ABSTRACT
Tick-borne encephalitis virus (TBEV) causes a severe and potentially fatal neuroinfection in humans. Despite its high medical relevance, no specific antiviral therapy is currently available. Here we demonstrate that treatment with a nucleoside analog, 7-deaza-2′-C-methyladenosine (7-deaza-2′-CMA), substantially improved disease outcomes, increased survival, and reduced signs of neuroinfection and viral titers in the brains of mice infected with a lethal dose of TBEV. To investigate the mechanism of action of 7-deaza-2′-CMA, two drug-resistant TBEV clones were generated and characterized. The two clones shared a signature amino acid substitution, S603T, in the viral NS5 RNA-dependent RNA polymerase (RdRp) domain. This mutation conferred resistance to various 2′-C-methylated nucleoside derivatives, but no cross-resistance was seen with other nucleoside analogs, such as 4′-C-azidocytidine and 2′-deoxy-2′-beta-hydroxy-4′-azidocytidine (RO-9187). All-atom molecular dynamics simulations revealed that the S603T RdRp mutant repels a water molecule that coordinates the position of a metal ion cofactor as 2′-C-methylated nucleoside analogs approach the active site. To investigate its phenotype, the S603T mutation was introduced into a recombinant TBEV strain (Oshima-IC) generated from an infectious cDNA clone and into a TBEV replicon that expresses a reporter luciferase gene (Oshima-REP-luc2A). The mutants were replication impaired, showing reduced growth and a small plaque size in mammalian cell culture and reduced levels of neuroinvasiveness and neurovirulence in rodent models. These results indicate that TBEV resistance to 2′-C-methylated nucleoside inhibitors is conferred by a single conservative mutation that causes a subtle atomic effect within the active site of the viral NS5 RdRp and is associated with strong attenuation of the virus.
IMPORTANCE This study found that the nucleoside analog 7-deaza-2′-C-methyladenosine (7-deaza-2′-CMA) has high antiviral activity against tick-borne encephalitis virus (TBEV), a pathogen that causes severe human neuroinfections in large areas of Europe and Asia and for which there is currently no specific therapy. Treating mice infected with a lethal dose of TBEV with 7-deaza-2′-CMA resulted in significantly higher survival rates and reduced the severity of neurological signs of the disease. Thus, this compound shows promise for further development as an anti-TBEV drug. It is important to generate drug-resistant mutants to understand how the drug works and to develop guidelines for patient treatment. We generated TBEV mutants that were resistant not only to 7-deaza-2′-CMA but also to a broad range of other 2′-C-methylated antiviral medications. Our findings suggest that combination therapy may be used to improve treatment and reduce the emergence of drug-resistant viruses during nucleoside analog therapy for TBEV infection.
KEYWORDS: antiviral agents, antiviral therapy, escape mutant, tick-borne encephalitis virus, tick-borne pathogens
INTRODUCTION
Nucleoside and nucleotide analogs represent an important class of therapeutic antiviral agents that are commonly used to treat many life-threatening viral diseases (1, 2). In particular, nucleoside/tide derivatives are a backbone of treatment for HIV/AIDS and for chronic hepatitis B virus (HBV) infection (3), with the nucleotide derivative sofosbuvir acting to revolutionize the treatment of chronic hepatitis C virus (HCV) infection (4, 5). The constant struggle to develop novel nucleoside/tide analogs as potential antiviral agents against HCV has yielded numerous interesting derivatives (6) that are potential treatments for diseases caused by other members of the Flaviviridae family. Indeed, 2′-methyl nucleoside derivatives act as polymerase reaction terminators (7, 8) and are active against HCV; they also have a profound antiviral effect against several medically important flaviviruses. For instance, 7-deaza-2′-C-methyladenosine (7-deaza-2′-CMA; also known as MK-608) (Fig. 1A), a 2′-methyl nucleoside analog originally developed as an inhibitor of HCV by Merck Research Laboratories (9), has strong antiviral effects against mosquito-borne members of the genus Flavivirus, such as dengue virus (DENV) (10) and Zika virus (11, 12). We recently found that 7-deaza-2′-CMA has inhibitory activity in vitro against tick-borne encephalitis virus (TBEV), the most medically important tick-borne flavivirus (13, 14).
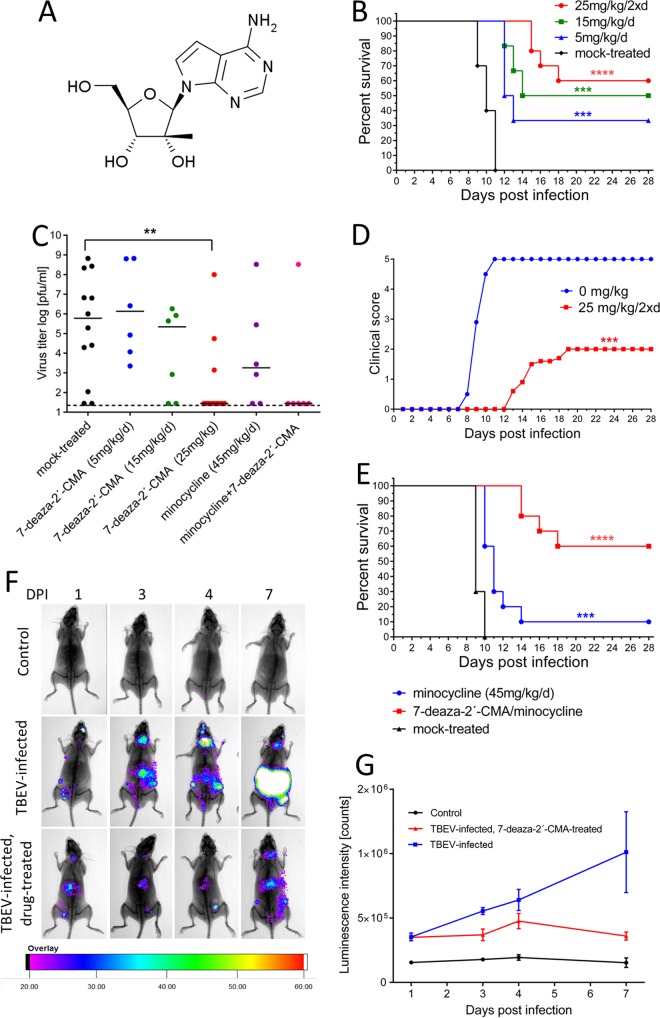
FIG 1.
7-Deaza-2′-CMA is effective at treating lethal TBEV infection in a mouse model. (A) Structure of 7-deaza-2′-CMA. (B) Groups of adult BALB/c mice were infected with a lethal dose of TBEV and treated intraperitoneally with the indicated doses of 7-deaza-2′-CMA or mock treated with PBS. Survival was monitored daily for 28 days. (C) Groups of adult BALB/c mice were infected with a lethal dose of TBEV and treated with the indicated dose of 7-deaza-2′-CMA, minocycline, a combination of 7-deaza-2′-CMA and minocycline, or PBS. On day 8 p.i., the brains of the mice were collected and homogenized, and the viral titers were determined using a plaque assay. (D) Illness signs were scored as follows: 0, no symptoms; 1, ruffled fur; 2, slowing of activity or hunched posture; 3, asthenia or mild paralysis; 4, lethargy, tremor, or complete paralysis of the limbs; and 5, death. (E) Groups of adult BALB/c mice were infected with a lethal dose of TBEV and treated intraperitoneally with minocycline, with a combination of minocycline and 7-deaza-2′-CMA, or with PBS. Survival was monitored daily for 28 days. (F and G) IFNβ+/Δβ-luc mice were infected with a lethal dose of TBEV and treated with 7-deaza-2′-CMA or mock treated with PBS. The bioluminescence signal indicating the IFN-β response to virus infection was scanned using a live-animal imaging device (F) and quantified (G). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
TBEV is the causative agent of tick-borne encephalitis (TBE), a potentially deadly neuroinfection that is prevalent in large areas of Europe and northern Asia. Despite available effective TBEV vaccines, the number of TBE cases continues to increase in Europe, and the risk areas continue to expand (15, 16). The severities of TBE clinical manifestations range from a mild flu-like infection to meningitis, encephalitis, and meningoencephalitis/radiculitis, with a 10% to 20% risk of long-lasting or permanent neuropsychiatric sequelae (17). Fatality rates range from <2% in Europe to up to 20% in Russia (17). No specific antiviral therapy is currently available; thus, there is an urgent need for efficient drugs to treat patients with TBE (17). Currently, 2′-methyl nucleoside derivatives, and 7-deaza-2′-CMA in particular, represent the lead candidates for further development of antivirals and their prodrug forms as effective therapeutics for this disease (13, 14).
Unfortunately, antiviral therapy based on chemical inhibitors of viral replication is often accompanied by the rapid emergence of drug-resistant “escape” mutants, substantially complicating therapy. Experience with the treatment of HIV, HBV, and HCV shows that resistance can develop rapidly with literally all of the direct-acting antiviral agents (18, 19). In the case of RNA viruses, including flaviviruses, the development of drug resistance is mainly related to the high mutation rates caused by the low fidelity of viral RNA-dependent RNA polymerases (RdRps), which introduce 10−4 to 10−6 error per nucleotide. This, together with a high viral replication rate, can generate highly variable viral populations that are termed quasispecies. Viral quasispecies represent dynamic mutant networks in which individual sequences are continuously diversified and regenerated by mutations, allowing the population to adapt quickly to changes in the environment, such as the presence of an antiviral drug (20, 21). Drug-resistant variants can rapidly become the dominant part of the viral population and lead to therapy failure. However, drug-resistant variants often also have a lower replicative capacity or “fitness” than wild-type viruses (22). If the therapy is stopped, the more virulent wild-type virus can again become dominant (22). Thus, it is crucial to identify specific mutations in all antiviral agents that lead to drug resistance, to characterize the replicative capacity of the resistant virus, and to evaluate the risk of potential therapeutic failure.
Here we evaluated the therapeutic effects of 7-deaza-2′-CMA in a lethal mouse model of TBE. We found that 7-deaza-2′-CMA treatment significantly reduced the development of signs of neuroinfection, reduced viral titers in the brain, and increased survival after the infection. We also generated a 7-deaza-2′-CMA-resistant TBEV variant by serial passage of the virus with increasing concentrations of the drug in vitro. The fitness of the drug-resistant virus was characterized both in vitro and in vivo in a mouse model. Whole-genome sequencing was used to identify a specific mutation associated with drug resistance and the phenotype change, and the effect of the identified mutation was confirmed by reverse genetics. The results revealed that a single conservative amino acid substitution in the sequence of the TBEV RNA-dependent RNA polymerase conferred resistance to a broad spectrum of 2′-methylated nucleoside inhibitors and strongly reduced viral replication fitness and viral neuroinvasiveness. In the absence of 7-deaza-2′-CMA, there was a rapid reversion to the wild-type genotype and phenotype in vitro but not in vivo. Taken together, these results have important implications for the development of drugs based on 2′-methylated nucleoside analogs as therapy for infections caused by flaviviruses.
