ABSTRACT
During primary HIV infection, the presence of minority drug resistance mutations (DRM) may be a consequence of sexual transmission, de novo mutations, or technical errors in identification. Baseline blood samples were collected from 24 HIV-infected antiretroviral-naive, genetically and epidemiologically linked source and recipient partners shortly after the recipient's estimated date of infection. An additional 32 longitudinal samples were available from 11 recipients. Deep sequencing of HIV reverse transcriptase (RT) was performed (Roche/454), and the sequences were screened for nucleoside and nonnucleoside RT inhibitor DRM. The likelihood of sexual transmission and persistence of DRM was assessed using Bayesian-based statistical modeling. While the majority of DRM (>20%) were consistently transmitted from source to recipient, the probability of detecting a minority DRM in the recipient was not increased when the same minority DRM was detected in the source (Bayes factor [BF] = 6.37). Longitudinal analyses revealed an exponential decay of DRM (BF = 0.05) while genetic diversity increased. Our analysis revealed no substantial evidence for sexual transmission of minority DRM (BF = 0.02). The presence of minority DRM during early infection, followed by a rapid decay, is consistent with the “mutation-selection balance” hypothesis, in which deleterious mutations are more efficiently purged later during HIV infection when the larger effective population size allows more efficient selection. Future studies using more recent sequencing technologies that are less prone to single-base errors should confirm these results by applying a similar Bayesian framework in other clinical settings.
IMPORTANCE The advent of sensitive sequencing platforms has led to an increased identification of minority drug resistance mutations (DRM), including among antiretroviral therapy-naive HIV-infected individuals. While transmission of DRM may impact future therapy options for newly infected individuals, the clinical significance of the detection of minority DRM remains controversial. In the present study, we applied deep-sequencing techniques within a Bayesian hierarchical framework to a cohort of 24 transmission pairs to investigate whether minority DRM detected shortly after transmission were the consequence of (i) sexual transmission from the source, (ii) de novo emergence shortly after infection followed by viral selection and evolution, or (iii) technical errors/limitations of deep-sequencing methods. We found no clear evidence to support the sexual transmission of minority resistant variants, and our results suggested that minor resistant variants may emerge de novo shortly after transmission, when the small effective population size limits efficient purge by natural selection.
KEYWORDS: minority drug resistance mutation, deep sequencing, Bayesian hierarchical framework, transmission, human immunodeficiency virus
INTRODUCTION
The transmission of drug resistance mutations (DRM) may limit therapy options for the newly infected individuals (1) and thwart efforts to control the HIV epidemic with antiretroviral therapy (ART) (2–5). Using ultrasensitive techniques, such as deep sequencing or allele-specific PCR, several studies have reported a high prevalence of “transmitted” minority DRM among the ART-naive population (6–8). The exact origin and clinical relevance of these minority variants carrying DRM are still debated (1, 5, 8–10). We previously performed deep sequencing to detect minority DRM in blood plasma collected from 32 HIV-infected individuals during primary infection, and we found that most low-frequency DRM in our cohort were likely identified because of methodological errors or were a consequence of error-prone HIV replication in the recipient (11). Our previous study and other similar studies have been limited by lack of characterization of the source partner viral population (6–8, 10, 11), lack of characterization of the primary source compartment of transmitted viral populations (e.g., genital secretion), and lack of longitudinal characterization of the viral population in recipients after transmission.
Here, we applied deep-sequencing techniques within a Bayesian hierarchical framework to a well-characterized cohort of 24 epidemiologically and genetically linked sexual transmission partners to investigate whether minority DRM detected shortly after transmission were the consequence of (i) sexual transmission from the source, (ii) de novo emergence shortly after infection followed by viral selection and evolution, or (iii) technical errors or analytical bias of deep-sequencing methods.
(Part of this study has been presented at the annual Conference on Retroviruses and Opportunistic Infections [CROI], Boston, MA, USA, 2016.)
RESULTS
Population characteristics and biological samples.
All study participants were men who have sex with men (MSM) infected with HIV-1 subtype B and without evidence of dual infection (12). Three identified source partners transmitted HIV to more than one recipient, leading to a total of 21 unique source partners (relationships between individuals are illustrated in Fig. 1). Among the 21 unique source partners, the median age was 32 years (interquartile range [IQR], 22 to 51), and the median CD4 T-cell count was 400 cells/mm3 (IQR, 270 to 821). The median HIV RNA levels in blood and seminal plasma were 4.7 log10/ml (IQR, 4.4 to 5.1) and 3.6 log10/ml (IQR, 2.7 to 5.5), respectively. All potential source partners were ART naive at the estimated time of transmission. Among the 24 recipients, baseline blood plasma samples were available within a median of 78 days (IQR, 18 to 130) from their estimated date of infection (EDI). The median age at baseline was 32 years (IQR, 26 to 41). The median HIV RNA level and CD4 T-cell count were 4.9 log10/ml (IQR, 4.2 to 5.8) and 507 cells/mm3 (IQR, 248 to 1,382), respectively. The median elapsed time between collection of paired source and recipient blood was 9 days (IQR, 2 to 29). Only 4 of the 21 source partners (19.0%) had paired seminal samples available, and longitudinal blood samples were available for 11 of the 24 recipients (45.8%), with a median of 3 time points (IQR, 2 to 3) collected over a median of 106 days (IQR, 56 to 219). Characteristics of the study populations are summarized in Table 1.
