ABSTRACT
Gut-homing α4β7high CD4+ T lymphocytes have been shown to be preferentially targeted by human immunodeficiency virus type 1 (HIV-1) and are implicated in HIV-1 pathogenesis. Previous studies demonstrated that HIV-1 envelope protein gp120 binds and signals through α4β7 and that this likely contributes to the infection of α4β7high T cells and promotes cell-to-cell virus transmission. Structures within the second variable loop (V2) of gp120, including the tripeptide motif LDV/I, are thought to mediate gp120-α4β7 binding. However, lack of α4β7 binding has been reported in gp120 proteins containing LDV/I, and the precise determinants of gp120-α4β7 binding are not fully defined. In this work, we report the novel finding that fibronectins mediate indirect gp120-α4β7 interactions. We show that Chinese hamster ovary (CHO) cells used to express recombinant gp120 produced fibronectins and other extracellular matrix proteins that copurified with gp120. CHO cell fibronectins were able to mediate the binding of a diverse panel of gp120 proteins to α4β7 in an in vitro cell binding assay. The V2 loop was not required for fibronectin-mediated binding of gp120 to α4β7, nor did V2-specific antibodies block this interaction. Removal of fibronectin through anion-exchange chromatography abrogated V2-independent gp120-α4β7 binding. Additionally, we showed a recombinant human fibronectin fragment mediated gp120-α4β7 interactions similarly to CHO cell fibronectin. These findings provide an explanation for the apparently contradictory observations regarding the gp120-α4β7 interaction and offer new insights into the potential role of fibronectin and other extracellular matrix proteins in HIV-1 biology.
IMPORTANCE Immune tissues within the gut are severely damaged by HIV-1, and this plays an important role in the development of AIDS. Integrin α4β7 plays a major role in the trafficking of lymphocytes, including CD4+ T cells, into gut lymphoid tissues. Previous reports indicate that some HIV-1 gp120 envelope proteins bind to and signal through α4β7, which may help explain the preferential infection of gut CD4+ T cells. In this study, we demonstrate that extracellular matrix proteins can mediate interactions between gp120 and α4β7. This suggests that the extracellular matrix may be an important mediator of HIV-1 interaction with α4β7-expressing cells. These findings provide new insight into the nature of HIV-1–α4β7 interactions and how these interactions may represent targets for therapeutic intervention.
KEYWORDS: α4β7 integrin, extracellular matrix proteins, fibronectin, gp120, human immunodeficiency virus
INTRODUCTION
Gut-associated lymphoid tissues (GALT) are preferentially targeted by human immunodeficiency virus type 1 (HIV-1) (1, 2). Soon after invading GALT, HIV-1 depletes the majority of intestinal CD4+ T lymphocytes and damages surrounding tissues through bystander effects (3). This is believed to contribute substantially to immune dysregulation, chronic inflammation, and the development of AIDS (4). CD4+ T cells expressing the α4β7 integrin have been proposed to play a role in HIV-1's apparent tropism for GALT. First, α4β7high cells traffic between peripheral mucosal sites and GALT, largely due to an interaction between α4β7 and mucosal vascular addressin cell adhesion molecule 1 (MAdCAM-1) expressed on gut vasculature (5, 6). For this reason, α4β7high cells may facilitate viral spread from mucosal portals of entry to intestinal sites. Consistent with this, the frequency of α4β7high lymphocytes within genital tissues correlates with infection risk in vivo (7, 8). Second, it has been demonstrated that the α4β7high memory CD4+ T cell subset is more susceptible to HIV-1 infection than α4β7− cells in vitro (9). This is also supported by studies that demonstrate preferential infection of α4β7high cells in vivo (10–12). Because these cells are present at high density in the gut (13) and are highly susceptible to HIV-1 infection, they may facilitate HIV-1 propagation throughout GALT.
Binding between α4β7 and the gp120 subunit of HIV-1 envelope protein has been described (14–20). This interaction has been proposed to enhance HIV-1 infection either by facilitating virus attachment to cells or by activating α4β7-mediated signaling. Notably, the monoclonal antibodies (MAbs) Act-1 and natalizumab, which block α4β7 and the α4 integrin chain, respectively, did not significantly inhibit HIV-1 infectivity in vitro (9, 21, 22). In contrast, targeting α4β7 with Act-1 in macaques infected with simian immunodeficiency virus (SIV) resulted in lower virus titers and significant improvements in CD4+ T cell numbers, as well as prevention of mucosal virus transmission (23–25). These different effects of α4β7 inhibition in vitro and in vivo argue against α4β7 functioning as a virus attachment factor. Furthermore, a recent study reported that a small-molecule inhibitor of α4β7 failed to inhibit infection in vivo, similarly to Act-1 (26). While both Act-1 and this small molecule blocked ligand binding to α4β7, the small molecule initiated α4β7-mediated signaling, whereas Act-1 blocked this signaling. This finding suggests that α4β7-mediated signaling may play an important role during infection in vivo and further argues against a role for α4β7 as a virus attachment factor. In support of this in vivo finding, gp120 has been demonstrated to initiate α4β7 signal transduction, leading to LFA-1 activation in vitro. This mechanism has been proposed to contribute to cell-to-cell transmission of HIV-1 and the preferential infection of α4β7high memory CD4+ T cells (14).
At present, the precise determinants of gp120-α4β7 interactions are not fully understood. The interaction is thought to be mediated by structures within the second variable loop (V2) of gp120, including the conserved tripeptide sequence LDV/I (14). However, the reactivity of gp120 to α4β7 is reported to vary considerably among gp120 proteins from different strains, and many gp120 molecules containing LDV/I have no measurable α4β7 binding (14, 27). It is thought that α4β7 reactivity may be strain specific and modulated by factors including the length of the V2 loop, the presence or absence of N-linked glycans (in variable regions 1, 2, and 4 and conserved region 3 of gp120), and the specific glycoforms present on gp120 (28). These factors are hypothesized to affect α4β7 binding by altering the conformation and exposure of LDV/I. More recently, additional determinants in both the V2 and V3 loops have been proposed to participate in α4β7 binding (18, 19). Although the nature of the interaction between gp120 and α4β7 is not fully understood, it is clear that any such interaction depends on more than the simple presence or absence of the LDV/I sequence.
In the present work, we sought to understand the nature of gp120-α4β7 binding in greater detail. We determined that CHO cells used to produce recombinant gp120 also produced cellular proteins with high α4β7 binding reactivity, and these proteins copurified with gp120 by lectin chromatography. Importantly, we discovered that these CHO cellular proteins were capable of mediating indirect interactions between α4β7 and HIV-1 envelope proteins. The proteins responsible for this indirect interaction included an α4β7-reactive isoform of fibronectin and other extracellular matrix components. These findings provide evidence that extracellular matrix proteins may mediate interactions between HIV-1 envelope proteins and α4β7 and offer new insight into the potential role of α4β7 in HIV-1 pathogenesis.
RESULTS
The α4β7 binding properties of 93MW959 gp120 depend on protein purification conditions.
Lack of gp120-α4β7 reactivity has been reported in studies that used recombinant gp120 purified by DEAE anion-exchange chromatography (27). In contrast, earlier reports of gp120-α4β7 reactivity utilized envelope proteins that were purified without the DEAE anion-exchange step (14, 28). Here, we examined the effects of DEAE chromatography in detail, using gp120 from the 93MW959 isolate (referred to below as MW959), which is reported to have strong α4β7 binding properties (28).
MW959 gp120 was produced by transient transfection of CHO cells and prepared with a three-step purification scheme. First, gp120-containing supernatant was purified on a Galanthus nivalis lectin affinity column (GNA). Second, DEAE chromatography was used to divide the GNA eluate into two fractions: DEAE flowthrough (material that did not bind to DEAE) and DEAE eluate (material that bound and was subsequently eluted from DEAE). Third, size exclusion chromatography (SEC) was used to analyze material recovered at each of these purification steps.
GNA eluate yielded 3 distinct peaks when analyzed by SEC (Fig. 1A, blue chromatogram), and these were numbered 1 to 3 in ascending order of their apparent sizes. DEAE chromatography separated peak 3 materials from peaks 1 and 2. DEAE flowthrough yielded two peaks that corresponded to peaks 1 and 2 of the GNA eluate (Fig. 1A, green chromatogram). DEAE eluate yielded a single peak on SEC that corresponded to peak 3 of the GNA eluate (Fig. 1A, red chromatogram).
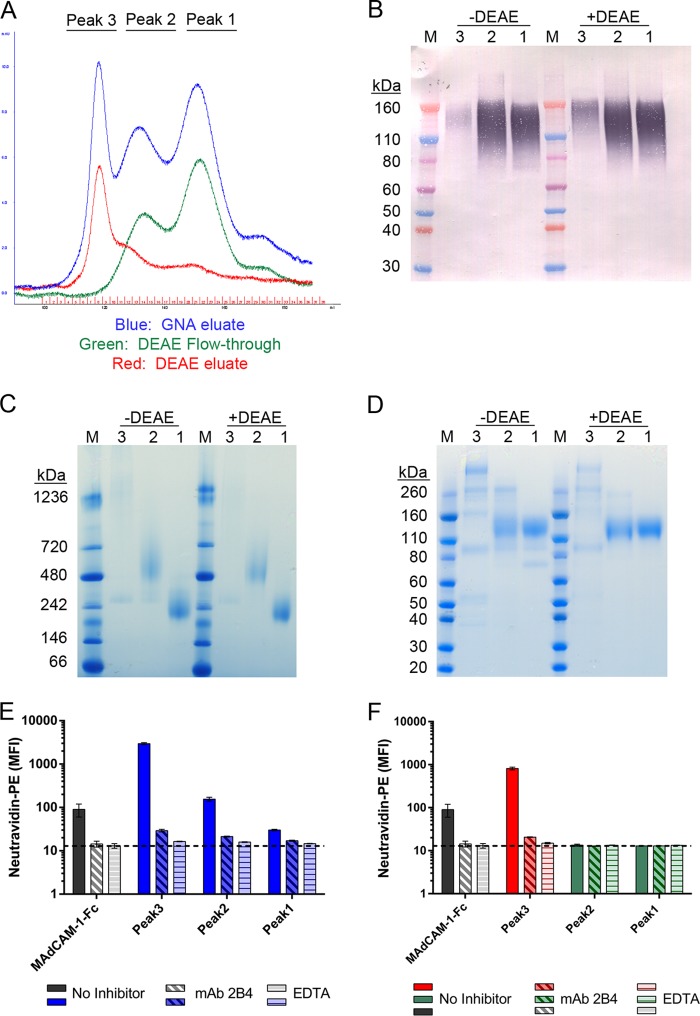
FIG 1.
Effect of DEAE purification on α4β7 reactivity of MW959 gp120. MW959 gp120 produced in CHO cells was analyzed at different stages of purification. (A) Overlay of UV chromatograms from size exclusion chromatography. (B to D) Peaks 1 to 3 were collected as indicated for panel A and analyzed by Western blotting (B), native-PAGE (C), and SDS-PAGE (D). Protein was loaded at 1 μg/lane for Western blotting and 5 μg/lane for native-PAGE and SDS-PAGE. Lanes: 1 to 3, peaks 1 to 3 from GNA eluate (−DEAE) or DEAE fractionation (+DEAE); M, molecular mass markers. Peak 3 +DEAE is the material recovered from DEAE eluate, and peaks 1 to 2 +DEAE are materials recovered from DEAE flowthrough. The Western blot was probed with HIV-1+ serum and detected with a chromogenic alkaline phosphatase substrate. (E and F) Materials collected for panel A were biotinylated and tested by a flow cytometry assay for binding to the α4β7-expressing cell line RPMI8866. (E) Cell binding of material in peaks 1 to 3 from GNA eluate. (F) Cell binding of material in peaks 1 to 3 from DEAE fractionation. Binding was measured without inhibitors (No Inhibitor), with α4β7-blocking antibody 2B4 (MAb 2B4), or with EDTA (EDTA). The dashed line indicates the background signal from cells stained with neutravidin-PE in the absence of biotinylated proteins. MAdCAM-1–Fc binding was included as a positive control for α4β7 binding. The cell binding results are the means and standard deviations of 3 replicate measurements from a single representative experiment.
The products of SEC purification were analyzed with polyacrylamide gel electrophoresis (PAGE) and Western blotting. The results from these analyses confirmed that peak 1 contained gp120 monomers and peak 2 contained gp120 dimers. Western blot analysis showed the majority of recombinant gp120 was present in peaks 1 and 2, whether or not DEAE was used (Fig. 1B). Native PAGE confirmed that peak 1 primarily contained gp120 monomers and peak 2 primarily contained gp120 dimers, whereas peak 3 contained a diffuse smear of high-molecular-weight material (Fig. 1C). Reduced and denatured samples analyzed by SDS-PAGE showed that peak 1 and peak 2 both contained a major band of 120 kDa, which was presumed to be gp120 (Fig. 1D). The same analysis revealed that peaks 1 and 2 prepared without DEAE contained additional contaminant bands that were not observed in material prepared with DEAE.
Material from each of these peaks was biotinylated and tested for α4β7 binding activity in a flow cytometry-based cell binding assay, using the α4β7-expressing cell line RPMI8866. Soluble MAdCAM-1–Fc fusion protein was used as a positive control for α4β7 binding. Specificity of binding was determined either by preblocking cells with the α4 integrin-blocking antibody 2B4 or by inhibiting integrins through removal of divalent cations with EDTA-containing buffer. All 3 peaks purified without DEAE exhibited α4β7 binding activity (Fig. 1E). Binding was highest in peak 3, lower in peak 2, and lowest in peak 1. In contrast, DEAE-purified gp120 monomers and dimers exhibited no α4β7 binding activity, but DEAE eluate exhibited high α4β7 reactivity (Fig. 1F). These results demonstrated that MW959 gp120 prepared without DEAE appeared to react with α4β7, whereas gp120 prepared with DEAE did not.
CHO produces α4β7-reactive cellular proteins.