RESULTS
7-Deaza-2′-CMA is effective at treating lethal TBEV infection in a mouse model.
The antiviral effects of 7-deaza-2′-CMA (Fig. 1A) were examined in vivo by use of a mouse model of lethal TBEV infection. Characteristic clinical signs of infection, such as ruffled fur, slowing of activity, asthenia, lethargy, tremor, and mild or complete paralysis of the limbs, typically appeared in TBEV-infected untreated adult BALB/c mice on days 8 to 11 postinfection (p.i.). Subcutaneous infection with 103 PFU was 100% lethal, with a mean survival time of 10 ± 1.4 days p.i. (Fig. 1B).
Treatment of TBEV-infected mice with 5 and 15 mg/kg of body weight of 7-deaza-2′-CMA once a day (initiated at the time of virus inoculation and ceased on day 17 p.i.) resulted in survival rates of 35% and 50% (P < 0.001) (Fig. 1B), respectively, and was associated with mean survival times of 12.5 ± 0.7 and 13 ± 1.4 days p.i., respectively. Treatment of TBEV-infected mice with 25 mg/kg of 7-deaza-2′-CMA that was initiated at the time of infection, administered twice daily, and ceased on day 17 p.i. resulted in a significantly higher survival rate (60%) (P < 0.0001) (Fig. 1B) and significantly reduced the development of clinical signs of neuroinfection (P < 0.001) (Fig. 1D). The mean survival time of infected and drug-treated mice was approximately 6.5 days longer than that of infected and mock-treated control mice. No apparent toxicity or other side effects were seen in mice treated with the highest dose of 7-deaza-2′-CMA, i.e., 25 mg/kg twice a day, during the entire 28-day monitoring period. Viral titers were significantly reduced in the brains of treated TBEV-infected mice compared to those in the brains of mock-treated TBEV-infected mice (P < 0.01) (Fig. 1C).
Mortality following TBEV infection is the result of both direct virus-mediated damage and immunopathology, so we tested whether treatment with a combination of 7-deaza-2′-CMA plus minocycline, a drug with immunomodulatory properties, would increase the survival of TBEV-infected mice. TBEV-infected mice were treated with 7-deaza-2′-CMA two times a day (25 mg/kg/dose) and/or with minocycline (45 mg/kg/day). Treatment with minocycline alone resulted in a significantly higher survival rate (P < 0.001) (Fig. 1E) and prolonged the mean survival time of the infected mice (11.8 ± 1.7 days). However, the survival rates and mean survival times after infection were comparable for mice treated with the drug combination and mice treated only with 7-deaza-2′-CMA, so the combination therapy showed no additive effects.
To monitor the antiviral effects of therapy with 7-deaza-2′-CMA in mice before the development of the first clinical signs of neuroinfection, we developed a bioluminescence mouse model of TBEV infection based on IFNβ+/Δβ-luc mice (23). This model allowed us to continuously monitor the development of TBEV infection and the effectiveness of antiviral therapy early in infection (Fig. 1F) by visualizing the host's response to the virus. Monitoring reporter activity in uninfected control mice revealed a background-level luminescence signal that was stable over the 7-day experimental period. In TBEV-infected mice, 2 to 3 times higher levels of luminescence signal were detected as early as day 1 p.i. A further increase in the luminescence signal was observed in the brain, lymph nodes, and peritoneal cavity (probably in the spleen and liver) on day 3 p.i. On day 4 p.i., the bioluminescence signal levels were approximately 5 times higher than those of uninfected controls. On day 7 p.i., when the mice showed the first clinical signs of infection, the signal was 7 times higher in the untreated TBEV-infected mice than in the uninfected controls (Fig. 1F).
Treatment of TBEV-infected IFNβ+/Δβ-luc mice with 7-deaza-2′-CMA (25 mg/kg two times a day) significantly reduced the bioluminescence signals at all investigated intervals. A low bioluminescence signal was detected primarily in the peritoneal cavity and/or lymph nodes; the signals in the brain had low values that were close to the background level (Fig. 1F and G).
TBEV resistance to 7-deaza-2′-CMA is associated with a single mutation in the NS5 gene.
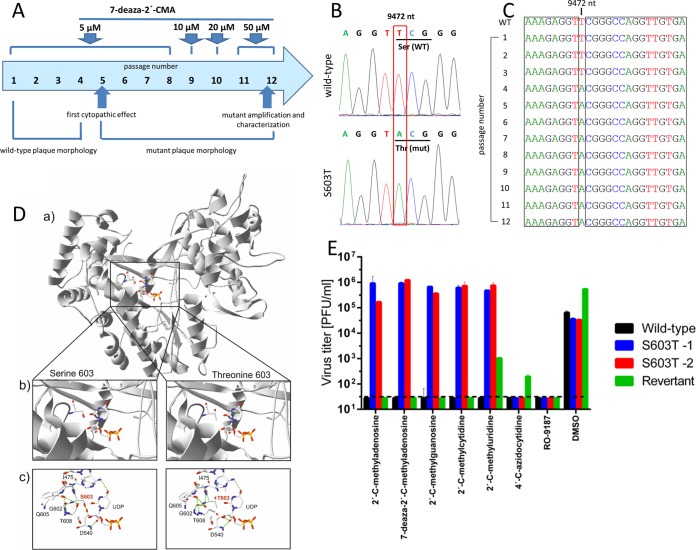
The generation of drug-resistant mutants is important for understanding how a drug works at the molecular level and can be helpful clinically in terms of guiding treatment. To select viruses that were resistant to 7-deaza-2′-CMA, TBEV was serially passaged in porcine kidney stable (PS) cells in the presence of increasing concentrations of 7-deaza-2′-CMA (Fig. 2A). Two drug-resistant clones were independently obtained after 12 passages with a maximal concentration of 50 μM 7-deaza-2′-CMA. Whole-genome sequence analysis (Sanger method) of the passaged viruses revealed a single mutation, at amino acid position 603 in the NS5 protein, that changed a serine residue to a threonine (Fig. 2B). This corresponded to a TCG → ACG nucleotide substitution in the NS5 gene. The same substitution was detected in both drug-resistant clones but not in the wild-type virus passaged in the absence of the drug. Sequencing of viruses after each passage revealed that the virus acquired the S603T mutation at passage 4 and that the mutation was retained until the end of the experiment (Fig. 2C). The S603T mutation mapped to the active site of the RNA-dependent RNA polymerase domain of the NS5 protein (Fig. 2D).
FIG 2.
TBEV resistance to 7-deaza-2′-CMA is associated with a single mutation in the NS5 gene. (A) Diagram showing the selection process for the generation of a TBEV strain resistant to 7-deaza-2′-CMA. TBEV was serially passaged in porcine kidney stable (PS) cells in the presence of increasing concentrations of 7-deaza-2′-CMA. (B and C) Whole-genome sequence analysis of the passaged virus revealed a single mutation, at amino acid position 603 in the NS5 protein, that changed a serine residue to a threonine. (D) The S603T mutation was mapped to the active site of the RNA-dependent RNA polymerase domain of the NS5 protein. Structural differences between the wild-type (S603) and mutated (T603) TBEV NS5 polymerases were examined. (a) Models of the wild-type and mutated TBEV NS5 polymerases were created by homology modeling using the structures of the West Nile virus (WNV) (PDB entry 2HFZ), dengue virus (DENV) (PDB entries 2J7U, 4V0Q, and 4V0R), and Japanese encephalitis virus (JEV) (PDB entry 4K6M) polymerases in MODELLER. (b) Closer view of the polymerase active sites. (c) Differences in the active site structures, showing changes in the hydrogen bond network. (E) The sensitivity or resistance of wild-type TBEV, the S603T TBEV mutant, and the revertant to a broad spectrum of diverse nucleoside inhibitors was evaluated in TBEV-infected PS cells in vitro. The S603T TBEV mutant showed high levels of resistance to a broad spectrum of 2′-C-methylated nucleoside inhibitors.
The S603T TBEV mutant is resistant to a broad spectrum of 2′-C-methylated nucleoside inhibitors.
To test whether the S603T mutation affects the sensitivity of TBEV to other nucleoside inhibitors, we tested a broad spectrum of nucleoside inhibitors to determine their inhibitory effects on TBEV-infected PS cells in vitro. A series of 2′-C-methyl-substituted nucleosides, 4′-C-azidocytidine, and RO-9187 were tested for inhibitory activity against both the S603T TBEV mutant and wild-type TBEV. The sensitivity of the S603T TBEV mutant to 7-deaza-2′-CMA (50% effective concentration [EC50] of >50 μM) was 46-fold lower than that of the wild-type virus (EC50 = 1.07 ± 0.03 μM) (Table 1). The S603T TBEV mutant showed high levels of cross-resistance to other 2′-C-methyl-modified nucleosides, such as 2′-C-methyladenosine, 2′-C-methylguanosine, 2′-C-methylcytidine, and 2′-C-methyluridine; their inhibitory effects were reduced dramatically (8- to 30-fold) in the S603T TBEV mutant compared to those against the wild-type virus (Fig. 2E). The inhibitory effects of 4′-C-azidocytidine and its stereochemical counterpart, RO-9187, were not affected in the S603T TBEV mutant, indicating an absence of cross-resistance between 4′-C-azido-modified and 2′-C-methylated nucleosides (Fig. 2E).
TABLE 1.
Sensitivities of wild-type and S603T mutant TBEV strains to 2′-C-methylated and 4′-C-azido-modified nucleosides
| Compound | EC50 (μM)a (fold increase compared to wild-type value) |
|
|---|---|---|
| Wild type | S603T mutant | |
| 7-Deaza-2′-C-methyladenosine | 1.07 ± 0.03 | >50 (>46.7) |
| 2′-C-Methyladenosine | 1.69 ± 0.19 | >50 (>29.5) |
| 2′-C-Methylguanosine | 1.98 ± 0.12 | >50 (>25.3) |
| 2′-C-Methylcytidine | 2.24 ± 0.27 | >50 (>22.3) |
| 2′-C-Methyluridine | 5.79 ± 0.15 | >50 (>8.6) |
| 4′-C-Azidocytidine | 5.45 ± 0.03 | 5.36 ± 0.21 (0.98) |
| RO-9187 | 0.99 ± 0.00 | 1.13 ± 0.21 (1.14) |
Determined from three independent experiments. EC50s were calculated as concentrations giving a 50% reduction of the viral titer by using the Reed-Muench method.