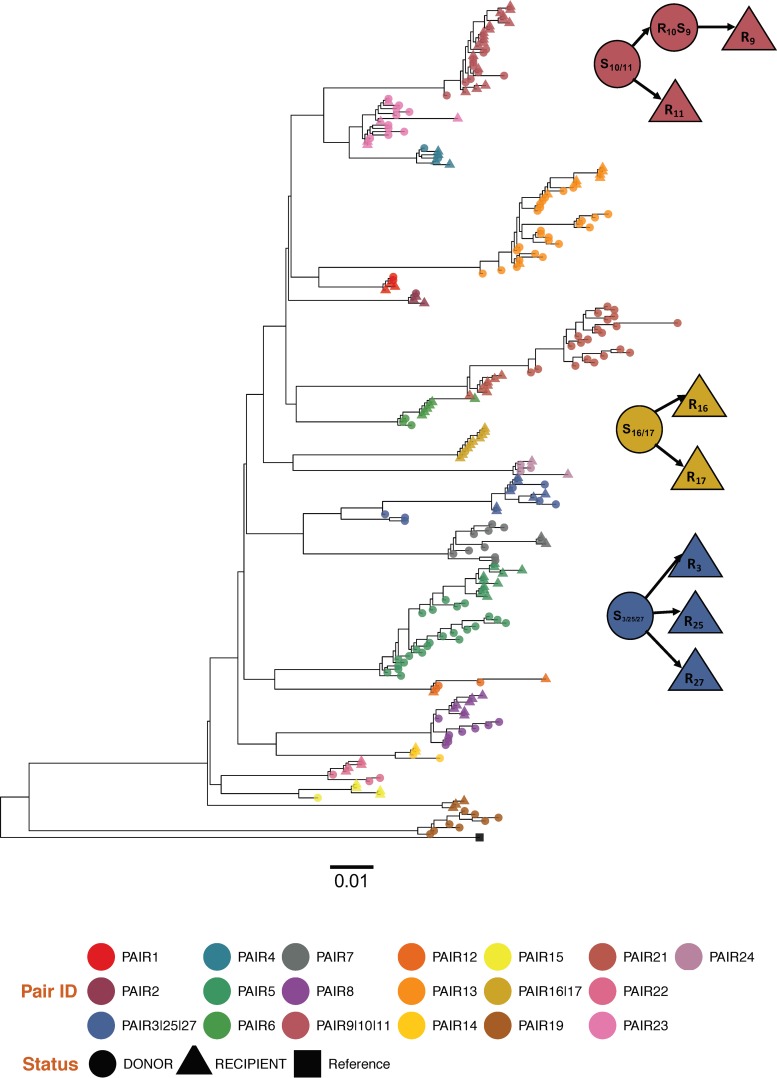
FIG 1.
Approximate maximum-likelihood phylogenetic tree of the partial pol regions for the 24 transmission pairs. A tree of the entire data set was reconstructed specifying a GTR+Gamma model. Each transmission pair clusters in a distinct monophyletic subtree (bootstrap support of >0.70) and is depicted in a different color. Shapes denote the status (i.e., source/recipient for each individual). The external group (black) is the subtype B HXB2 reference sequence. Pairs 3, 25, and 27, pairs 10 and 11, and pairs 16 and 17 shared a unique source partner. The recipient partner from pair 10 was also the source partner for pair 9 and is depicted with circles. S, source; R, recipient.
TABLE 1.
Population characteristics
| Characteristic at baseline (24 pairs) | Source | Recipient |
|---|---|---|
| No. of participants (no. with longitudinal data) | 21 (0) | 24 (11) |
| Median age, yr (IQR) | 32 (22–51) | 32 (20–59) |
| No. of paired seminal plasma sample | 4 | NAa |
| Median time from EDI (IQR) at baseline, days | NA | 78 (18–130) |
| Median CD4 cells/ml (IQR) | 400 (270–821) | 507 (248–1382) |
| Median HIV RNA level, log10 copies/ml (IQR) | ||
| Blood plasma | 4.7 (4.4–5.1) | 4.9 (4.2–5.8) |
| Seminal plasma | 3.6 (2.7–5.5) | NA |
| Median duration of follow-up, days (IQR) | NA | 106 (52–219) |
NA, not available.
Identification of DRM.
The median coverage for the pol/RT region in the blood and seminal plasma samples from the source and in blood plasma from recipients of 2,429 reads (IQR, 1,790 to 6,034), 2,478 reads (IQR, 1,711 to 4,969), and 5,248 reads (IQR, 3,840 to 6,648), respectively. Using these data, we determined the presence of DRM at each site and calculated the relative frequency in the source and recipient partners' samples. Minority variants with a prevalence below the background error rates were excluded. We found medians of 3.0 (IQR, 2.0 to 4.0) and 3.0 (IQR, 1.0 to 4.0) distinct DRM in the source and recipient partners, respectively. The median relative frequencies (DRMFreq) of the DRM in source and recipient partners were 2.3% (IQR, 0.5% to 7.1%) and 2.3% (IQR, 0.2% to 12.0%), respectively. Details of the identified DRM and their relative frequencies are summarized in Table 2.
TABLE 2.
Identification screening of NRTI and NNRTI minority DRM in source and recipient samples at baseline
| Pair | Source |
Recipient |
||||
|---|---|---|---|---|---|---|
| Origina | VLb | DRM (%)c | VL | Time from EDI (days) | DRM (%) | |
| 1 | BP | 4.4 | F227L (0.1), K103N (100) | 5.6 | 11 | K65N (1.3), D67N (2.4), K103N (100) |
| 2 | BP | 4.6 | K65R (5.6), L74V (0.1), Y115F (0.1), K219W (1.3), F227W (0.1) | 4.9 | 133 | K219W (3.7) |
| 3d | BP | 3.8 | Y188H (0.8), T215S (3.2), K219W (7.8), F227L (0.1) | 3.8 | 133 | K101Q (2.5) |
| 4 | BP | 4.4 | K219W (7.5), F227L (0.1) | 4.7 | 11 | K70R (5.7), T215V (62.6), K219W (21.8), F227L (0.2) |
| 5 | BP | 4.7 | T215V (31), K219W (6.9), F227L(0.1) | 6.4 | 11 | F227L (0.6) |
| 6 | BP | 5.0 | D67N (10.8), T215S (1.2), K219W (3.5), F227L (0.3), P236L (0.7) | 4.2 | 121 | G190S (0.3), T215V (13.9), K219W (62.5), F227L (0.1), P236L (0.1) |
| 7 | SP | 5.9 | D67N (2.0), E138A (95.6) | 5.9 | 27 | K70R (6.8), K219W(15), F227L(0.1), L324I (52) |
| BP | NAe | |||||
| 8 | BP | 4.2 | D67N (4.2), T215E (3.1), K219W (12), F227L (0.5) | 3.5 | 133 | K65N (3.2), D67N (2.1), K70R (8.6), L74I (0.1), V75L (0.2), K103R(11.4), Y188H (0.5), T215V (6.1), K219W (40.1), F227L (0.2) |
| 9 | BP | 3.5 | K103N (100), K219W (36), T215V (6.9) | 7.2 | 17 | K103N (100), T215V (8), F227L (1.4) |
| 10d | BP | 5.8 | K103N (100), T215V (1.3), F227L (1.0) | 6.1 | 88 | K103N (100), T215V (6.9), K219W (36.1) |
| 11d | BP | 5.8 | K103N (100), T215V (1.3), F227L (1.0) | 5.2 | 70 | K65N (8.1), K103N (100), F227L (0.2) |
| 12 | BP | 6.2 | F227L (0.8) | 5.9 | 19 | K65N (15), D67N (4.3), Y181S (12), M184I (1.6), T215I (1.0), K219W (5.1) |
| SP | 3.1 | D67N (2.3) | ||||
| 13 | BP | 3.7 | None | 3.9 | 14 | T215V (5.1), K219W (16.7), F227L (0.1) |
| 14 | BP | 5.2 | G190S (0.5), T215I (3.8), F227L (0.7) | 5.2 | 85 | None |
| 15 | BP | 5.5 | None | 3.7 | 87 | M184I (1.8), T215V (1.3), K219W (24.6), F227L (0.1) |
| 16d | BP | 4.7 | K65R (5.8), D67N (96.5) | 4.9 | 137 | K65R (8.8), D67N (91), T215S (100), K219E (88.4), F227L (0.1) |
| 17d | BP | 4.7 | K65R (5.8), D67N (96.5) | 5.2 | 32 | K65R (4.8), D67N (99), K219E (68.6) |
| 19 | BP | 4.7 | D67N (1.0) | 4.6 | 140 | K103N (1.4) |
| 21 | SP | 4.0 | D67N (2.8), K219W (77.4) | 4.6 | 85 | F227L (0.1), K219W (24.6) |
| BP | 4.9 | K101E (8.3), T215I (1.5), K219W (4.9), F227L (0.1) | ||||
| 22 | BP | 4.4 | None | 4.4 | 70 | T215V (5.7), F227L (0.1), K219W (32.4) |
| 23 | BP | 4.5 | D67N (5.3), K219W (4.4), F227L (0.3) | 5.1 | 114 | None |
| 24 | SP | 2.6 | F227L (1.1) | 4.9 | 96 | F227L (0.3) |
| BP | 2.6 | K65R (10.1), K70R (2.3), T215V (0.2), K219W (5.4) | ||||
| 25d | BP | 4.8 | Y188H (0.8), T215S (3.2), K219W (7.8), F227L (0.1) | 6.1 | 11 | F227L (1.5) |
| 27d | BP | 4.8 | Y188H (0.8), T215S (3.2), K219W (7.8), F227L (0.1) | 3.8 | 70 | None |
BP, blood plasma; SP, seminal plasma.
