ABSTRACT
Viruses display a wide range of genomic profiles and, consequently, a variety of gene expression strategies. Specific sequences associated with transcriptional processes have been described in viruses, and putative promoter motifs have been elucidated for some nucleocytoplasmic large DNA viruses (NCLDV). Among NCLDV, the Marseilleviridae is a well-recognized family because of its genomic mosaicism. The marseilleviruses have an ability to incorporate foreign genes, especially from sympatric organisms inhabiting Acanthamoeba, its main known host. Here, we identified for the first time an eight-nucleotide A/T-rich promoter sequence (AAATATTT) associated with 55% of marseillevirus genes that is conserved in all marseilleviruses lineages, a higher level of conservation than that of any giant virus described to date. We instigated our prediction about the promoter motif by biological assays and by evaluating how single mutations in this octamer can impact gene expression. The investigation of sequences that regulate the expression of genes relative to lateral transfer revealed that the promoter motifs do not appear to be incorporated by marseilleviruses from donor organisms. Indeed, analyses of the intergenic regions that regulate lateral gene transfer-related genes have revealed an independent origin of the marseillevirus intergenic regions that does not match gene-donor organisms. About 50% of AAATATTT motifs spread throughout intergenic regions of the marseilleviruses are present as multiple copies. We believe that such multiple motifs are associated with increased expression of a given gene or are related to incorporation of foreign genes into the mosaic genome of marseilleviruses.
IMPORTANCE The marseilleviruses draw attention because of the peculiar features of their genomes; however, little is known about their gene expression patterns or the factors that regulate those expression patterns. The limited published research on the expression patterns of the marseilleviruses and their unique genomes has led us to study the promoter motif sequences in the intergenic regions of the marseilleviruses. This work is the first to analyze promoter sequences in the genomes of the marseilleviruses. We also suggest a strong capacity to acquire foreign genes and to express those genes mediated by multiple copies of the promoter motifs available in intergenic regions. These findings contribute to an understanding of genomic expansion and plasticity observed in these giant viruses.
KEYWORDS: lateral gene transfer, Marseilleviridae, gene expression, promoter
INTRODUCTION
Since the discovery of the Acanthamoeba polyphaga mimivirus (APMV) in 2003 (1), new giant viruses, including Marseillevirus marseillevirus (MsV), have been isolated from clinical and environmental samples (2–12). According to phylogenies that are based on a set of core genes, the family Marseilleviridae encompasses four distinct lineages: lineage A, composed of the prototype MsV; lineage B, composed of Lausannevirus; lineage C, composed of Tunisvirus; and lineage D, composed of Brazilian marseillevirus (BR-MsV) (2, 4, 7, 8, 13, 14). MsV has an extensive genome, with circular, double-stranded DNA. It is approximately 370 kbp, it has a GC content of 45%, and it encodes 428 proteins, of which 187 were encoded by genes that originated from a putative lateral gene transfer (LGT) (2).
Some studies have described putative A/T-rich promoter sequences for nucleocytoplasmic large DNA viruses (NCLDV) from different families (15–21). In 2005, the mimivirus promoter sequence was identified. It showed a level of conservation unreported among eukaryotes, being present for about 45% of open reading frames (ORFs) in the mimivirus (22).
In this paper, we describe the identification of an octamer promoter motif, AAATATTT, present in 55% of marseillevirus genes. We used biological assays to confirm the prediction of a promoter sequence in the genome of a giant virus. We also performed an analysis of the distribution of motifs in the intergenic regions (IRs) that are associated with lateral gene transfer in marseilleviruses. Results showed that marseillevirus IRs are of an independent origin that is unrelated to respective donor organisms. Furthermore, multiple copies of available AAATATTT promoter motifs in IRs that are not associated with a downstream gene may help explain why the marseilleviruses have the ability to acquire foreign genes that contribute to its fitness and genome expansion.
