ABSTRACT
Immediate early proteins of human herpesvirus 6A (HHV-6A) are expressed at the outset of lytic infection and thereby regulate viral gene expression. Immediate early protein 2 (IE2) of HHV-6A is a transactivator that drives a variety of promoters. The C-terminal region of HHV-6A IE2 is shared among IE2 homologs in betaherpesviruses and is involved in dimerization, DNA binding, and transcription factor binding. In this study, the structure of the IE2 C-terminal domain (IE2-CTD) was determined by X-ray crystallography at a resolution of 2.5 Å. IE2-CTD forms a homodimer stabilized by a β-barrel core with two interchanging long loops. Unexpectedly, the core structure resembles those of the gammaherpesvirus factors EBNA1 of Epstein-Barr virus and LANA of Kaposi sarcoma-associated herpesvirus, but the interchanging loops are longer in IE2-CTD and form helix-turn-helix (HTH)-like motifs at their tips. The HTH and surrounding α-helices form a structural feature specific to the IE2 group. The apparent DNA-binding site (based on structural similarity with EBNA1 and LANA) resides on the opposite side of the HTH-like motifs, surrounded by positive electrostatic potential. Mapping analysis of conserved residues on the three-dimensional structure delineated a potential factor-binding site adjacent to the expected DNA-binding site. The predicted bi- or tripartite functional sites indicate a role for IE2-CTD as an adapter connecting the promoter and transcriptional factors that drive gene expression.
IMPORTANCE Human herpesvirus 6A (HHV-6A) and HHV-6B belong to betaherpesvirus subfamily. Both viruses establish lifelong latency after primary infection, and their reactivation poses a significant risk to immunocompromised patients. Immediate early protein 2 (IE2) of HHV-6A and HHV-6B is a transactivator that triggers viral replication and contains a DNA-binding domain shared with other betaherpesviruses such as human herpesvirus 7 and human cytomegalovirus. In this study, an atomic structure of the DNA-binding domain of HHV-6A IE2 was determined and analyzed, enabling a structure-based understanding of the functions of IE2, specifically DNA recognition and interaction with transcription factors. Unexpectedly, the dimeric core resembles the DNA-binding domain of transcription regulators from gammaherpesviruses, showing structural conservation as a DNA-binding domain but with its own unique structural features. These findings facilitate further characterization of this key viral transactivator.
KEYWORDS: HHV-6, IE2, structure
INTRODUCTION
Nine human herpesviruses have been identified and grouped into the alpha-, beta-, and gammaherpesvirus subfamilies (1). In all cases, the virions show a common architecture, and they all establish lifelong latency in the host, although pathogenesis and detailed characteristics are highly differentiated. Some core functional genes are shared across herpesviridae, but viruses in each subfamily also have distinctive genes. Human herpesvirus 6A (HHV-6A) and HHV-6B are members of the betaherpesviruses and were recently classified as different species according to their distinguishable characteristics and pathological properties (2–11). The betaherpesvirus subfamily also includes HHV-7 and human cytomegalovirus (HCMV) as well as a wide variety of animal herpesviruses. HHV-6B is a causative agent of exanthema subitum (roseola) in children (12), while HHV-6A has been shown to be linked to multiple sclerosis (13, 14), although the exact details of pathogenesis remain unclear. Reactivation of particularly HHV-6B in recipients receiving transplants under immune suppression can sometimes cause severe encephalitis (15, 16).
Gene expression in herpesviruses is highly regulated and proceeds in a cascade manner. After the virion enters a target cell, the viral genome is released, and it invades the nucleus, resulting in the expression of the first group of viral genes, known as immediate early (IE) genes, that encode proteins that promote the expression of the next gene group, the early genes, which are mainly involved in genome replication. Through this process, immediate early genes manage the onset of the lytic cycle of infection. The regulation of gene expression is important for both the transition to the lytic cycle and the maintenance of latency, and most immediate early gene products are believed to access the viral and/or host genome in the nucleus.
Immediate early protein 2 (IE2) of HHV-6A and HHV-6B is a transactivator that is thought to be important for the onset of lytic infection (17). IE2 is expressed at the immediate early phase of infection with IE1, and both proteins accumulate over the course of lytic infection (17). Both IE1 and IE2 are encoded at the IE-A locus in the viral genome (18, 19). The transcript for IE2, derived from U90 and U86/87, shares the 5′ region of U90 with IE1 (18–20). The IE2 transcript of HHV-6A strain U1102 encodes a protein of 1,500 amino acids (aa), although the lengths of the IE2 protein vary among strains of HHV-6A and HHV-6B (HHV-6A strain GS, 1,466 aa; HHV-6B HST, 1,513 aa; HHV-6B Z29, 1,520 aa) (17). Differences are attributed mainly to the lengths of the repeat sequences (SSRA/D) in the central region (21, 22) that are derived from repeat region R1 in open reading frame (ORF) U86. The C-terminal region of IE2 shares similarity with HCMV IE2, which is encoded by UL122, while the other regions of HHV-6A IE2 and HCMV IE2 are different (23, 24). HHV-6A IE2 recognizes a wide variety of promoters to transactivate gene expression, including its own HHV-6A IE promoter; the HHV-6A polymerase (POL) promoter; the cellular promoters CRE, NF-κB, and nuclear factor for activated T cell (NFAT); as well as the HIV-long terminal repeat (LTR) promoter (17, 24). Transactivation requires an intact C-terminal region, although the other regions are also needed for full activity (24). HCMV IE2 has been revealed to be critical for viral replication (25). Numerous studies have characterized the function of HCMV IE2 homologs, and the conserved C-terminal region is required for dimerization (23, 26, 27), DNA binding (23), and interactions with transcription factor II B (TFIIB) and TATA-binding protein (TBP) (28, 29). Therefore, the conserved C-terminal region of IE2 is expected to have several functional sites.
To further characterize the function of the C-terminal region of IE2, we purified and crystallized this domain (IE2 C-terminal domain [here called IE2-CTD]) from HHV-6A U1102 in a previous study (30). In the present work, the crystal structure was determined to a resolution of 2.5 Å, which revealed a dimeric structure similar to that of the gammaherpesvirus latency-associated factors EBNA1 of Epstein-Barr virus (EBV) and LANA of Kaposi sarcoma-associated herpesvirus (KSHV) albeit with some critical structural differences. Comparison with EBNA1 and LANA and a detailed structural analysis revealed the putative DNA-binding sites and transcription factor-binding sites and provided structural insights into the essential functions of HHV-6A IE2-CTD, particularly regarding transactivation activity.
RESULTS
Structural determination of HHV-6A IE2.
In our previous study, the conserved C-terminal region of HHV-6A U1102 IE2 (residues 1324 to 1500) (IE2-CTD) was purified and crystallized (30). X-ray diffraction data were collected from selenomethionine (SeMet)-labeled crystals, and the crystal structure was solved by using the single anomalous dispersion (SAD) method. Data collection statistics were reported previously (30). In the present study, the IE2-CTD structure was determined and refined at a resolution of 2.5 Å. Data collection and refinement statistics are shown in Table 1. The asymmetric unit contains four copies of IE2-CTD as two similar dimers (details of dimerization are discussed below). The two IE2-CTD dimers are similar, with a root mean square deviation (RMSD) of 0.51 or 1.20 Å for Cα atoms or all heavy atoms, respectively. Electron density was clearly discernible for residues 1334 to 1498, and only 10 and 2 residues from the N and C termini were disordered, respectively.
TABLE 1.