The above-mentioned results suggested to us that CHO cells may produce α4β7-reactive substances that are copurified with gp120. To test this, we prepared CHO-conditioned culture medium by growing nontransfected cells under conditions identical to those used for gp120 purification and then processed the resulting medium with GNA affinity chromatography, followed by SEC. The resulting CHO proteins yielded a major peak on SEC at the same elution time as peak 3 in MW959 purification. CHO proteins were collected in 3 separate fractions corresponding to the elution times of peaks 1 to 3 in the MW959 gp120 purification (Fig. 2A). These fractions were biotinylated and tested for α4β7 binding activity on RPMI8866 cells. All the fractions exhibited α4β7 binding, with the highest activity in fraction 3, lower activity in fraction 2, and the lowest activity in fraction 1 (Fig. 2B).
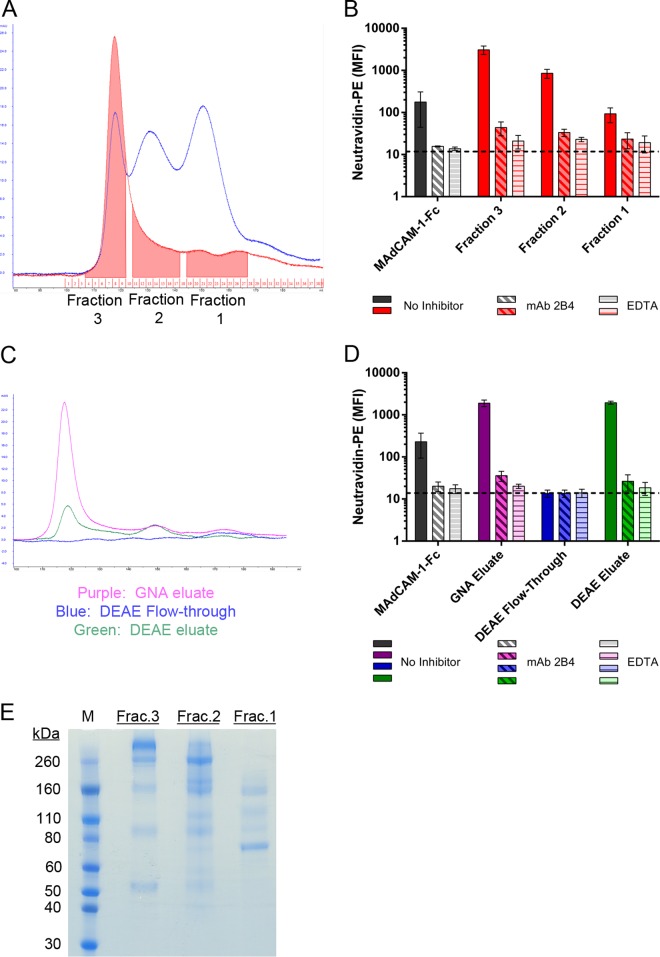
FIG 2.
CHO cells produce α4β7-reactive cellular proteins. Culture medium was conditioned by CHO cells and processed by GNA. (A) Comparison of GNA eluates from MW959 gp120 (blue) and CHO cell-conditioned medium (red) analyzed by size exclusion chromatography. CHO cell proteins were collected in 3 fractions, indicated by shaded areas in the red chromatogram. (B) Binding of protein fractions collected for panel A to RPMI8866 cells. The results are the means and standard deviations of 2 separate experiments. (C) CHO cell proteins were fractionated by DEAE and analyzed by size exclusion chromatography. (D) Binding of proteins collected for panel C to RPMI8866 cells. The results are the means and standard deviations of 3 separate experiments. (E) SDS-PAGE analysis of fractions collected for panel A.
These results demonstrated that CHO cells produce α4β7-reactive substances that are enriched by GNA and comigrate with gp120 monomers and dimers on SEC. Because α4β7 reactivity was not observed in DEAE-purified MW959 gp120, we tested the ability of DEAE to capture and remove these α4β7-reactive substances. Similar to gp120 purification, CHO-conditioned medium was processed by GNA and DEAE, followed by SEC. Minimal amounts of protein were recovered in the DEAE flowthrough (Fig. 2C, blue chromatogram), while the majority of proteins were bound by DEAE and could be recovered in the eluate (Fig. 2C, green chromatogram). CHO proteins recovered in the DEAE eluate had a SEC elution profile similar to that of CHO proteins captured by GNA (Fig. 2C, purple chromatogram). These fractions were assayed for α4β7 binding activity as described above. The DEAE flowthrough had no detectable α4β7 binding, whereas the DEAE eluate exhibited high α4β7 binding (Fig. 2D), and this indicated that DEAE is capable of capturing α4β7-reactive CHO proteins. The three CHO protein fractions, collected as shown in Fig. 2A, were analyzed by SDS-PAGE. Each fraction contained multiple protein bands (Fig. 2E), and they were similar to the contaminant bands observed in MW959 gp120 purified without DEAE (compare Fig. 1D). Overall, these results suggested that the α4β7 reactivity of MW959 gp120 in our assays depended on the presence of α4β7-reactive CHO proteins.
It has been reported that gp120 derived from human embryonic kidney 293 (HEK293) cells does not bind to α4β7, and we speculated that this may be due to a lack of α4β7-reactive proteins produced by 293 cells (28). Thus, we examined whether human embryonic kidney 293 cells produce α4β7-reactive cellular proteins. 293 cell culture medium was prepared and analyzed as described above. Surprisingly, α4β7 binding activity was again detected (Fig. 3). However, we also observed that 293 cells secreted fewer cellular proteins overall than CHO cells (91 ± 14 μg/liter for 293 cells versus 546 ± 210 μg/liter for CHO cells). Additionally, in separate experiments, we observed that recombinant gp120 yields were higher in 293 cells than in CHO cells (MW959 gp120 production, 1,024 μg/liter for 293 cells versus 444.6 ± 148.3 μg/liter for CHO cells). From this, we concluded that α4β7 reactivity may not be readily observed in 293 cell-derived gp120 because of a low relative abundance of cellular proteins, even without DEAE purification.
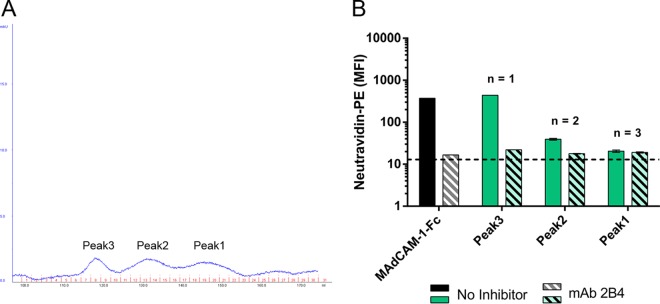
FIG 3.
293 cells produce α4β7-reactive cellular proteins. 293 cell-conditioned medium was processed by GNA and then analyzed by size exclusion chromatography. (A) UV chromatogram showing 3 peaks of 293 cellular proteins collected by size exclusion. (B) Binding of the proteins collected for panel A to RPMI8866 cells. The results presented are means and standard deviations; the number of replicates for each sample is indicated.
Inhibitors of α4 integrins block binding of CHO cellular proteins.
Several inhibitors have been used to demonstrate the specificity of gp120-α4β7 binding, and we asked if these agents inhibit CHO cell protein binding, as well. First, we tested the effects of the cyclic peptide CWLDVC. It is a competitive inhibitor of the LDV recognition site of α4 integrins and has been used to demonstrate LDV-dependent α4β7 binding of gp120 (14, 29). CWLDVC completely inhibited MAdCAM-1–Fc binding to RPMI8866 cells and inhibited the majority of CHO cell protein binding, as well (Fig. 4A). The scrambled control peptide CDLVWC had no effect on either protein. These results indicate that binding in our assays required the LDV recognition site of an α4 integrin. EDTA was found in previous assays to inhibit CHO cell protein binding (Fig. 2B), and it was included in these experiments as a control.
FIG 4.
Inhibition of CHO cellular protein binding by α4 integrin inhibitors. (A) Binding of MAdCAM-1–Fc (blue) or CHO cell proteins (green) to RPMI8866 cells in the presence of the cyclic peptide CWLDVC, scrambled control peptide CDLVWC, or buffer containing EDTA. (B) Binding in the presence of antibody 2B4 (α4 integrin), Act-1 (α4β7 integrin), P5D2 (β1 integrin), or a combination of Act-1 and P5D2. The results are the means and standard deviations of 3 separate experiments.
Next, we examined the effects of integrin-blocking antibodies. In previous assays, we established that CHO cell protein binding could be blocked by MAb 2B4, which inhibits the α4 integrin chain (Fig. 2B). We confirmed this result and also tested the effects of MAb Act-1, which is specific for the α4β7 heterodimer (30, 31). Surprisingly, Act-1 was found to inhibit only about half of the CHO cell protein binding signal but completely inhibited MAdCAM-1–Fc binding (Fig. 4B). The high degree of inhibition observed with MAb 2B4 and CWLDVC peptide, contrasted with the partial inhibition of MAb Act-1, suggested that CHO cell proteins may bind through both α4β7 and the related integrin α4β1. We tested this with MAb P5D2, which blocks the β1 integrin chain. As expected, P5D2 had no effect on MAdCAM-1–Fc binding. P5D2 did, however, inhibit about half of the CHO cell protein binding (Fig. 4B). When Act-1 and P5D2 were combined, the effects were additive, and CHO cell protein binding was inhibited to the same level as observed with MAb 2B4 (Fig. 4B).
These results suggest that CHO cell proteins are capable of binding to both α4β7 and α4β1. This is in contrast to other findings in which gp120-α4β7 binding is primarily through α4β7 and is significantly inhibited by Act-1 (14). One important caveat to these comparisons is that the present study used the RPMI8866 cell line while prior studies used primary human peripheral blood mononuclear cells (PBMCs). Because receptor expression or binding properties may differ between these cell types, direct comparisons cannot be made. In total, our experiments demonstrated that CHO cell protein binding was blocked by inhibitors of α4 integrins and that copurified CHO cell proteins can account for gp120-α4β7 binding signals under certain conditions.
CHO cellular proteins mediate indirect gp120-α4β7 binding.
Mixing experiments were carried out to better understand the reasons for lack of α4β7 reactivity in DEAE-purified MW959 gp120. In these experiments, labeled (biotinylated) and unlabeled (nonbiotinylated) components were mixed and tested for changes in binding activity of the labeled component in the presence of the unlabeled component. Because cell staining was carried out with neutravidin-R-phycoerythrin (PE), only biotinylated proteins in these mixtures were expected to produce a signal.
First, we ruled out the possibility that DEAE purification introduced α4β7-inhibitory substances. To test this, unlabeled DEAE-purified MW959 gp120 monomers were mixed with either labeled MAdCAM-1–Fc or labeled MW959 gp120 purified without DEAE. In both cases, DEAE-purified material did not inhibit the binding signals (Fig. 5A), which indicated that DEAE purification did not introduce α4β7-inhibitory activity.
FIG 5.
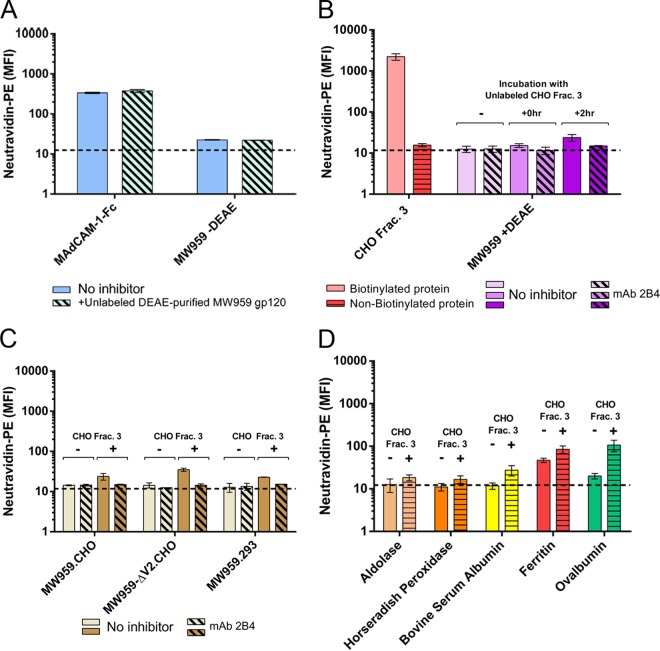
CHO cell proteins mediate indirect α4β7 binding. Biotinylated (labeled) and nonbiotinylated (unlabeled) proteins were mixed, and binding of the labeled component was measured on RPMI8866 cells by neutravidin-PE staining. (A) DEAE-purified MW959 gp120 was tested for α4β7-inhibitory activity. Binding of labeled MAdCAM-1–Fc or MW959 gp120 prepared without DEAE was measured in the absence (solid bars) or presence (hatched bars) of 10 μg unlabeled DEAE-purified gp120. (B) CHO cell proteins were tested for enhancement of α4β7 binding signals. Labeled CHO cell proteins produced binding signals (pink bar), but unlabeled proteins did not (red bar). Binding of DEAE-purified MW959 gp120 was measured without CHO cell proteins or with unlabeled CHO cell proteins combined with gp120 for 0 h or 2 h. (C) Wild-type MW959 gp120 was produced in CHO and 293 cells, and MW959-ΔV2 with a deleted V2 loop was produced in CHO cells. Binding of these proteins was measured with and without 2-h preincubation with unlabeled CHO cell proteins. (D) Binding of five non-HIV-1 proteins with and without 2-h preincubation with unlabeled CHO cell proteins. All the results are the means and standard deviations of 3 replicate measurements.
We next asked if CHO cell proteins could modify the binding behavior of DEAE-purified gp120. First, we verified that CHO cell fraction 3, which binds strongly to α4β7, produced no signal when not biotinylated (Fig. 5B). Labeled DEAE-purified MW959 gp120, with no measurable α4β7 binding, was then mixed with unlabeled CHO cell fraction 3. Surprisingly, we discovered a modest α4β7 binding signal when these proteins were incubated for 2 h prior to being added to cells (Fig. 5B). The same effect was not seen when the proteins were mixed directly on cells without preincubation (Fig. 5B). Our interpretation of these results was that CHO cell proteins mediated indirect gp120-α4β7 binding. Because an effect was seen only with 2 h of incubation, we concluded that the interaction between gp120 and CHO cell proteins was slow under these experimental conditions.