VL, viral load in log HIV RNA copies/ml.
DRM with frequencies above 20% are in bold.
Shared source partner for transmission pairs 3, 25, 27, pairs 16 and 17, and pairs 10 and 11. The recipient partner from pair 10 was also the source partner for pair 9 (also see Fig. 1).
NA, not available because of PCR failure.
Evidence against preferential transmission of viral populations harboring DRM.
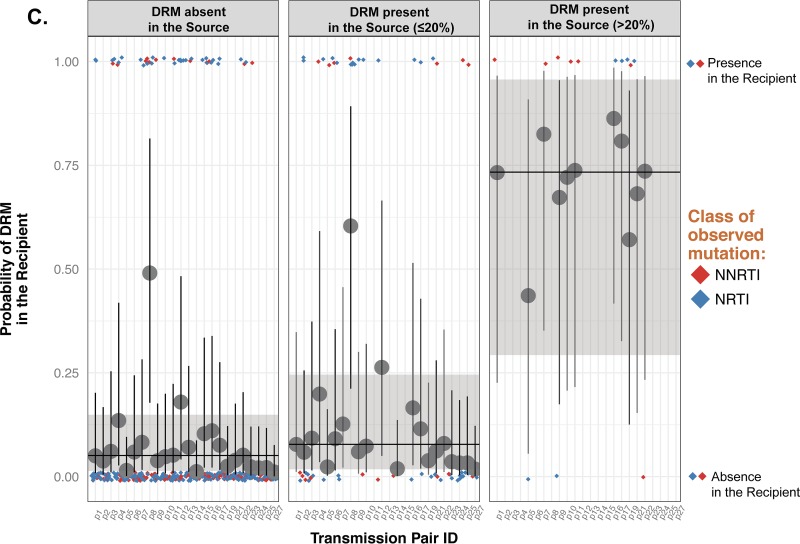
We sought to investigate the potential sexual transmission of minority DRM by comparing the identified DRM in confirmed source and recipient pairs. The probability of a DRM being present in the recipient at baseline when absent in the source (PR+∣S−) was low at 0.05, with a 95% credible interval of 0.01 to 0.13 (Fig. 2A, left panel). When a minority DRM was present in the source at a given site, the probability of detecting a minority DRM at the same site in the recipient (PR+∣Smin) was similarly low at 0.08 (0.02 to 0.24) (Fig. 2A, middle panel). As a comparison, the probability of DRM detection in the recipient when the source had a majority DRM (PR+∣Smaj) was much higher at 0.73 (0.29 to 0.96) (Fig. 2A, right panel). One example transmission pair (pair 9), where the individual dots are labeled with their respective mutations, is proposed in Fig. 2B. Altogether, Bayesian models confirmed that there was no increased probability of detecting a minority DRM in the recipient at a given site when present in the source (Bayes factor [BF] = 6.37 for PR+∣S− = PR+∣Smin) but were strongly indicative of an association between the presence of majority DRM in the source and in the recipients' partner PR+∣S− (BF ≈ 0). Due to the high frequency of the rare K219W mutation, which is likely not clinically relevant in contrast to other mutations at the same position (e.g., K219QE), we performed a new analysis to ensure that our conclusions were not affected by this mutation. Excluding K219W in fact did not qualitatively change other effects in the model; the percent changes in the regression coefficients were less than 5%, and all (non)significant effects remained so. The BFs remained above substantial as well (BF = 6.37 and 4.41 before and after exclusion of K219W, respectively) (Fig. 2C and Table 3).
FIG 2.
Probability of observing DRM in the recipient when the DRM in the source partner is absent (left panels), present at a frequency of ≤20% (middle panels), or present at a frequency of >20% (right panels) for the 24 transmission pairs (A), for pair 9 as an example (B), and after exclusion of mutation K219W (C). The colored diamonds indicate actual observations of DRM in the recipient at baseline (0, absence of mutation; 1, presence of mutation [right y axis]). Red diamonds, nonnucleoside reverse transcriptase inhibitors (NNRTIs); blue diamonds, nucleoside reverse transcriptase inhibitors (NRTIs). The horizontal line and the large dark gray dot indicate the median of the posterior distribution of the probability of DRM being present in the recipient for each scenario in the source (left y axis). The shaded area and the vertical line represent a 95% credible interval for each condition and each pair, respectively. (A) The 24 transmission pairs. (B) As an example, in pair 9, F227V was detected at baseline in the recipient but was absent in the source (left panel), T215V was present in the source (<20%) and in the recipient at baseline (middle panel), and K103N was present (>20%) in the source and in the recipient while K219 was detected only in the source (>20%) and was absent in the recipient at baseline (right panel). Mutations present in the source and/or in the recipient are indicated. (C) Similar data after exclusion of K219W. Excluding K219W did not qualitatively change other effects in the model, and the BFs remained above substantial (BF = 6.37 and 4.41 before and after exclusion of K219W, respectively).
TABLE 3.