Data collection and refinement statisticsa
Values in parentheses are for the outermost shell.
b Pearson correlation coefficient between two random half sets.
Structure of the IE2-CTD monomer and dimer.
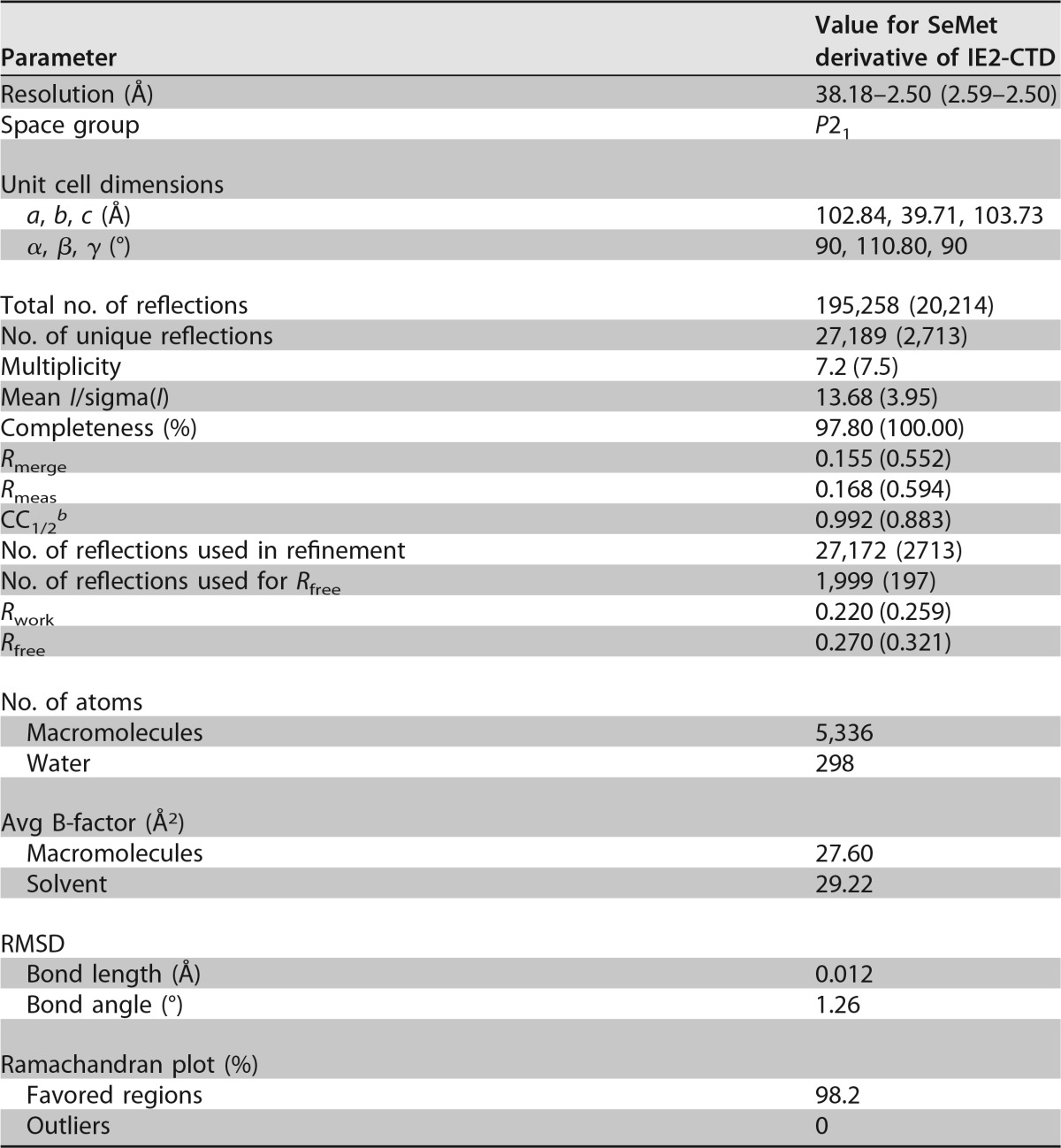
The crystal structure of IE2-CTD and its secondary structural topology are shown in Fig. 1. IE2-CTD has an α1β1α2β2β3α3α4β4α5β5α6 secondary structure (Fig. 1A and B), with the α1β1α2β2β3β4α5β5 portion forming a two-layered α/β fold with C-terminal α6 in a perpendicular direction. A helix-turn-helix (HTH)-like motif consisting of α3α4 is connected between β3 and β4 by a long coil region. The α/β fold is typical of the dimeric α-β-barrel superfamily (structural classification of proteins [SCOP] subfamily 54909 [31]). It was consistent with dimer formation via a central β-barrel (Fig. 1C).
FIG 1.
Crystal structures of HHV-6A IE2-CTD and the dimeric form. (A) The HHV-6A IE2-CTD monomer is shown in a ribbon representation with secondary structural elements assigned. The positions of N- and C-terminal residues are indicated. Only one monomer from the dimeric form is shown for clarity. (B) Topology diagram showing the secondary structure arrangement. α1β1α2β2β3β4α5β5 constitutes a two-layered α/β sandwich fold, while α3 and α4 are distant from the main globular domain and connected by a turn, resulting in a helix-turn-helix (HTH)-like motif. α-Helices and β-strands are represented by pink rectangles and yellow arrows, respectively. The turn is represented by a pink line. (C) Dimeric form of HHV-6A IE2-CTD shown as ribbon models from three different views. The two IE2-CTD molecules are shown in cyan and orange. The N- and C-terminal residues (K1334 and S1498) and the cysteine residues (C1471) (magenta) at the 2-fold axis are indicated. (D) Topology diagram of the two-layered α/β sandwich fold forming a closed barrel with an antiparallel β-sheet arrangement. Circles and trapeziums represent α-helices and β-strands, respectively. Long extrusions, including the HTH-like motif between β3 and β4, are omitted for clarity and represented by green dotted lines. Perpendicularly arranged α6-helices are shown as rectangles. (E) Size exclusion chromatography of purified IE2-CTD. The retention volumes of standards are indicated by arrows and molecular mass labels, and the expected retention volumes for the IE2-CTD dimer (40.8 kDa) and monomer (20.4 kDa) were calculated from the standard curve and are indicated by red arrows. IE2-CTD eluted as a single peak distinct from the void volume (∼45 ml), with a retention volume close to that expected for a dimer.
IE2-CTD forms a homodimer mainly through the β-sheets and interchanging loops (Fig. 1C and D). The interchanging loops of the two IE2-CTD monomers cross over so that HTH-like motifs at their tips form the outermost shell of the fold (Fig. 1C, bottom and lateral views). The β-barrel is formed by a combination of two antiparallel β-sheets of IE2-CTDs in an antiparallel orientation (Fig. 1D). The inside of the β-barrel is filled by hydrophobic/aromatic residues A1353, I1355, F1387, M1389 (SeMet in this structure), P1479, I1473, and I1439 and polar residues Y1367, Y1441, and C1471. A 2-fold axis runs perpendicularly to the top-to-bottom axis of the β-barrel (Fig. 1C, lateral view). The two Cys1471 residues are in close proximity to the 2-fold axis (Fig. 1C), but a disulfide bond is not observed, presumably because of the reducing agent used during purification and crystallization.
This dimeric structure of IE2-CTD in solution was also suggested by size exclusion chromatography, since purified IE2-CTD eluted earlier than would be expected for a monomer and close to the volume expected for a dimer (Fig. 1E). The molecular mass calculated from the elution volume was 33.8 kDa, which was slightly lower than the expected molecular mass of 40.8 kDa.