Because the V2 loop is implicated in gp120-α4β7 binding, we speculated that CHO cell proteins may modulate the activity of the V2 loop in a way that enhanced α4β7 reactivity. If this were the case, we reasoned that gp120 without a V2 loop should not produce an α4β7 binding signal in these mixing experiments. We examined this by creating an MW959 gp120 mutant in which the majority of the V2 loop (amino acid residues 161 to 185) was deleted. This mutant was designated MW959-ΔV2 and was produced in CHO cells and purified with DEAE as described above. MW959-ΔV2 gp120 exhibited no α4β7 binding activity by itself but did produce an α4β7 binding signal in mixing experiments with CHO cell fraction 3 (Fig. 5C). From this, we concluded that the binding signal seen in mixtures of gp120 and CHO cell proteins did not require the V2 loop.
Next, we examined whether gp120 produced in 293 cells interacted with CHO cell proteins. We speculated that 293 cell-derived gp120 may interact poorly with α4β7-reactive cellular proteins, and this could contribute to the lack of α4β7 binding signals. MW959 gp120 was produced in 293 cells and tested in mixing experiments with CHO cell proteins, as described above, and was found to react similarly to CHO cell-derived gp120 (Fig. 5C). This indicated that interaction between gp120 and cellular proteins was not dependent on specific glycosylation patterns or other posttranslational modifications that might vary between expression systems. It also further supported the hypothesis that low abundance of α4β7-reactive cellular proteins was the likely reason for lack of binding observed in 293 cell-derived gp120.
After determining that CHO cell proteins interact with these MW959 gp120 preparations, we tested the specificity of this interaction with five non-HIV-1 proteins (aldolase, horseradish peroxidase [HRP], bovine serum albumin, ferritin, and ovalbumin). These proteins were biotinylated and tested in mixing experiments as described above. CHO cell proteins did not significantly increase the binding of aldolase or horseradish peroxidase but did increase the binding of bovine serum albumin, ferritin, and ovalbumin (Fig. 5D). These results indicated that CHO cell proteins can interact with a variety of proteins, resulting in apparent α4β7 binding, and this interaction was not specific to gp120.
Collectively, these experiments showed that CHO cell proteins are not only capable of producing α4β7 binding signals by themselves, but also produce signals by mediating indirect interactions between α4β7 and gp120. The indirect gp120-α4β7 binding observed in the presence of CHO cell proteins seemed to be due to protein-protein interactions that did not require the V2 loop or CHO cell-specific posttranslational events, such as glycosylation.
CHO cell proteins mediate indirect gp120-α4β7 binding in a panel of diverse HIV-1 envelopes.
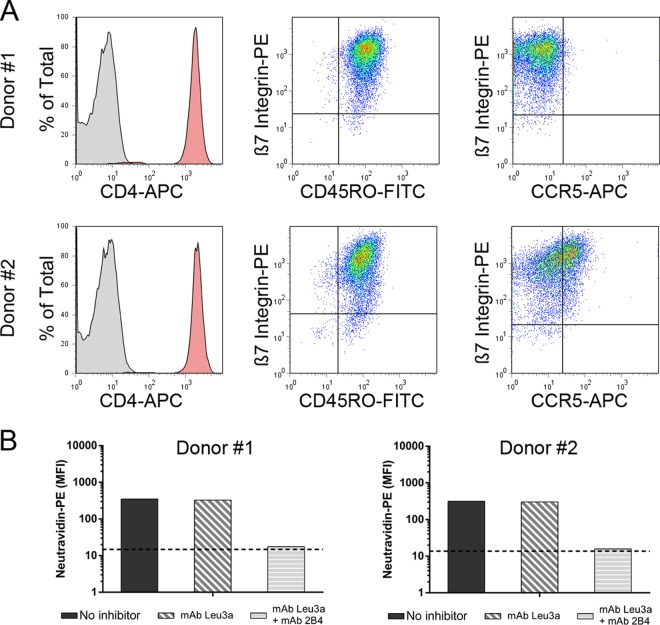
The above-described findings were based on gp120 from the MW959 isolate and assays that used the RPMI8866 cell line. We next asked whether these findings would extend to a variety of envelope proteins assayed with primary CD4+ α4β7+ T cells. Human PBMCs were acquired from two separate donors, and CD4+ cells were isolated from them. Primary CD4+ T cells were activated with anti-CD3 antibody and cultured in the presence of interleukin-2 and retinoic acid. The expression levels of relevant receptors were then measured by flow cytometry. Cells from both donors expressed high levels of CD4, α4β7, and the marker of cell activation CD45RO (Fig. 6A). CCR5 was not detected on cells from donor number 1 but was detected on approximately half of the cells from donor number 2 (Fig. 6A). MAdCAM-1–Fc bound to cells from both donors at similar levels. As expected, MAdCAM binding was not inhibited by the CD4-blocking antibody leu3a but was inhibited by a combination of leu3a and 2B4 antibodies (Fig. 6B).
FIG 6.
Characterization of primary CD4+ α4β7+ T cells. Human CD4+ cells were enriched from PBMCs of two separate donors and cultured in retinoic acid for 9 days prior to analysis. (A) Expression of CD4, β7 integrin, CD45RO, and CCR5 measured by flow cytometry. (B) Binding of MAdCAM-1–Fc to cells from both donors. Binding was measured without inhibitors, with the CD4-blocking antibody leu3a, or with a combination of leu3a and 2B4 antibodies.
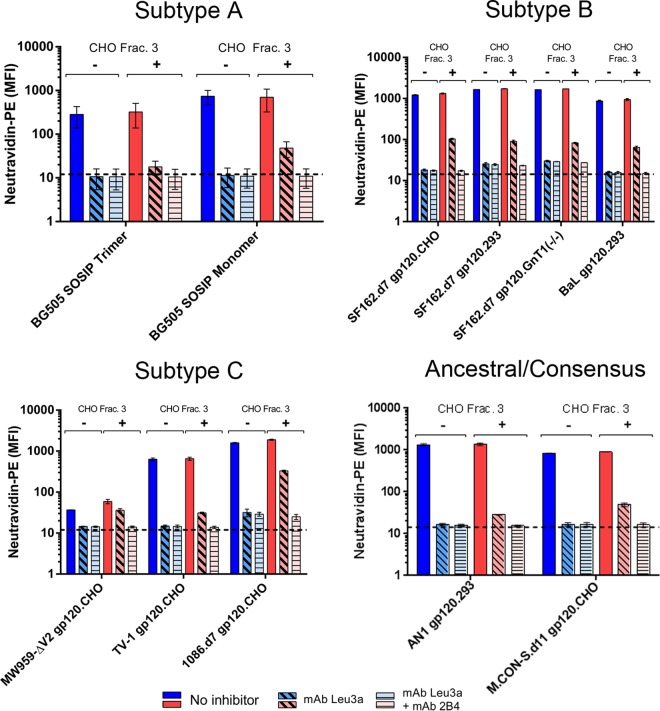
A panel of DEAE-purified envelope proteins was assembled and included representatives from HIV-1 subtypes A, B, and C. It also included two fully synthetic envelopes: the ancestral subtype B envelope AN1 (32) and an envelope based on a consensus of group M sequences (M.CON-S.d11) (33). We primarily included gp120 monomers, but also included the BG505.T332N.664 SOSIP trimer and corresponding BG505.T332N.664 SOSIP gp140 monomer (34). SF162 and 1086 gp120 proteins contained a deletion of the first 7 N-terminal amino acids of the mature protein (d7 mutation), and M.CON-S.d11 contained a deletion of the first 11 N-terminal amino acids (35). Additionally, the panel contained proteins produced in CHO and 293 cells, as well as one SF162 envelope produced in 293S GnT1−/− cells. All the proteins used in these experiments were prepared with a DEAE purification step.
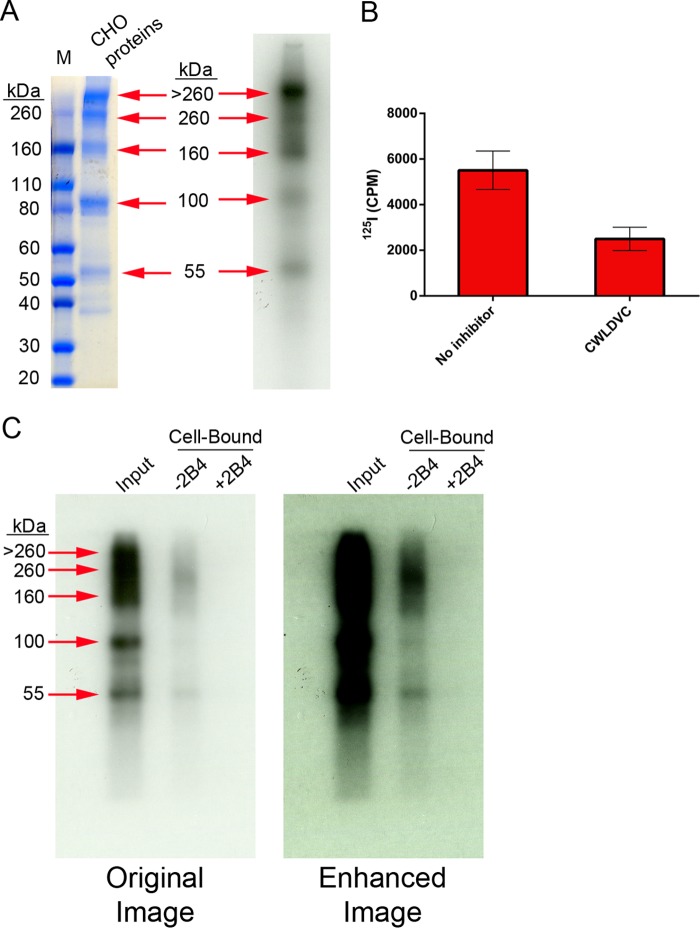
All of these envelope proteins displayed similar binding profiles on CD4+ α4β7+ cells (Fig. 7). In the absence of CHO cell proteins, all the envelopes produced a binding signal that was completely inhibited by CD4 antagonism with leu3a. In the presence of CHO cell proteins, the binding signal was mostly inhibited by leu3a but displayed residual CD4-independent binding that was inhibited with the addition of α4β7-blocking antibody. CHO cell proteins increased the total binding signal for all the envelopes studied. For most envelopes, binding was increased by 4 to 10%; for 1086.d7, binding was increased by 18%; and for MW959-ΔV2, binding was increased 45%. Increases in binding were statistically significant for SF162.d7.293, SF162.d7.GnT1−/−, MW959-ΔV2, 1086.d7.CHO, and M.CON-S.d11.CHO (unpaired t test; P < 0.05). These experiments showed that the majority of gp120 binding was through CD4 but that overall binding could be enhanced by CHO cell protein-mediated interactions with α4β7. In our assays, gp120-α4β7 binding required CHO cell proteins and was not apparently influenced by other envelope-specific factors (HIV-1 subtype, monomeric or trimeric envelope, and producer cell type).
FIG 7.
CHO cell proteins mediate α4β7 binding signals in a diverse panel of HIV-1 envelopes. Shown is binding of 11 different DEAE-purified envelope proteins to primary CD4+ α4β7+ T cells. The means and standard deviations of single measurements made on cells from both donors are presented. Binding was measured in the absence (blue bars) or presence (red bars) of unlabeled CHO cell proteins. For both of these conditions, binding was measured without inhibitors, with leu3a antibody, or with a combination of leu3a and 2B4 antibodies.
DEAE-purified envelopes display intact V2 epitopes.
Because the gp120-α4β7 interaction is hypothesized to involve LDV/I and possibly other structures within V2, we conducted experiments to verify that DEAE-purified envelopes display intact V2 epitopes. Additionally, several distinct conformations of the V2 loop have been described, and exposure of LDV/I may depend on these conformations (36–38). We therefore also asked if DEAE purification selects for any particular V2 conformation and if this could explain the lack of α4β7 reactivity in DEAE-purified envelopes.
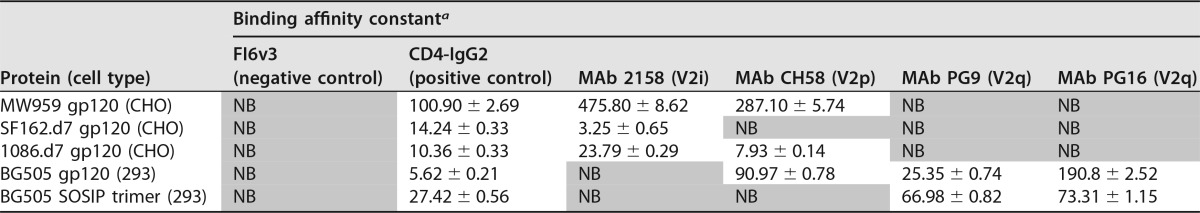
A biolayer interferometry assay was used to measure the binding of several envelope proteins to a panel of V2 antibodies. We included representatives from 3 antibody classes that are able to distinguish V2 conformations: PG9 and PG16, which bind V2q quaternary epitopes; CH58, which binds a V2p linear epitope; and 2158, which binds a V2i conformational epitope that includes LDV/I (39). The antibody FI6v3 to influenza virus hemagglutinin (HA) protein was included as a negative control, and CD4-IgG2 fusion protein was included as a positive control (40). No difference was observed in the reactivity of biotinylated versus nonbiotinylated envelopes to any of these antibodies, and aliquots of the same biotinylated envelopes used for cell binding assays were used for biolayer interferometry. The results of these experiments are summarized in Table 1.
TABLE 1.
Envelope protein binding to conformation-dependent V2 antibodies
a Binding affinity constants (KD) and associated measurement errors are provided for gp120-antibody pairs in which binding was observed. NB, no binding detected at 1 μM concentration of envelope protein (shaded).
All the envelopes bound to CD4 and were recognized by V2-specific antibodies, which indicated that they were correctly folded. Strain-specific binding to V2 antibodies was observed. Among the envelopes tested, there were representatives that bound to each of the V2 antibodies. As expected, none of the envelopes bound to PG9 and PG16, except for BG505 SOSIP trimer and BG505 gp120 monomer. PG9 and PG16 typically do not react with monomeric gp120 but have been reported previously to react with BG505 gp120 monomers (41).
These experiments confirmed that the DEAE-purified envelopes used in our assays displayed intact and accessible V2 epitopes. With the exception of BG505 gp120, all of them were the same envelope proteins used in our cell binding assays (Fig. 7). From this, we concluded that lack of α4β7 binding could not be attributed to a gross defect in the V2 loop or disruption of any particular V2 epitope by the additional DEAE purification step. However, the V2 domain can adopt multiple conformations, and we cannot exclude the possibility that specific V2 conformations required for α4β7 reactivity were underrepresented in our protein preparations.