Adjusted and unadjusted Bayes factor values for transmission of minority and majority DRM and covariates
| Model | Adjustmenta | BF |
||
|---|---|---|---|---|
| Minorityb | Majorityc | Covariate | ||
| Base | Unadjusted | 6.37 | <0.01 | NAd |
| Base without K219 | 4.41 | <0.01 | ||
| Base | Class of DRM | 6.86 | <0.01 | 4.84 |
| Sequencing coverage | 3.23 | <0.01 | 3.65 | |
| EDI | 5.36 | <0.01 | 5.61 | |
| Semen compartment | 4.94 | <0.01 | 0.88 | |
Unadjusted BFs are derived from models without covariates, while adjusted BFs are from models with covariates.
The BFs indicate substantial evidence against transmission of the minority DRM with or without adjustment (i.e., BF > 3).
The BFs indicate decisive evidence for transmission of the majority DRM with or without adjustment.
NA, not applicable.
To adjust for variations in HIV RNA levels, we investigated whether the absolute copy numbers of HIV RNA carrying a DRM (DRMcopies) in the source were predictive of the presence of DRM in the recipient at each specific site when DRM was not a majority population. Similarly to the case for DRMFreq, higher DRMcopies in the source also was not associated with increased probability of observing a DRM in the recipient (odds ratio [OR] [95% credible interval] = 1.03 [0.83 to 1.26] and BF = 48.7 for DRMFreq; OR = 1.04 [0.89 to 1.2] and BF = 60.2 for DRMcopies). Finally, to ensure that our initial results were not due to spurious detection of very low-level minority DRM, we performed sensitivity analyses with various DRM minimum thresholds by excluding minority DRM with frequencies below 10%, 5%, and 1% from our initial model. Bayesian models confirmed our observations regardless of the DRM minimum threshold (BFs for PR+∣S− = PR+∣Smin were 4.3, 5.1, and 3.6 after discarding minority DRM with frequencies below 10%, 5%, and 1%, respectively).
Sensitivity analysis.
Using the baseline model described above, we examined the potential confounding effects of relevant covariates, i.e., time from EDI to sampling, compartment of sampling (i.e., blood or seminal plasma), class of drug resistance mutation (i.e., nonnucleoside reverse transcriptase inhibitors [NNRTIs] versus nucleoside reverse transcriptase inhibitors [NRTIs], or depth of sequencing coverage (Table 3). None of these covariates significantly impacted the probability of the recipient having a minority DRM. That is, Bayes factors for minority DRM all remained >3 after adjustment, indicative of substantial evidence against transmission of the minority DRM with or without adjustment: EDI (OR = 1.71 [0.65 to 4.65], BF = 5.61, change in leave-one-out cross validation information criterion [ΔLOOIC] with standard error [SE] = −0.2 ± 2.3), compartment of sampling (OR = 51.6 [10.3 to 371.1], BF = 0.88, ΔLOOIC = 2.5 ± 3.4), class of DRM (OR = 1.19 [0.15 to 9.37], BF = 4.84), and sequencing coverage (OR = 1.78 [0.93 to 3.81], BF = 3.65, ΔLOOIC = 1.66 ± 3.0). Similarly, the BFs for majority DRM remained at <0.01, indicative of decisive evidence for transmission of the majority DRM with or without adjustment for the covariates mentioned above (Table 3).
No persistence of minority DRM during the course of infection.
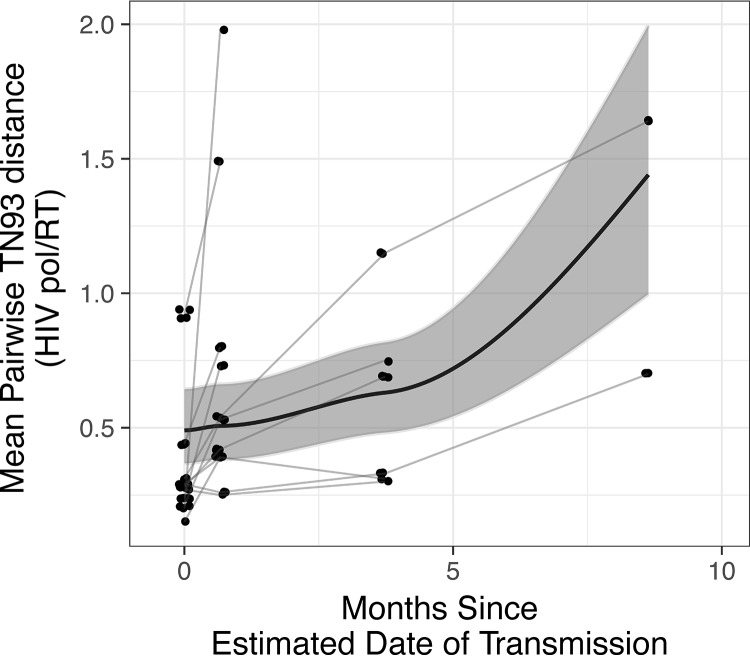
Finally, we evaluated the dynamics of DRM over a median duration of follow-up of 106 days (IQR, 56 to 219) among the 11 recipients with longitudinally collected samples. Using deep-sequence data from these samples, we fitted a mixed-effects exponential decay model. The decay rate was significantly greater than 0 (λ = 3.21 [1.19 to 11.64], BF = 0.05), indicating an exponential decay of the DRM frequency (Fig. 3). We found similar results after adjusting DRM frequencies to HIV RNA levels (i.e., by using DRMcopies). To further explore the relationship between decay and the EDI, we included EDI in the exponential decay model. EDI was not associated with either the baseline DRM frequency or the rate of decay, with the BFs not indicating strong enough evidence (BF = 6.91 and 0.9 for the baseline DRM frequency and the rate of decay, respectively). These results confirmed the significant decay of DRM over time regardless of the different sampling times from EDI across participants. Finally, the HIV pol/RT molecular diversity was modeled with a Bayesian hierarchical linear model with a log-normal link function. This model revealed a significant increase of the mean viral diversity over the study period (regression coefficient β = 0.67 [0.07 to 1.31], standard deviation = 0.2, BF = 0.02) (Fig. 4).
FIG 3.
Exponential decay of DRMFreq over time. The thin lines indicate means of DRMFreq across sites for each recipient. The thick lines and the gray shade represent the median and its 95% credible interval for the posterior distributions of the exponential-decay parameters. The Bayes factor indicates decisive evidence for exponential decay of DRMFreq over time (∼0.05).
FIG 4.
Increased HIV pol diversity over time. The thin lines indicate means of diversity across sites for each recipient. The thick lines and the gray shade represent the median and its 95% credible interval for the posterior distributions of the model parameters (Bayesian hierarchical linear model with a log-normal link function). The Bayes factor indicates very strong evidence for an increase in diversity over time (0.02).