Similarity with EBV EBNA1-DBD and KSHV LANA-DBD.
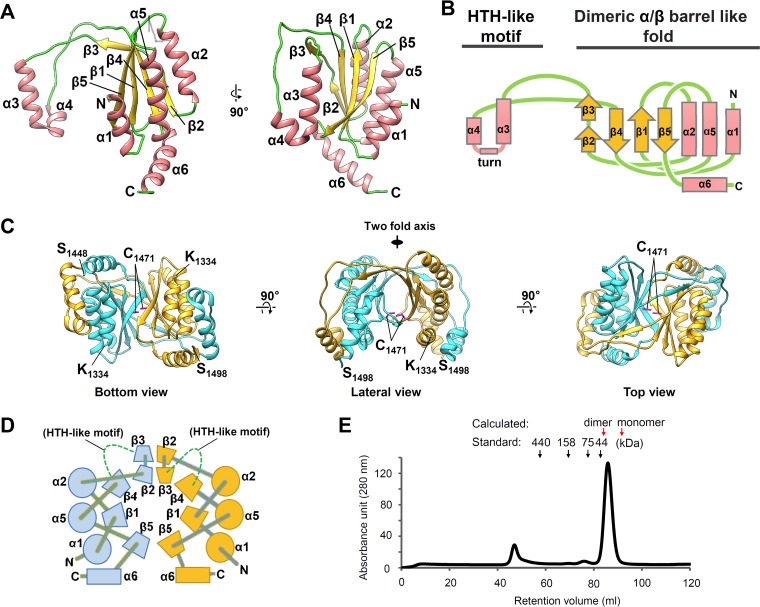
The dimeric form of IE2-CTD resembles the structure of the EBV EBNA1 DNA-binding domain (EBNA1-DBD) and KSHV LANA-DBD (Fig. 2A). EBV EBNA1 and KSHV LANA have the DBD at their C termini, like HHV-6A IE2, and all of them have repeat sequences at the N-terminal sides (Fig. 2B). The secondary structural topology of IE2-CTD in the central fold (α1β1α2β2β3β4α5β5) (Fig. 1B) is comparable to those of EBNA1-DBD (PDB accession number 1VHI) (32) and LANA1-DBD (PDB accession number 4K2J) (33) (Fig. 2C). Indeed, the IE2-CTD structure could be superposed on these structures (Fig. 2D), with RMSDs between 34 Cα atoms in the well-aligned β1β2β4β5 region and the connecting region between β2 and β3 of 1.20 Å and 1.33 Å for IE2-CTD/EBNA1-DBD and IE2-CTD/LANA-DBD, respectively. The mode of dimer formation is similar to that of EBNA1-DBD and LANA-DBD, in which the β-barrel forms the core. Dimers superposed with RMSD values of 1.43 Å and 1.53 Å for 68 β-barrel Cα atoms, indicating similar β-barrel structures.
FIG 2.
IE2-CTD has a fold similar to those of EBNA1-DBD and LANA-DBD. (A) Ribbon models of IE2-CTD, EBNA1-DBD (PDB accession number 1VHI [32]), and LANA-DBD (PDB accession number 4K2J [33]), shown in orange, cyan, and magenta, respectively. (B) Comparison of the domain structures among IE2, EBNA1, and LANA. EBNA1 and LANA have a C-terminal domain corresponding to IE2-CTD at the C-terminal ends (orange) and repeat regions at the N-terminal sides (gray). (C) Topology diagrams of EBNA1-DBD and LANA-DBD showing the secondary structure arrangement. The pink rectangles and yellow arrows represent α-helices and β-strands, as described in the legend of Fig. 1B. The regions in EBNA1-DBD and LANA-DBD comparable to α1β1α2β2β3β4α5β5 of IE2-CTD are indicated by purple boxes with dashed lines. (D) The two-layered α/β sandwich folds of IE2-CTD (orange) and EBNA1-DBD (cyan) or LANA-DBD (magenta) superposed to show similarity. For clarity, the unique IE2-CTD α1 and α6 regions and the corresponding regions in EBNA1-DBD and LANA-DBD are shown as transparent ribbons. (E) The three structure elements of IE2-CTD, α1, α6, and the internal loop, constitute a unique structure showing differences from EBNA1-DBD and LANA-DBD. The α-helices corresponding to IE2-CTD α1 are labeled α1′ for EBNA1-DBD and LANA-DBD. (F) Structure-based sequence alignment of IE2-CTD, EBNA1-DBD, and LANA-DBD. Secondary structure elements determined for IE2-CTD (Fig. 1A) are shown at the top. Just below, positions where at least 5% of the surface area is accessible to the solvent and considered exposed are designated by the letter e, while buried positions are labeled b. The positions occupied by strongly or weakly similar residues are highlighted by orange and gray, respectively. The conserved and buried residues, which are likely important for protein folding, are indicated by arrowheads above the sequence, and the positions where the side chains face the dimer interface are shown as red boxes.
In contrast, α1 of IE2-CTD and the corresponding helices of EBNA1-DBD and LANA-DBD are much less similar than the central fold region. α1 of IE2-CTD runs antiparallel alongside α2 of the central core, effectively extending the α-helical layer (Fig. 1A and B), whereas the corresponding helices in EBNA1-DBD and LANA-DBD reside on the outer face of the α-helical layer (Fig. 2E). Additionally, the HTH-like motifs and C-terminal α6 are specific features of IE2-CTD (Fig. 1B and 2E). These unique α1, α6, and HTH-like motif regions are spatially assembled at a distinct local area (Fig. 2E).
Similarity was further analyzed by a structure-based sequence alignment of the aligned α1β1α2β2β3β4α5β5 regions (Fig. 2F). There are several positions with similar residues, especially on the β-strands, although no fully conserved residues were found in the alignment. Most of the highly or weakly conserved residues are buried and likely contribute to protein folding (Fig. 2F, arrowheads), and five residues are buried in the dimer interface (Fig. 2F, red boxes). Therefore, structure conservation, but not sequence conservation, among IE2-CTD, EBNA1-DBD, and LANA-DBD was suggested.
Electrostatic potential on the IE2-CTD surface and putative DNA-binding site.
