ABSTRACT
Bovine herpesvirus 1 (BoHV-1), an important bovine pathogen, establishes lifelong latency in sensory neurons. Latently infected calves consistently reactivate from latency following a single intravenous injection of the synthetic corticosteroid dexamethasone. The immediate early transcription unit 1 (IEtu1) promoter, which drives bovine ICP0 (bICP0) and bICP4 expression, is stimulated by dexamethasone because it contains two glucocorticoid receptor (GR) response elements (GREs). Several Krüppel-like transcription factors (KLF), including KLF15, are induced during reactivation from latency, and they stimulate certain viral promoters and productive infection. In this study, we demonstrate that the GR and KLF15 were frequently expressed in the same trigeminal ganglion (TG) neuron during reactivation and cooperatively stimulated productive infection and IEtu1 GREs in mouse neuroblastoma cells (Neuro-2A). We further hypothesized that additional regions in the BoHV-1 genome are transactivated by the GR or stress-induced transcription factors. To test this hypothesis, BoHV-1 DNA fragments (less than 400 bp) containing potential GR and KLF binding sites were identified and examined for transcriptional activation by stress-induced transcription factors. Intergenic regions within the unique long 52 gene (UL52; a component of the DNA primase/helicase complex), bICP4, IEtu2, and the unique short region were stimulated by KLF15 and the GR. Chromatin immunoprecipitation studies revealed that the GR and KLF15 interacted with sequences within IEtu1 GREs and the UL52 fragment. Coimmunoprecipitation studies demonstrated that KLF15 and the GR were associated with each other in transfected cells. Since the GR stimulates KLF15 expression, we suggest that these two transcription factors form a feed-forward loop that stimulates viral gene expression and productive infection following stressful stimuli.
IMPORTANCE Bovine herpesvirus 1 (BoHV-1) is an important viral pathogen that causes respiratory disease and suppresses immune responses in cattle; consequently, life-threatening bacterial pneumonia can occur. Following acute infection, BoHV-1 establishes lifelong latency in sensory neurons. Reactivation from latency is initiated by the synthetic corticosteroid dexamethasone. Dexamethasone stimulates lytic cycle viral gene expression in sensory neurons of calves latently infected with BoHV-1, culminating in virus shedding and transmission. Two stress-induced cellular transcription factors, Krüppel-like transcription factor 15 (KLF15) and the glucocorticoid receptor (GR), cooperate to stimulate productive infection and viral transcription. Additional studies demonstrated that KLF15 and the GR form a stable complex and that these stress-induced transcription factors bind to viral DNA sequences, which correlates with transcriptional activation. The ability of the GR and KLF15 to synergistically stimulate viral gene expression and productive infection may be critical for the ability of BoHV-1 to reactivate from latency following stressful stimuli.
KEYWORDS: bovine herpesvirus 1, KLF15, gene regulation, glucocorticoids, latency, stress response
INTRODUCTION
Bovine herpesvirus 1 (BoHV-1) is an important pathogen of cattle that causes conjunctivitis and/or upper respiratory tract disease; consequently, erosion of mucosal surfaces occurs during acute infection (1, 2). BoHV-1 infections also suppress host immune responses (3), which can then increase the incidence of life-threatening bacterial pneumonia (4, 5). BoHV-1, stress, and other bovine viral pathogens contribute to the polymicrobial disease bovine respiratory disease complex (BRDC) (6). BRDC is the most important disease in cattle because it costs the U.S. cattle industry more than $1 billion in losses each year (1, 4, 7, 8). A BoHV-1 entry protein encoded by the poliovirus receptor-related 1 gene is a BRDC susceptibility gene for Holstein calves (9), confirming that BoHV-1 is an important BRDC cofactor.
Following acute infection, trigeminal ganglia (TG) are primary sites for lifelong latency (10, 11). Increased corticosteroid levels, due to food and water deprivation during shipping of cattle, weaning, and/or dramatic weather changes, increase the incidence of BoHV-1 reactivation from latency (11, 12). The synthetic corticosteroid dexamethasone (DEX) mimics the effects of stress, stimulates productive infection (13), and initiates reactivation from latency (14–21). Corticosteroids bind and activate the glucocorticoid receptor (GR) and mineralocorticoid receptor (MR) (22), suggesting that these nuclear hormone receptors mediate key aspects of reactivation from latency. Herpes simplex virus 1 (HSV-1) and presumably BoHV-1 genomes exist as “silent” chromatin during latency (23); conversely HSV-1 DNA is associated with unstable chromatin during productive infection (24, 25). Regardless of the reactivation stimulus, silent viral heterochromatin must be converted into an actively transcribing template in order for successful reactivation from latency to occur. BoHV-1 viral gene products can be readily detected within hours after DEX treatment (19, 26–28), suggesting that cellular transcription factors are pivotal for activating viral gene expression. DEX-induced cellular transcription factors were identified within the first 3 h in TG neurons following DEX treatment of latently infected calves, and these transcription factors stimulate certain viral promoters and productive infection (29). Several Krüppel-like transcription factors (KLF) were identified in this study, which is intriguing because these proteins bind GC- or CA-rich motifs (30), and the BoHV-1 genome is GC rich. Corticosteroids also have potent anti-inflammatory and immune-suppressive properties (22, 31–33), which would clearly enhance viral replication and spread during reactivation from latency.
BoHV-1 genes are expressed in three distinct phases during productive infection of cultured cells: immediate early (IE), early (E), and late (L) (14, 15). IE gene expression is stimulated by VP16, a tegument protein (34, 35). IE transcription unit 1 (IEtu1) encodes two transcriptional regulatory proteins, bovine ICP0 (bICP0) and bICP4, because a single IE transcript is differentially spliced and then translated into bICP0 or bICP4 (36–38). The bICP0 protein is also translated from an E mRNA (E2.6) because a separate E promoter drives expression of the bICP0 E transcript (36–39). IEtu1 promoter activity is stimulated by the GR and the synthetic corticosteroid DEX because there are two consensus GR response elements (GREs) located in the promoter (13), suggesting that this promoter is activated by stress-induced transcription factors during reactivation from latency.
In this study, we provide evidence that KLF15 and the GR were frequently detected in the same TG neurons during reactivation from latency but not during latency. Additional studies demonstrated that KLF15 and the GR cooperated to stimulate productive infection and transactivate a 280-bp fragment containing the IEtu1 GREs cloned upstream of a minimal simian virus 40 (SV40) early promoter. Since the BoHV-1 genome contains more than 100 putative GR binding sites (13), we also tested whether intergenic regions of the viral genome that contain binding sites for the GR and KLF family members were transactivated by stress-induced transcription factors. We provide evidence that KLF15 and the GR cooperate to transactivate a reporter construct containing a UL52 intergenic fragment. Additional intergenic regions functioned as transcriptional enhancers and were transactivated by KLF15. These findings provide evidence that KLF15 and the GR cooperate to stimulate viral transcription and productive infection following a stressful stimulus.
RESULTS
The GR and KLF15 are frequently present in the same neuron during reactivation and can stimulate productive infection in cultured cells.
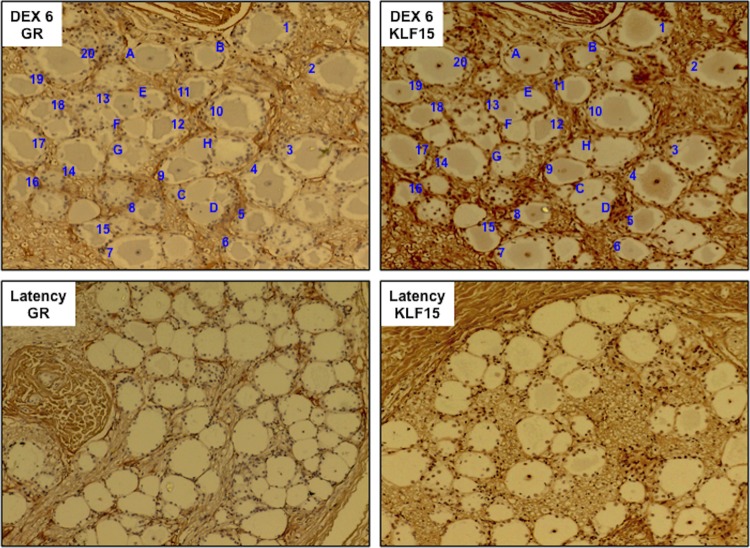
We hypothesized that KLF15 and the GR cooperate with each other to stimulate viral replication and gene expression during DEX-induced reactivation from latency. The rationale for this hypothesis is summarized below. The GR is expressed in a subset of neurons in the peripheral nervous system (40), GR-positive (GR+) neurons increase during reactivation (26, 27), and TG neurons that express bICP0 or VP16 during early stages of reactivation frequently express the GR (26, 27). Furthermore, KLF15 is a DEX-induced cellular transcription factor identified during reactivation from latency (29), and KLF15 stimulates the BoHV-1 IEtu1 promoter (29) as well as the HSV-1 ICP0 promoter (41). Finally, KLF15 and the GR were reported to be physically associated, which enhances the ability of corticosteroids to stimulate GR-dependent transcription (42–44). Initial studies tested whether the GR and KLF15 were expressed in the same TG neurons during DEX-induced reactivation from latency. Consecutive sections were cut from TG of latently infected calves that were treated with DEX for 6 h to initiate reactivation from latency. One section was stained with a GR antibody (Ab), and one was stained with a KLF15 antibody. We frequently observed GR+ neurons 6 h after DEX treatment that were also KLF15 positive (KLF15+) (Fig. 1, DEX 6 GR and DEX 6 KLF, respectively; blue numbers denote TG neurons stained with both antibodies). GR+ neurons that were not readily detected by the KLF15 antibody were also detected (Fig. 1, blue letters). Finally, there were several neurons that were not readily stained by either antibody (for example, the neuron to the right of neuron H in Fig. 1). As previously reported in bovine TG from latently infected calves or mouse TG explants (26, 41), the GR and KLF15 were detected only in a small subset of TG neurons during latency (Fig. 1, latency GR or KLF15 panel). Since we detected the GR and KLF15 in many TG neurons following DEX treatment for 6 h, we suggest that this is part of the normal stress response and that many GR+ and KLF15+ neurons are not latently infected.
FIG 1.
KLF15 and the GR are frequently found in the same TG neuron during DEX-induced reactivation from latency. Immunohistochemistry (IHC) of consecutive sections was examined for KLF15 and GR expression in TG neurons when latently infected calves were treated with DEX for 6 h to initiate reactivation from latency, as described in the Materials and Methods section. The numbers denote TG neurons that are positive for KLF15 and the GR. The letters denote GR+ KLF15-negative neurons. As a control, TG sections from latently infected calves were examined for GR and KLF15 expression. Since GR and KLF15 were not readily detected in TG neurons during latency, consecutive sections were not examined. These results are representative of TG sections from two different calves.