Monoclonal antibodies to V2 do not inhibit α4β7 binding signals.
V2 monoclonal antibodies, including MAb 2158, are reported to inhibit gp120-α4β7 binding in vitro (15, 18). Based on this, we asked whether V2 antibodies inhibit α4β7 binding signals seen in our assays with CHO cell proteins, perhaps by disrupting interactions between gp120 and CHO cell proteins. The two V2-directed antibodies CH58 and 2158 were tested in combination with gp120 in which antibody-gp120 interactions had been confirmed by biolayer interferometry. Antibodies and gp120 were incubated for 1 h prior to incubation with unlabeled CHO cell proteins, and then the mixtures were assayed for α4β7 binding on RPMI8866 cells. With all of the envelopes tested, the binding signal was increased by the addition of CHO cell proteins, but this was not inhibited with the addition of V2 antibodies (Fig. 8). The V2 antibodies were additionally tested against MAdCAM-1–Fc in a similar manner and did not inhibit MAdCAM binding (Fig. 8).
FIG 8.
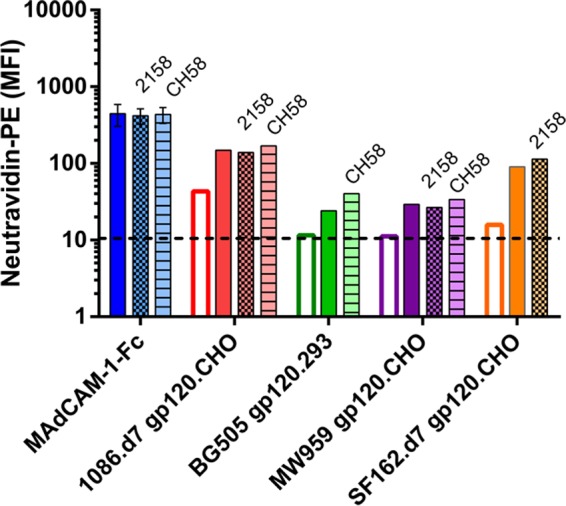
V2-directed antibodies do not inhibit α4β7 binding signals. Binding of gp120 without CHO cell proteins (open bars) was compared to binding with 2-h preincubation of gp120 and CHO cell proteins (solid and patterned bars). One hour prior to adding CHO cell proteins, 4 μg of gp120 was mixed with either 2 μg of MAb 2158 (cross-hatched bars), 2 μg MAb CH58 (hatched bars), or a matched volume of buffer without antibody (solid bars). MAdCAM-1–Fc was similarly preincubated with MAbs 2158 and CH58. The binding results are single measurements, except MAdCAM-1–Fc, which is the mean and standard deviation of 2 measurements.
Although these experiments were limited in scope, they indicated that under the conditions we employed, V2 antibodies do not inhibit in vitro gp120-α4β7 binding, as previously reported. Because the antibodies did not inhibit MAdCAM binding, we also concluded they do not have general α4β7-inhibitory activity. While we were unable to explain previous observations concerning V2 antibodies, these experiments indicate that the indirect binding mediated by CHO cell proteins does not involve the V2 loop and is not blocked by V2 antibodies.
CHO cells produce an α4β7-reactive isoform of fibronectin.
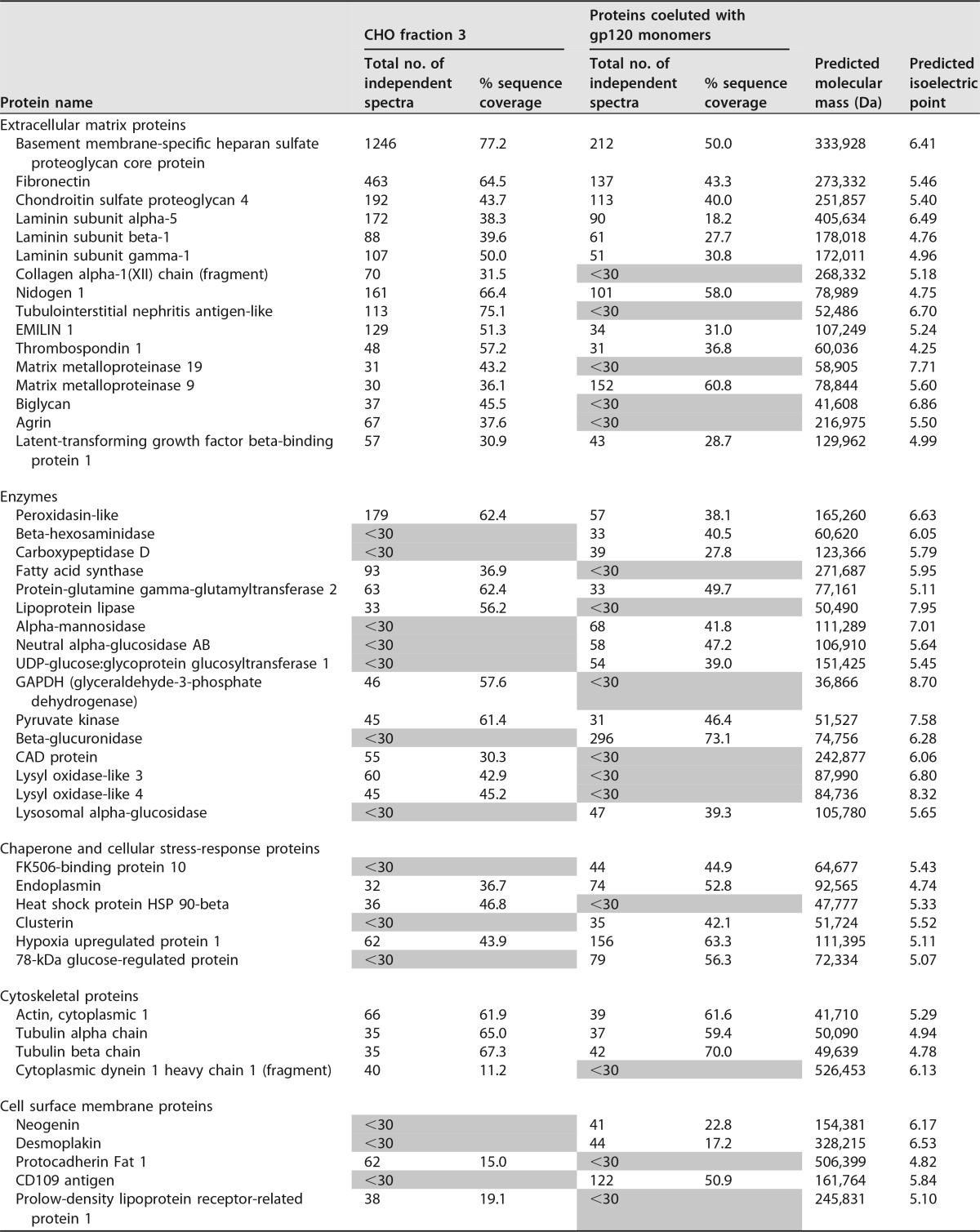
Because CHO cellular proteins were the main predictor of gp120-α4β7 binding in our assays, we sought to identify which proteins were responsible for the α4β7 binding activity. We began by generating lists of proteins present in CHO cell samples using a mass spectrometry (MS) shotgun sequencing approach. Proteins in CHO cell fraction 3, which were largely separated from gp120 monomers by SEC, were analyzed from a CHO cell-conditioned medium sample. We additionally analyzed the CHO cell proteins that were coeluted with MW959 gp120 monomers that could be removed by DEAE. For this, the order of purification steps was changed: GNA eluate was first separated by SEC, the resulting gp120 monomer fraction was passed through DEAE, and CHO cell proteins retained by DEAE were eluted and analyzed.
The results of these experiments are summarized in Table 2. For the sake of brevity, Table 2 lists only proteins that were identified by 30 or more independent spectra. Additionally, even though keratin was detected, it was presumed to be a contaminant of sample preparation and was excluded. Major components of CHO cell fraction 3 appeared to be extracellular matrix proteins, including the primary components of basement membrane (proteoglycans, fibronectin, collagen, laminin, and nidogen). CHO cell proteins coeluted with gp120 also contained many of these components, including very large proteins (proteoglycans, fibronectin, and laminin). This indicated there was likely a “trailing effect” with SEC, and not all of the high-molecular-weight components were fully excluded from the position where gp120 monomers elute.
TABLE 2.
Characterization of CHO proteins by mass spectrometrya
Shading, denotes proteins detected by 30 or fewer spectra.
Out of the proteins identified, fibronectin was the only protein with known α4β7 interactions. Cells produce several variants of fibronectin by alternative RNA splicing, and only some of them bind to α4β7 (42, 43). We therefore examined fibronectin in further detail and asked whether CHO cells produced α4β7-reactive fibronectins. Fibronectin-α4β7 binding occurs through the inclusion of the alternatively spliced type III connecting segment (IIICS). Within IIICS is a sequence designated the CS1 peptide that contains an LDV site required for α4β7 binding (44). We examined spectra from CHO cell fraction 3 and found 11 independent and high-quality spectra containing the complete fibronectin CS1 sequence (available via ProteomeXchange with identifier PXD006060 [https://www.ebi.ac.uk/pride/archive/projects/PXD006060]). These results confirmed that CHO cells produce an α4β7-reactive isoform of fibronectin. The experimentally determined amino acid sequence of CHO cell fibronectin CS1 was DELPQLVTLPHPNLHGPEILDVPST. Notably, this is 100% identical to the human fibronectin CS1 sequence (UniProtKB/Swiss-Prot accession no. P02751.4). Therefore, we concluded it was highly plausible that the CHO cell fibronectins in our samples reacted with human α4β7.
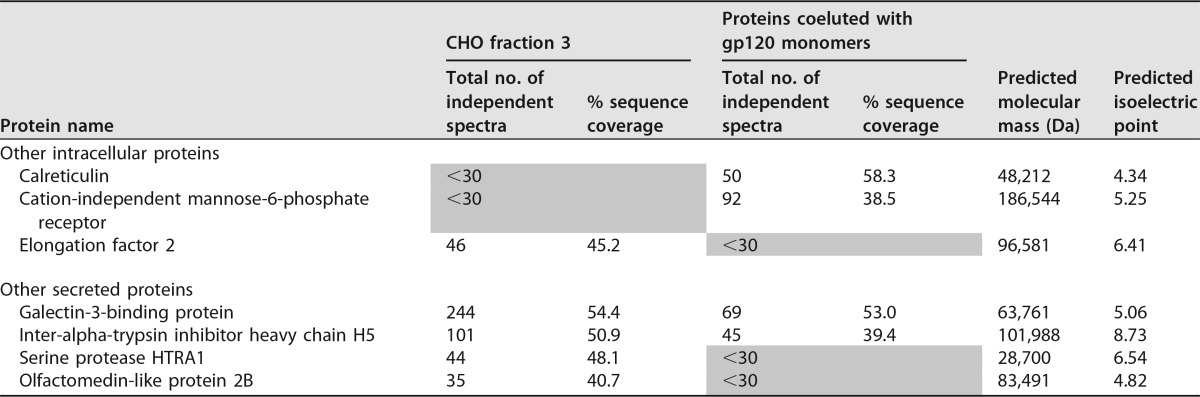
We next attempted to verify this with biochemical assays. First, CHO cell proteins were fractionated by DEAE using stepwise elution with 100 mM, 200 mM, 300 mM, and 400 mM sodium chloride. The 200 mM fraction was found to retain the relevant binding activity and was used for subsequent assays. Proteins were radiolabeled with 125I (Fig. 9A) and tested to ensure they retained α4β7 binding activity (Fig. 9B). Next, RPMI8866 cells were used to selectively capture the α4β7 binding components. For this, cells were incubated with 125I-labeled CHO cell proteins and washed, and then the cell-bound material was dissociated with buffer containing EDTA and CWLDVC peptide. The dissociated material was then separated by SDS-PAGE and visualized by autoradiography. The major component enriched on RPMI8866 cells appeared to be a 260-kDa band (Fig. 9C). Additionally, a band of approximately 55 kDa also appeared to be retained on RPMI8866 cells (Fig. 9C). A separate (nonradioactive) sample was separated by SDS-PAGE, and the 260-kDa and 55-kDa bands were excised, digested, and analyzed by liquid chromatography (LC)-MS. While these experiments were not carried out in a way that allowed quantitative analysis, they did identify components that appeared to be enriched relative to the unfractionated protein mixture. The two major components enriched in the 260-kDa band appeared to be fibronectin (664 peptides) and chondroitin sulfate proteoglycan (867 peptides) (available via ProteomeXchange with identifier PXD006063 [https://www.ebi.ac.uk/pride/archive/projects/PXD006063]). The major components enriched in the 55-kDa band appeared to be tubulointerstitial nephritis antigen-like protein (276 peptides) and calreticulin (186 peptides) (available via ProteomeXchange with identifier PXD006062 [https://www.ebi.ac.uk/pride/archive/projects/PXD006062]). Collectively, these findings were consistent with the identification of fibronectin as the CHO cell protein responsible for α4β7 reactivity.
FIG 9.
Identification of fibronectin as an α4β7 binding component of CHO cell proteins. The CHO cell proteins were labeled with 125I. (A) Comparison of proteins detected by autoradiography to 25 μg of proteins stained with Coomassie blue. Five major bands present in the Coomassie-stained gel were also identified by autoradiography, as indicated. (B) Proteins displayed α4β7-dependent binding to RPMI88666 cells after radiolabeling. (C) RPMI8866 cells were used to capture and identify α4β7 components. 125I-labeled protein was captured on cells in the presence or absence of MAb 2B4, and then the cell-bound material was dissociated and analyzed. Input protein was compared to cell-bound material by SDS-PAGE and autoradiography. Shown are the original autoradiograph and a contrast-enhanced image. The cells retained a major band of approximately 260 kDa and a minor band of 55 kDa.
Heparin sulfate inhibits fibronectin-mediated gp120-α4β7 binding.