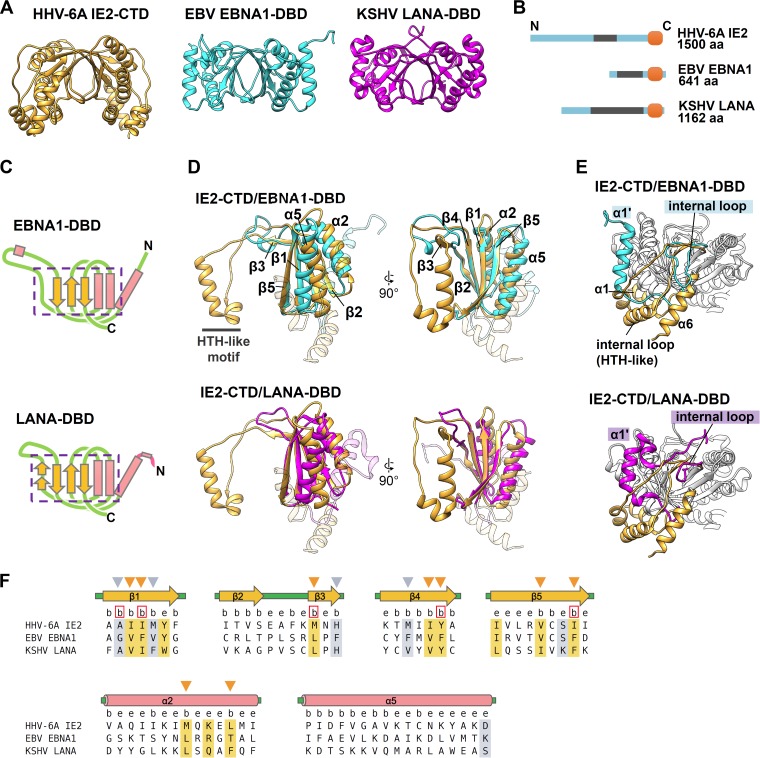
Next, the surface features possibly linked to IE2 function were investigated. The electrostatic potential of the IE2-CTD dimer is shown in Fig. 3A. There are three noticeable positively charged clusters, designated positively charged area 1 (PCA1), PCA2, and PCA3 (Fig. 3A). The symmetrically located PCA1 broadly covers the surface of the top side of IE2-CTD (Fig. 3A, top view), while PCA2 is located near the N terminus of IE2-CTD, including α1 and α5 (Fig. 3A, lateral view). PCA3 is formed from α3 of the HTH-like motif, specifically from the three lysine residues K1399, K1403, and K1410 (Fig. 3A, bottom and lateral views).
FIG 3.
Potential functional sites in IE2-CTD. (A) Electrostatic surface potential of IE2-CTD shown from the three views in Fig. 1C. Positively and negatively charged areas are shown in blue and red, respectively. The positions of residues involved in the three noticeable positively charged areas (PCA1, PCA2, and PCA3) are indicated. (B) Fully and highly conserved residues exposed on the dimer surface are shown in magenta and orange, respectively. The position of each residue is indicated. (C) Amino acid sequences of IE2 homologs from human and other betaherpesviruses were aligned by using Clustal Omega (57). Fully and highly conserved positions are shown in light purple and orange, respectively. The secondary structures are shown at the top, as described in the legend of Fig. 2F, and the gray bar indicates the region missing in the crystal structure. The letters e and b just below the structures stand for the exposed and buried residues, as described in the legend of Fig. 2F. Conserved and solvent-exposed residues are represented by purple and orange arrowheads, for fully and highly conserved residues, respectively. Abbreviations (accession numbers): HHV-6A, HHV-6A U1102 (gi∣205830876); HHV-6B, HHV-6B HST (gi∣4996073); HHV-7, HHV-7 UCL1 (gi∣541905786); HCMV, HCMV AD169, (GenBank accession number P19893.2); SaHV-4, saimirine herpesvirus 4 (gi∣360086591); PaHV-2, panine herpesvirus 2 (gi∣20026703); MaHV-3, macacine herpesvirus 3 (gi∣389889375); CeHV-5, cercopithecine herpesvirus 5 (gi∣359946547); MHV-8, murid herpesvirus 8 (GenBank accession number YP_007016515.1); MHV-1, murid herpesvirus 1 (gi∣330548).
The IE2-CTD dimer includes a conserved patch.
To further investigate the potential functional sites on IE2-CTD, we performed multiple-sequence alignment of IE2 homologs from betaherpesviruses for this region (Fig. 3B and C). In the folded region of IE2-CTD (residues 1334 to 1498), 20 residues are fully conserved between homologs, and 19 are highly conserved (Fig. 3B and C). These residues are spread throughout the aligned sequences. Among the fully conserved residues, 8 are buried in the fold, and 12 are at least partially exposed and accessible to the solvent. Regarding the highly conserved residues, 8 are buried, and 12 are solvent exposed. Mapping the exposed and conserved residues onto the IE2-CTD structure clearly delineated a localized patch on the lateral side (Fig. 3B, lateral view, magenta and orange). On the top side, Q1372, N1390, and K1436 surrounding the 2-fold axis are highly conserved (Fig. 3B, top view). On α4 of the HTH-like motif, W1417 is conserved and partially accessible to the solvent (Fig. 3B, lateral view), although most of the indole side chain is buried between the two lysine residues K1337 and K1453 on α1 and α5, respectively. K1453 is highly conserved, while residues at the position of K1337 are variable (Fig. 3C). The partially exposed and highly conserved L1336 residue resides at the peripheral region of the conserved patch described above and contacts W1417 (Fig. 3B, lateral view). These interactions between the HTH-like motif and the core fold likely fix the relative spatial arrangement of the HTH-like motif. G1412 at the turn of the HTH-like motif is also conserved (Fig. 3B and C, bottom and lateral views) and is presumably required for the tight turn (kink) between α3 and α4, since glycine residues at this position are a conserved feature of genuine HTH motifs (34).
The positively charged area and conserved patch are involved in the transactivation function of IE2.
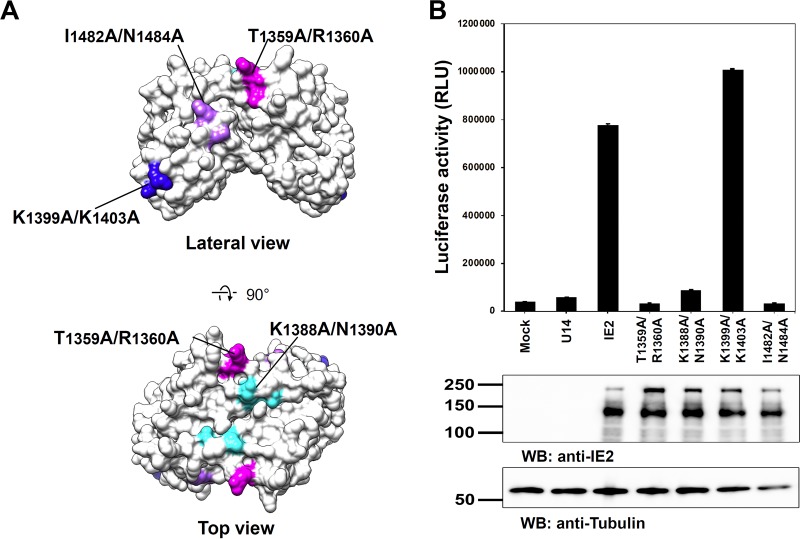
In order to assess the significance of the putative functional sites, we conducted a transactivation assay with IE2 mutants. Four pairs of residues at putative functional sites were replaced with alanine in the full-length IE2 expression plasmid (Fig. 4A). T1359 and R1360 are conserved among betaherpesvirus IE2 proteins (Fig. 3B and C) and constitute a part of the highly conserved patch (Fig. 3B and 4A). R1360 is also involved in PCA1 (Fig. 3A, top view). K1388 and N1390 are located around the 2-fold axis of IE2-CTD on the top view (Fig. 3A and B and 4A). K1388 is at the center of PCA1, and the adjacent residue N1390 is highly conserved (Fig. 3). K1399 and K1403 are involved in PCA3 and are located at the first α3 helix of the HTH-like motif (Fig. 3A and 4A). I1482 and N1484 are fully and highly conserved residues at the conserved patch, respectively (Fig. 3B and C and 4A).
FIG 4.