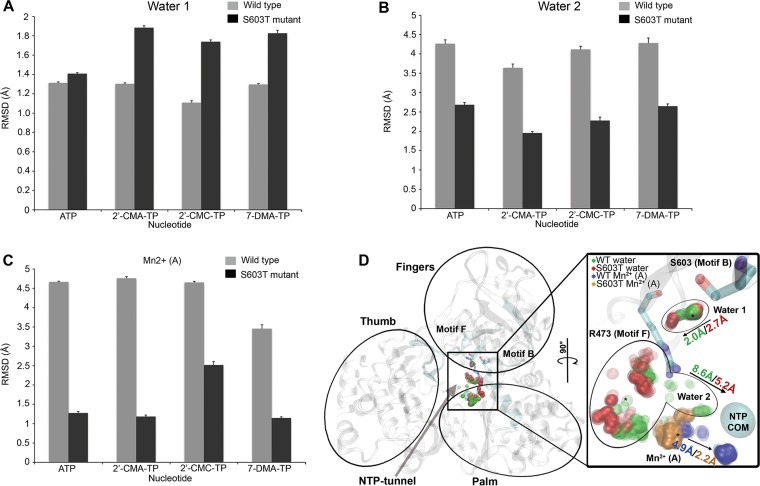
TBEV NS5 S603T escape mechanism from 7-deaza-2′-CMA as determined using all-atom molecular dynamics simulation.
All-atom stochastic software (24, 25) was used to determine how the NS5 polymerase from the S603T TBEV mutant helps the virus to escape 2′-C-methylated nucleoside triphosphates (NTPs). The starting position for each NTP (namely, ATP, 2′-C-methyladenosine, 2′-C-methylcytidine, and 7-deaza-2′-CMA) was ∼18 Å from the polymerase active site of the wild type and the S603T TBEV mutant via the NTP tunnel. Measurements were obtained for the root mean square deviation (RMSD) from the initial wild-type position for the following response variables: NTP, Mn2+ ion cofactor, and water molecule. A few water molecules were selected based on the following 2 criteria (since over 20 water molecules were included in the simulations): (i) the water molecule must be in close proximity to the NTP and substrate binding residues, i.e., the catalytic, priming, and interrogating residues (26); and (ii) the water molecule must have an RMSD of >0.5 Å from its initial position for all NTPs and strains.
Two-way analysis of variance (ANOVA) was performed (Table 2). The explanatory variables (NTP and strain) were both significant (P < 0.05) for each response variable, so the magnitude of the effect size was calculated as eta squared (η2) and was guided by Cohen's rule of thumb (27) (Table 2). Although small to large effect sizes were noted for the explanatory variable NTP and for the interaction between NTP and strain, we restricted our interpretation to response variables with large effect sizes caused by the strains. Figure 3A shows that one water molecule was farther from its initial position in the S603T TBEV mutant strain than in wild-type TBEV. In contrast, the second water molecule (Fig. 3B) showed a higher deviation in the wild-type virus than in the mutant strain.
TABLE 2.
Descriptive statistics for two-way ANOVA with RMSD data
| Molecule and source of variation | df | Sum of squares | Mean square | F value | P value | η2a |
|---|---|---|---|---|---|---|
| NTP N1 | ||||||
| NTP | 3 | 4,033.08 | 1,344.36 | 203.54 | <0.0001 | 0.211 |
| Strain | 1 | 382.39 | 382.39 | 57.89 | <0.0001 | 0.020 |
| NTP × strain | 3 | 1,112.68 | 370.89 | 56.15 | <0.0001 | 0.058 |
| Error | 2,064 | 13,632.73 | 6.61 | |||
| Water molecule 1 | ||||||
| NTP | 3 | 18.11 | 6.04 | 92.33 | <0.0001 | 0.062 |
| Strain | 1 | 113.28 | 113.28 | 1,732.35 | <0.0001 | 0.390b |
| NTP × strain | 3 | 23.96 | 7.99 | 122.15 | <0.0001 | 0.083 |
| Error | 2,064 | 134.97 | ||||
| Water molecule 2 | ||||||
| NTP | 3 | 197.36 | 65.79 | 40.17 | <0.0001 | 0.039 |
| Strain | 1 | 1,512.26 | 1,512.26 | 923.40 | <0.0001 | 0.296b |
| NTP × strain | 3 | 13.45 | 4.49 | 2.74 | 0.04 | 0.003 |
| Error | 2,064 | 3,380.22 | 1.64 | |||
| Water molecule 3 | ||||||
| NTP | 3 | 349.81 | 116.60 | 67.72 | <0.0001 | 0.070 |
| Strain | 1 | 397.69 | 397.69 | 230.97 | <0.0001 | 0.080 |
| NTP × strain | 3 | 679.31 | 226.44 | 131.51 | <0.0001 | 0.136 |
| Error | 2,064 | 3,553.90 | 1.72 | |||
| Mn2+ ion A | ||||||
| NTP | 3 | 385.21 | 128.40 | 171.78 | <0.0001 | 0.062 |
| Strain | 1 | 4,006.99 | 4,006.99 | 5,360.78 | <0.0001 | 0.649b |
| NTP × strain | 3 | 240.55 | 80.18 | 107.27 | <0.0001 | 0.038 |
| Error | 2,064 | 1,542.76 | 0.75 | |||
| Mn2+ ion B | ||||||
| NTP | 3 | 133.12 | 44.37 | 74.31 | <0.0001 | 0.062 |
| Strain | 1 | 128.06 | 128.06 | 214.44 | <0.0001 | 0.060 |
| NTP × strain | 3 | 643.12 | 214.37 | 358.99 | <0.0001 | 0.301 |
| Error | 2,064 | 1,232.52 | 0.60 |
Cohen's effect size guideline is as follows: small = 0.01, medium = 0.06, and large = 0.14.
Large effect size (for strain) between wild-type and S603T mutant TBEV NS5 polymerases.
FIG 3.
TBEV NS5 S603T mutant mechanism of escape from 7-deaza-2′-CMA as determined by all-atom molecular dynamics simulation. The bar charts show mean RMSD values with standard errors (y axes) for each NTP (x axes). Values for the two water molecules (A and B) and the Mn2+ ion (C) with large effect sizes (Table 2) are shown for the wild type (gray bars) and the S603T TBEV polymerase mutant (black bars). (D) TBEV polymerase structure, with the thumb, finger, and palm domains labeled. The B and F motifs are highlighted (cyan) and labeled to show the orientation of the NTP tunnel (arrow). The inset shows the active site turned 90°, which shows the positioning patterns of the two water molecules and the Mn2+ ion; dense clusters are shown in darker colors. The interrogating R473 residue of motif F, the S603 residue of motif B, and the active site center of mass (COM) are shown. The arrows indicate the direction of migration, with the largest RMSD shown for the wild type (WT) and the TBEV S603T mutant. The legend in the top left corner of the inset indicates the meaning of the colors for wild-type TBEV and the S603T TBEV mutant for the two water molecules and the Mn2+ ion. 2′-CMA, 2′-C-methyladenosine; 2′-CMC, 2′-C-methylcytidine; 7-DMA, 7-deaza-2′-C-methyladenosine; TP, triphosphate.
Two metal ions are bound within the RdRp catalytic site, with one ion (A) chelating with 3 catalytic aspartic residues and the other (B) chelating with 2 aspartic residues (26). Flavivirus RdRps are structurally homologous to other Flaviviridae RdRps (28), and their all-atom mechanism of escape from antiviral drugs is similar to that of the drug-resistant RdRp mutant of hepatitis C virus (29). The concerted positioning of the 2 water molecules and the Mn2+ A ion during the 7-deaza-2′-CMA exploration of TBEV polymerase in both strains is shown in Fig. 3D. The first water molecule is between the S603 residue and the conserved arginine, which is both an interrogating and a priming residue (26). The additional hydrophobic methyl group of threonine repels the water toward the arginine, leading to the higher deviation in the S603T TBEV mutant (Fig. 3A). The second water molecule is in proximity to the opening of the NTP tunnel and near the Mn2+ A ion. The positioning of the hydrophobic methyl group of threonine in the mutant prevents the second water molecule from migrating toward the positioning of the Mn2+ A ion, allowing it to maintain a lower RMSD than that for the wild type (Fig. 3B). The second wild-type TBEV water molecule forms large clusters at the Mn2+ A ion position, which may cause greater ion displacement than that in the S603T TBEV mutant (Fig. 3C).
The growth fitness of the S603T TBEV mutant is attenuated in cell culture.
To characterize the phenotype of the drug-resistant mutant, the plaque morphology and growth properties of the S603T TBEV mutant were assayed in PS cell cultures and compared to those of the wild-type virus (Fig. 4A). Wild-type virus produced plaques that ranged from 269 to 1,333 μm in diameter (mean, 730 ± 216 μm) and from 0.148 to 5.578 mm2 in area (mean, 1.814 ± 1.052 mm2); the plaques were mostly large, clear, round, and regular in shape (Fig. 4A to C). During passage of the virus with increasing concentrations of 7-deaza-2′-CMA, the wild-type-sized plaques gradually disappeared and were increasingly replaced by small and turbid plaques that ranged from 160 to 1,170 μm in diameter (mean, 540 ± 207 μm) and from 0.08 to 4.3 mm2 in area (mean, 1.051 ± 0.207 mm2) (Fig. 4A to C). The small-plaque phenotype became the dominant phenotype in the viral population starting at passage 5, and this was maintained during all subsequent passages.
FIG 4.
The S603T TBEV mutant has an attenuated phenotype. (A) (Top) The plaque morphology of the S603T TBEV mutant was assayed in a culture of porcine kidney stable (PS) cells and compared to that of the wild-type virus. (Bottom) Images of the plaques were scanned, and the plaque areas (B) and plaque diameters (C) of the wild-type virus and the S603T TBEV mutant were compared. (D) The growth kinetics of the S603T TBEV mutant was compared with the growth kinetics of the wild type and a genetic revertant to assess the replication efficacy of the mutant in PS cells. The neuroinvasiveness of the S603T TBEV mutant was studied in BALB/c mice and was compared to that of the wild-type virus. Adult BALB/c mice were infected subcutaneously with 103 PFU of either virus, and survival (E) and the clinical scores for neuroinfection symptoms (F) were monitored for 28 days. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The growth kinetics of the S603T TBEV mutant were compared to those of the wild-type virus to assess the replication efficacy of the mutant in PS cells. The wild-type virus reached a peak titer of 7.2 × 105 PFU/ml on day 2 p.i. Starting on day 4 p.i., the titer declined slightly and was 8.3 × 104 PFU/ml on day 7 p.i. (Fig. 4D). In contrast, the replication capacity of the drug-resistant virus (from the final stock after the 50 μM selection) was significantly lower than that of the wild type. The titers of the S603T TBEV mutant were approximately 103-fold lower for up to 3 days p.i. On days 4 to 6 p.i., the titers of the S603T TBEV mutant were 10- to 100-fold lower than the wild-type titers; on day 7 p.i., the difference in titers between the mutant and wild-type viruses was less pronounced, and both had titers of about 104 PFU/ml (Fig. 4D).