RESULTS AND DISCUSSION
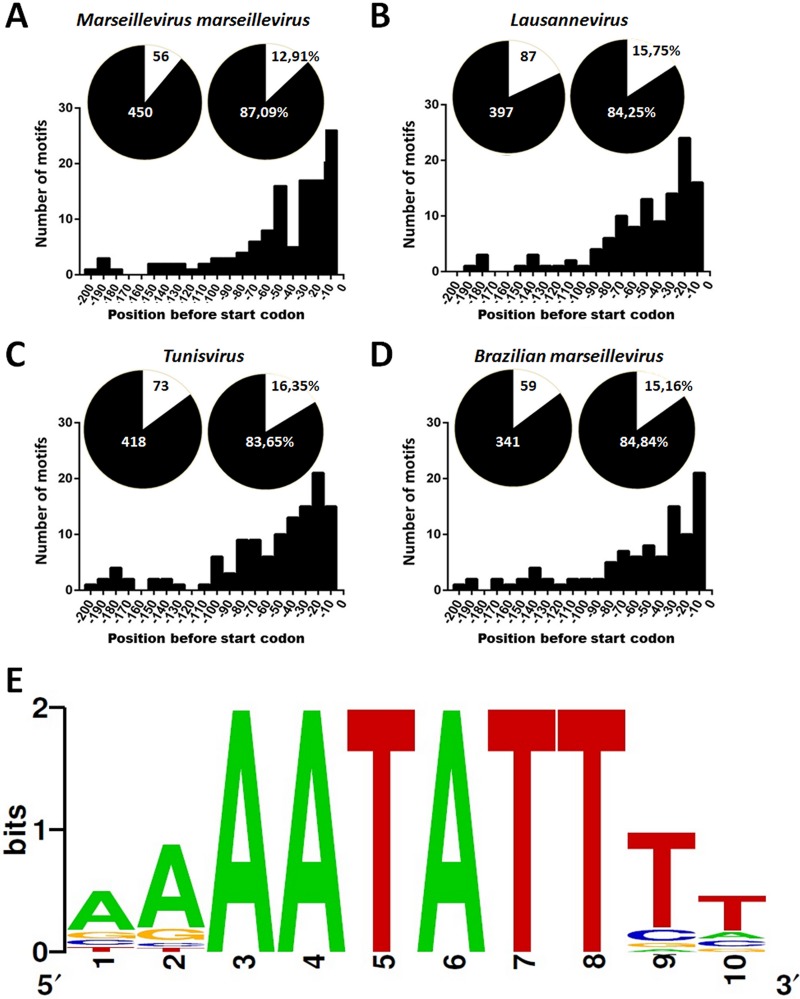
The AAATATTT motif is associated with more than half of the MsV genes.
The conserved AAATATTT motif (Fig. 1E) was found to be associated with 55.1% of MsV genes, which revealed an unprecedented conservation of the regulatory motif between viruses and eukaryotes. Similar conservation profiles were observed in all other marseilleviruses lineages: 49.5% in Lausannevirus, 41.9% in Tunisvirus, and 38.7% in BR-MsV (some overlapping genes in marseilleviruses seem to be regulated by a single motif). The AAATATTT motif occurred in 55.6% of MsV IRs, 49.4% of Lausannevirus IRs, 42.9% of Tunisvirus IRs, and 39.7% of BR-MsV IRs. These data reveal a strongly conserved motif in IRs across all lineages of the Marseilleviridae family. Such a high prevalence of motifs is not random; we found 350 to 700% more motifs in IRs than in coding regions (Fig. 1A to D). A total of 450 motifs were detected in MsV IRs, 397 in Lausannevirus IRs, 418 in Tunisvirus IRs, and 341 in BR-MsV IRs. In comparison, only 56, 87, 73, and 59 were found in coding regions of MsV, Lausannevirus, Tunisvirus, and BR-MsV, respectively (Fig. 1A to D). The presence of the motif upstream of marseillevirus genes was highly significant (P = 1.61 × 10−42 by Fisher exact test). These data are represented by the circles in Fig. 1A to D. Furthermore, the search for the AAATATTT motif in the Acanthamoeba castellanii genome showed that the motif is poorly represented; only about 35 AAATATTT motifs were found in the available genome. The presence of an A/T-rich promoter sequence has been described in the genomes of NCLDV, even in those presenting a high GC content, such as those in the following: phycodnaviruses (∼40% GC content) (23), iridoviruses (∼48% GC content) (24), and marseilleviruses (∼45% GC content) (2). As previously mentioned, bioinformatics studies on the mimivirus identified a nucleotide sequence (AAAATTGA) that occurs in about 45% of mimivirus open reading frames (22). In addition, the Cafeteria roenbergensis virus, another representative of the Mimiviridae family which is distantly related to mimiviruses, showed this promoter motif in the upstream region of 35% of its genes (25). Therefore, the AAATATTT promoter motif in marseilleviruses is the most representative and widespread motif considering members from different lineages of marseilleviruses.