Previous reports indicated that gp120 binds to the C-terminal heparin binding domain of fibronectin and that this can be inhibited with heparin sulfate (45). Based on this, we tested the ability of heparin to inhibit gp120-α4β7 interactions mediated by CHO cell fibronectin. Mixtures of 1086.d7 gp120 and CHO cell proteins were incubated for 2 h, and then various concentrations of heparin sulfate were added and binding to RPMI8866 cells was measured. Chondroitin sulfate, a glycosaminoglycan related to heparin, was used as a control in these experiments.
Heparin sulfate was able to inhibit fibronectin-mediated gp120-α4β7 binding in a concentration-dependent manner (Fig. 10). The half-maximal inhibitory concentration (IC50) of heparin was calculated as 0.25 μg/ml (95% confidence interval, 0.11 to 0.59 μg/ml). In contrast, chondroitin sulfate did not have a measurable inhibitory effect at the concentrations tested. These experiments supported the hypothesis that gp120 interacted with the heparin binding domain of CHO cell fibronectin.
FIG 10.
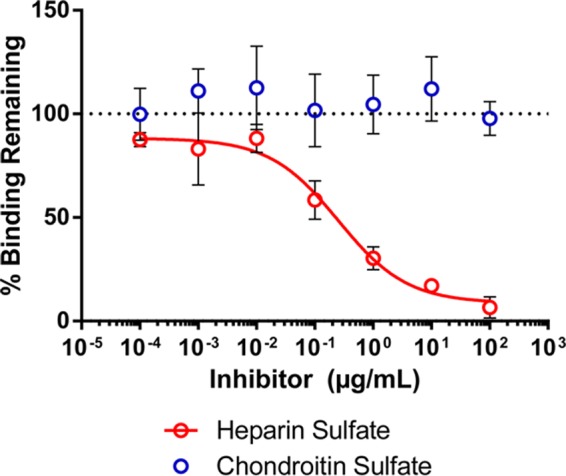
Heparin inhibits fibronectin-mediated gp120-α4β7 binding. 1086.d7 gp120 was incubated for 2 h with CHO cell proteins. Heparin sulfate or chondroitin sulfate was added at the indicated concentrations 10 min prior to incubation with RPMI8866 cells. The results are the means and standard deviations of three replicates; the red line is a best-fit curve of the heparin inhibition data.
A recombinant human fibronectin fragment reproduces CHO cell protein binding behavior.
Last, we asked if purified human fibronectin proteins could reproduce the binding phenomena observed with CHO cell proteins. Three fibronectin proteins were chosen for study. First was human plasma fibronectin, which was not expected to react with α4β7. Second was cellular fibronectins isolated from cultured human foreskin fibroblasts. These fibronectins were expected to be a heterogeneous mixture and potentially to contain α4β7-reactive isoforms. Last was retronectin, a chimeric recombinant human fibronectin fragment produced in Escherichia coli containing the cell binding domain, C-terminal heparin binding domain, and CS1 peptide of human fibronectin (TaKaRa) (46).
These three proteins were biotinylated and tested for α4β7 binding activity. As expected, plasma fibronectin did not bind to RPMI8866 cells. Cellular fibronectin produced a weak α4β7 binding signal, and retronectin produced a robust signal (Fig. 11A). The proteins were then tested for the presence of the CS1 peptide by Western blotting. All the proteins reacted with polyclonal anti-fibronectin antisera, but only retronectin reacted with monoclonal antibody to CS1 (Fig. 11B). Because retronectin bound strongly to α4β7 and was verified to contain CS1, it was used in subsequent assays. In mixing experiments, similar to those performed with CHO cell proteins, cell binding was observed in mixtures of unlabeled retronectin and labeled MW959 gp120 or MW959-ΔV2 gp120 (Fig. 11C). These results confirmed that human fibronectins can mediate indirect gp120-α4β7 interactions similarly to CHO cell proteins.
FIG 11.
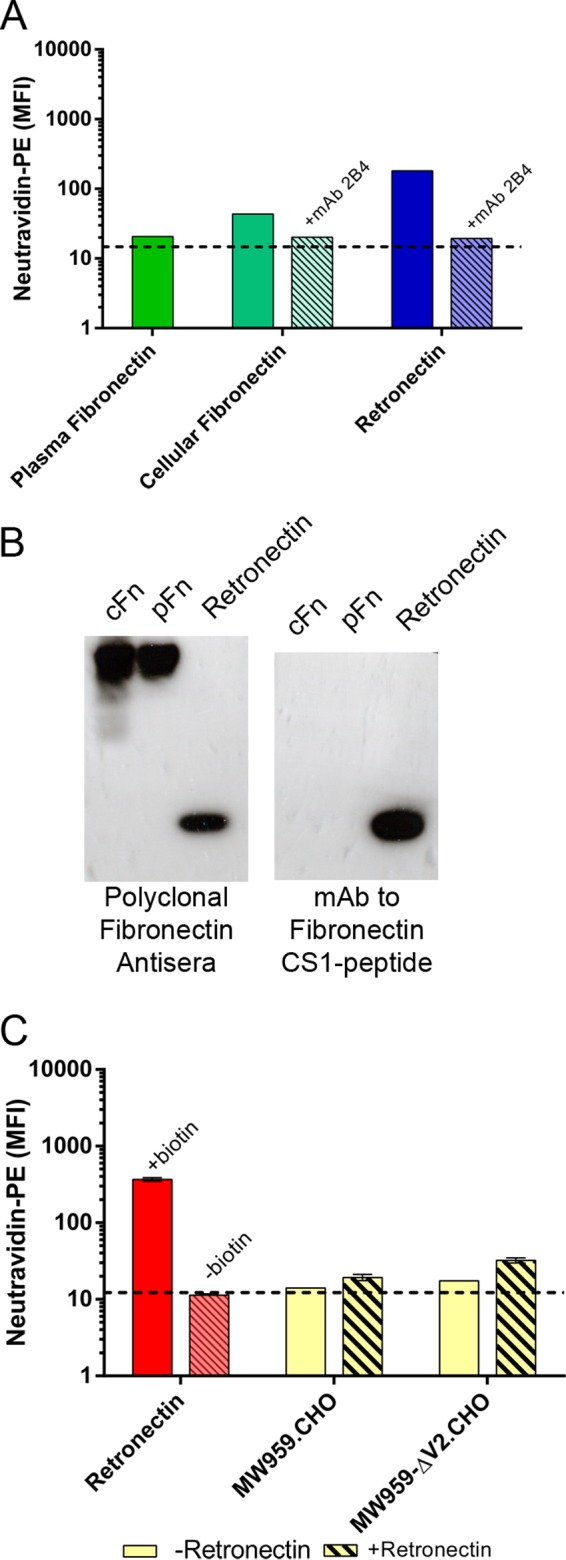
The recombinant human fibronectin fragment retronectin mediates gp120-α4β7 binding. (A) Plasma fibronectin, cellular fibronectin, and retronectin were biotinylated and tested for binding to RPMI8866 cells. The results of a single representative experiment are shown. (B) Western blot analysis of plasma fibronectin (pFn), cellular fibronectin (cFn), and retronectin. Shown are two parallel blots developed with either polyclonal fibronectin antiserum or monoclonal antibody to the fibronectin CS1 peptide; 1 μg of protein was loaded per lane. (C) Retronectin mediates gp120 binding to RPMI8866 cells. Biotinylated retronectin (+biotin) produces a binding signal, and nonbiotinylated retronectin (−biotin) does not. Binding of wild-type MW959 and MW959 with the V2 loop deleted was measured with (hatched bars) and without (solid bars) 2-h preincubation with retronectin. The results are the means and standard deviations of 3 replicate measures.
DISCUSSION
In this work, we provide evidence that extracellular matrix proteins, including fibronectin, can mediate indirect interactions between gp120 and α4β7. We initially observed that CHO cellular fibronectins, which copurified with gp120, could facilitate binding of gp120 to α4β7. We then extended this observation and verified that a recombinant human fibronectin fragment could also mediate gp120-α4β7 binding. These observations help reconcile conflicting reports concerning gp120-α4β7 interactions (14, 27) and at the same time raise the interesting possibility that the extracellular matrix may play a role in mediating HIV-1–α4β7 contact in vivo. This may have important implications with respect to α4β7 as a potential target for antiretroviral therapy and vaccines.
It has been reported that the LDV/I amino acid sequence within the V2 loop facilitates gp120-α4β7 interactions (14, 18, 19, 28). LDV/I is a well-conserved feature of V2 and has been hypothesized to be a structural mimic of the integrin binding motifs in MAdCAM-1 (LDT), VCAM-1 (IDS), and fibronectin (LDV) (14). In MAdCAM-1 and VCAM-1, these sites coordinate magnesium cations that bind to the α4 integrin metal ion-dependent adhesion site, while adjacent amino acid residues provide specificity that is critical for receptor binding (47, 48). Mimicry of integrin binding motifs has been reported in several distantly related virus families and may be a common strategy used to control host tropism (49); however, it is not known whether viruses use these motifs as integrin contact sites or as sites that accommodate divalent cations. Within HIV-1 envelopes, LDV/I has been proposed to coordinate divalent cations. However, similar to α4β7 binding with its native ligands, structures adjacent to the LDV/I site are likely required for receptor binding. In support of this, it has recently been reported that structures adjacent to LDV/I within short and cyclic V2 peptides promote binding to α4β7 (18, 19). Additionally, the conformation of the V2 loop may impact α4β7 reactivity. Our group (K. K. Lee and S.-L. Hu) has previously demonstrated that the V2 loop is highly dynamic and flexible (50). Cocrystallization studies with V2 MAbs have revealed both helical and β-strand conformations (51). These findings underscore the structural heterogeneity of the V2 loop. Presently, it is not known which conformations of V2 may mediate α4β7 binding. Among the recombinant envelope proteins employed in this study, all contained an LDV/I site. Additionally, these envelope proteins presented a diverse set of V2 conformations, as defined by their reactivity with a panel of conformation-dependent V2 antibodies. Despite this, we did not detect α4β7 reactivity in any of these proteins. This implies that LDV/I and other V2 structures within these envelope proteins were not presented in a way that was accessible to α4β7. Further studies are required to clarify if any specific V2 conformations may be required for α4β7 binding. Ideally, such studies would compare the α4β7 binding properties of DEAE-purified gp120 and V2 peptides matched to those proteins.
Vaccine-elicited V2-specific antibodies have been correlated with reduced risk of HIV-1 acquisition in the RV144 trial, but the underlying mechanism for this observation remains unclear (52). One possibility is that V2-directed antibodies may disrupt HIV-1 engagement with α4β7 and thus contribute to protection. In support of this, a small number of studies show gp120-α4β7 binding in vitro can be inhibited by V2-specific antibodies. Specifically, it has been reported that MAb 697D and MAb 2158 both inhibit binding of a clade A gp120 (18) and several mouse V2 MAbs inhibit binding of MN gp120 (15). In contrast to these findings, we failed to detect any inhibitory effect of V2 MAbs on fibronectin-mediated gp120 binding to α4β7. Thus, it remains unclear whether any vaccine-induced V2-specific antibody may offer protection through inhibiting HIV-1–α4β7 interaction. In this context, we note that an alternative role for V2-directed antibodies mediating antibody-dependent cellular cytotoxicity has also been proposed as a mechanism for protection in RV144 (53).
In this report, we identified an indirect interaction between gp120 and α4β7 that is mediated by the extracellular matrix. Specifically, we identified fibronectin as a protein that could mediate this interaction. Fibronectins are large glycoproteins of the extracellular matrix with functional roles in regulating cell migration, embryonic development, and wound healing (54). They have also been shown to interact with gp120 and enhance the infectivity of HIV-1 (55). Twenty fibronectin splice variants have been described in humans, and those containing the alternatively spliced IIICS support α4β7 binding through an LDV site (43, 44). In contrast, all fibronectin isoforms bind gp120 through the invariant C-terminal heparin binding domain (45). In our work, we observed that CHO cells produce a fibronectin variant containing the CS1 peptide within IIICS that supports α4β7 binding. CHO cell proteins, including fibronectin and the recombinant human fibronectin fragment retronectin, mediated indirect gp120-α4β7 binding. Based on these observations, we speculate that α4β7-reactive matrix fibronectins may colocalize with HIV-1 and α4β7+ cells in vivo and thus lead to the preferential infection of these cells. However, because our work is based on in vitro gp120 protein interactions, further studies are required to determine whether fibronectin plays any role in HIV-1 infection of α4β7+ cells.
Prior studies have demonstrated that fibronectin-gp120 contact occurs through the heparin binding domain of fibronectin (45). In agreement with this, we showed that fibronectin-mediated gp120-α4β7 binding in our assays could be inhibited by heparin sulfate. However, it is less clear which structures within gp120 are involved in this binding. The V3 loop and CD4 binding site have both been implicated in gp120-fibronectin interactions; however, neither has been definitively demonstrated to be sufficient or necessary for binding (45, 56). It is possible that specific structures within gp120 may engage fibronectin. Alternatively, the interaction may be less specific, involving interactions with glycans or charged amino acid residues. In studies reported here, we found the V2 loop is not likely to be involved in gp120-fibronectin interactions. However, more studies are needed to determine the nature of gp120-fibronectin interactions and whether these interactions may be inhibited by anti-HIV-1 antibodies.
At present, α4β7 antagonists are being investigated for use in HIV-1 treatment. In particular, the α4β7-blocking monoclonal antibody Act-1 has been extensively studied. A primatized version of Act-1 has been developed, and treatment with this antibody resulted in lower viral loads and higher CD4+ cell numbers in SIV-infected macaques (23–25). Additionally, treatment prior to vaginal challenge reduced the efficiency of SIV transmission (25). More recently, a combination regimen of antiretroviral drugs and Act-1 was shown to reconstitute gut CD4+ cells and induce a state of drug-free virological control in SIV-infected macaques (57). Currently, early-phase clinical trials are under way to test whether the humanized version of Act-1 (vedolizumab) can achieve similar results in humans (http://www.clinicaltrials.gov; identifier NCT02788175). Our studies raise an interesting scenario in which indirect HIV-1–α4β7 interactions, mediated by fibronectin, may be inhibited by Act-1. Further studies are needed to explore this as a possible mechanism of action for any therapeutic effect against HIV-1 attributable to Act-1.