The neuroinvasiveness of the S603T TBEV mutant is highly attenuated in mice.
The neuroinvasiveness of the S603T TBEV mutant was studied and compared to that of the wild-type virus in BALB/c mice. Adult BALB/c mice (n = 10 per group) were infected subcutaneously with 103 PFU of either virus, and clinical signs of neuroinfection and survival were monitored for 28 days. Mice infected with the wild-type virus showed severe clinical signs of infection (ruffled fur, hunched posture, and hind leg paralysis), with 90% mortality and a mean survival time of 10 ± 1 days (Fig. 4E). Mice infected with the S603T TBEV mutant showed significantly lower mortality (20%) (P < 0.001) and a significantly longer mean survival time (21 ± 2.8 days) (P < 0.001) than those of mice infected with the wild-type virus (Fig. 4E). There were also considerable differences in the onset of disease signs and clinical scoring outcomes (P < 0.0001) (Fig. 4F).
Brains were collected from mice that did not survive infection with the S603T TBEV mutant, viral RNA was isolated and amplified by PCR, and the NS5 gene was sequenced to exclude possible reversion of the S603T mutation to the wild-type genotype during replication in the mouse. We found that TBEV carrying the S603T mutation was present in all of the mouse brains we investigated, indicating that no reverse mutation occurred spontaneously during the infection (Table 3). However, a few additional mutations that led to amino acid substitutions were detected in the NS5 gene of the drug-resistant mutant following propagation in mice (Table 3).
TABLE 3.
Mutations in NS5 of the Hypr virus in the brains of S603T mutant-infected mice
| Mouse no. | Presence of mutationa |
|||||||
|---|---|---|---|---|---|---|---|---|
| nt 7967 (NS5 aa 101) |
nt 8967 (NS5 aa 434) |
nt 9022 (NS5 aa 453) |
nt 9131 (NS5 aa 498) |
nt 9409 (NS5 aa 582) |
nt 9410 (NS5 aa 582) |
nt 9472 (NS5 aa 603) |
nt 9524 (NS5 aa 620) |
|
| G → A (Arg → Lys) | C → T (Arg → His) | T → C (Tyr → His) | C → T (Ala → Val) | G → A (Ala → Thr) | C → T (Ala → Val) | T → A (Ser → Thr) | A → T (Gln → Leu) | |
| 1 | + | − | − | − | + | − | + | − |
| 2 | + | + | + | − | − | + | + | − |
| 3 | − | − | − | − | − | − | + | + |
| 4 | − | − | − | + | + | − | + | − |
Nucleotide numbering is based on the nucleotide sequence of the TBEV Hypr strain (GenBank accession no. U39292.1).
Rapid reversion of the S603T mutant to the wild-type genotype in vitro in the absence of 7-deaza-2′-CMA.
The stability of the S603T mutant genotype was investigated during serial passages in PS cells in the absence of 7-deaza-2′-CMA. After 5 passages, the S603T mutant genotype reverted to wild type. This correlated with a reemergence of the wild-type phenotype (i.e., large plaques and gross cytopathic effects in PS cell cultures and growth kinetics in PS cells that were comparable to those of the wild-type virus) (Fig. 4D). Moreover, the revertant was sensitive to all 2′-C-methylated nucleosides tested (Fig. 2E). The sensitivity of the virus to 4′-C-azidocytidine and RO-9187 was not affected during virus passaging (Fig. 2E).
Site-directed mutagenesis confirms the role of the S603T mutation in drug resistance and virus attenuation.
To examine the direct effects of the S603T mutation on TBEV characteristics, the mutation was introduced into a recombinant TBEV strain (Oshima-IC) generated from an infectious cDNA clone and a TBEV replicon that expresses a reporter luciferase gene (Oshima-REP-luc2A) (30, 31). In the absence of 7-deaza-2′-CMA, the introduced S603T mutation rapidly reverted to the parental sequence (Ser) in the recovered recombinant virus from cells transfected with the Oshima-IC S603T mRNA. The S603T mutant virus was recovered only in the presence of 7-deaza-2′-CMA, and the virus titer was <2 × 103 PFU/ml, while the titer was >106 PFU/ml in cells transfected with Oshima-IC wild-type mRNA. The focus size of the cells infected with the S603T mutant virus was noticeably smaller than that of cells infected with wild-type virus (Fig. 5A).
FIG 5.
Site-directed mutagenesis confirms the role of the S603T mutation in drug resistance and virus attenuation. (A) Focus formation in baby hamster kidney fibroblasts (BHK cells) infected with Oshima-IC. BHK cells were infected with the Oshima-IC wild-type (a) or S603T mutant (b) virus and stained with anti-TBEV E-specific antibodies at 36 or 72 h postinfection, respectively. (B) Luciferase activity in cells transfected with TBEV replicon RNA. BHK cells were transfected with Oshima-REP-luc2A wild-type or S603T mRNA, and the luciferase activity was examined at 72 h posttransfection. Luciferase activity is expressed in raw light units (RLU). (C) Effect of 7-deaza-2′-CMA on the replication of the TBEV replicon. BHK cells were transfected with Oshima-REP-luc2A wild-type or S603T mRNA, and the luciferase activity was examined at 72 h posttransfection. The luciferase activity in the absence of 7-deaza-2′-CMA (0 μM) was considered the 100% value. *, P < 0.05; **, P < 0.01 (relative to the value at 0 μM). (D) Stability of the S603T mutation. BHK cells were transfected with Oshima-IC S603T mRNA and incubated for 5 days in the presence of 0 to 50 μM 7-deaza-2′-CMA. Viral RNA was harvested from the cells, and the mutation site was sequenced. (E) Survival curve for mice following intracerebral infection with Oshima-IC. C57BL/6J mice were infected intracerebrally with 50 PFU of the Oshima-IC wild-type or S603T mutant virus and monitored for 21 days. (F) At the indicated number of days after infection, the virus titers in brains were determined using plaque assays. Error bars indicate standard deviations (n = 6). ***, P < 0.001.
The luciferase activity in cells transfected with Oshima-REP-luc2A was significantly reduced by the S603T mutation, indicating that the mutation affected the genomic replication of NS5 (Fig. 5B). While the luciferase activity of the wild-type replicon was reduced by 7-deaza-2′-CMA, in a dose-dependent manner, 7-deaza-2′-CMA did not affect the activity of the S603T replicon (Fig. 5C). This indicated that the S603T mutation was responsible for the resistance of the S603T TBEV mutant to 7-deaza-2′-CMA.
Mice were infected intracerebrally with the Oshima-IC wild-type or S603T TBEV strain. The two groups of mice had similar morbidity and mortality rates (Fig. 5E), but the survival time of mice infected with the S603T TBEV mutant was longer (14.75 days) than that of mice infected with wild-type TBEV (9.33 days) (P < 0.05). The virus titers in the brain were significantly lower for the mice infected with the S603T mutant than for those infected with the wild-type virus (Fig. 5F). Virus isolates recovered from the brain were subjected to NS5 sequence analysis, and most had the S603T mutation; notably, two additional amino acid substitutions were detected as well (Table 4).
TABLE 4.
Mutations in NS5 of the virus in the brains of S603T mutant-infected mice
| Mouse no. | Presence of mutationa |
Time of brain collection | ||
|---|---|---|---|---|
| nt 9471 (NS5 aa 603) |
nt 9523 (NS5 aa 620) |
nt 9660 (NS5 aa 666) |
||
| A → T (Thr → Ser) | A → T (Gln → Leu) | T → A (Cys → Ser) | ||
| 1 | − | + | + | Time of death |
| 2 | − | + | + | Time of death |
| 3 | + | + | + | Time of death |
| 4 | − | + | + | Time of death |
| 5 | − | − | + | 5 days p.i. |
| 6 | − | + | − | |
| 7 | − | + | + | |
| 8 | − | + | + | 8 days p.i. |
| 9 | − | + | + | |
| 10 | − | + | + | |
Nucleotide numbering is based on the nucleotide sequence of the TBEV Oshima strain (GenBank accession no. AB753012.1).
The stability of the S603T mutation was investigated in baby hamster kidney (BHK) cells transfected with Oshima-IC S603T mRNA. After incubation for 5 days in the presence of 7-deaza-2′-CMA, we observed that 7-deaza-2′-CMA at 25 μM and 50 μM acted to retain the S603T mutation, while mixed wild-type and mutated genotypes were detected when the concentration of 7-deaza-2′-CMA was 12.5 μM or lower. In the absence of 7-deaza-2′-CMA, the wild-type genotype dominated (Fig. 5D). This result confirmed that the mechanism underlying the generation of the drug-resistant TBEV mutant and the reversion to the wild-type genotype was due primarily to selection for a particular mutation, not only to a growth advantage of a particular viral clone that was present in the quasispecies.
DISCUSSION
TBEV NS5 is a candidate target for nucleoside analog inhibitors (2). We previously found that 2′-C-methylated nucleoside derivatives, and 7-deaza-2′-CMA in particular, have high antiviral activity against TBEV in vitro (13, 14). In the present study, we demonstrated the antiviral effect of 7-deaza-2′-CMA in a lethal rodent model of TBE. A dose of 25 mg/kg 7-deaza-2′-CMA administered twice a day starting on the day of infection showed the highest antiviral efficacy. This dose significantly increased mouse survival, decreased the viral titers in mouse brains, and reduced the clinical signs of neuroinfection in the infected animals (Fig. 1). The treatment started at the time of infection, which does not correlate with potential human drug use, since therapy is usually initiated during the neurological phase of the disease. This scenario is difficult to replicate because TBEV-infected mice usually die within 1 to 2 days after the initial onset of neurological signs of the infection. A transgenic IFNβ+/Δβ-luc mouse model was used to monitor the development of the infection in the mice by revealing the host beta interferon (IFN-β) response before the appearance of the first clinical signs of infection. Previous work showed that there is a good correlation between the bioluminescence signal and virus replication in these mice (32), allowing nondestructive and continuous monitoring of viral invasion and the level of virus replication in mouse organs. Indeed, this model is an excellent way to monitor the effectiveness of antiviral therapy in mice that exhibit no signs of disease. Using this model, we confirmed that the treatment of TBEV-infected mice with 7-deaza-2′-CMA reduced virus spread in the body and the level of virus replication in multiple mouse organs and tissues (Fig. 1F and G).