To test whether KLF15 and the activated GR cooperated to stimulate productive infection, mouse neuroblastoma cells (Neuro-2A) were cotransfected with genomic DNA of the gCblue virus, and the effects of KLF15 and the GR were examined. Neuro-2A cells were used for these studies because they have neuron-like properties (45), can be readily transfected, and are permissive for BoHV-1 (46). Neuro-2A cells were transfected with BoHV-1 gCblue DNA instead of infecting cells because VP16 and other regulatory proteins in the virion diminish the stimulatory effects of DEX on productive infection (data not shown). KLF15 and the GR stimulated the number of β-galactosidase-positive (β-Gal+) Neuro-2A cells more than 7-fold, which was significantly higher than treatment with GR plus DEX or with the GR or KLF15 alone (Fig. 2A). Cotransfection of gCblue and the GR plus KLF15 stimulated productive infection 4-fold even when DEX was not added to cultures, which was higher than the effects seen by GR and DEX treatment and significantly different than when cultures were transfected with just gCBlue DNA. Additional studies examined the effect of KLF15 and the GR on productive infection in rabbit skin (RS) cells (Fig. 2B). In general, the trends in RS cells were the same as in Neuro-2A cells except that the levels of induction by KLF15 and the GR were approximately 2-fold less in RS cells than in Neuro-2A cells.
FIG 2.
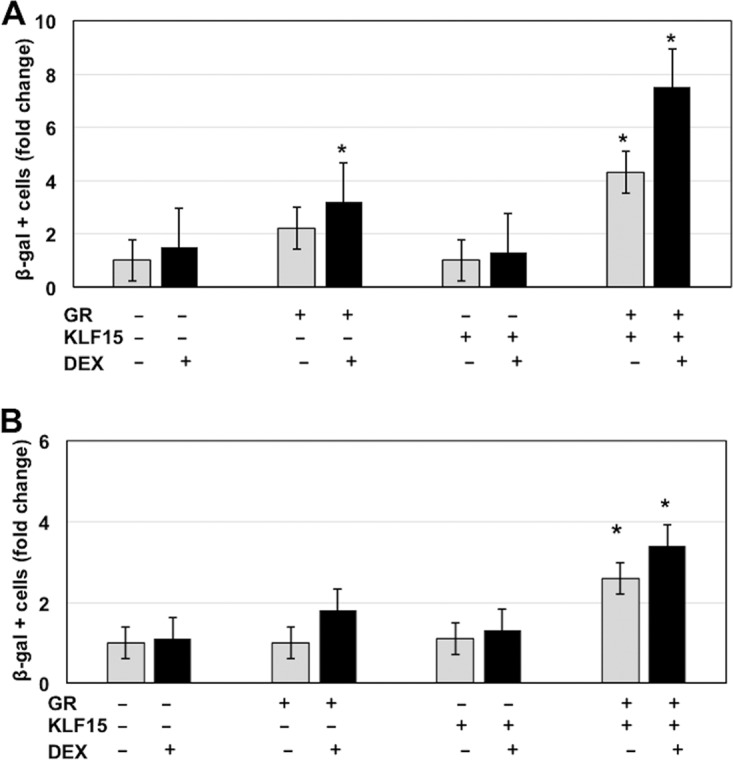
KLF15 and the GR cooperate to stimulate productive infection. Neuro-2A cells (A) or rabbit skin cells (B) were used for these studies. Twenty-four hours prior to transfection, 2% stripped fetal calf serum was added to the medium. Stripped fetal calf serum was used for these studies because normal serum contains steroid hormones, which activate the GR, as judged by nuclear localization of the GR following incubation with normal fetal calf serum (13). Cells incubated with stripped fetal calf serum for 24 h contain little or no nuclear GR. Cells were then transfected with 1.5 μg of BoHV-1 gCblue and, where indicated, a plasmid that expresses the mouse GR protein (1.0 μg DNA) and KLF15 (0.5 μg DNA). To maintain the same amount of DNA in each sample, empty vector was included in the samples. Designated cultures were then treated with water-soluble DEX (10 μM; Sigma). At 48 h after transfection, the number of β-Gal+ cells was counted. The value for the control (gCblue DNA treated with PBS after transfection) was set at 1. The results from DEX-treated cultures (black bars) were compared to those with the controls (white bars) and are an average of three independent studies. An asterisk denotes a significant difference between Neuro-2A cells transfected with BoHV-1 DNA (P < 0.05), using the Student's t test.
KLF15 and the GR cooperate to stimulate IEtu1 promoter activity.
To test whether KLF15 cooperated with the GR to stimulate IEtu1 promoter activity, transient-transfection studies were performed in Neuro-2A cells. A 280-bp fragment that contained both GREs within the IEtu1 promoter and flanking sequences was cloned upstream of the minimal SV40 early promoter and designated IEtu1 GREs (Fig. 3A and B). The IEtu1 GRE construct was used instead of the entire IEtu1 promoter because previous studies demonstrated that KLF15 transactivated an IEtu1 promoter construct that lacks the GREs (29), and the goal of this study was to test whether sequences adjacent to the GREs were stimulated by the GR and KLF15. We have consistently observed that KLF15 and the GR cooperated to stimulate the IEtu1 GRE construct approximately 40-fold whereas treatment with the GR plus DEX stimulated this construct approximately 8-fold (Fig. 3D).
FIG 3.
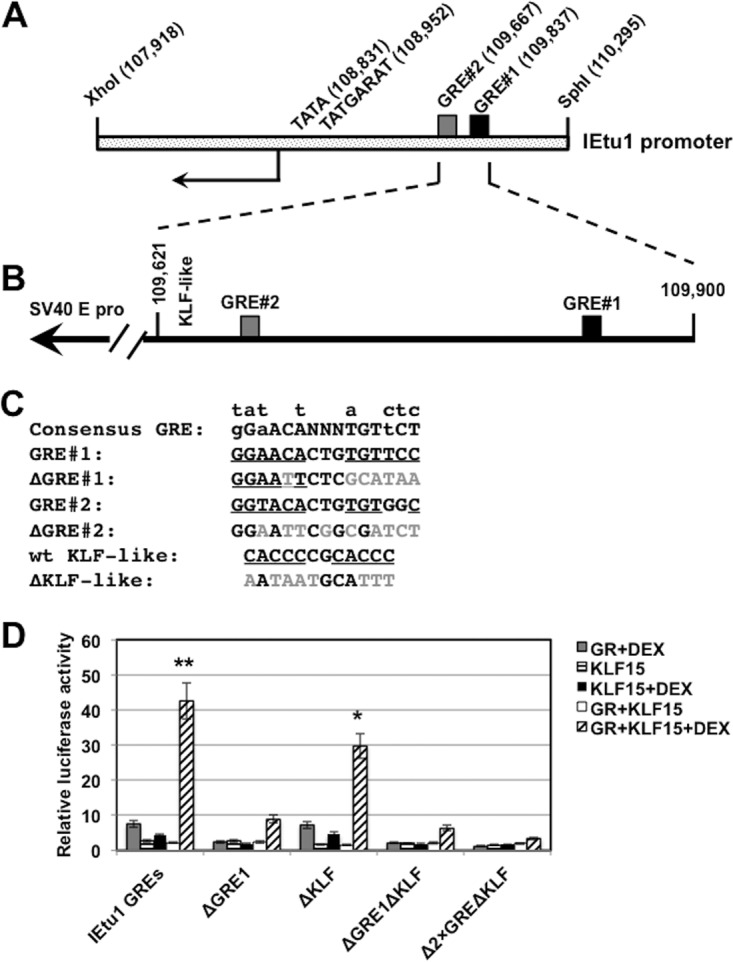
KLF15 and the GR cooperate to stimulate IEtu1 promoter activity. (A) Schematic of IEtu1 promoter and location of the TATA box, TAATGARAT motif, and the two GREs. Numbers in parentheses indicate the genomic location of the first nucleotide of each motif. (B) Schematic of the 280-bp fragment that contains the IEtu1 GREs and a KLF-like motif. This fragment was cloned upstream of the minimal SV40 early promoter in the luciferase vector (pGL3-promoter vector; Promega). (C) Nucleotide sequence of motifs in the IEtu1 GREs and mutations that were prepared. The mutations in GRE1 and GRE2 were previously shown to disrupt transactivation by the GR in transient-transfection studies (13). (D) Neuro-2A cells were treated with 2% stripped fetal calf serum 24 h prior to transfection. Neuro-2A cells were then transfected with the designated IEtu1 GRE plasmid (0.5 μg of DNA) and, where indicated, a plasmid that expresses the mouse GR protein (1.0 μg of DNA) and/or KLF15 (0.5 μg of DNA). To maintain the same amount of DNA in each sample, empty vector was included in certain samples. Designated cultures were then treated with water-soluble DEX (10 μM; Sigma) at 24 h after transfection. At 48 h after transfection, cells were harvested, and protein lysate was subjected to a dual-luciferase assay as described in the Materials and Methods section. Levels of promoter activity in the empty luciferase vector (pGL3-promoter vector) were normalized to a value of 1, and fold activation values for other samples are presented. The results are the average of three independent experiments, and error bars denote the standard errors. The double asterisks denote a significant difference (P < 0.05) between results for IEtu1 GREs relative to those for all mutant constructs following transfection with GR plus KLF15 and treatment with DEX, as determined by the Student t test. The single asterisk denotes a significant difference (P < 0.05) between results for the ΔKLF mutant relative to results for the other mutant constructs (ΔGRE1, ΔGRE1ΔKLF, and Δ2×GREΔKLF) when they are cotransfected with GR plus KLF15 and treated with DEX, as determined by the Student t test.
To identify sequences in the IEtu1 GREs that mediate transactivation by KLF15 and the GR, site-specific mutations of GRE1, GRE2, and KLF-like binding sites were prepared (Fig. 3C) and examined in Neuro-2A cells. A previous study demonstrated that the same GRE1 mutation interfered with transactivation mediated by GR plus DEX and was more important than GRE2 (13). Mutagenesis of both GREs abolished transactivation mediated by GR plus DEX (13). Disruption of GRE1 significantly inhibited transactivation by KLF15 and the GR (Fig. 3D). All KLF transcription factors contain similar DNA binding domains and thus can bind to a CACCC core motif (30, 47–49). Two KLF-like binding sites were identified in the IEtu1 GREs, and these motifs disrupted (Fig. 3C). Mutagenesis of the putative KLF-like binding sites reduced transactivation by KLF15 and the GR approximately 30%. Mutagenesis of the GRE1 and the KLF binding site (ΔGRE1ΔKLF) had results similar to those with the ΔGRE1 mutant. Mutagenesis of both GREs and the KLF binding site reduced KLF15- and GR-mediated transactivation to basal levels. In summary, mutagenesis of the GRE1 in the IEtu1 GRE fragment significantly reduced transactivation by KLF15 and the GRE.
Identification of intergenic regions in the BoHV-1 genome that contain putative GR and KLF binding sites.