Mutagenesis analysis of the putative functional sites found on the IE2-CTD structure. (A) Mutations in the putative functional sites analyzed in this study. The mutations T1359A/R1360A, K1388A/N1390A, K1399A/K1403A, and I1482A/N1484A are indicated on the IE2-CTD surface model and shown in magenta, cyan, blue, and light purple, respectively. (B) Transactivation activity of IE2 assessed by a luciferase reporter assay. Luciferase expression under the control of the HHV-6 IE promoter (35) was measured as relative luciferase units (RLU), and the averages of data from three replicate wells were plotted. The bars indicate the standard errors. Mock and U14 were used as controls and stand for the empty or U14-encoding vector instead of the IE2 vector. Representative results from at least three independent transfection experiments are shown. WB, Western blotting.
The expression of wild-type IE2 activated the expression of luciferase under the control of the major immediate early promoter (MIEp) of HHV-6, named 6MIEp-d6in1 (35) (Fig. 4B), compared with the cases without IE2 or with the expression of the HHV-6A tegument protein U14 as a control (Fig. 4B). The transactivation activity of IE2 was largely diminished by the substitutions T1359A/R1360A, K1388A/N1390A, and I1482A/N1484A, while the IE2 mutant with the K1399A/K1403A substitution maintained the transactivation activity (Fig. 4B). These results demonstrated that PCA1 and the conserved patch in IE2-CTD have significant functions in transactivation.
DISCUSSION
The C-terminal region of HHV-6A IE2 (IE2-CTD) was previously expressed, purified, and crystallized by our group (30). In the present study, the crystal structure of IE2-CTD was determined and analyzed in detail. The HHV-6A IE2-CTD structure adopts a well-organized homodimeric form (Fig. 1C and D). Dimer formation at the homologous C-terminal region has been reported for HCMV IE2 based on gel shift mobility assays (23, 27). Residues 1334 to 1498 of HHV-6A IE2-CTD correspond to residues 390 to 553 of HCMV IE2 and lie close to the region spanning residues 388 to 542, which includes residues previously predicted to contribute to dimerization (26). Although IE2-CTD is missing the N-terminal part (residues 1290 to 1333) of the homologous region (23, 30), the main features of IE2 are likely to be attributed to the folded part and are discussed below.
Importantly, the secondary structural topology and dimeric form of IE2-CTD revealed an unexpected similarity to gammaherpesvirus factors EBNA1 of EBV and LANA of KSHV, both of which possess DBDs at their C termini, as does HHV-6A IE2 and its homologs, with repeat sequences at the N-terminal sides (Fig. 2B). The structure-based sequence alignment indicated structural conservation, but not sequence conservation, at the core folding part (Fig. 2F); thus, they might show structural convergence as DNA-binding domains. EBV EBNA1 is expressed throughout all latent phases of EBV infection, and the DBD is the key domain that accesses double-stranded DNA (dsDNA) (36, 37). EBNA1 regulates the EBV latent origin of replication (oriP), genome synthesis, and the transactivation of viral genes and also suppresses the expression of particular viral promoters. KSHV LANA, the master regulator of KSHV latency (38), is expressed during latency and is associated mainly with DNA replication and the regulation of transcription. The common occurrence of this DNA-binding module at the C terminus may be indicative of a need for a latency-associated modulation of the DNA status in the nucleus. The DBDs found in EBNA1 and LANA are not limited to the Herpesviridae, since the transactivator E2 of human papillomavirus (HPV) and bovine papillomavirus (BPV) also possesses this feature at the C terminus (39, 40). This observed similarity suggests that the prevalence of DBDs is much higher than previously thought.
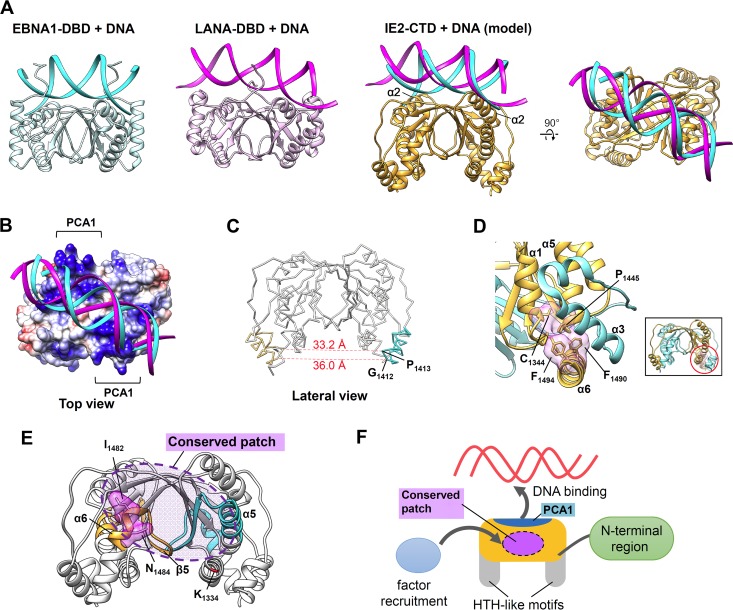
The structural similarity shared with EBNA1-DBD and LANA-DBD led us to predict a DNA-binding site in IE2-CTD. The crystal structures of DNA complexes are available for both EBNA1-DBD (PDB accession number 1B3T [41]) and KSHV LANA-DBD (PDB accession number 4UZB [42]). Both proteins bind DNA on the top side of IE2-CTD, and α-helices that correspond to α2 in IE2-CTD face the major groove of the bound dsDNA (Fig. 5A). On this top side, the β2-strand and the following coil region of IE2-CTD align well with these features in EBNA1-DBD and LANA-DBD (Fig. 2C); hence, a similar DNA backbone position would be anticipated and has been modeled. The predicted DNA-binding site of IE2-CTD includes the PCA1 region (Fig. 3A and 5B), which could interact with the negatively charged DNA backbone. This is consistent with the fact that EBNA1-DBD and LANA-DBD as well as BPV E2 and HPV E2 share similar positively charged patches at the DNA-binding interface (32, 33, 39–42). In a mutation assay, substitutions at K1388/N1390 within this putative DNA-binding site resulted in a loss of the transactivation function of IE2 (Fig. 4), and the functional significance of this area was suggested. IE2-CTD lacks some interaction sites that contribute to DNA binding in EBNA1-DBD and LANA-DBD, specifically the unique α1-helix and the loop between α5 and β5. The structural similarity and positively charged region of the top side of IE2-CTD are consistent with a DNA-binding site, although further experiments are needed to confirm this.
FIG 5.
Putative DNA-binding and functional sites of IE2-CTD. (A) DNA-binding mode of IE2-CTD predicted based on structural similarity with EBNA1-DBD and LANA-DBD. Structures of the EBNA1-DBD/DNA (PDB accession number 1B3T [41]) and LANA-DBD/DNA (PDB accession number 4UZB [42]) complexes are shown for reference. In the IE2-CTD/DNA model, cyan and magenta ribbons represent the expected DNA positions based on EBNA1-DBD and LANA-DBD, respectively. (B) Same model of IE2-CTD/DNA as in panel A, with the exception that the electrostatic potential on IE2-CTD is shown. PCA1 is located at the expected binding site. (C) The distances between the two HTH-like motifs were calculated as the distances between the Cα atoms of G1412 and P1413. (D) Conserved interaction among α6, α1, and α5. Fully conserved residues F1490 on α6, P1446 on α5, and C1344 on α1 constitute a hydrophobic cluster inside the fold. F1494 is the residue corresponding to Q548 of HCMV IE2, where substitution to arginine changed the IE2 activity (44), and resides on the adjacent region. The inset represents an overall view of IE2-CTD, and the red circle indicates the closeup region, although the observed angle was changed in the closeup view for clarity. (E) The region related to TFIIB binding in HCMV IE2 (28) was mapped on the IE2-CTD structure (cyan and orange). The location corresponds to the lateral side of IE2-CTD and overlaps the conserved patch shown in Fig. 3B. I1432/N1434, which is related to the transactivation of IE2 (Fig. 4), resides on this region and is shown as a surface model. The N-terminal position in the IE2-CTD structure near this site is indicated and shown in red. (F) Schematic illustration of the function of IE2-CTD. The top side houses the predicted DNA-binding site on PCA1, and the conserved patch (indicated by the dashed circle) on the lateral side is a possible site for the recruitment of transcription factors such as TFIIB. The HTH-like motif and N-terminal region may also be involved in these functions.