DISCUSSION
FDA-approved genotyping methods can reliably identify mutations present in >20% of the circulating HIV population (13, 14). The advent of more-sensitive sequencing platforms has led to an increased identification of minority DRM among ART-naive HIV-infected individuals (6–8). Similarly to previous reports (6, 15), in this analysis of epidemiologically and genetically linked partner pairs, the presence of majority DRM in the recipient was associated with the presence of the same DRM in the source, consistent with sexual transmission. Conversely, we found no increased probability of detecting a minority DRM in the recipient at any given site when the same minority DRM was detectable in the source (compared to no DMR in the source). This observation suggests that the detection of minority DRM in recently infected individual is not likely to be the consequence of sexual transmission of variants harboring DRM, consistent with our previous report (11).
In the 11 recipient partners with longitudinal sampling, we found a significant exponential decay of the DRM frequency while viral pol diversity increased over time. This rapid reversion of viruses harboring minority DRM to the wild type supports the hypothesis that the detected DRM at low levels are unlikely the consequence of false-positive detection due to technical errors. Indeed, this exponential decay in DRM frequency in the face of increasing diversity suggests the presence of negative selection. The presence of minority DRM mainly during early infection, when the effective population size (Ne) is low, is consistent with the mutation-selection balance hypothesis, in which deleterious mutations (i.e., DRM) are more efficiently purged later during HIV infection, when the larger effective population size allows selection to more efficiently remove slightly deleterious mutations. At early stages of infection, the inefficiency of selection under low-Ne conditions allows DRM to appear and persist at low frequencies. The rapid disappearance of minority DRM over the course of infection is consistent with the importance of Ne as a factor shaping the patterns of molecular evolution.
Although our study offers new and interesting data about the appearance of minority DRM, it has some important limitations. First, given the time elapsed between the EDI and sampling of the recipient partner's blood and between sampling of the recipient and the identified source, we cannot rule out that unobserved selective pressures and other sampling-related biases may have driven the emergence and/or disappearance of minority DRM. Second, while the genital tract is a known reservoir for distinct HIV variants (16–18), our analysis found no significant compartment effect, most likely because of the small samples size of 4 sources with genital secretion (19), which limited our ability to rule out transmission of minority DRM from the genital compartment.
When assessing viral variants related to HIV drug resistance at low levels, one must also consider the absolute level of HIV to assess template resampling and its effects on frequency determination and error associated with PCR amplification. In our study, HIV-1 RNA levels exceeded 1,000 copies/ml for all but one sample, limiting the risk of oversampling by deep sequencing. In addition, we confirmed our observations after adjusting for the level of HIV RNA and after excluding DRM at very low levels in our sensitivity analysis.
Another important limitation includes the use of the 454/Roche platform for sequencing: this high-throughput sequencing platform generates massively parallel short reads that are prone to homopolymer-associated, per-base errors (20). Variability between replicated deep-sequencing runs on the 454 platform has previously been reported (21, 22), with minority variants detected at levels between 1 and 5% in one replicate while being undetectable in another. In our previous work, we also observed discordance in the detection of amino acid residues in more than half of the replicate runs (11). To overcome these limitations, we apply rigorous quality control procedures for deep sequencing (23, 24). Our validated bioinformatics pipeline (25–27) includes strict quality filtering steps, and only quality-controlled reads were included in the iterative alignment procedure. Ideally, the relatedness of existing DRM variants within transmission pairs could be assessed in a phylogenetic framework. However, the 454 platform used here produced short individual reads that were not suitable for proper phylogenetic analysis. Therefore, we relied on a Bayesian-based statistical framework to model the sexual transmission and persistence of DRM. One major advantage of using this Bayesian framework is that it allows us to directly evaluate a null hypothesis (i.e., the probability of no transmission given the data). Evaluating the null hypothesis based on the more commonly used frequentist framework is almost impossible to do, since a P value (the probability of obtaining the data given the null hypothesis) of greater than the typical 5% alpha level does not support the null hypothesis.
Finally, our study assessed the putative transmission of DRM from treatment-naive source partners and did not include treatment-experienced sources who could have been more relevant/informative for study of transmitted DRM. However, most HIV transmission events occur when the source is still ART naive and often is unaware of their HIV status (28, 29), and investigating transmission of minority variants in this context remains enlightening for assessment of the transmission of minority variants and more broadly for understanding the biology concerning male-to-male sexual transmission of HIV.
In summary, in this study we applied deep-sequencing methods within a Bayesian framework in a uniquely characterized population of source and recipient HIV-infected partners. Despite the important limitations, we found no clear evidence to support the sexual transmission of minority resistant variants, although it should be noted that the absence of evidence does not rule out potential transmission of minority DRM via sexual exposure. Overall, our results suggested that minor resistant variants may emerge de novo shortly after transmission, when the small effective population size limits efficient purge by natural selection (30, 31). Future studies using recent sequencing technologies that provide longer reads and are less prone to single-base errors (MiSeq or PacBio with or without Primer ID) (32, 33) or single-genome amplification/Sanger sequencing in different settings (i.e., ART-experienced HIV-infected source partners) within a similar Bayesian framework would be informative for detecting clinically relevant minority variants and investigating their propensity for transmission.
MATERIALS AND METHODS
Ethics statement.
The UCSD Human Research Protections Program approved the study protocol, consent, and procedures for consent. All study participants provided written informed consent before any study procedures were undertaken.
Subject characteristics and data collection.
A total of 24 genetically and epidemiologically linked transmission pairs were included. Partnerships were identified through contact tracing of individuals newly diagnosed with primary or early HIV infection as part of the San Diego Primary Infection Resource Consortium (SD-PIRC). Each newly diagnosed participant was asked about his most recent sexual partners. HIV transmission was confirmed by sequence analysis, and phylogenetic linkage was inferred when the two viral strains were >98.5% similar in the HIV-1 pol/reverse transcriptase (pol/RT)-coding region (34). The direction of transmission (i.e., source versus recipient) was defined based on the estimated date of infection (EDI) for each individual (35), based on participant reporting and standardized serologic algorithms (36). Demographics, laboratory data (CD4 T-cell count and HIV RNA levels), and data on HIV risk factors (e.g., intravenous drug usage or sexually transmitted infections) and sexual behavior (receptive and insertive anal intercourse and number of sex partners) were collected. Blood plasma from both partners was collected at the time of recruitment; a subset of 11 recipients had additional longitudinal samples collected prior to ART initiation. Seminal plasma was collected at baseline for 4 out of the 21 (19.0%) unique source partners (37).
HIV RNA extraction and deep sequencing from blood and seminal plasma samples.