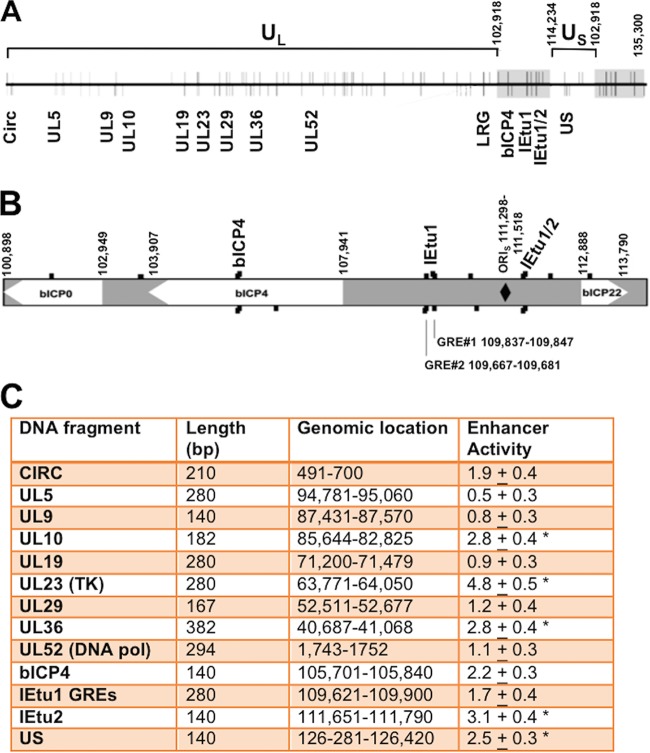
The BoHV-1 genome contains approximately 100 putative GREs (13) (Fig. 3A and B), suggesting that these sites play a role in stimulating viral gene expression following a stressful stimulus. We identified 13 regions in the viral genome that contained at least two putative GREs and a potential KLF binding site that were less than 400 bp apart (Fig. 4A to C). No preference was given to whether these sequences were contained in a known viral promoter because a GRE can be transactivated by the activated GR when the GR is located more than 5 kb from a start site of transcription (50). Sequences containing these regions were synthesized and cloned upstream of a minimal promoter (pGL3-promoter vector) to test whether these sequences were transactivated by stress-induced cellular transcription factors. The IEtu1 GRE construct was used for comparison with other viral fragments.
FIG 4.
Location of GREs on BoHV-1 and intergenic regions analyzed for stress-induced transcriptional activity. (A) Diagram of the linear BoHV-1 genome (horizontal line) with predicted GREs denoted by vertical lines. Lines above the genome represent GREs on the positive/forward DNA strand, while lines below indicate GREs on the negative strand. The terminal repeats are denoted by gray rectangles. Numbers denote genomic coordinates. Genomic regions that contain at least two putative GREs and potential KLF binding sites and are 400 bp or less are denoted by the gene in which they are located. LRG refers to the latency-related gene (10). The Circ gene encodes an IE transcript from the left end of the genome and spans covalently joined genome ends. These sequences were synthesized (Genescript), cloned into the pGL3-promoter vector, and then used for the studies described below. UL, unique long region. (B) Expanded genomic region that includes bICP0, bICP4, and bICP22. Genomic sequences located between bICP4 and bICP22 contain the origin of replication (ORIs, denoted by a black diamond) and IEtu1 and IEtu2 to the left and right of the ORIs, respectively (not highlighted). Black boxes denote enlarged GREs on positive (above) or negative (below) strands. GRE1 and GRE2 within the IEtu1 promoter are indicated, and the genomic region for the beginning and end of the sequences is given. (C) Neuro-2A cells were cotransfected with a plasmid containing the firefly luciferase gene downstream of the designated plasmid constructs (1 μg of DNA) and a plasmid encoding Renilla luciferase (0.05 μg of DNA) using Lipofectamine 3000. The level of promoter activity in the empty luciferase vector (pGL3-promoter vector) was normalized to a value of 1, and fold activation for other constructs is presented as enhancer activity. An asterisk denotes a significant difference between luciferase activities of the designated construct relative to the activity of the empty vector that contains only the SV40 early promoter (pGL3-promoter vector), using the Student's t test.
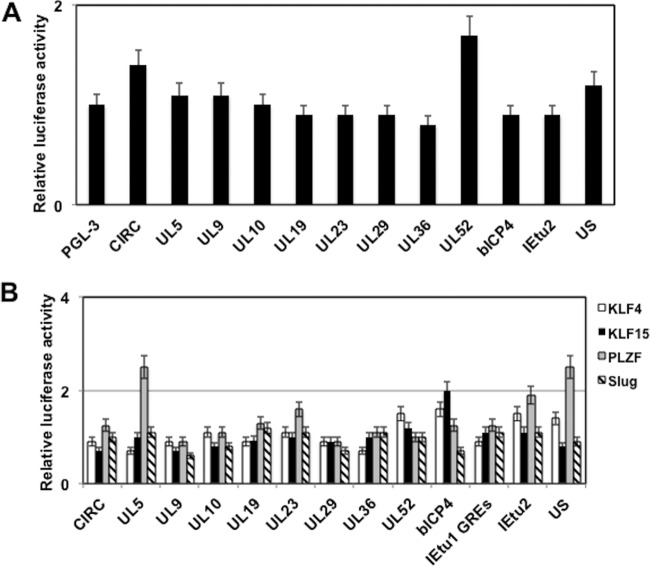
Initial studies tested whether any of these fragments functioned as a transcriptional enhancer in Neuro-2A cells (Fig. 4C). Relative to the empty vector (pGL3-promoter vector), the UL23 fragment consistently activated transcription 4.8-fold. Furthermore, the UL10, UL36, bICP4, IEtu2, and unique short region (US) fragments stimulated transcription by more than 2-fold compared to the level with the empty pGL3-promoter vector. In contrast, the UL5 fragment reduced promoter activity in Neuro-2A cells. In summary, this study suggested that certain intergenic fragments within the viral genome have weak enhancer activity.
Regulation of promoter activity by individual stress-induced transcription factors.
Although each fragment contained putative GREs, the ability of DEX to stimulate the respective fragments was not significantly different than that of the pGL3-promoter vector (Fig. 5A). Only the UL52 fragment was stimulated by DEX and the GR by approximately 2-fold.
FIG 5.
Identification of BoHV-1 intergenic regions that are regulated by stress-induced transcription factors. (A) Neuro-2A cells were cotransfected with the designated plasmid constructs (0.25 μg of DNA) containing the firefly luciferase gene that contains the BoHV-1 sequences, a plasmid that expresses the GR plasmid (1 μg of DNA), and a plasmid that expresses Renilla luciferase (0.05 μg of DNA). Following transfection, Neuro-2A cells were cultured in 2% stripped fetal calf serum after transfection. Twenty-four hours after transfection the designated Neuro-2A cultures were treated with water-soluble DEX (10 μM; Sigma). (B) Neuro-2A cells were cotransfected with the designated plasmid constructs (0.25 μg of DNA) containing the firefly luciferase gene and viral sequences, a plasmid encoding Renilla luciferase (0.05 μg of DNA), and a KLF transcription factor (KLF4, KLF15, or promyelocytic leukemia zinc finger [PLZF]) or a plasmid expressing Slug-1 (0.5 μg of DNA). The level of promoter activity in the empty luciferase vector (pGL3-promoter vector) was normalized to a value of 1, and fold activation values for other samples are presented. At 48 h after transfection, cells were harvested, and protein extracts were subjected to a dual-luciferase assay as described in the Materials and Methods section. The level of promoter activity in the empty luciferase vector (pGL3-promoter vector) was normalized to a value of 1, and fold activation values for other samples are presented. The results are the average of three independent experiments, and error bars denote the standard errors.
Microarray and immunohistochemistry studies revealed that three KLF family members were induced during early stages of reactivation from latency in TG neurons: KLF4, KLF15, and promyelocytic leukemia zinc finger (PLZF) (29). Slug was also examined because it is induced by DEX in TG during reactivation from latency and stimulated productive infection. PZLF stimulated the UL5 and US fragment more than 2-fold (Fig. 5B). KLF15 transactivated the promoter construct containing the bICP4 intergenic fragment approximately 2-fold. KLF4 and Slug essentially had no effect on any of the fragments that we examined. In summary, the individual stress-induced transcription factors identified in TG neurons did not dramatically transactivate any of the intergenic viral fragments inserted upstream of the SV40 early promoter compared to effect of the empty pGL3-promoter vector.
KLF15 cooperates with the GR to activate enhancer activity.
As discussed above, KLF15 and the GR were reported to interact with each other, and KLF15 is a coactivator of GR-dependent transcription (42–44). Thus, we examined the effect that KLF15 and the GR had on promoter activity of constructs containing intergenic regions. The UL52 intergenic region was stimulated 12-fold by treatment with GR and KLF15 plus DEX (Fig. 6A). Furthermore, the bICP4, IEtu2, and US fragments were stimulated more than 3-fold, which was higher than the effect of GR plus DEX and significantly different from the level with the empty vector control. In the absence of DEX, KLF15 and the GR had little effect on transcriptional activity of any construct, confirming that the transfection protocol did not dramatically activate the GR when cultures were incubated with stripped serum.
FIG 6.
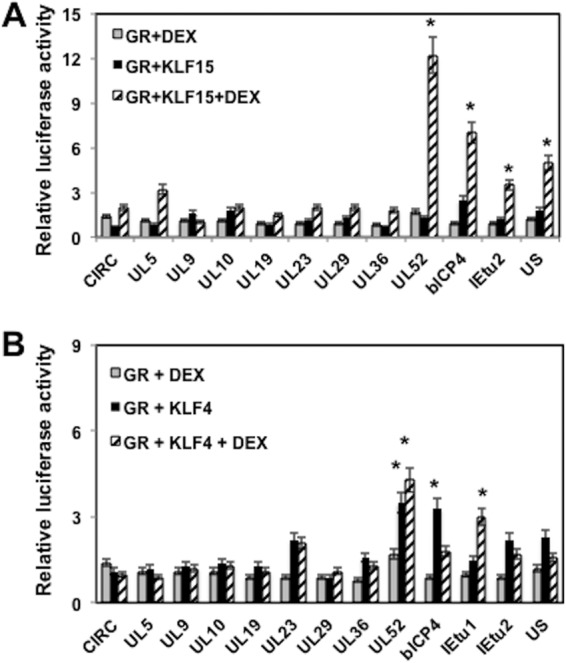
GR and KLF15 cooperate to transactivate certain BoHV-1 intergenic regions. Neuro-2A cells were cotransfected with the designated plasmid constructs (0.25 μg of DNA) containing the firefly luciferase gene downstream, a plasmid encoding Renilla luciferase (0.05 μg of DNA), KLF15 (A) or KLF4 (B) (0.5 μg of DNA), and the GR expression plasmid (1 μg of DNA). To maintain equal plasmid amounts in the transfection mixtures, the empty expression vector was added as needed. For these studies, Neuro-2A cells were cultured in 2% stripped fetal calf serum after transfection. Twenty-four hours after transfection the designated Neuro-2A cultures were treated with water-soluble DEX (10 μM; Sigma). At 48 h after transfection, cells were harvested, and protein extracts were subjected to a dual-luciferase assay as described in the Materials and Methods section. The results are the average of three independent experiments, and error bars denote the standard errors. An asterisk denotes a significant difference (P < 0.05) between the value for the empty control (pGL3-promoter vector), as determined by the Student t test. It should be noted that for UL52 the transactivation levels obtained with KLF4 plus GR were not significantly different when DEX was added.
As a control, we tested whether KLF4 cooperated with the GR to transactivate any of the fragments (Fig. 6B). The UL52 fragment was transactivated significantly more by KLF4 plus GR than by GR plus DEX or by the empty vector; however, addition of DEX to cultures transfected with KLF4 and the GR did not significantly increase promoter activity. Furthermore, the bICP4 fragment was transactivated more efficiently by KLF4 and the GR in the absence of DEX. Relative to treatment with DEX plus the GR, the IEtu1 GREs were transactivated approximately 3-fold by KLF4 and the GR in the presence of DEX; however, the difference was not as dramatic as that with KLF15 and the GR. In summary, these studies determined that KLF15 and the GR cooperated to transactivate promoter constructs that contained the UL52 and bICP4 intergenic fragments. Although KLF4 had an effect on GR-mediated transactivation, the effects were not as dramatic as those of KLF15.