The IE2 structure revealed how the dimeric fold shared with EBNA1 and LANA can be extended to include unique features. The degree of structural consistency among BPV E2, HPV E2, EBV EBNA1, and KSHV LANA in terms of secondary structural topology and the overall fold is higher than that for IE2-CTD. In the case of IE2-CTD, the extended long loop housing the HTH-like motif is appended between the second and third β-strands (β2/β3 and β4), which is a prominent difference (Fig. 2D and E). The existence of a putative helix-turn-helix motif in the region spanning residues 463 to 513 of HCMV was predicted previously (27), although the corresponding region of HHV-6A IE2 (residues 1408 to 1459) only partially overlaps the actual HTH-like motif region (residues 1400 to 1420) (Fig. 3C). G1412, located at this tight turn, is fully conserved among IE2 homologs (Fig. 3C), consistent with the presence of a glycine residue at the equivalent position of the canonical HTH motif (34). HTH motifs in dimeric proteins generally recognize dsDNA by inserting their second “recognition” helices into the major groove of the bound dsDNA. The distance between the two HTH-like motifs of IE2-CTD is approximately 33.2 to 36.0 Å, calculated for the symmetrical Cα atoms of P1413 and G1412 (Fig. 5C). This distance is comparable to the pitch of typical B-DNA (34 Å) and to the distance between typical HTH motifs in DNA-binding proteins (43). The HTH-like motifs and surrounding helices constitute PCA3 (Fig. 3A) and are presumably required for accessing DNA. However, the mutation assay revealed that substitutions of two lysine residues, K1399/K1403, in PCA3 (Fig. 3A) have no effect on the transactivation function of IE2 (Fig. 4). Therefore, some doubts remain about the functional significance of the HTH-like motif without further experimental evidence. The sequences of HTH-like motifs do not fully mirror the sequences observed for canonical HTH motifs (34), apart from the conserved G1412 residue at the tight turn, and the second α4-helix of the HTH-like motif is much shorter than that in canonical HTH motifs (34). Furthermore, α1 adjacent to the HTH-like motif appears to sterically hinder interactions with DNA. Thus, it was impossible for us to construct a DNA-binding model of this HTH-like motif without additional information.
C-terminal α6 of HHV-6A IE2-CTD is another region that is absent from EBNA1-DBD and LANA-DBD. Helix α6 includes the residue corresponding to Q548 in HCMV IE2, for which replacement with arginine caused a virus replication defect and altered transcriptional activity (44). In the HHV-6A IE2-CTD structure, the side chains of the corresponding residue F1494 and the adjacent residue F1490 on α6 protrude toward the cavity surrounded by α3 of the HTH-like motif, N-terminal α1, and α5 and thus help to fix the spatial arrangement of these structural elements (Fig. 5D). Although F1494 is not conserved in the IE2 group, F1490 is highly conserved as either F or Y and interacts with the fully conserved P1445 residue on α5 that in turn contacts the fully conserved C1344 residue on α1 (Fig. 5D). These highly conserved interactions suggest that the arrangement of α1, α5, and α6 is likely conserved among IE2 factors. Thus, the structural features specific to the IE2 group (α1, α6, and the HTH-like motif) and their spatial arrangement may be important for IE2-CTD, although the actual function and significance remain to be elucidated.
Besides the DBD, transactivators usually possess a transactivation domain (TAD) to recruit transcription factors. IE2 of HCMV has been revealed to include a TAD at the conserved C-terminal region (28, 29, 45). HCMV IE2 contains TBP- and TFIIB-binding sites at the regions spanning residues 290 to 504 and 290 to 542, respectively (28). The crystal structure of HHV-6A IE2-CTD includes residues 1334 to 1498, which correspond to residues 390 to 553 of HCMV IE2. The region spanning residues 290 to 390 of HCMV IE2 has been shown to be insufficient for binding to both TBP and TFIIB (28), and thus, the core folded domain revealed in the HHV-6A IE2-CTD structure is likely responsible for these interactions, at least in part. Regarding TFIIB, the region spanning residues 505 to 542 of HCMV IE2 is essential for this interaction because truncated HCMV IE2 spanning residues 290 to 504 exhibits a severely impaired binding ability (28). In the HHV-6A structure, the region spanning residues 505 to 542 of HCMV IE2 corresponds to residues 1450 to 1487, and it is located on the lateral side of the IE2-CTD dimer, overlapping the conserved patch (Fig. 5E, orange and cyan). The importance of the conserved patch was demonstrated by mutation analyses in which substitutions in I1482/N1484 caused a defect in the IE2 transactivation function (Fig. 4). This region includes the C-terminal half of α5 and β5 and the N-terminal half of α6 (Fig. 5E), and α5 and β5 are involved in the α/β layer that forms the core α/β fold. Therefore, it is also possible that the poor ability of the truncated HCMV IE2 construct spanning residues 290 to 504 to bind TFIIB was caused by a deletion in the core region and disrupted the overall fold of this structural domain. Although further experimental evidence is required, the local patch of conserved residues on the lateral side of IE2-CTD is a candidate binding site for these transcription factors. Because the N-terminal end of the IE2-CTD structure, including K1334, resides on the lateral side (Fig. 1C and 5E), the missing region spanning residues 1324 to 1333 (and the N-terminal region beyond that, which is conserved among betaherpesviruses) (Fig. 3C) (23, 30) may also contribute to the interaction.
The crystal structure of IE2-CTD, together with previously reported knowledge of this conserved region in IE2 homologs of betaherpesviruses, suggests locations of the putative dimeric DNA-binding and transcription factor-binding sites (Fig. 5F). The crystal structure will facilitate future mutational analyses aimed at confirming the predicted functional sites. In parallel, structural analysis of a complex between IE2-CTD and DNA will provide a clear view of how the DBD recognizes the promoter and drives gene expression.
MATERIALS AND METHODS
Cloning, expression, purification, crystallization, and X-ray diffraction of HHV-6A IE2-CTD.
Details of the expression, purification, crystallization, and X-ray diffraction of HHV-6A were reported previously (30). Briefly, the region spanning residues 1324 to 1500 of HHV-6A strain U1102 IE2 was subcloned into the pMAL-c2 vector (New England BioLabs) and expressed as a fusion protein with maltose-binding protein (MBP) at the N terminus. The vector was used to transform Escherichia coli strain B834, and protein expression was induced in M9 medium supplemented with SeMet by the addition of isopropyl-β-d-1-thiogalactopyranoside (IPTG). Cells were disrupted by ultrasonication, and the fusion protein was purified by using amylose resin (New England BioLabs). HRV3c was used to cleave the fusion protein, and IE2-CTD was further purified by SP Sepharose column chromatography. Crystals of IE2-CTD were obtained by the sitting-drop vapor diffusion method with a reservoir solution containing 24% polyethylene glycol 3350, 0.2 M sodium formate, 20 mM Tris-HCl (pH 8.8), 20 mM CaCl2, and 1 mM dithiothreitol (DTT). Because the SeMet derivative of IE2-CTD produced larger crystals, this was used throughout the study. X-ray diffraction data were collected by using synchrotron radiation at SPring-8 BL38B1.