HIV-1 coding regions of pol/RT (HXB2 coordinates 2708 to 3242) were amplified from blood and seminal plasma by PCR with region-specific primers (11). Deep sequencing was performed on a 454 GS FLX Titanium instrument (454 Life Sciences/Roche, Branford, CT) (12). For all but one seminal plasma sample, HIV-1 RNA levels exceeded 1,000 copies/ml.
Deep-sequencing processing and bioinformatics analysis.
Read (FASTA) and quality score files produced by the 454 instruments were analyzed using a purpose-built bioinformatics pipeline (25–27). The pipeline is available at https://github.com/veg/HIV-NGS, and the key steps are as follows. (i) The first filtering step included exclusion of low-quality reads (q score of <15) and correction for random sequencing errors or homopolymers (25). (ii) High-quality reads were aligned to the reference sequence (HXB2) using a codon-based algorithm (12). (iii) We applied a Bayesian inferences with Dirichlet multinomial mixtures model (38) to distinguish true low-frequency variants from sequencing errors (posterior probabilities of ≥99.99%). (iv) Representative reads were screened for evidence of recombination using GARD (39), APOBEC signatures, and frameshifts (40). (v) All sequences were screened for in-house cross-contamination using BLAST (41). Quality filtering parameters for deep-sequencing data are presented in Table 4. We computed the mean of all pairwise Tamura-Nei 93 distances between reads with at least 100 overlapping base pairs to quantify nucleotide diversity (42).
TABLE 4.
Quality filtering parameters for deep-sequencing data
| Parameter | Value |
|---|---|
| Minimum PHRED score for inclusion of sites | 15 |
| Minimum no. of sites with PHRED score higher than the minimum for inclusion of reads/fragments of reads | 75 |
| Minimum read length to be included in subsequent analyses | 100 |
| Minimum coverage | 250 |
| Window size for sliding-window estimation of nucleotide diversity | 150 |
| Length of sliding-window stride | 20 |
| Minimum no. of copies for a read to be considered a variant | 10 |
Identification of DRM.
We identified mutations associated with known resistance to nucleoside reverse transcriptase inhibitors (NRTIs) and nonnucleoside reverse transcriptase inhibitor (NNRTIs) according to the Stanford Drug Resistance Database (score of >35) (http://hivdb.stanford.edu). Filtered deep-sequencing reads were screened for the presence of amino acid mutations at these sites, and the relative frequency of each identified DRM was obtained as part of the bioinformatics pipeline (25, 26).
Estimation of lower limit of detection for DRM.
Since the lower limit of detection for each DRM is dependent on the sampling error and background error rate for each site, we included only DRM with residue frequencies greater than the background error rate, estimated using our published binomial mixture model (11) implemented in the online molecular sequence analysis server DataMonkey (43).
Statistical analyses.
Our objective was to evaluate the rate at which a DRM at a particular site was transmitted from source partners to their respective recipients and whether the transmission was affected by the relative frequency (DRMFreq) or the absolute copy numbers (DRMcopies [DRM frequency × HIV RNA copies/ml]) of each mutation. Minority DRM was defined as DRM with frequency below 20% (DRMFreq ≤ 20%). We tested whether the probability of observing a DRM in the recipient's blood plasma HIV RNA population sampled at baseline at a particular site differed across three conditions: (i) the source had no DRM (R+∣S−), (ii) a minority DRM was present in the source at a particular site (R+∣Smin), and (iii) a majority DRM (DRMFreq > 20%) was present in the source at a particular site (R+∣Smaj). Transmission of HIV variants harboring DRM can be affected by (i) between-pair variability (DRM might have transmitted within a particular pair more or less easily) and (ii) between-site variability (some DRM at a particular site might have transmitted more or less easily). To incorporate these sources of variability into a model, we fit Bayesian hierarchical Bernoulli logistic regression models with the three conditions as a within-pair fixed effect and two random intercepts for pairs and sites as crossed random effects. For modeling longitudinal data, we were mainly concerned with modeling two outcomes: DRM and diversity. For modeling DRM (DRMFreq and DRMcopies), we fit a Bayesian hierarchical exponential-decay model that is represented by the function μi = N0 exp(−λti), where t, μi, N0, and λ represent the time point, the mean at time point i, the baseline value at t = 0, and the rate of change, respectively. At each time point, the DRM was modeled with a normal distribution. Recipients were allowed to have their own random intercepts for N0, and λ. The mean of diversity at each time point was modeled with a linear model with a log-normal link function. Diversity was assumed to be log-normally distributed at each time point because of its positive skew, while its change in the mean at each time point was assumed to be linear. Again, each recipient had his own random intercept and time slope.
The model convergence was evaluated with R hat, which is the ratio of the average variance of 1,000 Markov-chain Monte Carlo (MCMC) samples within each of the 4 chains to the variance of the pooled MCMC samples across chains. An R hat of <1.1 is considered acceptable convergence (44). We evaluated whether relevant covariates improved the base model by comparing the differences in the leave-one-out cross validation information criterion and the standard error (ΔLOOIC ± 1 standard error [SE]) between two models. A positive ΔLOOIC value greater than 2 SE indicates an improvement in model fit. A weakly informative normal distribution with a mean of 0 and a standard deviation of 5 was used as a prior distribution for regression coefficients, and noninformative uniform distributions were used as prior distributions of the variances and random intercepts. While Bayesian modeling is not concerned with the traditional null-hypothesis significance testing, we provided the BF, a ratio of the probability of obtaining data given null and alternative hypotheses as a reference. BFs of 1 to 3, 3 to 10, 10 to 30, 30 to 100, and >100 are considered anecdotal, substantial, strong, very strong, and decisive evidence for a null hypothesis, respectively, while their reciprocals (1 to 1/3, etc.) indicate evidence against a null hypothesis (45). In other words, BFs of >3 are indicative of substantial evidence against transmission of the DRM, while BFs of ≈0 indicate decisive evidence for transmission of a variant harboring a DRM. To examine potential confounding effects of relevant covariates on DRM transmission, we computed BFs in a model that included those covariates separately (Table 3). We used the R statistical language (46) and the rstan package (47) to fit Bayesian models.
Accession number(s).
The sequencing data are available at the NCBI Sequence Read Archive under accession numbers SAMN04914210 to SAMN04914278.
ACKNOWLEDGMENTS
We are grateful to all the participants in the San Diego Primary Infection cohort (SD PIC) and to the CFAR Genomic, Translational Virology, and Flow Cytometry Cores. The HIV RNA quantification standard was obtained through the NIH AIDS Research and Reference Reagent Program, DAIDS, NIAID (HIV VQA RNA quantification standard from the DAIDS Virology Quality Assurance Program). We also acknowledge Matt Strain and Ben Murrell for insightful discussions.