Since TBE is an immunopathological disease that involves both virus damage and pathological immune reactions (33, 34), we tried combination therapy with both 7-deaza-2′-CMA and minocycline, a broad immunomodulatory compound that has anti-inflammatory, antiapoptotic, and neuroprotective properties (35). Minocycline is an antibiotic that is a tetracycline derivative, and it was reported previously that tetracycline could reduce the manifestations of the inflammatory response during TBE (36). Minocycline is effective in animal models of Japanese encephalitis (35, 37, 38) and is under clinical trials in humans with an acute form of viral encephalitis (39). The present study found that minocycline treatment of TBEV-infected mice slightly increased their survival and prolonged their mean survival time, but no additive effects were seen with combination therapy (Fig. 1E). Similar to our study, other studies have found that 7-deaza-2′-CMA is a potent inhibitor of virus replication and disease progression in mice infected with Zika virus (12) and that it reduces viremia in a dengue viremia mouse model (40). In human clinical trials for chronically HCV-infected patients, treatment with 7-deaza-2′-CMA failed due to adverse effects in patients after long-term treatment—possibly by mitochondrial toxicity (41). However, 7-deaza-2′-CMA remains a highly promising lead candidate for the treatment of acute infections caused not only by flaviviruses but also by other important emerging viruses (2, 11–14, 28, 42).
Low replicative fidelity and the high mutation rates of RNA viruses lead to viral resistance to antiviral drugs. Thus, for any antiviral agent, it is crucial to identify specific mutations that are responsible for drug resistance, to characterize the replicative capacity of the resistant virus, and to evaluate the risk of potential therapeutic failure. Here we selected 7-deaza-2′-CMA-resistant TBEV by serial passage of the virus with increasing 7-deaza-2′-CMA concentrations. Whole-genome sequencing revealed a single amino acid substitution, S603T, in the genome of the drug-resistant mutant compared to the genome of a wild-type virus. The mutation was located in the RdRp active site of viral NS5. Interestingly, serine and threonine are considered conservative amino acids, and the substitution caused only subtle changes in the structure of the NS5 RdRp active site (Fig. 2D). The signature high-level resistance mutation S603T conferred resistance not only to 7-deaza-2′-CMA but also to other 2′-C-methylated nucleoside derivatives. There was no cross-resistance of the S603T TBEV mutant to 4′-C-azidocytidine or RO-9187, suggesting that the mechanisms of resistance to 2′-C-methylated and 4′-C-azido compounds might be different (Fig. 2E). We speculate that the absence of the 2′-β-methyl moiety in 4′-C-modified nucleosides may allow these compounds to bind to NS5 and that their conformations are not hindered by the increased steric bulk of the S603T substitution in the NS5 active site (43, 44). Taken together, these results show that the S603T mutation-conferred resistance is directed toward the 2′-β-methyl-ribose moiety and does not involve other substitutions of the ribose ring or the heterobase identity/modifications (45).
Resistance to 2′-C-methylated nucleosides has been studied previously in other members of the Flaviviridae family. In HCV, the NS5B S282T amino acid change, which corresponds to an S603T substitution in motif B of TBEV RdRp, was first selected in the presence of 2′-C-methyladenosine (45). Recently, the S603T mutation, which is associated with additional amino acid substitutions in the NS5 active site (mainly the C666S and M644V mutations), was also reported for the 2′-C-methylcytidine-resistant Alkhurma hemorrhagic fever virus (46). The S604T substitution in motif B of the Zika virus recombinant NS5 polymerase corresponds to the S603T substitution in TBEV, and the S604T substitution confers resistance to the nucleoside inhibitor sofosbuvir triphosphate but not to the pyridoxine-derived nonnucleoside small-molecule inhibitor DMB213 (47). In contrast, two substitutions in the methyltransferase domain of NS5 of dengue virus type 2, A60T and Y201H, confer resistance to 7-deaza-2′-CMA (48).
The all-atom escape from 2′-C-methylated nucleosides by Flaviviridae serine mutant polymerases uses a concerted mechanism within the active site that involves substrate binding residues, metal ions, and water molecules. The 2′-C-methylated nucleoside analogs use water molecules as camouflage during active site exploration. The hydrophobic methyl group in the serine-to-threonine mutant repels the water molecules surrounding the analogs, reducing its interrogation by the conserved arginine in motif F (29). This conserved arginine is essential for NTP incorporation (49). During the initial exploration of 7-deaza-2′-CMA, the S603T TBEV mutant polymerase maintains the position of a water molecule near the NTP tunnel opening and the Mn2+ A ion. This may explain the reduced replication of the S603T TBEV mutant, since water molecules are used to coordinate the NTP phosphate groups with the metal ions in wild-type polymerases (50) (Fig. 3).
In the Zika virus recombinant NS5 polymerase, the S604T substitution reduces enzyme activity compared to that of the wild-type polymerase (47). However, to the best of our knowledge, no studies have investigated the biological properties of flaviviruses that are resistant to 2′-C-methylated nucleoside analogs. Here we showed that the drug-resistant TBEV strain produced small turbid plaques and showed reduced growth fitness in vitro (Fig. 4A to D). This was confirmed by use of a drug-resistant virus that was selected by cell culture passages under the pressure of 7-deaza-2′-CMA as well as by use of an infectious cDNA TBEV clone and replicon with the introduced S603T substitution (Fig. 5A and B). During passaging of the virus under pressure of 7-deaza-2′-CMA, the slowly replicating S603T TBEV mutant probably had a selective advantage in terms of outcompeting other variants with a higher replication rate (20). Reduced replication fitness might limit the spread of the resistant virus in cell cultures and may closely correlate with the altered plaque morphology seen with the mutant clone (51–53). This would explain why rapidly replicating wild-type viruses formed large plaques, whereas the S603T TBEV mutant resulted in small plaques.
The lower replication capacity of the TBEV mutant may explain its rapid reversion to a drug-sensitive phenotype when 7-deaza-2′-CMA is no longer present. Under nonselective conditions, the replication capacity of the S603T TBEV mutant is probably not sufficient to give the mutant a replicative advantage over fitter variants in the viral quasispecies, resulting in elimination of the S603T TBEV mutant from the infected cell culture. This process might be considerably accelerated by spontaneous reverse mutations that contribute to a rapid increase in the wild-type genotypes in the progeny of a viral population when the selective agent is absent (54).
The resistance of the S603T TBEV mutant to 2′-C-methylated nucleoside analogs was associated with strong virus attenuation in mice. We found that the S603T mutation significantly reduced viral neuroinvasiveness after subcutaneous inoculation as well as neurovirulence after intracerebral administration (Fig. 4E and F and 5E and F). After subcutaneous inoculation, TBEV isolates from the brains of dead or sacrificed moribund mice were subjected to sequence analysis and found to contain the S603T mutation. With one exception, the same thing was observed in the brains of mice inoculated intracerebrally. A couple of additional mutations in the NS5 gene were identified in viruses recovered from mice infected with the S603T TBEV mutant, but it is unknown whether these substitutions represent compensatory mutations or random mutations. The Q620L mutation was detected in almost all virus clones that were recovered from mouse brains after intracerebral inoculation with Oshima-IC S603T and from the brain of one mouse following subcutaneous inoculation with the Hypr S603T TBEV mutant (Tables 3 and 4). The effects of these mutations on the biological properties of the virus and on antiviral resistance merit further study in the future.
In conclusion, we found that 7-deaza-2′-CMA is a strong TBEV inhibitor in vivo, significantly increases the survival of TBEV-infected mice, and reduces virus titers and the clinical signs of neuroinfection in brains. A single conservative mutation, S603T, in the NS5 RdRp caused only subtle changes in the structure of the polymerase active site but was associated with cross-resistance to various 2′-C-methylated nucleoside analogs and with strong reductions in viral fitness both in vitro and in a mouse model. The S603T TBEV mutant was not coresistant to 4′-C-azidocytidine or RO-9187, suggesting that combination therapy might improve treatment and reduce the emergence of drug-resistant viruses during therapy with 2′-C-methylated nucleoside analogs.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in strict accordance with Czech and Japanese laws and guidelines for the use of experimental animals and the protection of animals against cruelty. The animal care and use protocols adhered to the fundamental guidelines for proper conduct of animal experiments and related activities in academic research institutions by the Ministry of Education, Culture, Sports, Science and Technology (notice 71, 1 June 2006), the standards relating to the care and management of laboratory animals and relief of pain of the Ministry of the Environment (notice 84, 30 August 2013), and the Act on Welfare and Management of Animals (revised 5 September 2012) in Japan and to Animal Welfare Act 246/1992 Coll. in the Czech Republic. All procedures were reviewed by local ethics committees and approved by the president of Hokkaido University after review by the Animal Care and Use Committee of Hokkaido University and by the Ministry of Agriculture of the Czech Republic (permit no. MZe 1650).
Virus, cells, and antiviral compounds.
The well-characterized, low-passage-number TBEV strain Hypr, a member of the European TBEV subtype, was passaged 6 times in the brains of suckling mice and used in this study (the virus was obtained from the virus collection of the National Institute of Public Health, Prague, Czech Republic). This strain was originally isolated from the blood of a 10-year-old child who died because of TBE in 1953 (55).
PS cells (56) used for plaque assay, viral subculture, and construction of virus growth curves (see below) were obtained from the National Cell Culture Collection, National Institute of Public Health, Prague, Czech Republic. The cells were cultured at 37°C in Leibovitz (L-15) medium supplemented with 3% newborn calf serum plus a 1% mixture of penicillin and glutamine (Sigma-Aldrich). BHK cells (obtained from the American Type Culture Collection [ATCC]) were used for transfection of the Oshima-IC and Oshima-REP-luc2A plasmids and for infection with the Oshima-IC virus.
The following antiviral compounds were purchased: 2′-C-methyl-substituted nucleosides from Carbosynth (Compton, United Kingdom) and 4′-azidocytidine and 2′-deoxy-2′-beta-hydroxy-4′-azidocytidine (RO-9187) from Medchemexpress (Stockholm, Sweden). For in vitro studies, the test compounds were solubilized in 100% dimethyl sulfoxide (DMSO) to yield 10 mM stock solutions.
Plaque assay.
Virus titers were evaluated by plaque assay as described previously (13, 14, 57). Briefly, 10-fold dilutions of TBEV in L-15 medium were prepared in 24-well tissue culture plates, and PS cells were added in suspension (0.6 × 105 to 1.5 × 105 cells per well). Following incubation for 4 h, the infected PS cell suspension was overlaid with 1.5% (wt/vol) carboxymethyl cellulose in L-15 medium. After a 5-day cultivation at 37°C, the infected plates were washed extensively with phosphate-buffered saline (PBS), and the cell monolayers were stained with naphthalene black. Plaque numbers were counted, and the virus titer was expressed as the number of PFU per milliliter.