Structure determination and refinement.
During the preliminary structural analysis, experimental phasing was performed by using the SAD method in Phenix.Autosol (46, 47). The coordinates and individual B-factors of atoms in the structure were further refined by using Phenix.Refine (48). Coot (49) was used to validate and correct the structural model. A composite omit map was calculated and used to verify the final model. Structural analysis and figure preparation were conducted by using UCSF Chimera (50) and MolMol (51).
Size exclusion column chromatography.
Size exclusion column chromatography was performed by using procedures similar to those described previously for the HHV-6B U14 N-terminal domain (52). Briefly, purified HHV-6A IE2-CTD was loaded onto a Superdex 200-pg column (GE Healthcare) equilibrated with running buffer (20 mM Tris-HCl [pH 8.0], 1 M NaCl, 20 mM MgCl2, 1 mM EDTA, and 1 mM DTT). The molecular size was estimated from the elution volume of IE2-CTD and the calibration curve obtained by using the HWM calibration kit (GE Healthcare).
Antibodies.
An anti-IE2 antibody was prepared according to the following procedures. The codon-optimized IE2 gene was used as a template, and the C-terminal region spanning residues 994 to 1500 was amplified by PCR. The PCR product was digested with BamHI and XhoI and inserted in frame into the pMAL-c2 bacterial expression vector digested with BamHI and SalI. The purified protein (MBP-AIE2 residues 994 to 1500) was used as an antigen for the immunization of mice. The obtained monoclonal antibody against HHV-6A IE2 was named AIE2-1. The animal experiment was approved by the president of Kobe University after review by the Institutional Animal Care and Use Committee (permission number P140105) and carried out according to the Kobe University Animal Experimentation Regulations.
Plasmid construction.
IE2 expression vectors were constructed by using the pCAGGS-MCS plasmid (53). The IE2 expression vector was used as the template for KOD-Plus mutagenesis (Toyobo). The whole IE2 sequence and the introduced substitutions were verified by sequencing on a 3130 genetic analyzer (Applied Biosystems). The pCAGGS plasmid with the HHV-6A U14 gene was described previously (54) and used as a control. The 6MIEp-d6in1 plasmid (pGL3-basic vector), which includes the HHV-6B MIEp followed by a firefly luciferase gene, was described previously (35).
Luciferase reporter assay.
293T cells were plated onto 12-well plates at a density of 2.0 × 105 cells per well before transfection and were transfected with 0.5 μg of the 6MIEp-d6in1/pGL3 plasmid (35) and 2 μg of the IE2 plasmid (wild type or mutants) by using PEImax (Invitrogen). Twenty-four hours after transfection, firefly luciferase activity was measured by using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's protocols, using a Trister LB941 multimode reader (Berthold Technologies). The expression of IE2 was confirmed by Western blotting with the AIE2-1 antibody as described previously (55, 56). Tubulin was detected by an α-tubulin antibody (Sigma-Aldrich).
Accession number(s).
Structure factors and coordinates of HHV-6A IE2-CTD have been deposited in the Protein Data Bank under accession number 5WX8.
ACKNOWLEDGMENTS
We thank Huamin Tang (Nanjing Medical University) and Bochao Wang (Kobe University Graduate School of Medicine) for technical assistance. We thank Jun-ichi Miyazaki for providing the pCAGGS-MCS plasmid (53). Synchrotron radiation experiments were performed at BL38B1 in SPring-8 with the approval of RIKEN (proposal no. 2016A2556).
This study was supported in part by a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (JSPS) with KAKENHI grant number 17859322.
REFERENCES
- 1.Davison AJ. 2010. Herpesvirus systematics. Vet Microbiol 143:52–69. doi: 10.1016/j.vetmic.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, DiLuca D, Flamand L, Frenkel N, Gallo R, Gompels UA, Hollsberg P, Jacobson S, Luppi M, Lusso P, Malnati M, Medveczky P, Mori Y, Pellett PE, Pritchett JC, Yamanishi K, Yoshikawa T. 2014. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol 159:863–870. doi: 10.1007/s00705-013-1902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams MJ, Carstens EB. 2012. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012). Arch Virol 157:1411–1422. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aubin JT, Collandre H, Candotti D, Ingrand D, Rouzioux C, Burgard M, Richard S, Huraux JM, Agut H. 1991. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J Clin Microbiol 29:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume G, Guerrini S, Liu X, Foa-Tomasi L. 1993. Monoclonal antibodies to glycoprotein B differentiate human herpesvirus 6 into two clusters, variants A and B. J Gen Virol 74(Part 10):2257–2262. doi: 10.1099/0022-1317-74-10-2257. [DOI] [PubMed] [Google Scholar]
- 6.Jasirwan C, Furusawa Y, Tang H, Maeki T, Mori Y. 2014. Human herpesvirus-6A gQ1 and gQ2 are critical for human CD46 usage. Microbiol Immunol 58:22–30. doi: 10.1111/1348-0421.12110. [DOI] [PubMed] [Google Scholar]
- 7.Mori Y. 2009. Recent topics related to human herpesvirus 6 cell tropism. Cell Microbiol 11:1001–1006. doi: 10.1111/j.1462-5822.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- 8.Tang H, Mori Y. 2015. Determinants of human CD134 essential for entry of human herpesvirus 6B. J Virol 89:10125–10129. doi: 10.1128/JVI.01606-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang H, Serada S, Kawabata A, Ota M, Hayashi E, Naka T, Yamanishi K, Mori Y. 2013. CD134 is a cellular receptor specific for human herpesvirus-6B entry. Proc Natl Acad Sci U S A 110:9096–9099. doi: 10.1073/pnas.1305187110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang H, Wang J, Mahmoud NF, Mori Y. 2014. Detailed study of the interaction between human herpesvirus 6B glycoprotein complex and its cellular receptor, human CD134. J Virol 88:10875–10882. doi: 10.1128/JVI.01447-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyatt LS, Balachandran N, Frenkel N. 1990. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J Infect Dis 162:852–857. doi: 10.1093/infdis/162.4.852. [DOI] [PubMed] [Google Scholar]
- 12.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065–1067. [DOI] [PubMed] [Google Scholar]
- 13.Behzad-Behbahani A, Mikaeili MH, Entezam M, Mojiri A, Pour GY, Arasteh MM, Rahsaz M, Banihashemi M, Khadang B, Moaddeb A, Nematollahi Z, Azarpira N. 2011. Human herpesvirus-6 viral load and antibody titer in serum samples of patients with multiple sclerosis. J Microbiol Immunol Infect 44:247–251. doi: 10.1016/j.jmii.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Voumvourakis KI, Kitsos DK, Tsiodras S, Petrikkos G, Stamboulis E. 2010. Human herpesvirus 6 infection as a trigger of multiple sclerosis. Mayo Clin Proc 85:1023–1030. doi: 10.4065/mcp.2010.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogata M, Fukuda T, Teshima T. 2015. Human herpesvirus-6 encephalitis after allogeneic hematopoietic cell transplantation: what we do and do not know. Bone Marrow Transplant 50:1030–1036. doi: 10.1038/bmt.2015.76. [DOI] [PubMed] [Google Scholar]
- 16.Ongradi J, Ablashi DV, Yoshikawa T, Stercz B, Ogata M. 2017. Roseolovirus-associated encephalitis in immunocompetent and immunocompromised individuals. J Neurovirol 23:1–19. doi: 10.1007/s13365-016-0473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gravel A, Tomoiu A, Cloutier N, Gosselin J, Flamand L. 2003. Characterization of the immediate-early 2 protein of human herpesvirus 6, a promiscuous transcriptional activator. Virology 308:340–353. doi: 10.1016/S0042-6822(03)00007-2. [DOI] [PubMed] [Google Scholar]