Localization of sequences in the UL52 region that are responsive to KLF15 and the GR.
The UL52 intergenic region contains two GRE half-binding sites, a cluster of Sp1 binding sites, and a KLF-like binding site (Fig. 7A). GRE half-binding sites can be bound and transactivated by GR monomers, but adjacent transcription factor binding sites play a bigger role than “whole” GREs (22, 50–52). Mutations in each of these sites were synthesized (Fig. 7B), and their ability to be transactivated by KLF15 and the GR examined in Neuro-2A cells. Mutations in the GRE1 half-binding site and Sp1 binding sites reduced transactivation to approximately 3-fold (Fig. 7C). Although the ΔGRE2 half-binding site and ΔKLF mutants were not transactivated as efficiently by KLF15 and the GR, the level of transactivation was more than 4-fold.
FIG 7.
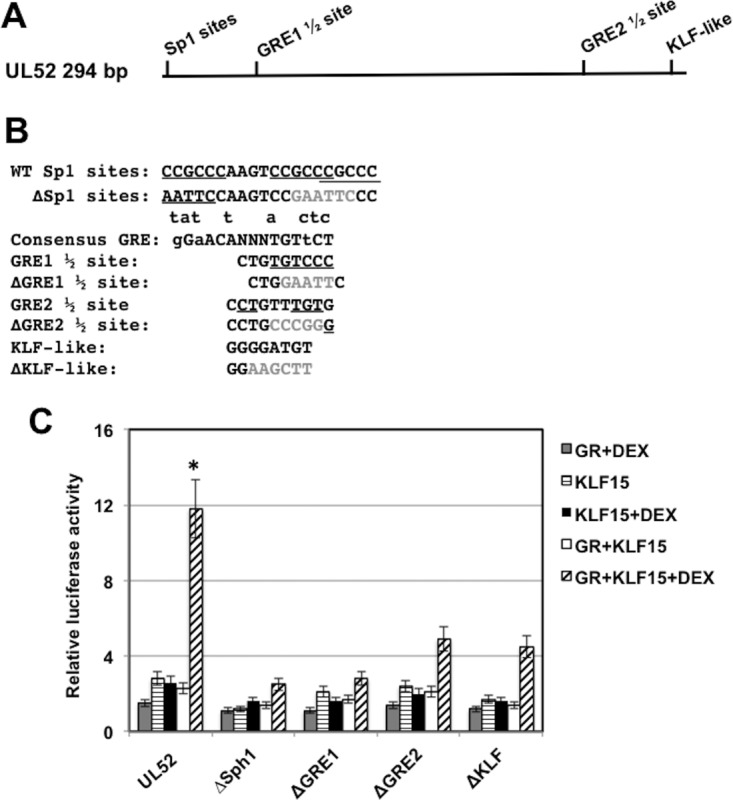
Identification of UL52 intergenic sequences that are important for transactivation by KLF15 and the GR. (A) Location of the GRE half-binding sites (GRE1 1/2 and GRE2 1/2) and potential KLF binding sites in the 294-bp UL52 fragment. (B) UL52 sequences contain three closely linked and partially overlapping Sp1 binding sites that are underlined. The ΔSp1 binding site mutant replaced the Sp1 binding site with an EcoRI restriction enzyme site (GAATTC). The consensus GRE binding site is shown (small letters above consensus are nucleotides that can be part of a functional GRE). The GRE1 half-binding site is shown, and the underlined nucleotides match the consensus. The ΔGRE1 half-binding site contains an EcoRI restriction enzyme site (GAATTC) that replaced the GRE sequences. The GRE2 half-binding site is shown, and the underlined nucleotides match the consensus. The ΔGRE2 half-binding site contains an SmaI restriction enzyme site (CCCGGG) that replaced GRE sequence. The KLF-like binding site is shown, and the ΔKLF mutant contains a HindIII restriction enzyme site (AAGCTT) that replaced the KLF-like binding motif. (C) Neuro-2A cells were cotransfected with the designated plasmid constructs (0.25 μg of DNA) containing the firefly luciferase gene downstream, a plasmid encoding Renilla luciferase (0.05 μg of DNA), KLF15 (0.5 μg DNA), and the GR-expressing plasmid (1 μg of DNA). To maintain equal plasmid amounts in the transfection mixtures, the empty expression vector was added as needed. For these studies, Neuro-2A cells were cultured in 2% stripped fetal calf serum after transfection. Twenty-four hours after transfection the designated Neuro-2A cultures were treated with water-soluble DEX (10 μM; Sigma). At 48 h after transfection, cells were harvested, and protein extracts were subjected to a dual-luciferase assay as described in the Materials and Methods section. The results are the average of three independent experiments, and error bars denote the standard errors. The asterisk denotes a significant difference (P < 0.05) between results with UL52 and the mutant constructs when they are cotransfected with GR plus KLF15 and treated with DEX, as determined by a Student t test.
The GR is associated with KLF15 in transfected Neuro-2A cells.
To confirm whether the GR is associated with KLF15, Neuro-2A cells were cotransfected with plasmids that express KLF15 and the GR, and coimmunoprecipitation (co-IP) studies were performed. Following IP with the GR antibody, we consistently detected KLF15 in the immunoprecipitate when cultures were treated with DEX (Fig. 8A) and when cultures were not treated with DEX after transfection (Fig. 8B). KLF15 was also immunoprecipitated by the GR antibody when the GR was not overexpressed (data not shown). When the IP was performed with the KLF15 antibody, the GR was not readily detected in the immunoprecipitate (Fig. 8A and B) unless the blots were overexposed (lanes IP KLF15*). The KLF15 antibody used for this study was produced in goats using a 13-amino-acid peptide, suggesting that this epitope was masked when KLF15 interacted with other cellular proteins or that this epitope does not yield antibodies that efficiently immunoprecipitate the protein. However, the KLF15 antibody specifically recognized KLF15 in Western blot studies.
FIG 8.
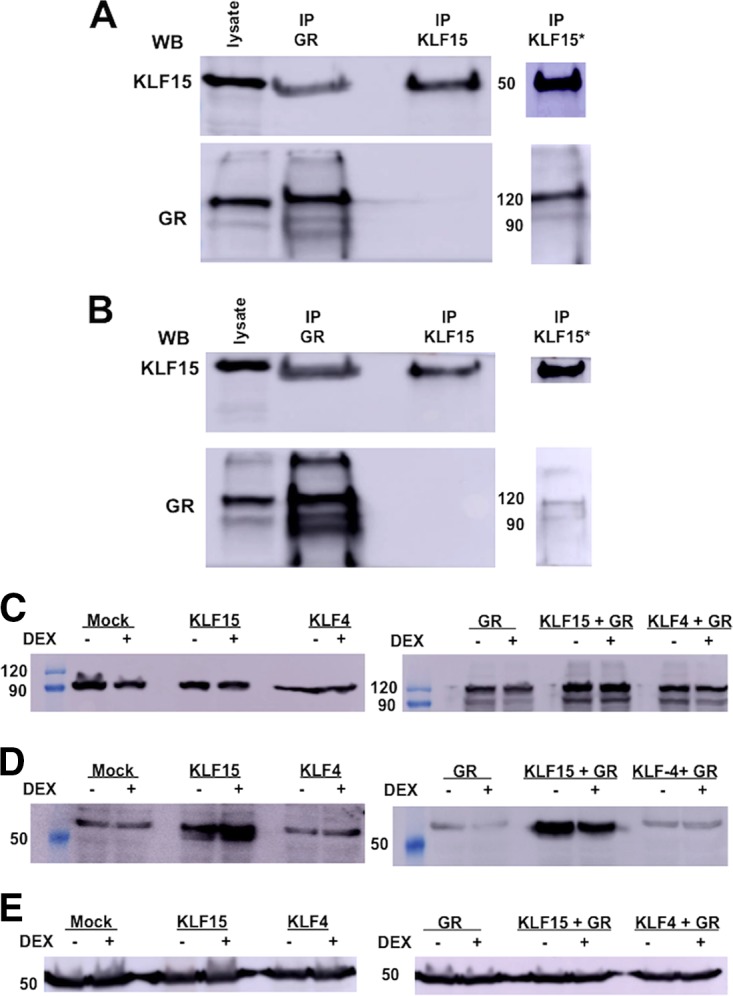
The GR interacts with KLF15. Neuro-2A cells were grown to 80% confluence on 100-mm dishes. Cells were cotransfected with plasmids that express KLF15 (1.5 μg) and the GR (2 μg). Cultures were treated with DEX (10 μM) in 2% stripped-serum medium for 4 h (A) before cell lysate was harvested or treated with DEX (B). Whole-cell lysate was prepared, and co-IP studies were performed using the GR or KLF15 antibody as described in Materials and Methods. Following IP with the designated antibody, the GR or KLF15 was detected in the immunoprecipitate by Western blotting (WB). Input lysate (50 μg of protein) was used as a positive control. (C) Neuro-2A cells were cotransfected with plasmids that express KLF15 (0.5 μg), KLF4 (0.5 μg), and the GR (1 μg) as indicated in the figure. Whole-cell lysate was prepared using RIPA buffer, proteins were separated by SDS-PAGE (50 μg in each lane), and GR expression was detected by Western blotting. Cell lysate from nontransfected Neuro-2A cells was used to examine endogenous GR expression. (D) Neuro-2A cells were cotransfected with plasmids that express KLF15 (0.5 μg), KLF4 (0.5 μg), and the GR (1 μg) as indicated on the figure. Whole-cell lysate was prepared using RIPA buffer, proteins (50 μg in each lane) were separated by SDS-PAGE, and Western blot analysis was performed using anti-KLF15. Cell lysate from nontransfected Neuro-2A cells was used to examine endogenous KLF15 expression. (E) As a loading control, tubulin levels were examined. For each lane, 50 μg was loaded. Values on the sides of the blots indicate molecular mass in kilodaltons.
Additional studies were performed to test whether interactions between KLF15 and the GR increased the stability of either protein. Endogenous levels of the GR were not altered dramatically when Neuro-2A cells were transfected with the KLF15 or KLF4 expression plasmid. When the GR expression plasmid was transfected into Neuro-2A cells, there was an additional GR-specific band that migrated at approximately 120 kDa, whereas the endogenous GR-specific band expressed in Neuro-2A cells migrated at approximately 90 kDa (Fig. 8 C, right panel). It is well established that the GR primary transcript is alternatively spliced and that different GR protein isoforms are expressed that have different activities (51). Thus, we suggest that the endogenous GR protein expressed in Neuro-2A cells is translated from an alternatively spliced transcript that has little transactivating activity or that the endogenous GR is prone to proteolysis. When the GR and KLF15 were cotransfected into Neuro-2A cells, we observed KLF15 protein levels similar to those in cells transfected with just the KLF15 expression plasmid, suggesting that the 90- and 120-kDa GR proteins did not dramatically affect KLF15 protein levels (Fig. 8D, right panel). As expected, similar levels of the loading control, tubulin, were expressed in Neuro-2A cells regardless of which plasmid was used to transfect these cells (Fig. 8E). In summary, these studies revealed that the GR interacted with KLF15 and that this interaction was independent of DEX treatment.
KLF15 and the GR interact with sequences located in the IEtu1 GREs and UL52 fragment.