In summary, our work demonstrates that an indirect gp120-α4β7 interaction can be mediated by extracellular matrix proteins, such as fibronectin. Our findings further suggest that studies of the in vivo effects of α4β7 antagonists may need to take into consideration the role of extracellular matrix proteins, such as fibronectin, in mediating indirect interactions between HIV-1 and α4β7. Further studies are needed to clarify whether and how the extracellular matrix in relevant species and tissues may play a role in HIV-1 infection of gut-homing cells.
MATERIALS AND METHODS
Reagents and cell lines.
The following fluorophores were used in flow cytometry analysis: allophycocyanin (APC), phycoerythrin (PE), and fluorescein isothiocyanate (FITC). Fluorescent antibodies to CD4 (clone RPA-T4; APC), CCR5 (clone 2D7; APC), and β7 integrin (clone FIB504; PE) were from Becton Dickinson. Fluorescent antibody to CD45RO (clone UCHL1; FITC) was from Biolegend. Neutravidin conjugated to PE was from ThermoFisher Scientific. The CD4-blocking antibody leu3a (clone SK3) was from Becton Dickinson. Integrin-blocking antibodies 2B4 and P5D2 were from R&D Systems. Human V2 monoclonal antibody 2158 was a gift from Susan Zolla-Pazner. Human V2 monoclonal antibodies PG9 and PG16 were from the International AIDS Vaccine Initiative. FI6v3 influenza virus HA monoclonal antibody was produced in house from a stable HEK293F cell line and purified from culture supernatants by protein A affinity chromatography. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS (DAIDS), NIAID, NIH: CD4-IgG2 fusion protein from Progenics Pharmaceuticals, Human V2 monoclonal antibody CH58 from Barton F. Haynes and Hua-Xin Liao, and anti-human α4β7 integrin antibody Act-1 from A. A. Ansari. The CWLDVC and CLDVWC peptides were purchased from GenScript. These peptides were cyclized by disulfide bonds, and purity was 98% or greater as assessed by high-pressure liquid chromatography and mass spectrometry. The MAdCAM-1–Fc fusion protein was from R&D Systems. Biotinylated HRP was from Pierce. Aldolase, ovalbumin, and ferritin were from GE Life Sciences. Bovine serum albumin (BSA), plasma fibronectin, cellular fibronectin, heparin sulfate derived from porcine intestinal mucosa (189 U/mg), and chondroitin sulfate B derived from porcine intestinal mucosa were from Sigma-Aldrich. Retronectin was from TaKaRa. RPMI8866 cells were from Sigma-Aldrich and were maintained at 0.2 × 106 to 1.0 × 106 cells/ml at 37°C and 5% CO2 in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Freestyle CHO-S and Freestyle 293-F cells used to produce recombinant proteins were purchased from ThermoFisher Scientific and cultured in serum-free and antibiotic-free media according to the manufacturer's instructions in spinner cultures maintained at 37°C and 8% CO2.
Human CD4+ α4β7+ cells.
Used leukocyte filters from healthy donors were purchased from Bloodworks Northwest (Seattle, WA) and used to isolate human PBMCs. Cells were recovered from the filters by back flushing with 200 ml of phosphate-buffered saline (PBS) containing 0.2 g/liter EDTA and 10,000 U/liter heparin. PBMCs were separated by Ficoll Paque, and magnetic cell separation was used to enrich CD4+ cells by negative selection (Miltenyi Biotec). The cells were activated for 2 days with 250 ng/ml OKT3 antibody (BioLegend) and cultured continuously with 20 U/ml interleukin-2 (Roche) and 10 nM all-transretinoic acid (Sigma-Aldrich). The culture medium was RPMI 1640 supplemented with fetal bovine serum, glutamine, and antibiotics as described above. The cells were used for binding studies 9 days after establishing cultures.
Source of HIV-1 gp120 expression plasmids and proteins.
An expression plasmid for MW959 gp120 was produced by Genewiz. The nucleotide sequence for 93MW959.18 gp120 (GenBank accession no. U08453.1) was codon optimized for expression in CHO cells, synthesized de novo, and inserted by topoisomerase cloning into the pcDNA3.1/V5-His TOPO vector (Invitrogen). The first 30 amino acids of the native 93MW959.18 sequence were replaced with the human tissue plasminogen activator leader peptide (amino acid sequence MDAMKRGLCCVLLLCGAVFVSAS). This plasmid was used to generate the MW959-ΔV2 gp120 mutant by site-directed mutagenesis (QuikChange Lightning kit; Agilent) using a primer with the following sequence: ATGAAGAACTGCTCCTTCAACGAGAAGGACAACAGC. Expression plasmids for BG505.T332N.664 SOSIP and furin were gifts from John Moore (34). The expression plasmid for BG505 gp120 was generated from the BG505.T332N.664 SOSIP plasmid by introducing a stop codon. The expression plasmid for AN1 gp120 was from James Arthos, as previously described (14).
The following proteins were prepared in the laboratory of Shiu-Lok Hu at the University of Washington: MW959 produced in CHO and 293 cells, MW959-ΔV2 produced in CHO cells, BG505 SOSIP produced in 293 cells, BG505 gp120 produced in 293 cells, and AN1 gp120 produced in 293 cells. The following proteins were produced at the Duke Human Vaccine Institute, Duke University Medical Center, and provided as a gift by David Montefiori: SF162.d7 gp120 produced in CHO, 293, and 293S GnT1−/− cells and 1086.d7 gp120 produced in CHO cells. TV-1 gp120 was produced in CHO cells by Novartis and provided as a gift by David Montefiori. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: BaL gp120 produced in 293 cells from DAIDS, NIAID, and M.CON-S.d11 gp120 produced in 293 cells from the Duke Human Vaccine Institute, Duke University Medical Center. Unless otherwise specified, purification of all these proteins included DEAE chromatography.
Production and purification of recombinant HIV-1 envelopes.
HIV-1 envelope expression plasmids were prepared with an endotoxin-free plasmid megaprep kit (Qiagen). The transfection procedure was the same for CHO-S and 293-F cells; 1.0 × 109 cells were suspended in 50 ml of medium containing 1 mg of plasmid and 2 mg of polyethyleneimine (Polysciences, Inc.). Cultures were incubated for 4 h and then diluted to 1 liter and maintained for 3 days. The culture supernatants were clarified by centrifugation and passed through a 0.2-μm filter, and protease inhibitors (aprotinin, leupeptin, and pepstatin A, all from Sigma-Aldrich) were added. For BG505 SOSIP production, cells were cotransfected with 750 μg BG505 SOSIP expression vector and 250 μg furin expression vector to ensure complete furin-mediated cleavage of the SOSIP trimer. For cell-conditioned media without recombinant protein expression, 1.0 × 109 cells were diluted to 1 liter and maintained for 3 days before harvesting as described above.
Purification was carried out with an ÄKTA 10/100 purifier (GE Life Sciences) maintained at 4°C. A 10-ml GNA column (G. nivalis lectin conjugated to agarose; Vector Laboratories) was prepared and preequilibrated with binding buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.5). Culture supernatant was loaded onto GNA at 1 ml/min. The column was then washed with 10 column volumes (CV) of high-salt wash buffer (500 mM NaCl, 20 mM Tris-HCl, pH 7.5), followed by 10 CV of binding buffer, and bound protein was eluted with binding buffer containing 1 M methyl-α-d-mannopyranoside (Sigma-Aldrich). Peak fractions were pooled and exchanged into DEAE binding buffer (100 mM NaCl, 20 mM Tris-HCl, pH 8.0) by diafiltration with Centricon centrifugal concentrators (Millipore) using 1,500 or greater diavolumes. The sample was then loaded at 1 ml/min onto a prepacked 5-ml DEAE Sepharose column (GE Life Sciences). Column flowthrough was collected, and the column was washed with 2 CV DEAE binding buffer. Material bound to DEAE was eluted by applying a gradient of 0 to 100% DEAE elution buffer (1 M NaCl, 20 mM Tris-HCl, pH 8.0) over 5 CV. Samples were concentrated with Amicon centrifugal concentrators (Millipore) and then purified by SEC on a 320-ml HighLoad 26/600 Superdex 200 column (GE Life Sciences). SEC was carried out at 1.2 ml/min with PBS (10 mM sodium phosphate, 150 mM NaCl, pH 7.4). Peak fractions were pooled and concentrated with Amicon centrifugal concentrators, and protein concentrations were determined by a bicinchoninic acid assay (Pierce).
Polyacrylamide gel electrophoresis and Western blot analysis.
Reagents used for protein gels were obtained from ThermoFisher Scientific. For SDS-PAGE, protein samples were reduced and denatured in NuPage lithium dodecyl sulfate buffer containing dithiothreitol (DTT) at 70°C for 10 min. Proteins were then separated by electrophoresis on a 4 to 12% Bis-Tris gel using Novex Sharp prestained protein standards as molecular weight markers. SimplyBlue Coomassie G-250 stain was used to visualize proteins according to the manufacturer's instructions. For Native PAGE, nondenatured proteins were loaded on a 3 to 12% Bis-Tris native gel using NativeMark unstained protein standards as molecular weight markers. The gel was run and stained according to the manufacturer's instructions.
For Western blotting, proteins separated by SDS-PAGE were transferred to polyvinylidene difluoride (PVDF) membranes with an iBlot device (Invitrogen). The membranes were incubated for 1 h in blocking buffer (PBS containing 5% nonfat milk and 0.05% Tween 20) and then incubated overnight at 4°C with primary antibodies diluted in blocking buffer. For gp120 detection, pooled plasma from 131 Zambian women infected with clade C HIV-1 (a gift from Charles Wood) was diluted 1:2,000. For fibronectin detection, rabbit polyclonal anti-fibronectin (Abcam) was diluted 1:1,000. For fibronectin CS1 detection, mouse monoclonal IgM against human fibronectin CS1 peptide (Santa Cruz Biotechnology; clone P1F11) was diluted 1:200. The membranes were washed three times in PBS containing 0.05% Tween 20 and then incubated for 1 h with secondary antibodies diluted in blocking buffer. For gp120 detection, alkaline phosphatase-conjugated goat anti-human IgG (Sigma-Aldrich) was diluted 1:10,000. For fibronectin detection, HRP-conjugated goat anti-rabbit IgG (Calbiochem) was diluted 1:10,000. For fibronectin CS1 detection, HRP-conjugated goat anti-mouse IgM (Santa Cruz Biotechnology) was diluted 1:2,000. The membranes were washed extensively with PBS containing 0.05% Tween 20 and then incubated with BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium (NBT) substrate (Sigma-Aldrich) for alkaline phosphatase detection or chemiluminescence substrate (Pierce) for HRP detection. HRP signals were captured on film (Amersham Hyperfilm ECL; GE Life Sciences).
Protein biotinylation.
Proteins were biotinylated with EZ-Link N-hydroxysuccinimide (NHS) biotin reagent (Pierce). The proteins were combined with NHS biotin in a 1:20 molar ratio and incubated for 30 min at room temperature. Samples from cell-conditioned media were labeled with the same biotin/protein ratio as was used for gp120 labeling. Reactions were quenched by adding Tris-HCl, pH 8.0, and then samples were dialyzed overnight against HEPES-buffered saline (150 mM NaCl, 10 mM HEPES, pH 7.4) at 4°C, followed by an additional 3 h in fresh buffer. The concentration of biotinylated proteins was measured by a bicinchoninic acid assay (Pierce) or by absorbance at 280 nm on a NanoDrop 1000 spectrophotometer (ThermoFisher Scientific). Proteins were put into single-use aliquots and stored at −80°C until they were used in cell binding assays.
Flow cytometry-based α4β7 binding assay.
Binding assays were performed as previously described (14). All binding and washing steps were carried out at 4°C in either manganese-containing buffer (150 mM NaCl, 10 mM HEPES, pH 7.4, 1 mM MnCl2, 100 μM CaCl2, 0.5% BSA, 0.09% sodium azide) or EDTA-containing buffer without divalent cations (150 mM NaCl, 10 mM HEPES, pH 7.4, 5 mM EDTA, 0.5% BSA, 0.09% sodium azide). The cells were either RPMI8866, which expresses α4β7 but not CD4, or primary CD4+ α4β7+ T cells. The cells were washed with binding buffer and plated on 96-well plates at 1.0 × 106 cells/ml, 200 μl per well. Fc receptors were blocked with 15 μg/ml normal mouse IgG and 5 μg/ml normal human IgG. Some wells received 2 μg of the following: integrin-blocking antibody 2B4, P5D2, or Act-1 or CD4-blocking antibody leu3a. In wells in which multiple antibodies were combined, 2 μg of each antibody was used. Other wells received 5 μg of the integrin-blocking peptide CWLDVC or the scrambled control CDLVWC. Cells were incubated for 10 min with the inhibitors, and then biotinylated proteins were added and the cells were incubated for an additional 20 min at 4°C; 4 μg of biotinylated protein was added to each well, except for MAdCAM-1–Fc, which was added at 100 ng per well. For mixing experiments, 4 μg of unlabeled proteins was combined with 4 μg of biotinylated proteins at 4°C for 0 or 2 h prior to adding to cells. For experiments with V2 monoclonal antibodies, 4 μg of biotinylated gp120 was combined with 2 μg of antibody and incubated for 1 h at room temperature, and then, 4 μg of unlabeled proteins (or a matched volume of buffer without protein) was added, and the mixtures were incubated at 4°C for an additional 2 h prior to adding them to cells. For heparin sulfate and chondroitin sulfate competition experiments, 4 μg of biotinylated gp120 was incubated for 2 h with 4 μg of unlabeled proteins, followed by 10 min incubation with 100 μg/ml to 100 pg/ml inhibitor prior to adding it to cells. Following protein binding, the cells were washed twice and stained for 20 min at 4°C in binding buffer containing fluorescent probes. RPMI8866 cells were stained with 2.5 μg/ml neutravidin-PE, and primary cells were stained with a mixture of neutravidin-PE and 1:100 dilutions of CD45RO-FITC and CCR5-APC antibodies. Aliquots of cells without biotinylated proteins were carried through the preceding wash/bind steps and stained with 1:100 dilutions of CD4-APC and integrin β7-PE antibodies. The cells were washed twice and then fixed in PBS containing 1% formaldehyde prior to analysis. Data were acquired on a BD FACSCalibur instrument with CellQuest Pro software (Becton Dickinson; version 6.0). A total of 20,000 events were recorded for each sample. The data were analyzed with FlowJo software (Treestar; version 7.2.2). The neutravidin-PE binding signal was reported in units of median fluorescence intensity (MFI).