FIG 1.
AAATATTT motif in intergenic regions of marseilleviruses lineages. (A to D) The distribution of the AAATATTT motif in intergenic and coding regions is represented by a circle graphic with black and white colors, respectively. The bar graphs represent the distribution −200 bp upstream of the ATG start codon in all marseilleviruses lineages. (E) The promoter motif sequence logo that was generated using the Berkeley Logo platform.
The AAATATTT motifs are located mainly at −1 to −100 bp upstream of ATG.
The AAATATTT motifs are mainly distributed in positions that are −1 to −100 bp upstream of translation start codons (ATG) for all lineages of marseilleviruses (Fig. 1A to D). This predominant localization is consistent with the promoter patterns demonstrated for most organisms, including NCLDV members (21). In eukaryotes, one classic element of the core promoter is the TATA box, which is represented by the consensus sequence TATAAAT that is located at 25 to 30 bp upstream from the transcription start sites (26). Bacterial genomes present short conserved motifs such as the hexamers TATAAT and TTGACA, which are located at 10 and 35 bp upstream of the transcription start site, respectively (27). Base composition analyses of the IRs of the poxviruses, asfarviruses, phycodnaviruses, iridoviruses, and mimiviruses show that NCLDV members present A/T-rich motifs associated with transcription that is located, in general, within a 150-bp region upstream from the start codon (16, 17, 22, 28, 29). These results reinforce our findings for marseilleviruses.
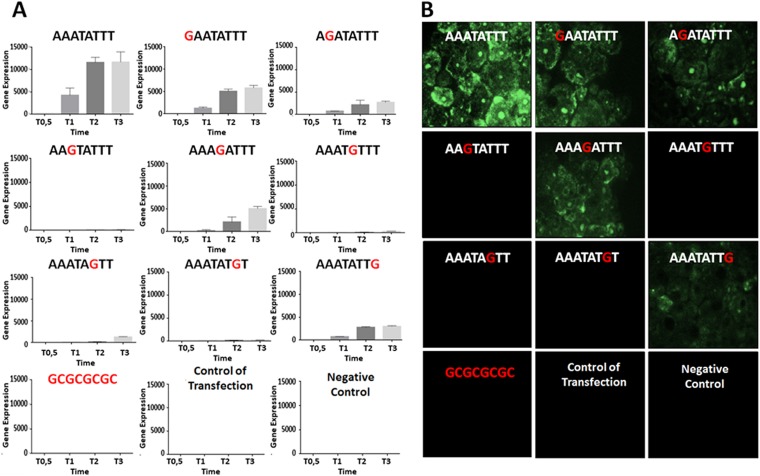
Biological validation confirmed the importance of the AAATATTT motif for MsV gene expression.
Acanthamoeba castellanii cells infected with MsV and transfected with plasmids containing the wild-type promoter motif (AAATATTT) showed high expression of enhanced green fluorescent protein (eGFP) transcripts at 1 h, 2 h, and 3 h postinfection (Fig. 2A). However, considerable reductions in eGFP transcript levels were detected for all mutant versions of this motif (including GAATATTT, AGATATTT, AAGTATTT, AAAGATTT, AAATGTTT, AAATAGTT, AAATATGT, and AAATATTG), even though the expression levels were not completely abolished when some motifs (GAATATTT, AGATATTT, AAAGATTT, and AAATATTG) were used in the assays. eGFP transcripts were not detected at 0.5 h postinfection in all cases, which may be due to the time needed for viral RNA polymerases to encounter the plasmids and trigger gene expression or the absence of complete RNA polymerase subunits in the virion (3). Furthermore, cells infected with MsV and then transfected with the plasmid containing the GC-rich sequence showed no transcript expression, which is consistent with the lack of expression observed for the controls. eGFP expression, which was analyzed by fluorescence microscopy, corroborated the importance of the AAATATTT motif (Fig. 2B). Higher eGFP expression was observed in cells transfected with plasmids containing the sequence AAATATTT, while a negative effect on gene expression was observed for GAATATTT, AGATATTT, AAAGATTT, and AAATATTG sequences. For the remaining sequences, expression was completely abolished, similar to what was demonstrated for eGFP transcripts. Because substitution of these eight nucleotides can result in a decline or loss of promoter activity, these results confirmed that the AAATATTT motif can act as a promoter sequence during the MsV replication cycle.