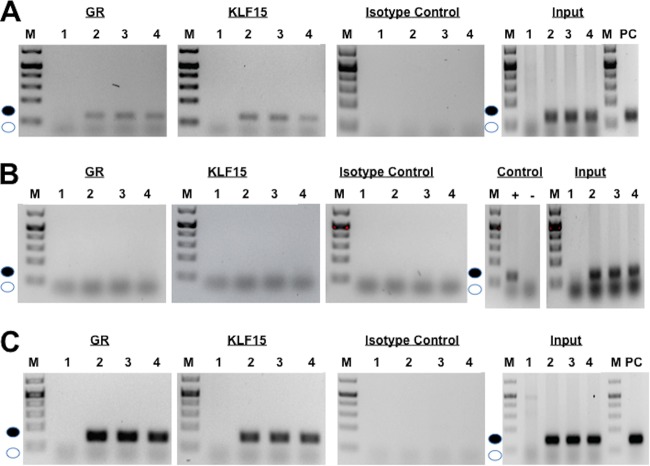
To test whether KLF15 and the GR interact with sequences located in the IEtu1 GREs and the UL52 fragment, chromatin immunoprecipitation (ChIP) studies were performed. For this study, Neuro-2A cells were transfected with the promoter construct containing the IEtu1 GREs (Fig. 9A, lane 2) and a plasmid expressing the KLF15 and GR without DEX (lane 3) or with DEX treatment (lane 4). As previously reported (13), the GR was bound to IEtu1 promoter sequences containing the GREs whether cells were transfected with the GR expression vector or treated with DEX, as judged by amplification of the 107-bp fragment (Fig. 9A, GR panel, lanes 2 to 4, filled circle). Additional studies demonstrated that KLF15 interacted with sequences in the IEtu1 GREs in transfected Neuro-2A cells whether these cells were transfected with a plasmid expressing the mouse GR or treated with DEX (Fig. 9A, KLF15 panel, lanes 2 to 4). In contrast, the 107-bp PCR product spanning the IEtu1 GREs was not detected when the IP was performed using an isotype control antibody (Fig. 9A). All input samples (prior to IP) yielded the specific 107-bp PCR product except Neuro-2A cells not transfected with the IEtu1 GRE construct (Fig. 9A, input panel, lane 1). Since the mutant lacking the two GREs and KLF (Δ2×GREΔKLF) was not transactivated by KLF15 and the GR (Fig. 3D), we tested whether the GR and KLF15 were bound to Δ2×GREΔKLF sequences. In contrast to results with a wild-type (wt) IEtu1 GRE construct, we were unable to detect binding of KLF15 and the GR to sequences in the Δ2×GREΔKLF plasmid following transfection of Neuro-2A cells (Fig. 9B). As expected, the primers amplified the positive control for Δ2×GREΔKLF and the input samples that contained the Δ2×GREΔKLF plasmid.
FIG 9.
Interaction between GR and KLF15 with IEtu1 GREs and UL52 sequences. Neuro-2A cells were cotransfected with the IEtu1 GRE construct (A; 4 μg of DNA), the Δ2×GREΔKLF (B; 4 μg of DNA), or the UL52 plasmid (C; 4 μg of DNA) and KLF15 and/or the GR plasmid (1 μg of DNA). Empty vector was added to maintain the same concentration of DNA in each transfection assay. Designated cultures were treated with DEX as described above. (A) Neuro-2A cells were transfected with no plasmid (lanes 1), with the IEtu1 GRE construct (lanes 2), with IEtu1 GREs, KLF15, and the GR (no DEX treatment) (lanes 3), and with IEtu1 GREs, KLF15, and the GR (lanes 4). Cultures were treated with DEX (10 μM) in 2% stripped-serum medium for 4 h before cells were harvested. (B) Neuro-2A cells were transfected with no plasmid (lanes 1), with the Δ2×GREΔKLF construct (lanes 2), with Δ2×GREΔKLF, KLF15, and the GR (no DEX treatment) (lanes 3), and with Δ2×GREΔKLF, KLF15, and the GR (lanes 4). Cultures were treated with DEX (10 μM) in 2% stripped-serum medium for 4 h before cells were harvested. (C) Neuro-2A cells were transfected with no plasmid (lanes 1), with a UL52 construct (lanes 2), with UL52, KLF15, and the GR (no DEX treatment) (lanes 3), and with UL52, KLF15, and the GR (lanes 4). Cultures were treated with DEX (10 μM) in 2% stripped-serum medium for 4 h before cells were harvested. Transfected cells were processed for ChIP as described in the Materials and Methods section, and immunoprecipitation (IP) was conducted using the GR antibody, KLF15 antibody, or isotype control antibody. Input was 10% of the total DNA-protein complexes that were used for IP, and then PCR was performed using PCR primers described in the Materials and Methods section. The filled circle denotes the specific PCR fragment of 107 bp for the IEtu1 GREs, 107 bp for Δ2×GREΔKLF, or 132 bp for UL52, and the open circle denotes the position of primer dimers. These results are representative of five independent studies for the GR ChIP and of two for the KLF15 ChIP and Δ2×GREΔKLF. M, molecular mass marker.
KLF15 (Fig. 9C, KLF15, lanes 2 to 4) and the GR (Fig. 9C, GR, lanes 2 to 4) were also bound to the UL52 fragment, as judged by specific amplification of a 132-bp fragment. When the IP was performed with the isotype control antibody, the 132-bp PCR product was not detected (Fig. 9C, isotype panel, lanes 1 to 4). As expected, the input samples yielded the specific 132-bp PCR product in all samples initially transfected with the UL52 promoter construct (Fig. 9C, input panels). In summary, these studies revealed that KLF15 and the GR specifically interacted with the IEtu1 GREs and UL52 fragment, which correlated with their ability to be transactivated by these two stress-inducible transcription factors.
DISCUSSION
In this study, we provided evidence that two stress-induced cellular transcription factors (KLF15 and GR) cooperated to stimulate productive infection and transactivate the IEtu1 GREs as well as several intergenic fragments (UL52, bICP4, IEtu2, and US). DEX, which mimics the effects of stress, consistently induces BoHV-1 reactivation from latency, in part because lytic cycle viral RNA and proteins are detected in TG neurons within hours after treatment (17, 18, 26–28, 53). In response to stress, the GR and KLF15 regulate gene expression dynamics and integrate signals by a feed-forward loop (42, 43, 54). The hallmark of a feed-forward loop is a primary factor (GR in this case) that stimulates expression of a second factor, KLF15 (29, 41–43, 54–56), and the two factors activate expression of genes in a specific pathway, for example, genes that regulate adipogenesis (57) and amino acid metabolizing enzymes (42, 43). Our studies suggest that activation of BoHV-1 gene expression following a stressful stimulus is, in part, regulated by a feed-forward loop containing the GR and KLF15.
The IEtu1 promoter is a unique viral promoter because the TATA box and single TAATGARAT motif are located more than 700 bases from the first GRE (Fig. 2A, GRE2). Sequences containing the two GREs are not required for VP16-mediated activation of the IEtu1 promoter (34) or transactivation by KLF4 and KLF15 (29). Thus, it was not surprising that the fragment spanning the IEtu1 GREs was not transactivated efficiently by KLF15 alone. Based on these observations, we suggest that the GREs are not required for regulating bICP0 and bICP4 IE expression during productive infection. Consequently, it should be possible to construct a mutant virus lacking the IEtu1 GREs that grows efficiently in cultured cells. Thus, the precise role that the IEtu1 GREs play during stress-mediated reactivation from latency in calves can be directly assessed.
KLF15, like other KLF family members, resembles the Sp1 transcription factor family, and both transcription factor families interact with GC-rich motifs (30, 47). It is well established that the BoHV-1 genome, as well as other Alphaherpesvirinae subfamily members, are GC rich, and many viral promoters contain Sp1 consensus binding sites as well as additional GC-rich motifs (30). Mutating the GREs within the IEtu1 GREs and UL52 construct was crucial for transactivation by the GR and KLF15, indicating that the GRE is required. Overlapping Sp1 binding sites and GREs were important for transactivation of the UL52 fragment by KLF15 and GR; however, the IEtu1 GREs lacked similar motifs. Several KLF15 binding sites have been reported (58–61), suggesting that sequence requirements for cooperative transactivation by KLF15 and the GR are flexible and may be difficult to identify by merely analyzing the sequence. It is also possible that when the GR is associated with KLF15, the GR initially binds to a GRE, and KLF15 DNA binding properties are not as specific.
The finding that the BoHV-1 genome contains 100 putative GREs (13) suggests that additional GREs play a role in stimulating productive infection and viral gene expression following stressful stimuli. Although many of the intergenic GREs were not transactivated by the GR or KLF15 alone, the GR and KLF15 cooperated to transactivate fragments derived from UL52, the bICP4 open reading frame (ORF), IEtu2, and US. Certain GREs in cellular chromatin can stimulate transcription from more than 5 kb to the nearest promoter (50). Furthermore, the activated GR can specifically bind transcriptionally silent chromatin (62–64) and form a nuclease-hypersensitive site, culminating in transcriptional activation (65, 66). These properties are consistent with the GR being termed a “pioneer transcription factor” (67, 68), which can activate transcription of certain viral genes when the GRE is not adjacent to core promoter sequences.
The ability of corticosteroids to interfere with immune responses (31, 32, 69) is also likely to be a contributing factor for increasing the incidence of reactivation from latency. For example, the activated GR interacts with two cellular transcription factors, activating protein 1 (AP-1) and nuclear factor kappa light-chain enhancer of activated B cells (NF-κB) that promote inflammation (reviewed in references 22, 33, and 70). AP-1 and NF-κB are required for activating the beta interferon promoter/enhancer following virus infection (71), which is significant because beta interferon has anti-herpesvirus activity, including interfering with reactivation from latency (72–77). Furthermore, infiltrating lymphocytes in TG undergo apoptosis following corticosteroid treatment (31, 32, 69, 78), and soluble factors produced by T cells (79, 80) and/or dendritic cells (81, 82) were reported to interfere with reactivation from latency. A previous study revealed that DEX treatment of latently infected calves leads to viral gene expression prior to apoptosis of infiltrating lymphocytes in TG (27). These observations suggest that the immune-inhibitory functions of corticosteroids are not critical during the early burst of viral gene expression after a reactivation stimulus but are likely to be important for restricting virus shedding and spread at later stages of reactivation from latency.
MATERIALS AND METHODS
Cells and virus.
Mouse neuroblastoma cells (Neuro-2A), rabbit skin (RS) cells, and bovine kidney cells (CRIB) were grown in Eagle's minimal essential medium (EMEM) supplemented with 10% fetal calf serum (FCS), penicillin (10 U/ml), and streptomycin (100 μg/ml).
A BoHV-1 mutant containing the β-Gal gene in place of the viral gC gene was obtained from S. Chowdury (LSU School of Veterinary Medicine) (gCblue virus), and stocks of this virus were grown in CRIB cells. The gCblue virus grows to similar titers as the wt parental virus and expresses the LacZ gene. Procedures for preparing genomic DNA were described previously (83).
Infection of calves and immunohistochemistry studies.
All TG samples from calves used for this study were previously described (27, 29, 84, 85). In brief, BoHV-1-free crossbred calves (∼200 kg) were inoculated with 107 PFU of BoHV-1 into ocular and nasal cavities as described previously (17, 83, 86–89). At 60 days postinfection (dpi), calves were not shedding virus and were operationally defined as being latently infected. Certain latently infected calves were injected intravenously in their jugular vein with 100 mg of water-soluble DEX (D2915; Sigma) to initiate reactivation from latency. Experiments were performed in accordance with American Association of Laboratory Animal Care and University of Nebraska IACUC committee guidelines (A3459).
