagen K, Aiden EL, Kornberg R. 2017. Polycomb-mediated chromatin loops revealed by a sub-kilobase resolution chromatin interaction map. bioRxiv. doi: 10.1101/099804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujioka M, Mistry H, Schedl P, Jaynes JB. 2016. Determinants of chromosome architecture: insulator pairing in cis and in trans. PLoS Genet 12:e1005889. doi: 10.1371/journal.pgen.1005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kellum R, Schedl P. 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol 12:2424–2431. doi: 10.1128/MCB.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kellum R, Schedl P. 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64:941–950. doi: 10.1016/0092-8674(91)90318-S. [DOI] [PubMed] [Google Scholar]
- 10.Vazquez J, Schedl P. 1994. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J 13:5984–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigrist CJA, Pirrotta V. 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. 1999. The Mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153:1333–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 14.Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. 2010. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem 285:4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belote JM, Lucchesi JC. 1980. Male-specific lethal mutations of Drosophila melanogaster. Genetics 96:165–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelbart ME, Kuroda MI. 2009. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller CI, Akhtar A. 2015. The MSL complex: juggling RNA-protein interactions for dosage compensation and beyond. Curr Opin Genet Dev 31:1–11. doi: 10.1016/j.gde.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Meller VH, Rattner BP. 2002. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J 21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith ER, Allis CD, Lucchesi JC. 2001. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem 276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- 20.Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. 2011. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98:513–522. doi: 10.1016/S0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- 22.Kageyama Y, Mengus G, Gilfillan G, Kennedy HG, Stuckenholz C, Kelley RL, Becker PB, Kuroda MI. 2001. Association and spreading of the Drosophila dosage compensation complex from a discrete roX1 chromatin entry site. EMBO J 20:2236–2245. doi: 10.1093/emboj/20.9.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, Kuroda MI. 2008. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. 2006. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorchakov AA, Alekseyenko AA, Kharchenko P, Park PJ, Kuroda MI. 2009. Long-range spreading of dosage compensation in Drosophila captures transcribed autosomal genes inserted on X. Genes Dev 23:2266–2271. doi: 10.1101/gad.1840409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adkins NL, Hagerman T, Georgel P. 2006. GAGA protein: a multi-faceted transcription factor. Biochem Cell Biol 84:559–567. doi: 10.1139/o06-062. [DOI] [PubMed] [Google Scholar]
- 27.Meller VH, Kuroda MI. 2002. Sex and the single chromosome. Adv Genet 46:1–24. [DOI] [PubMed] [Google Scholar]
- 28.Oh H, Park Y, Kuroda MI. 2003. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev 17:1334–1339. doi: 10.1101/gad.1082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramírez F, Lingg T, Toscano S, Lam KC, Georgiev P, Chung HR, Lajoie BR, de Wit E, Zhan Y, de Laat W, Dekker J, Manke T, Akhtar A. 2015. High-affinity sites form an interaction network to facilitate spreading of the MSL complex across the X chromosome in Drosophila. Mol Cell 60:146–162. doi: 10.1016/j.molcel.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larschan E, Soruco MML, Lee OK, Peng S, Bishop E, Chery J, Goebel K, Feng J, Park PJ, Kuroda MI. 2012. Identification of chromatin-associated regulators of MSL complex targeting in Drosophila dosage compensation. PLoS Genet 8:e1002830. doi: 10.1371/journal.pgen.1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soruco MML, Chery J, Bishop EP, Siggers T, Tolstorukov MY, Leydon AR, Sugden AU, Goebel K, Feng J, Xia P, Vedenko A, Bulyk ML, Park PJ, Larschan E. 2013. The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev 27:1551–1556. doi: 10.1101/gad.214585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuzu G, Kaye EG, Chery J, Siggers T, Yang L, Dobson JR, Boor S, Bliss J, Liu W, Jogl G, Rohs R, Singh ND, Bulyk ML, Tolstorukov MY, Larschan E. 2016. Expansion of GA dinucleotide repeats increases the density of CLAMP binding sites on the X-chromosome to promote Drosophila dosage compensation. PLoS Genet 12:e1006120. doi: 10.1371/journal.pgen.1006120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenberg AJ, Yanowitz JL, Schedl P. 2004. The Drosophila GAGA factor is required for dosage compensation in males and for the formation of the male-specific-lethal complex chromatin entry site at 12DE. Genetics 166:279–289. doi: 10.1534/genetics.166.