Images of the plaques were scanned on a BioVendor C-series array reader specially modified for plaque assays. Imaging was done sequentially well by well, with illumination from the bottom of the sample and with a full high-definition (HD) camera system. Every image was then processed by BioVendor Analytics software, which automatically counts plaques based on specialized image processing algorithms developed for this task.
Selection of drug-resistant viruses.
PS cells were seeded into 96-well plates (approximately 2 × 104 cells per well) and incubated for 24 h to form a confluent monolayer. Following incubation, the medium was aspirated from the wells and replaced with 200 μl of fresh medium containing 5 μM 7-deaza-2′-CMA. Simultaneously, the cells were infected with the Hypr TBEV strain at a multiplicity of infection (MOI) of 0.1. The formation of cytopathic effect (CPE) was monitored visually using an Olympus BX-5 microscope, yielding a 40 to 50% CPE in virus-infected cultures. The culture medium was then harvested and used for infection of fresh cell monolayers. A 5 μM concentration of 7-deaza-2′-CMA was used during the initial 8 passages. The subsequent 4 passages were performed with a gradual increase in drug concentration to 50 μM (Fig. 2A). In parallel, a control TBEV isolate was not passaged with 7-deaza-2′-CMA but in the presence of 0.5% (vol/vol) DMSO and was used as a mock-selected wild-type virus. After passage 12, the drug-resistant and control TBEV strains were harvested and subjected to an additional subculture to make a virus stock for further testing (titer of about 106 PFU/ml for the wild type and 104 to 105 PFU/ml for the drug-resistant TBEV strain). The in vitro selection of drug-resistant TBEV strains was performed in duplicate, resulting in two independent mutant clones, denoted S603T-1 and S603T-2.
In order to reinduce the wild-type virus (revertant) from the S603T TBEV mutant population, the S603T TBEV mutant was repeatedly passaged on PS cells in the absence of 7-deaza-2′-CMA. The sensitivity of the obtained viral progeny to 7-deaza-2′-CMA was determined after each passage by cultivation of the virus in the presence of 7-deaza-2′-CMA (50 μM). After passage 7, the obtained revertant was subjected to one more passage to prepare a virus stock for further testing (virus titer of about 106 PFU/ml). Obtained viruses were subjected to full-length sequence analysis (both mutants as well as the parental virus were sequenced), virulence characterization, and assessment of sensitivity to different classes of antivirals.
Antiviral assays.
A viral titer inhibition assay was used to measure the antiviral efficacy of the nucleoside analogs in cell culture and was performed as described previously (13). Briefly, PS cells were seeded in 96-well plates at approximately 2 × 104 cells/well and incubated for 24 h to form a confluent monolayer. The growth medium was then aspirated from the wells and replaced with 200 μl of fresh medium containing 50 μM test compound (3 wells per compound) and simultaneously infected with TBEV at an MOI of 0.1. For the mock-treated controls, DMSO was added to virus-infected and mock-infected cells, to a final concentration of 0.5% (vol/vol). The infected cell monolayers were monitored by use of a microscope to yield 70% to 90% CPE. Viral titers were determined by plaque assays and expressed as numbers of PFU per milliliter.
To evaluate the growth kinetics of the drug-resistant TBEV mutant, PS cell monolayers cultured for 24 h in 96-well plates were treated with 200 μl of medium containing 50 μM 7-deaza-2′-CMA and infected with TBEV at an MOI of 0.1. For the wild-type and revertant TBEV strains, medium containing 0.5% (vol/vol) DMSO was used. The medium was collected from the wells daily (days 1 to 7 p.i.; three wells at each time point), and the viral titers were determined by plaque assay. The virus titer values were used to construct TBEV growth curves.
For the dose-response studies with 2′-C-methylated and 4′-C-azido-modified nucleoside analogs, PS cell monolayers were cultured with 200 μl of medium containing individual test compounds at concentrations ranging from 0 to 50 μM and were simultaneously infected with TBEV at an MOI of 0.1. The medium was collected from the wells on day 3 or 4 p.i. (wild type) or day 6 or 7 p.i. (mutant), and the viral titers were determined by plaque assay and used to construct TBEV dose-response curves and to estimate the 50% effective concentration (EC50).
RNA isolation, PCR, and whole-genome sequencing.
RNA was isolated from growth medium or brain tissue homogenate by use of a QIAmp viral RNA minikit (Qiagen). Reverse transcription was performed using a ProtoScript First Strand cDNA kit (New England BioLabs) according to the manufacturer's instructions for the synthesis of first-strand cDNA, which was subsequently used for PCR amplification using 1 μl of the antisense primer (0.5 μM). To cover the entire TBEV genome, 35 overlapping DNA fragments were amplified by PCR as described previously (58), using PPP master mix (Top-Bio, Czech Republic), 1 μl of each primer (stock concentration, 0.01 mM; primer sequences are available upon request), and 1 μl of template. The cycling conditions were as follows: (i) denaturation (5 min at 95°C) and (ii) 30 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 1 min 30 s. The PCR products were visualized in a 1.7% agarose gel in Tris-acetate-EDTA buffer. DNA was purified using a High Pure PCR product purification kit (Roche) according to the manufacturer's recommendations. The PCR products were subsequently directly sequenced (Sanger method) by a commercial service (SEQme, Czech Republic). Both the nucleotide sequence and the deduced amino acid sequence were analyzed using BioEdit Sequence Alignment Editor, version 7.2.0.
Viruses from cDNA clones and replicons.
The TBEV Oshima-IC and Oshima-REP-luc2A replicons were prepared from an infectious cDNA clone (28) and a replicon expressing a firefly luciferase gene (29), using the Oshima 5-10 strain (accession no. AB062003), which was isolated in Hokkaido, Japan, in 1995 (59). Standard PCR mutagenesis techniques were used to construct Oshima-IC and Oshima-REP-luc2A S603T, in which a T-to-A mutation at nucleotide position 9471 was introduced.
RNAs were transcribed from the Oshima-IC and Oshima-REP-luc2A plasmids by use of an mMESSAGE mMACHINE SP6 kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and then transfected into BHK cells by use of TransIT-mRNA (Mirus Bio, Madison, WI, USA) as described previously (60).
For the focus assay, monolayers of BHK cells were infected with the Oshima-IC virus, overlaid with medium containing 2% fetal calf serum (FCS) and 1.5% carboxymethyl cellulose, and incubated for the indicated time. After incubation, cells were fixed with 4% (wt/vol) paraformaldehyde and permeabilized with 0.2% (vol/vol) Triton X-100. After blocking with 2% (wt/vol) bovine serum albumin (BSA), the cells were incubated with polyclonal mouse ascites fluid against TBEV. After extensive washing, the cells were incubated with Alexa 488-conjugated anti-mouse IgG antibodies (Thermo Fisher Scientific, Inc.). The cells were observed using a BZ-9000 fluorescence microscope (Keyence, Osaka, Japan).
To prepare cell extracts for the luciferase assays, BHK cells were washed with PBS, lysed using cell culture reporter lysis buffer (Promega, Madison, WI, USA), and incubated at room temperature for 10 min. Luciferase assays were carried out using a luciferase assay system (Promega) according to the manufacturer's instructions.
Infection of mice with TBEV.
To evaluate the anti-TBEV effects of 7-deaza-2′-CMA in vivo, four groups of 6-week-old female BALB/c mice were injected subcutaneously with TBEV (1,000 PFU/mouse) and immediately treated intraperitoneally with 200 μl of 7-deaza-2′-CMA, as follows: group 1 mice (n = 10) were nontreated control animals, group 2 mice (n = 6) were treated with 5 mg/kg once a day, group 3 mice (n = 6) were treated with 15 mg/kg once a day, and group 4 mice (n = 10) were treated with 25 mg/kg twice a day at 8-h intervals. 7-Deaza-2′-CMA was freshly solubilized in sterile saline buffer before each injection and was administered to the animals for 17 days p.i. The clinical scores and survival rates of the TBEV-infected mice were monitored daily for 28 days. Illness signs were scored as follows: 0, no symptoms; 1, ruffled fur; 2, slowing of activity or hunched posture; 3, asthenia or mild paralysis; 4, lethargy, tremor, or complete paralysis of the limbs; and 5, death.
In order to assess in vivo the antiviral efficacy of minocycline, 3 groups (n = 10/group) of 6-week-old female BALB/c mice were infected subcutaneously with TBEV (1,000 PFU/mouse) and immediately treated as follows: group 1 mice were nontreated control animals, group 2 mice were treated with 45 mg/kg minocycline, and group 3 mice were treated with a combination of 45 mg/kg minocycline and 25 mg/kg 7-deaza-2′-CMA (33). Whereas minocycline was injected into the mice once daily for 8 days, 7-deaza-2′-CMA was administered twice daily at 8-h intervals over the 17-day period. The clinical scores and survival rates were monitored daily.
To determine the TBEV titers in mouse brains, groups of 6-week-old BALB/c (n = 6 or 12 mice per group) mice were infected subcutaneously with 1,000 PFU of TBEV and treated intraperitoneally with 7-deaza-2′-CMA once daily at concentrations ranging from 0 to 15 mg/kg or twice daily with 25 mg/kg of 7-deaza-2′-CMA. On day 8 p.i., the brains were collected, weighed, and homogenized using Precellys 24 (Bertin Technologies), and 20% (wt/vol) suspensions were prepared in L-15 medium containing 3% newborn calf serum. Each homogenate was clarified by centrifugation at 5,000 × g, and the supernatant medium was used for plaque assays.
The anti-TBEV activity of 7-deaza-2′-CMA was further evaluated using in vivo bioluminescence imaging of IFNβ+/Δβ-luc mice (23). The IFNβ+/Δβ-luc mice were obtained by crossing the mouse strain IFNβΔβ-luc/Δβ-luc (ifnb1tm2.2lien) with albino (Tyrc2J) C57BL/6 mice. Six-week-old IFNβ+/Δβ-luc mice were divided into 3 groups (n = 3/group) and treated as follows: group 1 mice were mock-infected nontreated control animals, group 2 mice were injected subcutaneously with 1,000 PFU/ml TBEV, and group 3 mice were infected subcutaneously with 1,000 PFU/ml TBEV and injected intraperitoneally with 25 mg/kg of 7-deaza-2′-CMA twice a day for the 8-day experimental period. In vivo imaging was performed on days 1, 3, 4, and 7 p.i. Animals were anesthetized by intramuscular administration of a mixture of 10% ketamine (Narkamon) and 2% xylazine (Rometar). Following anesthesia, mice were injected intraperitoneally with 150 mg/kg of d-luciferin (Promega) in PBS. Bioluminescence imaging was carried out using an In-Vivo Xtreme imager (Bruker). The exposure time was 60 s, and 4 × 4 binning was used to obtain the optimal signal intensity.