Immunohistochemistry (IHC) studies were performed using an ABC kit (Vector Laboratories) according to specifications of the manufacturer as previously described (83). Thin sections (4 to 5 μm) of TG were cut, and the consecutive sections were numbered, mounted on glass slides, and processed as described previously (41, 84, 90). One slide was stained with anti-GR antibody (3660S; Cell Signaling) using a 1:100 antibody dilution, and a consecutive slide was incubated with the KLF15 antibody (ab2647; Abcam) using a 1:250 antibody dilution overnight in a humidified chamber at 4°C. The next day, slides were washed in 1× Tris-buffered saline (TBS) and incubated in biotinylated goat anti-rabbit IgG (PK-6101; Vector Laboratories) for 30 min at room temperature in a humidified chamber. Avidin-biotinylated enzyme complex was added to the slides for 30 min of incubation at room temperature. After three washes in 1× TBS, slides were incubated with freshly prepared substrate (SK-4800; Vector Laboratories), rinsed with distilled water, and counterstained with hematoxylin.
Quantification of β-Gal-positive cells.
Neuro-2A cells and RS cells grown in 60-mm plates were cotransfected with 1.5 μg of the gCblue viral genome and the designated amounts of plasmid expressing GR or KLF15, as stated in the legend to Fig. 5, using Lipofectamine 3000 (catalog no. L3000075; Invitrogen). At 48 h after transfection, cells were fixed with a solution containing 2% formaldehyde and 0.2% glutaraldehyde in phosphate-buffered saline (PBS) and then stained with a solution containing 1% Bluo-Gal, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 0.5 M MgCl2 in PBS. The number of β-galactosidase (β-Gal)-positive cells was determined as described previously (17, 83, 91, 92). In brief, the number of β-Gal-positive cells in cultures expressing the blank vector was set at 1 for each experiment. To calculate fold change of β-Gal-positive cells, the number of blue cells in cultures transfected with the plasmids of interest were divided by the number of blue cells in cultures transfected with the blank vector. The effect that KLF15, DEX, and overexpression of the GR had on productive infection is expressed as fold induction relative to the control level. This representation of the data minimized the differences in cell density, Lipofectamine 3000 lot variation, and transfection efficiency.
Plasmids.
A 280-bp fragment that contains the two GREs located in the IEtu1 promoter was synthesized by Genescript and cloned into the pGL3-promoter vector at the unique KpnI and XhoI restriction sites (IEtu1 GREs). Mutants of the IEtu1 GREs were synthesized by Genescript and cloned into the pGL3-promoter vector at KpnI and XhoI restriction enzyme sites. These mutants are ΔKLF, ΔGRE1, ΔGRE1ΔKLF, Δ2×GREΔKLF and are described in Fig. 3C. The UL52 mutant constructs were synthesized by Genescript and were cloned into the pGL3 -promoter vector at KpnI and XhoI restriction enzyme sites (Fig. 7).
A mouse GR expression vector was obtained from Joseph Cidlowski, NIH. The KLF4 expression vector was obtained from Jonathan Katz (University of Pennsylvania). The KLF15 expression vector was obtained from Deborah Otteson (University of Houston). The PLZF expression vector was obtained from Derek Sant'Angelo (Sloan-Kettering Cancer Center). The Slug expression vector was obtained from Paul Wade (NIEHS, Research Triangle Park, NC). All plasmids were prepared from bacterial cultures by alkaline lysis and two rounds of cesium chloride centrifugation.
Transfection and dual-luciferase reporter assay.
Neuro-2A cells (8 × 105) were seeded into 60-mm dishes containing EMEM with 10% FCS at 24 h prior to transfection. Two hours before transfection, medium was replaced with fresh growth medium lacking any antibiotics. Cells were cotransfected with the designated plasmid containing the firefly luciferase gene downstream of the SV40 early promoter (0.25 μg of plasmid DNA) and a plasmid encoding Renilla luciferase under the control of a minimal herpesvirus thymidine kinase (TK) promoter (50 ng of DNA) as described in the legends to Fig. 3, 4C, and 5 to 7 and Table 1. To maintain equal plasmid amounts in the transfection mixtures, the empty expression vector was added as needed. Neuro-2A cells were incubated in 2% charcoal-stripped fetal calf serum after transfection. At 24 h after transfection, Neuro-2A cultures were treated with water-soluble DEX (10 μM [D2915; Sigma]). Forty-eight hours after transfection, cells were harvested, and protein extracts were subjected to a dual-luciferase assay using a commercially available kit (E1910; Promega). Luminescence was measured by using a GloMax 20/20 luminometer (E5331; Promega).
Coimmunoprecipitation studies and Western blot analysis.
Neuro-2A cells grown on 100-mm dishes were cotransfected with plasmids that express KLF15 (1.5 μg of DNA) and the mouse GR (2 μg of DNA). Cultures were treated with DEX (10 μM) in 2% stripped-serum medium for 4 h before harvesting of transfected cultures. Whole-cell extracts were prepared with radioimmunoprecipitation assay (RIPA) lysis buffer with 1× protease inhibitor cocktail (Thermo Fisher Scientific), and protein concentration was quantified. Protein lysate (500 μg) was combined with anti-GR and/or anti-KLF15 (5 μg) antibody, and the reaction mixtures were incubated overnight at 4°C on a rotator. Protein A Dynabeads (catalog number 10001D; Life Technologies) were added and incubated for 2 h at 4°C with rotation. Immunoprecipitates were collected using a magnet DynaMag, (catalog no. 12321D; Life Technologies), the supernatants were removed, and the Dynabeads-antigen (Ag)-Ab complexes were washed three times with 1 ml of washing buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS). Proteins were eluted from Dynabeads by incubation with 30 μl of elution buffer (1% SDS, 100 mM NaHCO3); the samples were vortexed and then incubated in a water bath at 42°C for 30 min. The eluent was loaded onto an SDS-PAGE gel and Western blotting was performed as described below.
At 48 h after transfection, whole-cell lysate was prepared. Cultures were washed with phosphate-buffered saline (PBS) and suspended in RIPA lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 mM EDTA [pH 8.0], 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and one tablet of complete protease inhibitor (Roche Molecular Biochemicals) in 10 ml of buffer. Cell lysate was incubated on ice for 30 min, sonicated, and then clarified by centrifugation at 13,000 rpm at 4°C for 15 min. Protein concentrations were quantified by the Bradford assay. For SDS-PAGE, proteins were mixed with an equal amount of 2× sample loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 50 mM dithiothreitol, 0.1% bromophenol blue, 10% glycerol) and boiled for 5 min. Proteins were separated in an SDS–10% PAGE gel. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore) and blocked for 1 h in 5% (wt/vol) nonfat dry milk with 1× Tris-buffered saline–0.1% Tween 20 (TBS-T). Membranes were then incubated with the designated primary antibody at 4°C as described for Fig. 8 with gentle shaking overnight. The primary antibody was diluted 1:1,000 in the blocking solution. An antibody directed against tubulin was used as a loading control. After 45 min of washing with TBS-T, the blots were incubated with secondary antibodies (peroxidase-conjugated immunoglobulin G [Amersham Biosciences]), which were diluted 1:2,000 in 5% nonfat milk in TBS-T for 1 h. Blots were washed 45 min with TBS-T and exposed to Amersham ECL reagents, and then autoradiography was performed. Primary antibodies were purchased from Cell Signaling (anti-GR [D8H2], 3660S) and Abcam (anti-KLF4 antibody [ab72543] and anti-KLF15 [ab2647]). The secondary donkey anti-rabbit antibody (NA9340V) was purchased from GE Healthcare, secondary sheep anti-mouse antibody was purchased from GE Healthcare, and the secondary donkey anti-goat (sc-2020) was purchased from Santa Cruz Biotechnology.
ChIP assay.
Chromatin immunoprecipitation (ChIP) studies were performed, as previously described, in Neuro-2A cells (13), except that Lipofectamine 3000 (L3000075; Invitrogen) was used as a transfection reagent. Neuro-2A cells were grown on 100-mm dishes and cotransfected with the designated promoter constructs as described in the legend to Fig. 9. Cultures were treated with DEX (10 μM) in 2% stripped-serum medium for 4 h before cells were harvested. Neuro-2A cells were washed twice with phosphate-buffered saline (PBS). A volume of 0.5 ml of 16% paraformaldehyde was added for cross-linking with gentle shaking at 20°C for 10 min. Cross-linking was stopped by addition of 0.5 ml of 2.5 M glycine with shaking for 5 min. Cells were pelleted by centrifugation at 1,000 × g at 4°C followed by two washes with ice-cold PBS. The final pellet was suspended in 0.75 ml of cell lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8.0, 1% Triton X, 0.1% sodium deoxycholate, 0.1% SDS) with 1× protease inhibitors (catalogue no. 78425; Thermo Fisher Scientific) and incubated on ice for 10 min. Cells were vortexed every 2 min to promote lysis. The suspension was then sonicated 10 times for 10 s on ice. Sonicated samples were centrifuged at 8,000 × g for 1 min at 4°C, and the supernatant was divided into two tubes. Samples were precleared by adding 75 μl of agarose-salmon sperm DNA protein A beads (catalog no. 16-201; Millipore) and incubation for 1 h at 4°C. Agarose beads were removed by centrifugation. Samples were diluted in RIPA buffer (50 mM Tris HCl [pH 8.0], 150 mM NaCl, 2 mM EDTA [pH 8.0], 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) with 1× protease inhibitors, and 2 μg of anti-GR antibody (3600S; Cell Signaling) or anti-KLF15 antibody (ab2647; Abcam) was added. A tube that contained an isotype control IgG (I8140; Sigma) was used as a control for specific binding to the GR or KLF15 antibodies. Tubes were incubated overnight at 4°C, and samples were continuously rotated. Twenty microliters of protein A Dynabeads (catalog no. 10001D; Life Technologies) was added the next morning and allowed to incubate for 2 h at 4°C with rotation. Tubes were placed on a magnet (DynaMag, catalogue no. 12321D; Life Technologies), and the supernatant was removed. The Dynabeads-Ab-Ag-DNA complexes were washed three times with 1 ml of washing buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS). DNA-protein complexes were eluted from Dynabeads by incubation with 120 μl of elution buffer (1% SDS, 100 mM NaHCO3), vortexed gently, and then incubated in a water bath for 20 min at 30°C. Dynabeads were removed, and the supernatant was transferred to another tube. RNase (0.5 mg/ml) and proteinase K (20 mg/ml) were added, and then the mixture was incubated in a water bath at 65°C overnight to de-cross-link the protein from DNA. Samples were extracted twice with phenol-chloroform-isoamyl alcohol. A 0.1 volume of 3 M sodium acetate and 2 μl of glycogen (20 mg/ml) were added to samples, and then DNA was precipitated with 2.5 volumes of 100% cold ethanol, washed with 70% ethanol, dried in a Savant ISS110 SpeedVac concentrator (Thermo Scientific), and then suspended in 30 μl of nuclease-free water. PCR was performed using primers that amplify the IEtu1 GREs and Δ2×GREΔKLF (forward primer, 5′-CCCACTTTTGCCTGTGTG-3′; reverse primer, 5′-TTTTCCTCCTCCTTCCCC-3′). These primers yield a product of 107 bp. The UL52 primers are 5′-GCCTGTGTCCCCAACAAG-3′ (forward primer) and 5′-ATTTCACCCTGGTTCCCG-3′ (reverse primer). These primers yield a 132-bp product.