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urban J, Doherty C, Rieder L, Jordan W, Tsiarli M, Larschan E. 2017. The essential Drosophila CLAMP protein differentially regulates non-coding roX RNAs in male and females. Chromosome Res 25:101–113. doi: 10.1007/s10577-016-9541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtsuki S, Levine M. 1998. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev 12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belozerov VE, Majumder P, Shen P, Cai HN. 2003. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J 22:3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolle D, Cleard F, Aoki T, Deshpande G, Schedl P, Karch F. 2015. Functional requirements for Fab-7 boundary activity in the bithorax. Mol Cell Biol 35:3739–3752. doi: 10.1128/MCB.00456-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweinsberg SE, Schedl P. 2004. Developmental modulation of Fab-7 boundary function. Development 131:4743–4749. doi: 10.1242/dev.01343. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Barolo S, Szymanski P, Levine M. 1996. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev 10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]
- 40.Rodin S, Kyrchanova O, Pomerantseva E, Parshikov A, Georgiev P. 2007. New properties of Drosophila Fab-7 insulator. Genetics 177:113–121. doi: 10.1534/genetics.107.075887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagstrom K, Muller M, Schedl P. 1997. A polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics 146:1365–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cleard F, Wolle D, Taverner AM, Aoki T, Deshpande G, Andolfatto P, Karch F, Schedl P. 2017. Different evolutionary strategies to conserve chromatin boundary function in the bithorax complex. Genetics 205:589–603. doi: 10.1534/genetics.116.195586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyrchanova O, Chetverina D, Maksimenko O, Kullyev A, Georgiev P. 2008. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res 36:7019–7028. doi: 10.1093/nar/gkn781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai HN, Shen P. 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 46.Gilfillan GD, König C, Dahlsveen IK, Prakoura N, Straub T, Lamm R, Fauth T, Becker PB. 2007. Cumulative contributions of weak DNA determinants to targeting the Drosophila dosage compensation complex. Nucleic Acids Res 35:3561–3572. doi: 10.1093/nar/gkm282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. 2002. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- 48.Kyrchanova O, Mogila V, Wolle D, Magbanua JP, White R, Georgiev P, Schedl P. 2015. The boundary paradox in the Bithorax complex. Mech Dev 138:122–132. doi: 10.1016/j.mod.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorn R, Krauss V. 2003. The modifier of mdg4 locus in Drosophila: functional complexity is resolved by trans splicing. Genetica 117:165–177. doi: 10.1023/A:1022983810016. [DOI] [PubMed] [Google Scholar]
- 50.Bonchuk A, Denisov S, Georgiev P, Maksimenko O. 2011. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J Mol Biol 412:423–436. doi: 10.1016/j.jmb.2011.07.052. [DOI] [PubMed] [Google Scholar]
- 51.Villa R, Schauer T, Smialowski P, Straub T, Becker PB. 2016. PionX sites mark the X chromosome for dosage compensation. Nature 537:244–248. doi: 10.1038/nature19338. [DOI] [PubMed] [Google Scholar]
- 52.Aoki T, Schweinsberg S, Manasson J, Schedl P. 2008. A stage-specific factor confers Fab-7 boundary activity during early embryogenesis in Drosophila Mol Cell Biol 28:1047–1060. doi: 10.1128/MCB.01622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamada FN, Park PJ, Gordadze PR, Kuroda MI. 2005. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev 19:2289–2294. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuda NJ, Guertin MJ, Sharma S, Danko CG, Martins AL, Siepel A, Lis JT. 2015. GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet 11:e1005108. doi: 10.1371/journal.pgen.1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. 2007. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.