To evaluate the virulence of the Hypr S603T mutant in a mouse model of TBEV infection, 2 groups (n = 10/group) of 6-week-old BALB/c female mice were challenged subcutaneously with wild-type virus or with the S603T TBEV mutant (1,000 PFU/mouse). Symptoms of illness, clinical scores, and survival rates of TBEV-infected mice were monitored daily during the 28-day experimental period.
To assess the virulence of the Oshima S603T TBEV mutants by using cDNA clones and replicons, 5-week-old female C57BL/6J mice (Charles River Laboratories Japan, Inc., Yokohama, Japan) were anesthetized and inoculated intracerebrally with ∼50 PFU of the Oshima-IC virus. The mice were weighed daily and checked for clinical signs of illness for 21 days. Morbidity was defined as >10% weight loss. To analyze viral titers and gene expression levels, 3 mice were sacrificed on days 5 and 8 p.i., and brain samples were collected following perfusion with cold PBS and stored at −80°C.
Stochastic molecular simulations.
A homology-generated model of the TBEV NS5 polymerase was used for simulations as reported previously (13) and was optimized using Schrödinger's Maestro Protein Preparation Wizard tool (61). The S603T TBEV mutant was generated using Schrödinger's Maestro software. Stochastic molecular simulations were performed using the Metropolis Monte Carlo-based Protein Energy Landscape Exploration (PELE) server (24; https://pele.bsc.es/). The PELE software applications are explained elsewhere (24, 25). PELE software implements the optimized potentials for liquid simulations (OPLS-2005) (62) and was previously shown to be suitable for metals (63); however, metals are treated as ions and the bonds as ionic rather than covalent. For the simulations, the bonds between the metal ions (Mn2+) and the associated residues from the polymerase were bridged and converted to zero-order bonds.
Modifications to the ready-made PELE script are available upon request. A total of 24 CPUs were used for each simulation. The top 3 CPUs that explored the region close to the TBEV NS5 active site were combined, resulting in ∼250 frames per simulation. Since our data set represents a large sample size, the central limit theorem (CLT) validates normal-based methods (64, 65). The data were collected using Visual Molecular Dynamics (VMD) software (66). Two-way ANOVA was used to test the differences in the categorical explanatory variables nucleotide triphosphate (or NTP) and strain (wild type or S603T TBEV mutant) for each response variable. The VMD software generated the structural representations.
Statistical analyses.
Data are expressed as means ± standard deviations (SD), and the significance of differences between groups was evaluated using the Mann-Whitney U test or ANOVA. Survival rates were analyzed using the log rank Mantel-Cox test. All tests were performed using GraphPad Prism 5.04 (GraphPad Software, Inc., USA). P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We thank Jan Karasek from BioVendor Instruments, Czech Republic, for help with the scanning and analysis of plaques by use of a prototype instrument and software analytics. We thank Ivana Huvarova for providing excellent technical assistance.
This study was supported by a grant from the Ministry of Health of the Czech Republic (grant 16-34238A); by Project FIT (Pharmacology, Immunotherapy, nanoToxicology), which was funded by the European Regional Development Fund (to D.R.); by grants-in-aid for scientific research (grants 26660220 and 16K15032) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and by a grant provided by the Ichiro Kanehara Foundation (to K.Y.).
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
REFERENCES
- 1.De Clercq E, Li G. 2016. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev 29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldescu V, Behnam MAM, Vasilakis N, Klein CD. 2017. Broad-spectrum agents for flaviviral infections: dengue, Zika and beyond. Nat Rev Drug Discov 16:565–586. doi: 10.1038/nrd.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cihlar T, Ray AS. 2010. Nucleoside and nucleotide HIV reverse transcriptase inhibitors: 25 years after zidovudine. Antiviral Res 85:39–58. doi: 10.1016/j.antiviral.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR, Jacobson IM, Kowdley KV, Nyberg L, Subramanian GM, Hyland RH, Arterburn S, Jiang D, McNally J, Brainard D, Symonds WT, McHutchison JG, Sheikh AM, Younossi Z, Gane EJ. 2013. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med 368:1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 5.Schinazi R, Halfon P, Marcellin P, Asselah T. 2014. HCV direct-acting antiviral agents: the best interferon-free combinations. Liver Int 34(Suppl 1):S69–S78. doi: 10.1111/liv.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sofia MJ, Chang W, Furman PA, Mosley RT, Ross BS. 2012. Nucleoside, nucleotide, and non-nucleoside inhibitors of hepatitis C virus NS5B RNA-dependent RNA-polymerase. J Med Chem 55:2481–2531. doi: 10.1021/jm201384j. [DOI] [PubMed] [Google Scholar]
- 7.Hercík K, Kozak J, Šála M, Dejmek M, Hřebabecký H, Zborníková E, Smola M, Ruzek D, Nencka R, Boura E. 2017. Adenosine triphosphate analogs can efficiently inhibit the Zika virus RNA-dependent RNA polymerase. Antiviral Res 137:131–133. doi: 10.1016/j.antiviral.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Potisopon S, Ferron F, Fattorini V, Selisko B, Canard B. 2017. Substrate selectivity of dengue and Zika virus NS5 polymerase towards 2′-modified nucleotide analogues. Antiviral Res 140:25–36. doi: 10.1016/j.antiviral.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Olsen DB, Eldrup AB, Bartholomew L, Bhat B, Bosserman MR, Ceccacci A, Colwell LF, Fay JF, Flores OA, Getty KL, Grobler JA, LaFemina RL, Markel EJ, Migliaccio G, Prhavc M, Stahlhut MW, Tomassini JE, MacCoss M, Hazuda DJ, Carroll SS. 2004. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob Agents Chemother 48:3944–3953. doi: 10.1128/AAC.48.10.3944-3953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YL, Yokokawa F, Shi PY. 2015. The search for nucleoside/nucleotide analog inhibitors of dengue virus. Antiviral Res 122:12–19. doi: 10.1016/j.antiviral.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Eyer L, Nencka R, Huvarová I, Palus M, Joao Alves M, Gould EA, De Clercq E, Rùžek D. 2016. Nucleoside inhibitors of Zika virus. J Infect Dis 214:707–711. doi: 10.1093/infdis/jiw226. [DOI] [PubMed] [Google Scholar]
- 12.Zmurko J, Marques RE, Schols D, Verbeken E, Kaptein SJ, Neyts J. 2016. The viral polymerase inhibitor 7-deaza-2′-C-methyladenosine is a potent inhibitor of in vitro Zika virus replication and delays disease progression in a robust mouse infection model. PLoS Negl Trop Dis 10:e0004695. doi: 10.1371/journal.pntd.0004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eyer L, Valdés JJ, Gil VA, Nencka R, Hřebabecký H, Šála M, Salát J, Černý J, Palus M, De Clercq E, Rùžek D. 2015. Nucleoside inhibitors of tick-borne encephalitis virus. Antimicrob Agents Chemother 59:5483–5493. doi: 10.1128/AAC.00807-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eyer L, Šmídková M, Nencka R, Neča J, Kastl T, Palus M, De Clercq E, Rùžek D. 2016. Structure-activity relationships of nucleoside analogues for inhibition of tick-borne encephalitis virus. Antiviral Res 133:119–129. doi: 10.1016/j.antiviral.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 15.Dumpis U, Crook D, Oksi J. 1999. Tick-borne encephalitis. Clin Infect Dis 28:882–890. doi: 10.1086/515195. [DOI] [PubMed] [Google Scholar]
- 16.Heinz FX, Mandl CW. 1993. The molecular biology of tick-borne encephalitis virus. Review article. APMIS 101:735–745. doi: 10.1111/j.1699-0463.1993.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 17.Růzek D, Dobler G, Donoso Mantke O. 2010. Tick-borne encephalitis: pathogenesis and clinical implications. Travel Med Infect Dis 8:223–232. doi: 10.1016/j.tmaid.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Poveda E, Wyles DL, Mena A, Pedreira JD, Castro-Iglesias A, Cachay E. 2014. Update on hepatitis C virus resistance to direct-acting antiviral agents. Antiviral Res 108:181–191. doi: 10.1016/j.antiviral.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Menéndez-Arias L, Álvarez M, Pacheco B. 2014. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: mechanism of action and resistance. Curr Opin Virol 8:1–9. doi: 10.1016/j.coviro.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Lauring AS, Andino R. 2010. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog 6:e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauring AS, Frydman J, Andino R. 2013. The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol 11:327–336. doi: 10.1038/nrmicro3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger KL, Scherer J, Ranga M, Sha N, Stern JO, Quinson AM, Kukolj G. 2015. Baseline polymorphisms and emergence of drug resistance in the NS3/4A protease of hepatitis C virus genotype 1 following treatment with faldaprevir and pegylated interferon alpha 2a/ribavirin in phase 2 and phase 3 studies. Antimicrob Agents Chemother 59:6017–6025. doi: 10.1128/AAC.00932-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lienenklaus S, Cornitescu M, Zietara N, Łyszkiewicz M, Gekara N, Jabłónska J, Edenhofer F, Rajewsky K, Bruder D, Hafner M, Staeheli P, Weiss S. 2009. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J Immunol 183:3229–3236. doi: 10.4049/jimmunol.0804277. [DOI] [PubMed] [Google Scholar]
- 24.Madadkar-Sobhani A, Guallar V. 2013. PELE web server: atomistic study of biomolecular systems at your fingertips. Nucleic Acids Res 41(Web Server issue):W322–W328. doi: 10.1093/nar/gkt454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borrelli KW, Vitalis A, Alcantara R, Guallar V. 2005. PELE: protein energy landscape exploration. A novel Monte Carlo based technique. J Chem Theory Comput 1:1304–1311. doi: 10.1021/ct0501811. [DOI] [PubMed] [Google Scholar]
- 26.Bressanelli S, Tomei L, Rey FA, De Francesco R. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J Virol 76:3482–3492. doi: 10.1128/JVI.76.7.3482-3492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen J. 1988. Statistical power analysis for the behavioral sciences. Erlbaum, Hillsdale, NJ. [Google Scholar]
- 28.Valdés JJ, Gil VA, Butterill PT, Rùžek D. 2016. An all-atom, active site exploration of antiviral inhibitors that target Flaviviridae polymerases. J Gen Virol 97:2552–2565. doi: 10.1099/jgv.0.000569. [DOI] [PubMed] [Google Scholar]