ACKNOWLEDGMENTS
This research was supported by grants from the USDA-NIFA Competitive Grants Program (13-01041 and 16-09370), funds derived from the Sitlington Endowment, and support from the Oklahoma Center for Respiratory and Infectious Diseases (National Institutes of Health Centers for Biomedical Research Excellence grant P20GM103648). Fouad S. El-mayet was supported by a fellowship from the Egyptian Ministry of Higher Education, Mission Sector (JS-3541).
REFERENCES
- 1.Hodgson PD, Aich P, Manuja A, Hokamp K, Roche FM, Brinkman FSL, Potter A, Babiuk LA, Griebel PJ. 2005. Effect of stress on viral-bacterial synergy in bovine respiratory disease: novel mechanisms to regulate inflammation. Comp Funct Genomics 6:244–250. doi: 10.1002/cfg.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones C, Chowdhury S. 2010. Bovine herpesvirus type 1 (BHV-1) is an important cofactor in the bovine respiratory disease complex. Vet Clin North Am Food Anim Pract 26:303–321. doi: 10.1016/j.cvfa.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Jones C. 2009. Regulation of innate immune responses by bovine herpesvirus 1 and infected cell protein 0. Viruses 1:255–275. doi: 10.3390/v1020255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell J. 2005. Bovine respiratory disease. University of Arkansas, Division of Agriculture Research and Extension, document FSA3082. University of Arkansas Cooperative Extension Service, Little Rock, AR. [Google Scholar]
- 5.Rice Carrasco-Medina JAL, Hodgins DC, Shewen PE. 2007. Mannheimia haemolytica and bovine respiratory disease. Anim Health Res Rev 8:117–128. doi: 10.1017/S1466252307001375. [DOI] [PubMed] [Google Scholar]
- 6.Jones C, Chowdhury S. 2007. A review of the biology of bovine herpesvirus type 1 (BHV-1), its role as a cofactor in the bovine respiratory disease complex, and development of improved vaccines. Adv Anim Health 8:187–205. doi: 10.1017/S146625230700134X. [DOI] [PubMed] [Google Scholar]
- 7.Edwards AJ. 1996. Respiratory diseases of feedlot cattle in central USA. Bovine Pract 30:5–7. [Google Scholar]
- 8.Frank GH. (ed). 1984. Bacteria as etiologic agents in bovine respiratory disease. Texas A&M University Press, College Station, TX. [Google Scholar]
- 9.Neibergs HL, Seabury CM, Wojtowicz AJ, Wang Z, Scraggs E, Kiser JN, Neupane M, Womack JE, Van Eenennaam A, Hagevortm GR, Lehenbauer TW, Aly S, Davis J, Taylor JF, the Bovine Respiratory Disease Complex Coordinated Agricultural Project Research Team. 2014. Susceptibility loci revealed for bovine respiratory disease complex in pre-weaned Holstein calves. BMC Genomics 15:1164. doi: 10.1186/1471-2164-15-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones C, da Silva LF, Sinani D. 2011. Regulation of the latency-reactivation cycle by products encoded by the bovine herpesvirus 1 (BHV-1) latency-related gene. J Neurovirol 17:535–545. doi: 10.1007/s13365-011-0060-3. [DOI] [PubMed] [Google Scholar]
- 11.Jones C. 2013. Bovine herpes virus 1 (BHV-1) and herpes simplex virus type 1 (HSV-1) promote survival of latently infected sensory neurons, in part by inhibiting apoptosis. J Cell Death 6:1–16. doi: 10.4137/JCD.S10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones C. 2014. Reactivation from latency by α-herpesvirinae subfamily members: a stressful situation. Curr Topics Virol 12:99–118. [Google Scholar]
- 13.Kook I, Henley C, Meyer F, Hoffmann F, Jones C. 2015. Bovine herpesvirus 1 productive infection and the immediate early transcription unit 1 are stimulated by the synthetic corticosteroid dexamethasone. Virology 484:377–385. doi: 10.1016/j.virol.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Jones C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv Virus Res 51:81–133. doi: 10.1016/S0065-3527(08)60784-8. [DOI] [PubMed] [Google Scholar]
- 15.Jones C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin Microbiol Rev 16:79–95. doi: 10.1128/CMR.16.1.79-95.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones C, Geiser V, Henderson G, Jiang Y, Meyer F, Perez S, Zhang Y. 2006. Functional analysis of bovine herpesvirus 1 (BHV-1) genes expressed during latency. Vet Microbiol 113:199–210. doi: 10.1016/j.vetmic.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Inman M, Lovato L, Doster A, Jones C. 2002. A mutation in the latency-related gene of bovine herpesvirus 1 disrupts the latency reactivation cycle in calves. J Virol 76:6771–6779. doi: 10.1128/JVI.76.13.6771-6779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones C, Newby TJ, Holt T, Doster A, Stone M, Ciacci-Zanella J, Webster CJ, Jackwood MW. 2000. Analysis of latency in cattle after inoculation with a temperature sensitive mutant of bovine herpesvirus 1 (RLB106). Vaccine 18:3185–3195. doi: 10.1016/S0264-410X(00)00106-7. [DOI] [PubMed] [Google Scholar]
- 19.Rock D, Lokensgard J, Lewis T, Kutish G. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J Virol 66:2484–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheffy BE, Davies DH. 1972. Reactivation of a bovine herpesvirus after corticosteroid treatment. Proc Soc Exp Biol Med 140:974–976. doi: 10.3181/00379727-140-36592. [DOI] [PubMed] [Google Scholar]
- 21.Shimeld C, Hill TJ, Blyth WA, Easty DL. 1990. Reactivation of latent infection and induction of recurrent herpetic eye disease in mice. J Gen Virol 71:397–404. doi: 10.1099/0022-1317-71-2-397. [DOI] [PubMed] [Google Scholar]
- 22.Oakley RH, Cidlowski JA. 2013. The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J Allergy Clin Immunol 132:1033–1044. doi: 10.1016/j.jaci.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 24.Lacasse JL, Schang LM. 2012. Herpes simplex virus 1 DNA is in unstable nucleosomes throughout the lytic infection cycle, and the instability of the nucleosomes is independent of DNA replication. J Virol 86:11287–11300. doi: 10.1128/JVI.01468-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacasse JL, Schang LM. 2010. During lytic infection, herpes simplex virus type 1 DNA is in complexes with the properties of unstable nucleosomes. J Virol 84:1920–1933. doi: 10.1128/JVI.01934-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frizzo da Silva L, Kook I, Doster A, Jones C. 2013. Bovine herpesvirus 1 regulatory proteins, bICP0 and VP16, are readily detected in trigeminal ganglionic neurons expressing the glucocorticoid receptor during the early stages of reactivation from latency. J Virol 87:11214–11222. doi: 10.1128/JVI.01737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kook I, Doster A, Jones C. 2015. Bovine herpesvirus 1 regulatory proteins are detected in trigeminal ganglionic neurons during the early stages of stress-induced escape from latency. J Neurovirol 21:585–591. doi: 10.1007/s13365-015-0339-x. [DOI] [PubMed] [Google Scholar]
- 28.Winkler MT, Doster A, Jones C. 2000. Persistence and reactivation of bovine herpesvirus 1 in the tonsil of latently infected calves. J Virol 74:5337–5346. doi: 10.1128/JVI.74.11.5337-5346.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Workman A, Eudy J, Smith L, Frizzo da Silva L, Sinani D, Bricker H, Cook E, Doster A, Jones C. 2012. Cellular transcription factors induced in trigeminal ganglia during dexamethasone-induced reactivation from latency stimulate bovine herpesvirus 1 productive infection and certain viral promoters. J Virol 86:2459–2473. doi: 10.1128/JVI.06143-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaczynski J, Cook T, Urrutia R. 2003. Sp1- and Krüppel-like transcription factors. Genome Biol 4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnes PJ. 1998. Anti-inflammatory actions of glucocorticoids: molecular mechanisms. Clin Sci 94:557–572. doi: 10.1042/cs0940557. [DOI] [PubMed] [Google Scholar]
- 32.Schoneveld OJ, Gaemers IC, Lamers WH. 2004. Mechanisms of glucocorticoid signalling. Biochim Biophys Acta 1680:114–128. doi: 10.1016/j.bbaexp.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Smoak KL, Cidlowski JA. 2004. Mechanisms of glucocorticoid receptor signaling during inflammation. Mech Ageing Dev 125:697–706. doi: 10.1016/j.mad.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Misra V, Bratanich AC, Carpenter D, O'Hare P. 1994. Protein and DNA elements involved in transactivation of the promoter of the bovine herpesvirus (BHV) 1 IE-1 transcription unit by the BHV alpha gene trans-inducing factor. J Virol 68:4898–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misra V, Walker S, Hayes S, O'Hare P. 1995. The bovine herpesvirus alpha gene trans-inducing factor activates transcription by mechanisms different from those of its herpes simplex virus type 1 counterpart VP16. J Virol 69:5209–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirth UV, Fraefel C, Vogt B, Vlcek C, Paces V, Schwyzer M. 1992. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J Virol 66:2763–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wirth UV, Gunkel K, Engels M, Schwyzer M. 1989. Spatial and temporal distribution of bovine herpesvirus 1 transcripts. J Virol 63:4882–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wirth UV, Vogt B, Schwyzer M. 1991. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J Virol 65:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraefel C, Zeng J, Choffat Y, Engels M, Schwyzer M, Ackermann M. 1994. Identification and zinc dependence of the bovine herpesvirus 1 transactivator protein BICP0. J Virol 68:3154–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeLeon M, Covenas R, Chadi G, Narvaez JA, Fuxe K, Cintra A. 1994. Subpopulations of primary sensory neurons show coexistence of neuropeptides and glucocorticoid receptors in the rat spinal and trigeminal ganglia. Brain Res 14:338–342. doi: 10.1016/0006-8993(94)91034-0. [DOI] [PubMed] [Google Scholar]