- 18.Gompels UA, Nicholas J, Lawrence G, Jones M, Thomson BJ, Martin ME, Efstathiou S, Craxton M, Macaulay HA. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 19.Isegawa Y, Mukai T, Nakano K, Kagawa M, Chen J, Mori Y, Sunagawa T, Kawanishi K, Sashihara J, Hata A, Zou P, Kosuge H, Yamanishi K. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J Virol 73:8053–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiewe U, Neipel F, Schreiner D, Fleckenstein B. 1994. Structure and transcription of an immediate-early region in the human herpesvirus 6 genome. J Virol 68:2978–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholas J. 1994. Nucleotide sequence analysis of a 21-kbp region of the genome of human herpesvirus-6 containing homologues of human cytomegalovirus major immediate-early and replication genes. Virology 204:738–750. doi: 10.1006/viro.1994.1589. [DOI] [PubMed] [Google Scholar]
- 22.Papanikolaou E, Kouvatsis V, Dimitriadis G, Inoue N, Arsenakis M. 2002. Identification and characterization of the gene products of open reading frame U86/87 of human herpesvirus 6. Virus Res 89:89–101. doi: 10.1016/S0168-1702(02)00126-0. [DOI] [PubMed] [Google Scholar]
- 23.Chiou CJ, Zong J, Waheed I, Hayward GS. 1993. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J Virol 67:6201–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomoiu A, Gravel A, Flamand L. 2006. Mapping of human herpesvirus 6 immediate-early 2 protein transactivation domains. Virology 354:91–102. doi: 10.1016/j.virol.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 25.Marchini A, Liu H, Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J Virol 75:1870–1878. doi: 10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn JH, Chiou CJ, Hayward GS. 1998. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene 210:25–36. doi: 10.1016/S0378-1119(98)00056-0. [DOI] [PubMed] [Google Scholar]
- 27.Furnari BA, Poma E, Kowalik TF, Huong SM, Huang ES. 1993. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J Virol 67:4981–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caswell R, Hagemeier C, Chiou CJ, Hayward G, Kouzarides T, Sinclair J. 1993. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol 74(Part 12):2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 29.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol 66:4452–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Nishimura M, Wakata A, Mori Y. 2017. Purification, crystallization and X-ray diffraction study of the C-terminal domain of human herpesvirus 6A immediate early protein 2. Kobe J Med Sci 62:E142–E149. [PMC free article] [PubMed] [Google Scholar]
- 31.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. 2008. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res 36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bochkarev A, Barwell JA, Pfuetzner RA, Furey W Jr, Edwards AM, Frappier L. 1995. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell 83:39–46. doi: 10.1016/0092-8674(95)90232-5. [DOI] [PubMed] [Google Scholar]
- 33.Domsic JF, Chen HS, Lu F, Marmorstein R, Lieberman PM. 2013. Molecular basis for oligomeric-DNA binding and episome maintenance by KSHV LANA. PLoS Pathog 9:e1003672. doi: 10.1371/journal.ppat.1003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brennan RG, Matthews BW. 1989. The helix-turn-helix DNA binding motif. J Biol Chem 264:1903–1906. [PubMed] [Google Scholar]
- 35.Matsuura M, Takemoto M, Yamanishi K, Mori Y. 2011. Human herpesvirus 6 major immediate early promoter has strong activity in T cells and is useful for heterologous gene expression. Virol J 8:9. doi: 10.1186/1743-422X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frappier L. 2012. EBNA1 and host factors in Epstein-Barr virus latent DNA replication. Curr Opin Virol 2:733–739. doi: 10.1016/j.coviro.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Frappier L. 2012. The Epstein-Barr virus EBNA1 protein. Scientifica (Cairo) 2012:438204. doi: 10.6064/2012/438204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uppal T, Banerjee S, Sun Z, Verma SC, Robertson ES. 2014. KSHV LANA—the master regulator of KSHV latency. Viruses 6:4961–4998. doi: 10.3390/v6124961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegde RS, Androphy EJ. 1998. Crystal structure of the E2 DNA-binding domain from human papillomavirus type 16: implications for its DNA binding-site selection mechanism. J Mol Biol 284:1479–1489. doi: 10.1006/jmbi.1998.2260. [DOI] [PubMed] [Google Scholar]
- 40.Hegde RS, Grossman SR, Laimins LA, Sigler PB. 1992. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature 359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 41.Bochkarev A, Bochkareva E, Frappier L, Edwards AM. 1998. The 2.2 A structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1. J Mol Biol 284:1273–1278. doi: 10.1006/jmbi.1998.2247. [DOI] [PubMed] [Google Scholar]
- 42.Hellert J, Weidner-Glunde M, Krausze J, Lunsdorf H, Ritter C, Schulz TF, Luhrs T. 2015. The 3D structure of Kaposi sarcoma herpesvirus LANA C-terminal domain bound to DNA. Proc Natl Acad Sci U S A 112:6694–6699. doi: 10.1073/pnas.1421804112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alberts B. 2002. Molecular biology of the cell, 4th ed Garland Science, New York, NY. [Google Scholar]
- 44.Burgdorf SW, Clark CL, Burgdorf JR, Spector DH. 2011. Mutation of glutamine to arginine at position 548 of IE2 86 in human cytomegalovirus leads to decreased expression of IE2 40, IE2 60, UL83, and UL84 and increased transcription of US8-9 and US29-32. J Virol 85:11098–11110. doi: 10.1128/JVI.05315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. 1994. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J 13:2897–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. 2009. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr 65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Koradi R, Billeter M, Wuthrich K. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14:51–55, 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 52.Wang B, Nishimura M, Tang H, Kawabata A, Mahmoud NF, Khanlari Z, Hamada D, Tsuruta H, Mori Y. 2016. Crystal structure of human herpesvirus 6B tegument protein U14. PLoS Pathog 12:e1005594. doi: 10.1371/journal.ppat.1005594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 54.Mori J, Kawabata A, Tang H, Tadagaki K, Mizuguchi H, Kuroda K, Mori Y. 2015. Human herpesvirus-6 U14 induces cell-cycle arrest in G2/M phase by associating with a cellular protein, EDD. PLoS One 10:e0137420. doi: 10.1371/journal.pone.0137420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akkapaiboon P, Mori Y, Sadaoka T, Yonemoto S, Yamanishi K. 2004. Intracellular processing of human herpesvirus 6 glycoproteins Q1 and Q2 into tetrameric complexes expressed on the viral envelope. J Virol 78:7969–7983. doi: 10.1128/JVI.78.15.7969-7983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mori Y, Akkapaiboon P, Yonemoto S, Koike M, Takemoto M, Sadaoka T, Sasamoto Y, Konishi S, Uchiyama Y, Yamanishi K. 2004. Discovery of a second form of tripartite complex containing gH-gL of human herpesvirus 6 and observations on CD46. J Virol 78:4609–4616. doi: 10.1128/JVI.78.9.4609-4616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]