A.C. participated in the study design and data analysis and wrote the primary version of the manuscript; M.N. performed the primary data analysis and wrote the primary version of the manuscript; J.O.W. participated in the study design and revised the manuscript; S.J.L. and D.M.S. contributed to the study design, enrolled all participants, and revised the manuscript; S.R.M. participated in the study design and revised the manuscript; and S.G. participated in the study design, participated in the data analyses, and wrote the primary version of the manuscript.
A.C., M.N., S.J.L., S.R.M., and S.G. do not have any commercial or other associations that might pose a conflict of interest. D.M.S. has received grant support from ViiV Pharmaceuticals and consultant fees from Gen-Probe and Testing Talent Services. J.O.W. and S.J.L. have received grant support from Gilead Science Inc.
This work was supported primarily by a grant from the National Institutes of Health, University of California, San Francisco-Gladstone Institute of Virology and Immunology Center for AIDS Research (P30-AI027763) (CNIHR), California HIV Research Program Ideal Awards to S.G. and J.O.W. (ID15-SD-063 and ID15-SD-052), the Department of Veterans Affairs, the James B. Pendleton Charitable Trust, and additional grants from the National Institutes of Health: AI100665, MH100974, MH097520, DA034978, AI007384, AI027763, AI106039, AI43638, AI074621, AI036214, MH101012, UL1TR000100, CARE U19 AI096113, and AI068636-09.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Paredes R, Lalama CM, Ribaudo HJ, Schackman BR, Shikuma C, Giguel F, Meyer WA III, Johnson VA, Fiscus SA, D'Aquila RT, Gulick RM, Kuritzkes DR. 2010. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis 201:662–671. doi: 10.1086/650543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett DE. 2006. The requirement for surveillance of HIV drug resistance within antiretroviral rollout in the developing world. Curr Opin Infect Dis 19:607–614. doi: 10.1097/QCO.0b013e3280109ff1. [DOI] [PubMed] [Google Scholar]
- 3.Lee GQ, Bangsberg DR, Muzoora C, Boum Y, Oyugi JH, Emenyonu N, Bennett J, Hunt PW, Knapp D, Brumme CJ, Harrigan PR, Martin JN. 2014. Prevalence and virologic consequences of transmitted HIV-1 drug resistance in Uganda. AIDS Res Hum Retroviruses 30:896–906. doi: 10.1089/aid.2014.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Vaerenbergh K. 2001. Study of the impact of HIV genotypic drug resistance testing on therapy efficacy. Verh K Acad Geneeskd Belg 63:447–473. [PubMed] [Google Scholar]
- 5.Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Metzner KJ, Kozal MJ, Hullsiek KH, Balduin M, Jakobsen MR, Geretti AM, Thiebaut R, Ostergaard L, Masquelier B, Johnson JA, Miller MD, Kuritzkes DR. 2011. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 305:1327–1335. doi: 10.1001/jama.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakobsen MR, Tolstrup M, Søgaard OS, Jørgensen LB, Gorry PR, Laursen A, Ostergaard L. 2010. Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin Infect Dis 50:566–573. doi: 10.1086/650001. [DOI] [PubMed] [Google Scholar]
- 7.Metzner KJ, Rauch P, Walter H, Boesecke C, Zöllner B, Jessen H, Schewe K, Fenske S, Gellermann H, Stellbrink H-J. 2005. Detection of minor populations of drug-resistant HIV-1 in acute seroconverters. AIDS 19:1819–1825. doi: 10.1097/01.aids.0000189878.97480.ed. [DOI] [PubMed] [Google Scholar]
- 8.Johnson JA, Li J-F, Wei X, Lipscomb J, Irlbeck D, Craig C, Smith A, Bennett DE, Monsour M, Sandstrom P, Lanier ER, Heneine W. 2008. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy. PLoS Med 5:e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lataillade M, Chiarella J, Yang R, Schnittman S, Wirtz V, Uy J, Seekins D, Krystal M, Mancini M, McGrath D, Simen B, Egholm M, Kozal M. 2010. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naïve subjects in the CASTLE study. PLoS One 5:e10952. doi: 10.1371/journal.pone.0010952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzner KJ, Scherrer AU, Preiswerk B, Joos B, von Wyl V, Leemann C, Rieder P, Braun D, Grube C, Kuster H, Böni J, Yerly S, Klimkait T, Aubert V, Furrer H, Battegay M, Vernazza PL, Cavassini M, Calmy A, Bernasconi E, Weber R, Günthard HF, Swiss HIV Cohort Study. 2013. Origin of minority drug-resistant HIV-1 variants in primary HIV-1 infection. J Infect Dis 208:1102–1112. doi: 10.1093/infdis/jit310. [DOI] [PubMed] [Google Scholar]
- 11.Gianella S, Delport W, Pacold ME, Young JA, Choi JY, Little SJ, Richman DD, Kosakovsky Pond SL, Smith DM. 2011. Detection of minority resistance during early HIV-1 infection: natural variation and spurious detection rather than transmission and evolution of multiple viral variants. J Virol 85:8359–8367. doi: 10.1128/JVI.02582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pacold M, Smith D, Little S, Cheng PM, Jordan P, Ignacio C, Richman D, Pond SK. 2010. Comparison of methods to detect HIV dual infection. AIDS Res Hum Retroviruses 26:1291–1298. doi: 10.1089/aid.2010.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eshleman SH, Hackett J Jr, Swanson P, Cunningham SP, Drews B, Brennan C, Devare SG, Zekeng L, Kaptue L, Marlowe N. 2004. Performance of the Celera Diagnostics ViroSeq HIV-1 Genotyping System for sequence-based analysis of diverse human immunodeficiency virus type 1 strains. J Clin Microbiol 42:2711–2717. doi: 10.1128/JCM.42.6.2711-2717.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. 2007. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res 17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panichsillapakit T, Smith DM, Wertheim JO, Richman DD, Little SJ, Mehta SR. 2016. Prevalence of transmitted HIV drug resistance among recently infected persons in San Diego, CA 1996-2013. J Acquir Immune Defic Syndr 71:228–236. doi: 10.1097/QAI.0000000000000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith DM, Wong JK, Shao H, Hightower GK, Mai SH, Moreno JM, Ignacio CC, Frost SD, Richman DD, Little SJ. 2007. Long-term persistence of transmitted HIV drug resistance in male genital tract secretions: implications for secondary transmission. J Infect Dis 196:356–360. doi: 10.1086/519164. [DOI] [PubMed] [Google Scholar]