- 41.Sinani D, Cordes E, Workman A, Thunuguntia P, Jones C. 2013. Stress induced cellular transcription factors expressed in trigeminal ganglionic neurons stimulate the herpes simplex virus type 1 (HSV-1) infected cell protein 0 (ICP0) promoter. J Virol 87:13042–130471192. doi: 10.1128/JVI.02783-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasse S, Zuo Z, Kadiyala V, Zhang L, Pufall MA, Jain MK, Phang TL, Stormo GD, Gerber AN. 2015. Response element composition governs correlations between binding site affinity and transcription in glucocorticoid receptor feed-forward loops. J Biol Chem 290:19756–19769. doi: 10.1074/jbc.M115.668558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasse S, Mailoux CM, Barczak AJ, Wang Q, Altonsy MO, Jain MK, Haldar SM, Gerber AN. 2013. The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol Cell Biol 33:2104–2115. doi: 10.1128/MCB.01474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knoedler JR, Denver RJ. 2014. Kruppel-like factors are effectors of nuclear receptor signaling. Gen Comp Endocrinol 203:49–59. doi: 10.1016/j.ygcen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tremblay R, Sikorska M, Sandhu JK, Lanthier P, Ribecco-Lutkiewicz M, Bani-Yaghoub M. 2010. Differentiation of mouse Neuro-2A cells into dopamine neurons. J Neurosci Methods 186:60–67. doi: 10.1016/j.jneumeth.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Thunuguntla P, El-mayet FS, Jones C. 2017. Bovine herpesvirus 1 can efficiently infect the human (SH-SY5Y) but not the mouse neuroblastoma cell line (Neuro-2A). Virus Res 232:1–5. doi: 10.1016/j.virusres.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Bieker JJ. 2001. Kruppel-like factors: three fingers in many pies. J Biol Chem 276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 48.Black AR, Black JD, Azizkhan-Clifford J. 2001. Sp1 and Kruppel-like transcription factor family of transcription factors in cell growth and cancer. J Cell Physiol 188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 49.McConnell BB, Yang VW. 2010. Mammalian Kruppel-like factors in health and diseases. Physiol Rev 90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polman JAE, Welten JE, Bosch DS, de Jonge RT, Balog J, van der Maarel SM, de Kloet ER, Datson NA. 2012. A genome-wide signature of glucocorticoid receptor binding in neuronal PC12 cells. BMC Neurosci 13:118. doi: 10.1186/1471-2202-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu NZ, Cidlowski JA. 2006. Glucocorticoid receptor isoforms generate transcription specificity. Trends Cell Biol 16:301–307. doi: 10.1016/j.tcb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 52.Kassel O, Herrlich P. 2007. Crosstalk between the glucocorticoid receptor and other transcription factors: molecular aspects. Mol Cell Endocrinol 275:13–29. doi: 10.1016/j.mce.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Winkler MT, Doster A, Sur JH, Jones C. 2002. Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet Microbiol 86:139–155. doi: 10.1016/S0378-1135(01)00498-9. [DOI] [PubMed] [Google Scholar]
- 54.Mangan S, Alon U. 2003. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A 100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alon U. 2007. Network motifs: theory and experimental approaches. Nat Rev Genet 8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 56.Masuno K, Haldar SM, Jeyaraji D, Mailoux CM, Huang X, Panettieri RA Jr, Jain MK, Gerber AN. 2011. Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. Am J Respir Cell Mol Biol 45:642–649. doi: 10.1165/rcmb.2010-0369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asada M, Rauch A, Shimizu H, Maruyama H, Miaki S, Shigamori M, Kawasome H, Ishiyama H, Tuckermann J, Asahara H. 2011. DNA-binding dependent glucocorticoid receptor activity promotes adipogenesis via Kruppel-like factor 15 gene expression. Lab Invest 91:203–215. doi: 10.1038/labinvest.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uchida S, Tanaka Y, Ito H, Saitoh-Ohara F, Inazawa J, Yokoyama KK, Sasaki S, Marumo F. 2000. Transcriptional regulation of the CLC-K1 by myc-associated zinc finger protein, a novel zinc finger repressor. Mol Cell Biol 20:7319–7331. doi: 10.1128/MCB.20.19.7319-7331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeuchi K, Yahagi N, Aita Y, Murayama Y, Sawada Y, Piao X, Toya N, Oya Y, Shikama A, Takarada A, Masuda Y, Nishi M, Kuobota M, Izumida Y, Yamamoto T, Sekiya M, Matsuzaka T, Nakagawa Y, Urayama O, Kawakami Y, Iizuka Y, Gotoda T, Itaka K, Kataoka K, Nagai R, Kadowaki T, Nyamada Lin Y, Jain MK, Shimano H. 2016. KLF15 enables switching between lipogenesis and gluconeogenesis during fasting. Cell Rep 16:2373–2386. doi: 10.1016/j.celrep.2016.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du X, Rosenfield RL, Qin K. 2009. KLF15 is a transcriptional regulator of the human 17β-hydroxysteroid dehydrogenase type 5 gene. A potential link between regulation of testosterone production and fat stores in women. J Clin Endocrinol Metab 94:2594–2601. doi: 10.1210/jc.2009-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Otteson DC, Lai H, Liu Y, Zack DJ. 2005. Zinc-finger domains of the transcriptional repressor KLF15 binds multiple sites in rhodopsin and IRBP promoters including the CRS-1 and G-rich elements. BMC Mol Biol 6:15. doi: 10.1186/1471-2199-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perlmann T. 1992. Glucocorticoid receptor DNA-binding specificity is increased by the organization of DNA in nucleosomes. Proc Natl Acad Sci U S A 89:3884–3888. doi: 10.1073/pnas.89.9.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perlmann T, Wrange O. 1988. Specific glucocorticoid receptor binding to DNA reconstituted in nucleosome. EMBO J 7:3073–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin Q, Wrange O. 1993. Translational positioning of a nucleosome: glucocorticoid response element modulates glucocorticoid receptor affinity. Genes Dev 7:2471–2482. doi: 10.1101/gad.7.12a.2471. [DOI] [PubMed] [Google Scholar]
- 65.Richard-Foy H, Hager GL. 1987. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J 6:2321–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zaret KS, Yamamoto KR. 1984. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell 258:1780–1784. [DOI] [PubMed] [Google Scholar]
- 67.Zaret KS, Carrol JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iwafuchi-Doi M, Zaret KS. 2014. Pioneer transcription factors in cell reprogramming. Genes Dev 28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Funder JW. 1997. Glucocorticoids and mineralocorticoid receptors: biology and clinical relevance. Annu Rev Med 48:231–240. doi: 10.1146/annurev.med.48.1.231. [DOI] [PubMed] [Google Scholar]
- 70.Rhen T, Cidlowski JA. 2005. Antiinflammatory action of glucocorticoids—new mechanisms of old drugs. N Engl J Med 353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 71.Goodbourn S, Zinn K, Maniatis T. 1985. Human beta-interferon gene expression is regulated by an inducible enhancer element. Cell 41:509–520. doi: 10.1016/S0092-8674(85)80024-6. [DOI] [PubMed] [Google Scholar]
- 72.Hendricks RL, Weber PC, Taylor JL, Koumbis A, Tumpey TM, Glorioso JC. 1991. Endogenously produced interferon alpha protects mice from herpes simplex virus type 1 corneal disease. J Gen Virol 72:1601–1610. doi: 10.1099/0022-1317-72-7-1601. [DOI] [PubMed] [Google Scholar]
- 73.Katze MG, Heng Y, Gale M. 2002. Viruses and interferon: fight for supremacy. Nat Rev Immunol 2:675–686. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 74.Leib DA, Harrison TE, Laslo KM, Machalek MA, Moorman NJ, Virgin HW. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J Exp Med 189:663–672. doi: 10.1084/jem.189.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu T, Khanna KM, Carriere BN, Hendricks RL. 2001. Gamma interferon can prevent herpes simplex virus type 1 reactivation from latency in sensory neurons. J Virol 75:11178–11184. doi: 10.1128/JVI.75.22.11178-11184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Melroe GT, DeLuca NA, Knipe DM. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J Virol 78:8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mikloska Z, Cunningham AL. 2001. Alpha and gamma interferons inhibit herpes simplex virus type 1 infection and spread in epidermal cells after axonal transmission. J Virol 75:11821–11826. doi: 10.1128/JVI.75.23.11821-11826.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Migliorati G, Nicoletti I, D'Adamio F, Spreca A, Pagliacci C, Riccardi C. 1994. Dexamethasone induces apoptosis in mouse natural killer cells and cytotoxic T lymphocytes. Immunology 81:21–26. [PMC free article] [PubMed] [Google Scholar]
- 79.Decman V, Freeman ML, Kinchington PR, Hendricks RL. 2005. Immune control of HSV-1 latency. Viral Immunol 18:466–473. doi: 10.1089/vim.2005.18.466. [DOI] [PubMed] [Google Scholar]
- 80.Knickelbein JE, Khanna KM, Yee MB, Baty CJ, Kinchington PR, Hendricks RL. 2008. Noncytotoxic lytic granule-mediated CD8+ T cell inhibition of HSV-1 reactivation from neuronal latency. Science 322:268–272. doi: 10.1126/science.1164164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matundan H, Mott KR, Ghiasi H. 2014. Role of CD8+ T cells and lymphoid dendritic cells in protection from ocular herpes simplex virus 1 challenge in immunized mice. J Virol 88:8016–8027. doi: 10.1128/JVI.00913-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mott KR, Allen SJ, Zandian M, Ghiasi H. 2014. Coregulatory interactions among CD8α dendritic cells, the latency-associated transcript, and programmed death 1 contribute to higher levels of herpes simplex virus 1 latency. J Virol 88:6599–6610. doi: 10.1128/JVI.00590-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Inman M, Lovato L, Doster A, Jones C. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J Virol 75:8507–8515. doi: 10.1128/JVI.75.18.8507-8515.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Y, Hancock M, Workman A, Doster A, Jones C. 2016. Beta-catenin, a transcription factor activated by canonical Wnt signaling, is expressed in sensory neurons of calves latently infected with bovine herpesvirus 1. J Virol 90:3148–3159. doi: 10.1128/JVI.02971-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu L, Workman A, Jones C. 2017. A potential role for a beta-catenin coactivator (high-mobility group AT-hook 1 protein) during the latency-reactivation cycle of bovine herpesvirus 1. J Virol 91:e02132-16. doi: 10.1128/JVI.02132-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lovato L, Inman M, Henderson G, Doster A, Jones C. 2003. Infection of cattle with a bovine herpesvirus 1 (BHV-1) strain that contains a mutation in the latency related gene leads to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J Virol 77:4848–4857. doi: 10.1128/JVI.77.8.4848-4857.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perez S, Inman M, Doster A, Jones C. 2005. Latency-related gene encoded by bovine herpesvirus 1 promotes virus growth and reactivation from latency in tonsils of infected calves. J Clin Microbiol 43:393–401. doi: 10.1128/JCM.43.1.393-401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perez S, Lovato L, Zhou J, Doster A, Jones C. 2006. Comparison of inflammatory infiltrates in trigeminal ganglia of cattle infected with wild type BHV-1 versus a virus strain containing a mutation in the LR (latency-related) gene. J Neurovirol 12:392–397. doi: 10.1080/13550280600936459. [DOI] [PubMed] [Google Scholar]
- 89.Perez S, Meyer F, Saira K, Doster A, Jones C. 2008. Premature expression of the latency-related RNA encoded by bovine herpesvirus 1 correlates with higher levels of beta interferon RNA expression in productively infected cells. J Gen Virol 89:1338–1345. doi: 10.1099/vir.0.83481-0. [DOI] [PubMed] [Google Scholar]
- 90.Sinani D, Frizzo da Silva L, Jones C. 2013. A bovine herpesvirus 1 protein expressed in latently infected neurons (ORF2) promotes neurite sprouting in the presence of activated Notch1 or Notch3. J Virol 87:1183–1192. doi: 10.1128/JVI.02783-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inman M, Zhang Y, Geiser V, Jones C. 2001. The zinc ring finger in the bICP0 protein encoded by bovine herpes virus-1 mediates toxicity and activates productive infection. J Gen Virol 82:483–492. doi: 10.1099/0022-1317-82-3-483. [DOI] [PubMed] [Google Scholar]
- 92.Geiser V, Inman M, Zhang Y, Jones C. 2002. The latency related (LR) gene of bovine herpes virus 1 (BHV-1) can inhibit the ability of bICP0 to activate productive infection. J Gen Virol 83:2965–2971. doi: 10.1099/0022-1317-83-12-2965. [DOI] [PubMed] [Google Scholar]