- 29.Valdés JJ, Butterill PT, Rùžek D. 2017. Flaviviridae viruses use a common molecular mechanism to escape nucleoside analogue inhibitors. Biochem Biophys Res Commun 2017:S0006-291X(17)30535-1. doi: 10.1016/j.bbrc.2017.03.068. [DOI] [PubMed] [Google Scholar]
- 30.Hayasaka D, Gritsun TS, Yoshii K, Ueki T, Goto A, Mizutani T, Kariwa H, Iwasaki T, Gould EA, Takashima I. 2004. Amino acid changes responsible for attenuation of virus neurovirulence in an infectious cDNA clone of the Oshima strain of tick-borne encephalitis virus. J Gen Virol 85:1007–1018. doi: 10.1099/vir.0.19668-0. [DOI] [PubMed] [Google Scholar]
- 31.Yoshii K, Ikawa A, Chiba Y, Omori Y, Maeda J, Murata R, Kariwa H, Takashima I. 2009. Establishment of a neutralization test involving reporter gene-expressing virus-like particles of tick-borne encephalitis virus. J Virol Methods 161:173–176. doi: 10.1016/j.jviromet.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Weber E, Finsterbusch K, Lindquist R, Nair S, Lienenklaus S, Gekara NO, Janik D, Weiss S, Kalinke U, Överby AK, Kröger A. 2014. Type I interferon protects mice from fatal neurotropic infection with Langat virus by systemic and local antiviral responses. J Virol 88:12202–12212. doi: 10.1128/JVI.01215-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Růzek D, Salát J, Palus M, Gritsun TS, Gould EA, Dyková I, Skallová A, Jelínek J, Kopecký J, Grubhoffer L. 2009. CD8+ T-cells mediate immunopathology in tick-borne encephalitis. Virology 384:1–6. doi: 10.1016/j.virol.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 34.Palus M, Vojtíšková J, Salát J, Kopecký J, Grubhoffer L, Lipoldová M, Demant P, Rùžek D. 2013. Mice with different susceptibility to tick-borne encephalitis virus infection show selective neutralizing antibody response and inflammatory reaction in the central nervous system. J Neuroinflammation 10:77. doi: 10.1186/1742-2094-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra MK, Basu A. 2008. Minocycline neuroprotects, reduces microglial activation, inhibits caspase 3 induction, and viral replication following Japanese encephalitis. J Neurochem 105:1582–1595. doi: 10.1111/j.1471-4159.2008.05238.x. [DOI] [PubMed] [Google Scholar]
- 36.Atrasheuskaya AV, Fredeking TM, Ignatyev GM. 2003. Changes in immune parameters and their correction in human cases of tick-borne encephalitis. Clin Exp Immunol 131:148–154. doi: 10.1046/j.1365-2249.2003.02050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishra MK, Dutta K, Saheb SK, Basu A. 2009. Understanding the molecular mechanism of blood-brain barrier damage in an experimental model of Japanese encephalitis: correlation with minocycline administration as a therapeutic agent. Neurochem Int 55:717–723. doi: 10.1016/j.neuint.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Dutta K, Mishra MK, Nazmi A, Kumawat KL, Basu A. 2010. Minocycline differentially modulates macrophage mediated peripheral immune response following Japanese encephalitis virus infection. Immunobiology 215:884–893. doi: 10.1016/j.imbio.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Kumar R, Basu A, Sinha S, Das M, Tripathi P, Jain A, Kumar C, Atam V, Khan S, Singh AS. 2016. Role of oral minocycline in acute encephalitis syndrome in India—a randomized controlled trial. BMC Infect Dis 16:67. doi: 10.1186/s12879-016-1385-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schul W, Liu W, Xu HY, Flamand M, Vasudevan SG. 2007. A dengue fever viremia model in mice shows reduction in viral replication and suppression of the inflammatory response after treatment with antiviral drugs. J Infect Dis 195:665–674. doi: 10.1086/511310. [DOI] [PubMed] [Google Scholar]
- 41.Arnold JJ, Sharma SD, Feng JY, Ray AS, Smidansky ED, Kireeva ML, Cho A, Perry J, Vela JE, Park Y, Xu Y, Tian Y, Babusis D, Barauskus O, Peterson BR, Gnatt A, Kashlev M, Zhong W, Cameron CE. 2012. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog 8:e1003030. doi: 10.1371/journal.ppat.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu R, Smidansky ED, Oh HS, Takhampunya R, Padmanabhan R, Cameron CE, Peterson BR. 2010. Synthesis of a 6-methyl-7-deaza analogue of adenosine that potently inhibits replication of polio and dengue viruses. J Med Chem 53:7958–7966. doi: 10.1021/jm100593s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klumpp K, Kalayanov G, Ma H, Le Pogam S, Leveque V, Jiang WR, Inocencio N, De Witte A, Rajyaguru S, Tai E, Chanda S, Irwin MR, Sund C, Winqist A, Maltseva T, Eriksson S, Usova E, Smith M, Alker A, Najera I, Cammack N, Martin JA, Johansson NG, Smith DB. 2008. 2′-Deoxy-4′-azido nucleoside analogs are highly potent inhibitors of hepatitis C virus replication despite the lack of 2′-alpha-hydroxyl groups. J Biol Chem 283:2167–2175. doi: 10.1074/jbc.M708929200. [DOI] [PubMed] [Google Scholar]
- 44.Klumpp K, Lévêque V, Le Pogam S, Ma H, Jiang WR, Kang H, Granycome C, Singer M, Laxton C, Hang JQ, Sarma K, Smith DB, Heindl D, Hobbs CJ, Merrett JH, Symons J, Cammack N, Martin JA, Devos R, Nájera I. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J Biol Chem 281:3793–3799. doi: 10.1074/jbc.M510195200. [DOI] [PubMed] [Google Scholar]
- 45.Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, Bartholomew L, Bosserman MR, Ceccacci A, Colwell LF, Cortese R, De Francesco R, Eldrup AB, Getty KL, Hou XS, LaFemina RL, Ludmerer SW, MacCoss M, McMasters DR, Stahlhut MW, Olsen DB, Hazuda DJ, Flores OA. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem 278:49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- 46.Flint M, McMullan LK, Dodd KA, Bird BH, Khristova ML, Nichol ST, Spiropoulou CF. 2014. Inhibitors of the tick-borne, hemorrhagic fever-associated flaviviruses. Antimicrob Agents Chemother 58:3206–3216. doi: 10.1128/AAC.02393-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu HT, Hassounah SA, Colby-Germinario SP, Oliveira M, Fogarty C, Quan Y, Han Y, Golubkov O, Ibanescu I, Brenner B, Stranix BR, Wainberg MA. 2017. Purification of Zika virus RNA-dependent RNA polymerase and its use to identify small-molecule Zika inhibitors. J Antimicrob Chemother 72:727–734. doi: 10.1093/jac/dkw514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mateo R, Nagamine CM, Kirkegaard K. 2015. Suppression of drug resistance in dengue virus. mBio 6:e01960-15. doi: 10.1128/mBio.01960-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curti E, Jaeger J. 2013. Residues Arg283, Arg285, and Ile287 in the nucleotide binding pocket of bovine viral diarrhea virus NS5B RNA polymerase affect catalysis and fidelity. J Virol 87:199–207. doi: 10.1128/JVI.06968-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steitz TA, Steitz JA. 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A 90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobson BM, Procter DJ, Hollett NA, Flesch IE, Newsome TP, Tscharke DC. 2014. Vaccinia virus F5 is required for normal plaque morphology in multiple cell lines but not replication in culture or virulence in mice. Virology 456–457:145–156. doi: 10.1016/j.virol.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 52.Mateu CG, Recalde MP, Artuso MC, Hermida G, Linero FN, Scolaro LA, Damonte EB, Pujol CA, Carlucci MJ. 2011. Emergence of herpes simplex virus-1 syncytial variants with altered virulence for mice after selection with a natural carrageenan. Sex Transm Dis 38:555–561. [PubMed] [Google Scholar]
- 53.Rubio MP, López-Bueno A, Almendral JM. 2005. Virulent variants emerging in mice infected with the apathogenic prototype strain of the parvovirus minute virus of mice exhibit a capsid with low avidity for a primary receptor. J Virol 79:11280–11290. doi: 10.1128/JVI.79.17.11280-11290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCown MF, Rajyaguru S, Le Pogam S, Ali S, Jiang WR, Kang H, Symons J, Cammack N, Najera I. 2008. The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob Agents Chemother 52:1604–1612. doi: 10.1128/AAC.01317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pospisil L, Jandasek L, Pesek J. 1954. Isolation of new strains of meningoencephalitis virus in the Brno region during the summer of 1953. Lek List 9:3–5. [PubMed] [Google Scholar]
- 56.Kozuch O, Mayer V. 1975. Pig kidney epithelial (PS) cells: a perfect tool for the study of flaviviruses and some other arboviruses. Acta Virol 19:498. [PubMed] [Google Scholar]
- 57.De Madrid AT, Porterfield JS. 1969. A simple micro-culture method for the study of group B arboviruses. Bull World Health Organ 40:113–121. [PMC free article] [PubMed] [Google Scholar]
- 58.Růzek D, Gritsun TS, Forrester NL, Gould EA, Kopecký J, Golovchenko M, Rudenko N, Grubhoffer L. 2008. Mutations in the NS2B and NS3 genes affect mouse neuroinvasiveness of a Western European field strain of tick-borne encephalitis virus. Virology 374:249–255. doi: 10.1016/j.virol.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 59.Takashima I, Morita K, Chiba M, Hayasaka D, Sato T, Takezawa C, Igarashi A, Kariwa H, Yoshimatsu K, Arikawa J, Hashimoto N. 1997. A case of tick-borne encephalitis in Japan and isolation of the virus. J Clin Microbiol 35:1943–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshii K, Igarashi M, Ito K, Kariwa H, Holbrook MR, Takashima I. 2011. Construction of an infectious cDNA clone for Omsk hemorrhagic fever virus, and characterization of mutations in NS2A and NS5. Virus Res 155:61–68. doi: 10.1016/j.virusres.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 61.Schrödinger LLC. 2016. Release, 2016-3. Maestro. Schrödinger LLC, New York, NY. [Google Scholar]
- 62.Jorgensen WL, Tirado-Rives J. 1988. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc 110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 63.Giannotti MI, Cabeza de Vaca I, Artés JM, Sanz F, Guallar V, Gorostiza P. 2015. Direct measurement of the nanomechanical stability of a redox protein active site and its dependence upon metal binding. J Phys Chem B 119:12050–12058. doi: 10.1021/acs.jpcb.5b06382. [DOI] [PubMed] [Google Scholar]
- 64.Läärä E. 2009. Statistics: reasoning on uncertainty, and the insignificance of testing null. Ann Zool Fennici 46:138–157. [Google Scholar]
- 65.Zuur AF, Ieno EN, Elphick CS. 2010. A protocol for data exploration to avoid common statistical problems. Methods Ecol Evol 1:3–14. doi: 10.1111/j.2041-210X.2009.00001.x. [DOI] [Google Scholar]
- 66.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]