FIG 2.
Biological assays confirm that the AAATATTT motif is a promoter sequence of MsV. (A) Enhanced green fluorescent protein (eGFP) expression was measured by quantitative PCR (qPCR). Acanthamoeba castellanii cells were infected with MsV and transfected with plasmids that contained the GFP gene under the control of the AAATATTT motif and other similarly mutated motifs (position −10 bp from ATG). Cells were collected at different times postinfection (0.5 h, 1 h, 2 h, and 3 h), and expression of eGFP transcripts was measured by qPCR. The data were calculated using the 2−ΔΔCT method and were represented as the standard deviations from two independent biological assays. (B) eGFP expression was detected by fluorescence microscopy. Acanthamoeba castellanii cells were infected with MsV (multiplicity of infection of 10) and then transfected with the indicated construct plasmids. eGFP expression was visualized at 3 h postinfection.
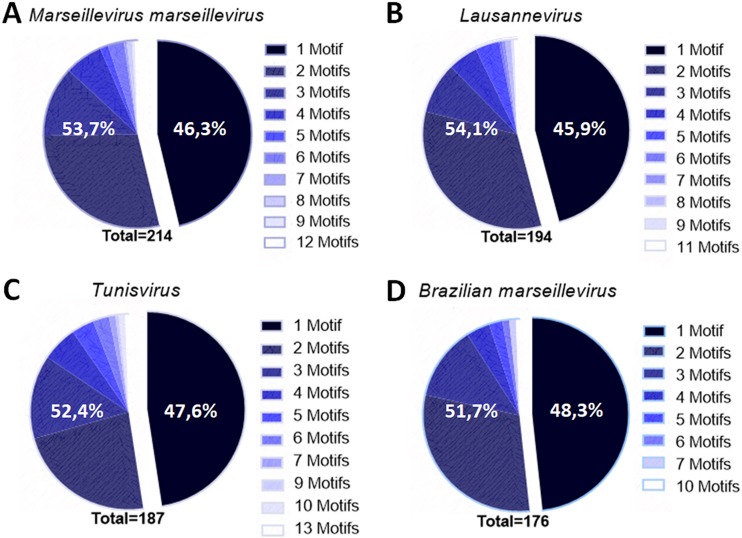
Marseillevirus intergenic regions display multiple repetitions of the AAATATTT motif.
More than 50% of the IRs of the marseilleviruses contain more than one copy of the AAATATTT motifs (Fig. 3). We found 450, 397, 418, and 341 motif repeats in a total of 214, 194, 187, and 176 IRs in MsV, Lausannevirus, Tunisvirus and BR-MsV, respectively. All marseilleviruses lineages presented more than one promoter motif sequence in approximately 50% of their IRs that contained the motif. Interestingly, some IRs harbored large numbers of up to 13 such repeats (Fig. 3). In MsV, more than one motif copy was observed in 53.7% of the IRs (between 2 and 12 repeats) (Fig. 3A). In Lausannevirus, more than one repeat was observed in 54.1% of the IRs (between two and 11 repeats) (Fig. 3B). In Tunisvirus and BR-MsV, more than one motif was found in 52.4% (with up to 13 repeats) and 51.7% of the IRs, respectively (Fig. 3C and D). These noteworthy findings motivated us to investigate the implications of repetitions in the MsV genome.
FIG 3.
Most marseillevirus intergenic regions present more than one promoter motif. Shown is a graphic representation of the number of repetitions of the motifs in different lineages of marseilleviruses.
The marseillevirus intergenic regions seem to have a different origin than those of respective genes acquired by LGT.