- 17.Eron JJ, Vernazza PL, Johnston DM, Seillier-Moiseiwitsch F, Alcorn TM, Fiscus SA, Cohen MS. 1998. Resistance of HIV-1 to antiretroviral agents in blood and seminal plasma: implications for transmission. AIDS 12:F181–F189. doi: 10.1097/00002030-199815000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Tirado G, Jove G, Kumar R, Noel RJ, Reyes E, Sepulveda G, Yamamura Y, Kumar A. 2004. Differential virus evolution in blood and genital tract of HIV-infected females: evidence for the involvement of drug and non-drug resistance-associated mutations. Virology 324:577–586. doi: 10.1016/j.virol.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 19.Rouder JN, Speckman PL, Sun D, Morey RD, Iverson G. 2009. Bayesian t tests for accepting and rejecting the null hypothesis. Psychon Bull Rev 16:225–237. doi: 10.3758/PBR.16.2.225. [DOI] [PubMed] [Google Scholar]
- 20.Gilles A, Meglecz E, Pech N, Ferreira S, Malausa T, Martin JF. 2011. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics 12:245. doi: 10.1186/1471-2164-12-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varghese V, Wang E, Babrzadeh F, Bachmann MH, Shahriar R, Liu T, Mappala SJM, Gharizadeh B, Fessel WJ, Katzenstein D, Kassaye S, Shafer RW. 2010. Nucleic acid template and the risk of a PCR-induced HIV-1 drug resistance mutation. PLoS One 5:e10992. doi: 10.1371/journal.pone.0010992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poon AF, Swenson LC, Dong WW, Deng W, Kosakovsky Pond SL, Brumme ZL, Mullins JI, Richman DD, Harrigan PR, Frost SD. 2010. Phylogenetic analysis of population-based and deep sequencing data to identify coevolving sites in the nef gene of HIV-1. Mol Biol Evol 27:819–832. doi: 10.1093/molbev/msp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson SJ, Welkers MR, Depledge DP, Coulter E, Breuer JM, de Jong MD, Kellam P. 2013. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos Trans R Soc Lond B Biol Sci 368:20120205. doi: 10.1098/rstb.2012.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher RG, Smith DM, Murrell B, Slabbert R, Kirby BM, Edson C, Cotton MF, Haubrich RH, Kosakovsky Pond SL, Van Zyl GU. 2015. Next generation sequencing improves detection of drug resistance mutations in infants after PMTCT failure. J Clin Virol 62:48–53. doi: 10.1016/j.jcv.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter CC, Wagner GA, Hightower GK, Caballero G, Phung P, Richman DD, Pond SL, Smith DM. 2015. HIV-1 neutralizing antibody response and viral genetic diversity characterized with next generation sequencing. Virology 474:34–40. doi: 10.1016/j.virol.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner GA, Pacold ME, Kosakovsky Pond SL, Caballero G, Chaillon A, Rudolph AE, Morris SR, Little SJ, Richman DD, Smith DM. 2014. Incidence and prevalence of intrasubtype HIV-1 dual infection in at-risk men in the United States. J Infect Dis 209:1032–1038. doi: 10.1093/infdis/jit633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall HI, Holtgrave DR, Maulsby C. 2012. HIV transmission rates from persons living with HIV who are aware and unaware of their infection. AIDS 26:893–896. doi: 10.1097/QAD.0b013e328351f73f. [DOI] [PubMed] [Google Scholar]
- 29.Skarbinski J, Rosenberg E, Paz-Bailey G, Hall HI, Rose CE, Viall AH, Fagan JL, Lansky A, Mermin JH. 2015. Human immunodeficiency virus transmission at each step of the care continuum in the United States. JAMA Intern Med 175:588–596. doi: 10.1001/jamainternmed.2014.8180. [DOI] [PubMed] [Google Scholar]
- 30.Lynch M, Conery JS. 2003. The origins of genome complexity. Science 302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 31.Yi S, Streelman JT. 2005. Genome size is negatively correlated with effective population size in ray-finned fish. Trends Genet 21:643–646. doi: 10.1016/j.tig.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Dudley DM, Bailey AL, Mehta SH, Hughes AL, Kirk GD, Westergaard RP, O'Connor DH. 2014. Cross-clade simultaneous HIV drug resistance genotyping for reverse transcriptase, protease, and integrase inhibitor mutations by Illumina MiSeq. Retrovirology 11:122. doi: 10.1186/s12977-014-0122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keys JR, Zhou S, Anderson JA, Eron JJ Jr, Rackoff LA, Jabara C, Swanstrom R. 2015. Primer ID informs next-generation sequencing platforms and reveals preexisting drug resistance mutations in the HIV-1 reverse transcriptase coding domain. AIDS Res Hum Retroviruses 31:658–668. doi: 10.1089/aid.2014.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oster AM, Wertheim JO, Hernandez AL, Ocfemia MC, Saduvala N, Hall HI. 2015. Using molecular HIV surveillance data to understand transmission between subpopulations in the United States. J Acquir Immune Defic Syndr 70:444–451. doi: 10.1097/QAI.0000000000000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butler DM, Delport W, Kosakovsky Pond SL, Lakdawala MK, Cheng PM, Little SJ, Richman DD, Smith DM. 2010. The origins of sexually transmitted HIV among men who have sex with men. Sci Transl Med 2:18re1. doi: 10.1126/scitranslmed.3000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris SR, Little SJ, Cunningham T, Garfein RS, Richman DD, Smith DM. 2010. Evaluation of an HIV nucleic acid testing program with automated Internet and voicemail systems to deliver results. Ann Intern Med 152:778–785. doi: 10.7326/0003-4819-152-12-201006150-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gianella S, Strain MC, Rought SE, Vargas MV, Little SJ, Richman DD, Spina CA, Smith DM. 2012. Associations between virologic and immunologic dynamics in blood and in the male genital tract. J Virol 86:1307–1315. doi: 10.1128/JVI.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye X, Yu YK, Altschul SF. 2010. Compositional adjustment of Dirichlet mixture priors. J Comput Biol 17:1607–1620. doi: 10.1089/cmb.2010.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 40.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for inferring selection. Mol Biol Evol 30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. [DOI] [PubMed] [Google Scholar]
- 43.Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gelman A, Carlin JB, Stern HS. 2013. Bayesian data analysis, 3rd ed Chapman and Hall/CRC, London, United Kingdom. [Google Scholar]
- 45.Wetzels R, Matzke D, Lee MD, Rouder JN, Iverson GJ, Wagenmakers EJ. 2011. Statistical evidence in experimental psychology: an empirical comparison using 855 t tests. Perspect Psychol Sci 6:291–298. doi: 10.1177/1745691611406923. [DOI] [PubMed] [Google Scholar]
- 46.R Core Team. 2016. R: a language and environment for statistical computing. http://www.r-project.org/.
- 47.Stan Development Team. 2016. Stan: a C++ library for probability and sampling, version 2.12.0. http://mc-stan.org/.