V2 antibody binding by biolayer interferometry.
An Octet RED96 instrument (Fortebio) was used to perform biolayer interferometry. The assay buffer was PBS with 1% BSA, 0.03% Tween 20, and 0.02% sodium azide. Antibodies were diluted in assay buffer to 10 μg/ml, and serial dilutions of envelope proteins were prepared in assay buffer (1,000 nM, 500 nM, 250 nM, 125 nM, and 62.5 nM). The regeneration buffer was 0.1 M glycine, pH 1.5; 200 μl of each solution per well was used in a low-binding 96-well plate. All assay steps were carried out at 32°C with constant mixing at 1,000 rpm. Anti-human IgG Fc biosensors (Fortebio) were preequilibrated in assay buffer and then transferred to antibody-containing wells for 300 s. After antibody capture, the sensors were washed in assay buffer, and baseline signals were recorded for 60 s. Antibody-coated tips were transferred to wells containing envelope proteins, and association curves were recorded for 180 s. The tips were then transferred to wells containing assay buffer, and dissociation curves were recorded for 300 s. The tips were regenerated by cycling 3 times between regeneration and assay buffers for 5 s in each buffer, and the tips were reused up to 3 times. A baseline signal was measured in every assay from an antibody-coated tip carried through association and dissociation steps in wells containing assay buffer without envelope proteins. Data were analyzed with ForteBio data analysis software (version 7.1). Data processing was as follows: baseline signals were subtracted, baselines were aligned to the y axis, and interstep correction and Savitzky-Golay filtering were applied. Response curves were fitted to processed data with a 1:1 global fit model and were used to compute association (Ka), dissociation (Kd), and binding affinity (KD) constants.
Mass spectrometry protein identification.
For in-solution digestion, 1-μg samples of proteins were vacuum dried in siliconized tubes with a Savant SpeedVac concentrator (ThermoFisher). The proteins were reconstituted in denaturation buffer (7 M urea, 100 mM ammonium bicarbonate, 5 mM DTT) and incubated at 56°C for 45 min. Cysteines were alkylated with 14 mM iodoacetamide for 30 min at room temperature. The iodoacetamide was quenched with an additional 5 mM DTT, and the solution was diluted 1:4 with 100 mM ammonium bicarbonate. Calcium chloride and sequencing-grade trypsin (Promega) were added to 1 mM and 4 ng/μl, respectively, and samples were incubated overnight at 37°C. Trypsin digestion was stopped with 0.4% (vol/vol) trifluoroacetic acid. Samples were centrifuged to remove precipitates, and acidified supernatants were vacuum concentrated to 20 μl. Tryptic peptides were desalted with C18 ZipTips (Millipore) according to the manufacturer's instructions.
For in-gel digestion, proteins were separated by SDS-PAGE and visualized with Coomassie blue as described above. Individual protein bands were excised and dehydrated by 3 washes with acetonitrile. Gel pieces were rehydrated in buffer containing 25 mM ammonium bicarbonate and 5 mM DTT and incubated for 45 min at 56°C. Cysteines were alkylated in 25 mM ammonium bicarbonate and 80 mM iodoacetamide for 30 min. Gel slices were rinsed with water, dehydrated in acetonitrile, and then rehydrated in 50 mM ammonium bicarbonate containing 12.5 ng/μl trypsin and incubated overnight at 37°C. Tryptic peptides were extracted by one wash with water and three washes with 50% acetonitrile containing 5% formic acid and were vacuum concentrated to 10 μl.
Samples were analyzed on an Orbitrap Fusion Lumos mass spectrometer (ThermoFisher Scientific) equipped with a nano-Acquity ultraperformance liquid chromatography (UPLC) system (Waters) and an in-house-developed nanospray ionization source. Thirty-centimeter columns were prepared in house from fused silica capillary tubing (75-μm inner diameter and 360-μm outer diameter; Kinesis Scientific) with laser-pulled tips and packed with 5-μm C18 resin with a 120-Å pore size (Reprosil-Pur C18-AQ; ESI Source Solutions). The mobile phases were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). Five microliters of sample was loaded onto the column from the autosampler and desalted for 40 min with 2% mobile phase B at 300 nl/min. Peptides were separated using a linear gradient from 5 to 30% mobile phase B over 80 min and a flow rate of 300 nl/min. The column was then washed with 80% mobile phase B for 10 min, followed by equilibration with 2% mobile phase B for 20 min prior to subsequent injections. Eluted peptides were detected using a data-dependent acquisition method. Survey scans of peptide precursors were performed in the Orbitrap mass analyzer from 375 to 1,575 m/z at 120k resolution (at 200 m/z) with a 7 × 105 ion count target and a maximum injection time of 50 ms. After survey scans, tandem MS (MS-MS) was performed on the most abundant precursors exhibiting a charge state of 2 to 4 and greater than 1 × 104 intensity by isolating them in the quadrupole with an isolation width of 1.6 m/z. High-energy collisional dissociation fragmentation was applied with normalized collision energy of 30%, and the resulting fragments were detected using the rapid scan rate in the ion trap. The ion count target for MS-MS was set to 1 × 104, and the maximum injection time was limited to 100 ms. Dynamic exclusion was set to 30 s with 10 ppm mass tolerance around the precursor and its isotopes.
Thermo .raw files were converted to the mzXML format using the ReAdW (version 2016.1.0) converter. The mzXML files were searched against a Cricetulus griseus protein sequence database downloaded from UniProt appended with common contaminant sequences (24,000 total sequence entries). A database search was performed using Comet (version 2016.01 revision 2) with the following search parameters: 20 ppm precursor tolerance, concatenated target-decoy search, tryptic digest allowing 2 missed cleavages, oxidized methionine variable modification, and carboxyamidomethylation static modification on cysteine. The Comet search results were then processed with PeptideProphet and ProteinProphet tools from the Trans-Proteomic Pipeline software suite (version 5.0.0; Typhoon).
Protein radiolabeling, cell binding, and autoradiography studies.
CHO cell proteins captured on DEAE and eluted with 200 mM NaCl were used for radiolabeling. Iodine-125 (44.4 MBq; PerkinElmer; catalog number NEZ033010MC) was diluted in 70 μl of PBS. One iodobead (Pierce) was rinsed with PBS and added to the 125I solution for 5 min at room temperature. Then, 70 μg of protein diluted to 230 μl in PBS was added to the 125I-iodobead solution and incubated for 5 min at room temperature. The reaction was stopped by rapidly transferring protein to a clean tube. Unreacted 125I was removed by passing the sample through a disposable PD-10 desalting column (GE Life Sciences) with PBS and manually collecting 500-μl fractions. Protein-containing fractions were pooled and concentrated to 100 μl with Amicon centrifugal concentrators (Millipore). Aliquots of the sample were counted in a Packard Cobra II gamma counter, and the ratio of radiolabeled protein to unreacted 125I was assessed by instant thin-layer chromatography (iTLC). For this, 1 μl of sample was dried onto an iTLC strip (Agilent), and the strip was developed for 10 min in PBS with 20% methanol and then dried. 125I-labeled protein remained at the bottom of the strip, and unreacted 125I migrated to the top. The strip was cut in half, and the radioactivity in both halves was measured by gamma counting. The total activity of the final product was 46.6 kBq/μl, and 97.9% of it was protein associated. Five microliters of radiolabeled protein was reduced, denatured, and separated by SDS-PAGE as described above, and the gel was exposed for 30 min to film (Amersham Hyperfilm MP; GE Life Sciences) at room temperature for autoradiography.
The procedure for cell binding studies with 125I-labeled protein was similar to the flow cytometry binding assays described above. RPMI8866 cells were washed with manganese-containing binding buffer, put in 200-μl aliquots in 1.5-ml tubes at 1.0 × 106 cells/ml, and incubated with or without 5 μg of CWLDVC peptide. Radiolabeled protein (46 kBq [2.76 × 106 dpm]) was added to each tube and incubated for 20 min at 4°C, and then cells were washed twice and counted in a Packard Cobra II gamma counter. Tubes without cells were carried through the preceding wash/binding steps, and counts from them were subtracted from the cell counts to determine cell-associated radioactivity.
The binding procedure was modified to enrich and identify α4β7-reactive proteins using RPMI8866 cells. The cells were washed in manganese-containing buffer and separated into two 500-μl aliquots at 2.0 × 106 cells/ml. Ten micrograms of 2B4 antibody was added to one tube, and samples were incubated for 10 min; 168 kBq (1.0 × 107 dpm) of radiolabeled protein was added, and the cells were incubated for 30 min at 4°C. The cells were washed 3 times in 1 ml of manganese-containing buffer without added BSA and transferred to a clean tube at each wash. The cells were then incubated for 30 min at 37°C in 300 μl of dissociation buffer (150 mM NaCl, 10 mM HEPES, pH 7.4, 5 mM EDTA, and 250 μg/ml CWLDVC peptide). The dissociation buffer was collected; concentrated to 30 μl with Amicon centrifugal concentrators (Millipore); and then reduced, denatured, and separated by SDS-PAGE as described above. Input sample (140 Bq [8,400 dpm]) was included on the gel to compare the protein compositions of input and cell-dissociated samples. Autoradiography was performed by exposing the gel to film, as described above, with a BioMax HE intensifying screen (Kodak) at −80°C for 11 days.
Statistical analysis.
GraphPad Prism software (version 6.03) was used for all graphing and statistical analyses. Binding of gp120 in the presence or absence of CHO cell proteins was compared using an unpaired t test, with a significance level (α) of 0.05. Nonlinear regression was used to fit heparin inhibition data and to calculate the IC50.
Accession number(s).
Mass spectrometry data sets have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifiers PXD006060 (https://www.ebi.ac.uk/pride/archive/projects/PXD006060; CHO cell peak 3), PXD006061 (https://www.ebi.ac.uk/pride/archive/projects/PXD006061; proteins coeluted with gp120), PXD006062 (https://www.ebi.ac.uk/pride/archive/projects/PXD006062; 55-kDa CHO proteins), and PXD006063 (https://www.ebi.ac.uk/pride/archive/projects/PXD006063; 260-kDa CHO proteins) (58).
ACKNOWLEDGMENTS
We thank Scott Wilbur and Don Hamlin of the Department of Radiation Oncology, University of Washington, for assistance with radiolabeling experiments. We also thank David Montefiori, Susan Zolla-Pazner, and Charles Wood for generously sharing reagents that were used in our experiments. Finally, we thank James Williams for producing and purifying the FI6v3 antibody used in this work.
This work is supported by the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery OPP1033102 (W.G., B.C., M.G., K.K.L., S.L.H.), National Institutes of Health grant P51 OD010425 (S.L.H.), and University of Washington Proteomics Resource grant UWPR95794 (P.V.H., J.K.E.). D.P. was supported by a Pharmaceutical Sciences Training Grant (T32 GM007750) from the National Institute of General Medical Sciences, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Brenchley JM, Douek DC. 2008. HIV infection and the gastrointestinal immune system. Mucosal Immunol 1:23–30. doi: 10.1038/mi.2007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siewe B, Landay A. 2012. Key concepts in the early immunology of HIV-1 infection. Curr Infect Dis Rep 14:102–109. doi: 10.1007/s11908-011-0235-3. [DOI] [PubMed] [Google Scholar]
- 3.Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, Dandekar S. 2003. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol 77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picker LJ. 2006. Immunopathogenesis of acute AIDS virus infection. Curr Opin Immunol 18:399–405. doi: 10.1016/j.coi.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Gorfu G, Rivera-Nieves J, Ley K. 2009. Role of beta7 integrins in intestinal lymphocyte homing and retention. Curr Mol Med 9:836–850. doi: 10.2174/156652409789105525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denucci CC, Mitchell JS, Shimizu Y. 2009. Integrin function in T-cell homing to lymphoid and nonlymphoid sites: getting there and staying there. Crit Rev Immunol 29:87–109. doi: 10.1615/CritRevImmunol.v29.i2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu A, Yang Y, Liu L, Meng Z, Li L, Qiu C, Xu J, Zhang X. 2014. Differential compartmentalization of HIV-targeting immune cells in inner and outer foreskin tissue. PLoS One 9:e85176. doi: 10.1371/journal.pone.0085176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKinnon LR, Nyanga B, Chege D, Izulla P, Kimani M, Huibner S, Gelmon L, Block KE, Cicala C, Anzala AO, Arthos J, Kimani J, Kaul R. 2011. Characterization of a human cervical CD4+ T cell subset coexpressing multiple markers of HIV susceptibility. J Immunol 187:6032–6042. doi: 10.4049/jimmunol.1101836. [DOI] [PubMed] [Google Scholar]
- 9.Cicala C, Martinelli E, McNally JP, Goode DJ, Gopaul R, Hiatt J, Jelicic K, Kottilil S, Macleod K, O'Shea A, Patel N, Van Ryk D, Wei D, Pascuccio M, Yi L, McKinnon L, Izulla P, Kimani J, Kaul R, Fauci AS, Arthos J. 2009. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc Natl Acad Sci U S A 106:20877–20882. doi: 10.1073/pnas.0911796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krzysiek R, Rudent A, Bouchet-Delbos L, Foussat A, Boutillon C, Portier A, Ingrand D, Sereni D, Galanaud P, Grangeot-Keros L, Emilie D. 2001. Preferential and persistent depletion of CCR5+ T-helper lymphocytes with nonlymphoid homing potential despite early treatment of primary HIV infection. Blood 98:3169–3171. doi: 10.1182/blood.V98.10.3169. [DOI] [PubMed] [Google Scholar]
- 11.Byrareddy SN, Sidell N, Arthos J, Cicala C, Zhao C, Little DM, Dunbar P, Yang GX, Pierzchalski K, Kane MA, Mayne AE, Song B, Soares MA, Villinger F, Fauci AS, Ansari AA. 2015. Species-specific differences in the expression and regulation of alpha4beta7 integrin in various nonhuman primates. J Immunol 194:5968–5979. doi: 10.4049/jimmunol.1402866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girard A, Vergnon D, Depince-Berger AE, Roblin X, Lutch F, Lambert C, Rochereau N, Bourlet T, Genin C, Paul S. 2016. A high rate of beta7+ gut homing lymphocytes in HIV infected immunogical non responders is associated with poor CD4 T cell recovery during suppressive HAART. J Acquir Immune Defic Syndr doi: 10.1097/qai.0000000000000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farstad IN, Halstensen TS, Lien B, Kilshaw PJ, Lazarovits AI, Brandtzaeg P. 1996. Distribution of beta 7 integrins in human intestinal mucosa and organized gut-associated lymphoid tissue. Immunology 89:227–237. doi: 10.1046/j.1365-2567.1996.d01-727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. 2008. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol 9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura GR, Fonseca DP, O'Rourke SM, Vollrath AL, Berman PW. 2012. Monoclonal antibodies to the V2 domain of MN-rgp120: fine mapping of epitopes and inhibition of alpha4beta7 binding. PLoS One 7:e39045. doi: 10.1371/journal.pone.0039045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Pauza CD. 2011. HIV envelope-mediated, CCR5/alpha4beta7-dependent killing of CD4-negative gammadelta T cells which are lost during progression to AIDS. Blood 118:5824–5831. doi: 10.1182/blood-2011-05-356535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Pauza CD. 2012. The alpha4beta7 integrin binds HIV envelope but does not mediate bystander killing of gammadelta T cells. Blood 120:698–699. doi: 10.1182/blood-2012-03-420117. [DOI] [PubMed] [Google Scholar]
- 18.Tassaneetrithep B, Tivon D, Swetnam J, Karasavvas N, Michael NL, Kim JH, Marovich M, Cardozo T. 2014. Cryptic determinant of alpha4beta7 binding in the V2 loop of HIV-1 gp120. PLoS One 9:e108446. doi: 10.1371/journal.pone.0108446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peachman KK, Karasavvas N, Chenine AL, McLinden R, Rerks-Ngarm S, Jaranit K, Nitayaphan S, Pitisuttithum P, Tovanabutra S, Zolla-Pazner S, Michael NL, Kim JH, Alving CR, Rao M. 2015. Identification of new regions in HIV-1 gp120 variable 2 and 3 loops that bind to alpha4beta7 integrin receptor. PLoS One 10:e0143895. doi: 10.1371/journal.pone.0143895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Jin W, Du T, Wu B, Liu Y, Shattock RJ, Hu Q. 2014. Binding of HIV-1 virions to alpha4beta 7 expressing cells and impact of antagonizing alpha4beta 7 on HIV-1 infection of primary CD4+ T cells. Virol Sin 29:381–392. doi: 10.1007/s12250-014-3525-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, Salazar MG, Decker JM, Parrish EH, Berg A, Hopper J, Hora B, Kumar A, Mahlokozera T, Yuan S, Coleman C, Vermeulen M, Ding H, Ochsenbauer C, Tilton JC, Permar SR, Kappes JC, Betts MR, Busch MP, Gao F, Montefiori D, Haynes BF, Shaw GM, Hahn BH, Doms RW. 2012. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Pathog 8:e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauls E, Ballana E, Moncunill G, Bofill M, Clotet B, Ramo-Tello C, Este JA. 2009. Evaluation of the anti-HIV activity of natalizumab, an antibody against integrin alpha4. AIDS 23:266–268. doi: 10.1097/QAD.0b013e328320a7f8. [DOI] [PubMed] [Google Scholar]
- 23.Pereira LE, Onlamoon N, Wang X, Wang R, Li J, Reimann KA, Villinger F, Pattanapanyasat K, Mori K, Ansari AA. 2009. Preliminary in vivo efficacy studies of a recombinant rhesus anti-alpha(4)beta(7) monoclonal antibody. Cell Immunol 259:165–176. doi: 10.1016/j.cellimm.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ansari AA, Reimann KA, Mayne AE, Takahashi Y, Stephenson ST, Wang R, Wang X, Li J, Price AA, Little DM, Zaidi M, Lyles R, Villinger F. 2011. Blocking of alpha4beta7 gut-homing integrin during acute infection leads to decreased plasma and gastrointestinal tissue viral loads in simian immunodeficiency virus-infected rhesus macaques. J Immunol 186:1044–1059. doi: 10.4049/jimmunol.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrareddy SN, Kallam B, Arthos J, Cicala C, Nawaz F, Hiatt J, Kersh EN, McNicholl JM, Hanson D, Reimann KA, Brameier M, Walter L, Rogers K, Mayne AE, Dunbar P, Villinger T, Little D, Parslow TG, Santangelo PJ, Villinger F, Fauci AS, Ansari AA. 2014. Targeting alpha4beta7 integrin reduces mucosal transmission of simian immunodeficiency virus and protects gut-associated lymphoid tissue from infection. Nat Med 20:1397–1400. doi: 10.1038/nm.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arrode-Bruses G, Goode D, Kleinbeck K, Wilk J, Frank I, Byrareddy S, Arthos J, Grasperge B, Blanchard J, Zydowsky T, Gettie A, Martinelli E. 2016. A small molecule, which competes with MAdCAM-1, activates integrin alpha4beta7 and fails to prevent mucosal transmission of SHIV-SF162P3. PLoS Pathog 12:e1005720. doi: 10.1371/journal.ppat.1005720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez LG, Chen H, Liao HX, Montefiori DC. 2014. Envelope glycoprotein binding to the integrin alpha4beta7 is not a general property of most HIV-1 strains. J Virol 88:10767–10777. doi: 10.1128/JVI.03296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nawaz F, Cicala C, Van Ryk D, Block KE, Jelicic K, McNally JP, Ogundare O, Pascuccio M, Patel N, Wei D, Fauci AS, Arthos J. 2011. The genotype of early-transmitting HIV gp120s promotes alpha (4) beta(7)-reactivity, revealing alpha (4) beta(7) +/CD4+ T cells as key targets in mucosal transmission. PLoS Pathog 7:e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanderslice P, Ren K, Revelle JK, Kim DC, Scott D, Bjercke RJ, Yeh ET, Beck PJ, Kogan TP. 1997. A cyclic hexapeptide is a potent antagonist of alpha 4 integrins. J Immunol 158:1710–1718. [PubMed] [Google Scholar]
- 30.Lazarovits AI, Moscicki RA, Kurnick JT, Camerini D, Bhan AK, Baird LG, Erikson M, Colvin RB. 1984. Lymphocyte activation antigens. I. A monoclonal antibody, anti-Act I, defines a new late lymphocyte activation antigen. J Immunol 133:1857–1862. [PubMed] [Google Scholar]
- 31.Schweighoffer T, Tanaka Y, Tidswell M, Erle DJ, Horgan KJ, Luce GE, Lazarovits AI, Buck D, Shaw S. 1993. Selective expression of integrin alpha 4 beta 7 on a subset of human CD4+ memory T cells with hallmarks of gut-trophism. J Immunol 151:717–729. [PubMed] [Google Scholar]
- 32.Doria-Rose NA, Learn GH, Rodrigo AG, Nickle DC, Li F, Mahalanabis M, Hensel MT, McLaughlin S, Edmonson PF, Montefiori D, Barnett SW, Haigwood NL, Mullins JI. 2005. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J Virol 79:11214–11224. doi: 10.1128/JVI.79.17.11214-11224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho Z, Ma BJ, Li Y, Decker JM, Nabel GJ, Montefiori DC, Hahn BH, Korber BT, Gao F, Haynes BF. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alam SM, Liao HX, Tomaras GD, Bonsignori M, Tsao CY, Hwang KK, Chen H, Lloyd KE, Bowman C, Sutherland L, Jeffries TL Jr, Kozink DM, Stewart S, Anasti K, Jaeger FH, Parks R, Yates NL, Overman RG, Sinangil F, Berman PW, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Karasavva N, Rerks-Ngarm S, Kim JH, Michael NL, Zolla-Pazner S, Santra S, Letvin NL, Harrison SC, Haynes BF. 2013. Antigenicity and immunogenicity of RV144 vaccine AIDSVAX clade E envelope immunogen is enhanced by a gp120 N-terminal deletion. J Virol 87:1554–1568. doi: 10.1128/JVI.00718-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayr LM, Cohen S, Spurrier B, Kong XP, Zolla-Pazner S. 2013. Epitope mapping of conformational V2-specific anti-HIV human monoclonal antibodies reveals an immunodominant site in V2. PLoS One 8:e70859. doi: 10.1371/journal.pone.0070859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang ZY, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. 2013. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spurrier B, Sampson J, Gorny MK, Zolla-Pazner S, Kong XP. 2014. Functional implications of the binding mode of a human conformation-dependent V2 monoclonal antibody against HIV. J Virol 88:4100–4112. doi: 10.1128/JVI.03153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laursen NS, Wilson IA. 2013. Broadly neutralizing antibodies against influenza viruses. Antiviral Res 98:476–483. doi: 10.1016/j.antiviral.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffenberg S, Powell R, Carpov A, Wagner D, Wilson A, Kosakovsky Pond S, Lindsay R, Arendt H, Destefano J, Phogat S, Poignard P, Fling SP, Simek M, Labranche C, Montefiori D, Wrin T, Phung P, Burton D, Koff W, King CR, Parks CL, Caulfield MJ. 2013. Identification of an HIV-1 clade A envelope that exhibits broad antigenicity and neutralization sensitivity and elicits antibodies targeting three distinct epitopes. J Virol 87:5372–5383. doi: 10.1128/JVI.02827-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruegg C, Postigo AA, Sikorski EE, Butcher EC, Pytela R, Erle DJ. 1992. Role of integrin alpha 4 beta 7/alpha 4 beta P in lymphocyte adherence to fibronectin and VCAM-1 and in homotypic cell clustering. J Cell Biol 117:179–189. doi: 10.1083/jcb.117.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gutman A, Kornblihtt AR. 1987. Identification of a third region of cell-specific alternative splicing in human fibronectin mRNA. Proc Natl Acad Sci U S A 84:7179–7182. doi: 10.1073/pnas.84.20.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mould AP, Wheldon LA, Komoriya A, Wayner EA, Yamada KM, Humphries MJ. 1990. Affinity chromatographic isolation of the melanoma adhesion receptor for the IIICS region of fibronectin and its identification as the integrin alpha 4 beta 1. J Biol Chem 265:4020–4024. [PubMed] [Google Scholar]
- 45.Bozzini S, Falcone V, Conaldi PG, Visai L, Biancone L, Dolei A, Toniolo A, Speziale P. 1998. Heparin-binding domain of human fibronectin binds HIV-1 gp120/160 and reduces virus infectivity. J Med Virol 54:44–53. doi:. [DOI] [PubMed] [Google Scholar]
- 46.Pollok KE, Hanenberg H, Noblitt TW, Schroeder WL, Kato I, Emanuel D, Williams DA. 1998. High-efficiency gene transfer into normal and adenosine deaminase-deficient T lymphocytes is mediated by transduction on recombinant fibronectin fragments. J Virol 72:4882–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green N, Rosebrook J, Cochran N, Tan K, Wang JH, Springer TA, Briskin MJ. 1999. Mutational analysis of MAdCAM-1/alpha4beta7 interactions reveals significant binding determinants in both the first and second immunoglobulin domains. Cell Adhes Commun 7:167–181. doi: 10.3109/15419069909010800. [DOI] [PubMed] [Google Scholar]
- 48.Clements JM, Newham P, Shepherd M, Gilbert R, Dudgeon TJ, Needham LA, Edwards RM, Berry L, Brass A, Humphries MJ. 1994. Identification of a key integrin-binding sequence in VCAM-1 homologous to the LDV active site in fibronectin. J Cell Sci 107:2127–2135. [DOI] [PubMed] [Google Scholar]
- 49.Triantafilou K, Takada Y, Triantafilou M. 2001. Mechanisms of integrin-mediated virus attachment and internalization process. Crit Rev Immunol 21:311–322. doi: 10.1615/CritRevImmunol.v21.i4.10. [DOI] [PubMed] [Google Scholar]
- 50.Liang Y, Guttman M, Davenport TM, Hu SL, Lee KK. 2016. Probing the impact of local structural dynamics of conformational epitopes on antibody recognition. Biochemistry 55:2197–2213. doi: 10.1021/acs.biochem.5b01354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang X, Totrov M, Li W, Sampson JM, Williams C, Lu H, Wu X, Lu S, Wang S, Zolla-Pazner S, Kong XP. 2016. Rationally designed immunogens targeting HIV-1 gp120 V1V2 induce distinct conformation-specific antibody responses in rabbits. J Virol 90:11007–11019. doi: 10.1128/JVI.01409-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonsignori M, Pollara J, Moody MA, Alpert MD, Chen X, Hwang KK, Gilbert PB, Huang Y, Gurley TC, Kozink DM, Marshall DJ, Whitesides JF, Tsao CY, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Kim JH, Michael NL, Tomaras GD, Montefiori DC, Lewis GK, DeVico A, Evans DT, Ferrari G, Liao HX, Haynes BF. 2012. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol 86:11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pankov R, Yamada KM. 2002. Fibronectin at a glance. J Cell Sci 115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 55.Greco G, Pal S, Pasqualini R, Schnapp LM. 2002. Matrix fibronectin increases HIV stability and infectivity. J Immunol 168:5722–5729. doi: 10.4049/jimmunol.168.11.5722. [DOI] [PubMed] [Google Scholar]
- 56.Tellier MC, Greco G, Klotman M, Mosoian A, Cara A, Arap W, Ruoslahti E, Pasqualini R, Schnapp LM. 2000. Superfibronectin, a multimeric form of fibronectin, increases HIV infection of primary CD4+ T lymphocytes. J Immunol 164:3236–3245. doi: 10.4049/jimmunol.164.6.3236. [DOI] [PubMed] [Google Scholar]
- 57.Byrareddy SN, Arthos J, Cicala C, Villinger F, Ortiz KT, Little D, Sidell N, Kane MA, Yu J, Jones JW, Santangelo PJ, Zurla C, McKinnon LR, Arnold KB, Woody CE, Walter L, Roos C, Noll A, Van Ryk D, Jelicic K, Cimbro R, Gumber S, Reid MD, Adsay V, Amancha PK, Mayne AE, Parslow TG, Fauci AS, Ansari AA. 2016. Sustained virologic control in SIV+ macaques after antiretroviral and alpha4beta7 antibody therapy. Science 354:197–202. doi: 10.1126/science.aag1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:11033. doi: 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]