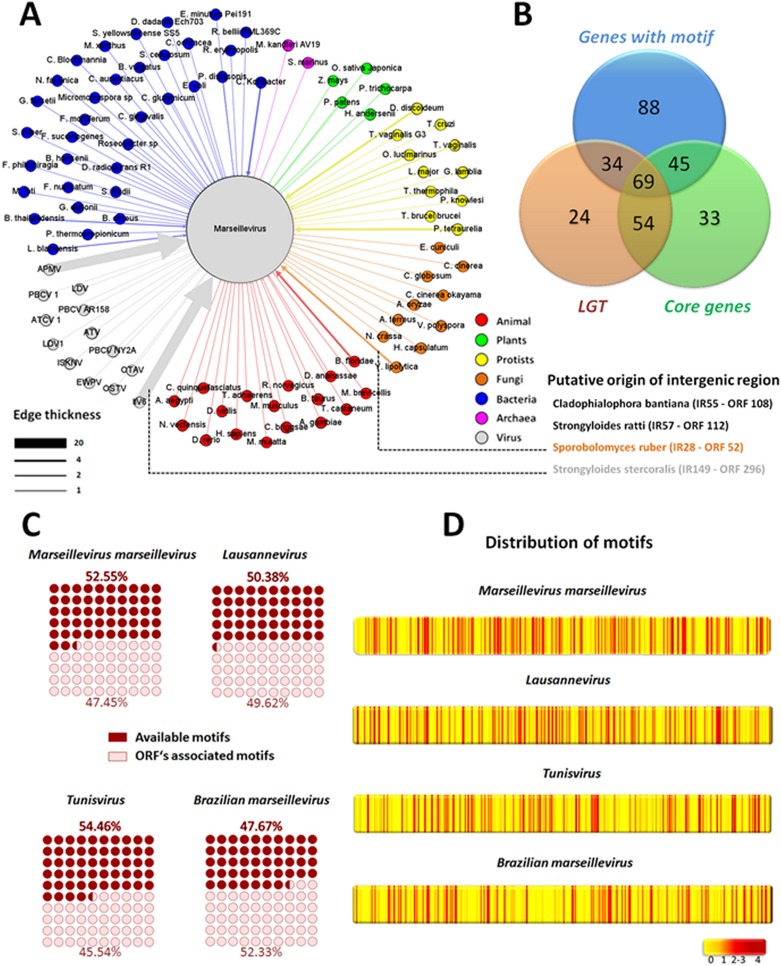
As previously mentioned, some research suggests that lateral gene transfer events are common among the marseilleviruses (2, 30). In our research, we observed that approximately 60% of genes that have been involved in LGT are regulated by IRs that present the AAATATTT motif (Fig. 4B). We decided to verify if the AAATATTT motif could have been acquired from donor organisms at the same time as the gene during LGT (that is, the transfer of the promoter plus gene). To investigate this, we analyzed the upstream promoter regions of all LGT donor organisms. Interestingly, the AAATATTT motif was present in only 14 of the 179 (7.8%) upstream regions in the LGT-associated donor genes that we evaluated (P = 4.17 × 10−25 by Fisher exact test) (data not shown). In addition, the search for sequences with similar IRs (cutoffs were an E value of <0.001 and coverage of >40%) revealed that only 4 of 317 (1.3%) IRs were similar to any organisms in the NCBI GenBank database (P = 1.85 × 10−47 by Fisher exact test) (Fig. 4A). These results suggest that neither the motif nor the IRs originated from LGT-related donors.
FIG 4.
Intergenic regions upstream of genes acquired by lateral gene transfer were not acquired from LGT donor organisms. (A) The network of LGTs in MsV. Gene donor organisms are represented as colored circles connected to MsV. The thickness of the edges is proportional to the number of open reading frames transferred from an organism to MsV. The putative origins of intergenic regions found in MsV are depicted. (B) Venn diagram demonstrating the relationship among core genes, LGT genes, and genes regulated by the AAATATTT motif. (C) Representation of the number of motifs that control genes or available motifs that are present but do not control any genes. (D) Heat map showing that the AAATATTT promoter motif is homogeneously distributed in the genomes of marseillevirus lineages. APMV, Acanthamoeba polyphaga mimivirus; ATCV1, Acantocisto turfacea virus 1; ATV, Ambystoma tigrinum virus; EWPV, Erwinia phage; ISKNV, infectious spleen and kidney necrosis virus; IIV6, invertebrate iridescent virus 6; LDV, lymphocystis disease virus; OTAV, Ostreococcus tauri virus; OSTV, Ostreococcus virus; PBCV, Paramecium bursaria chlorella virus.
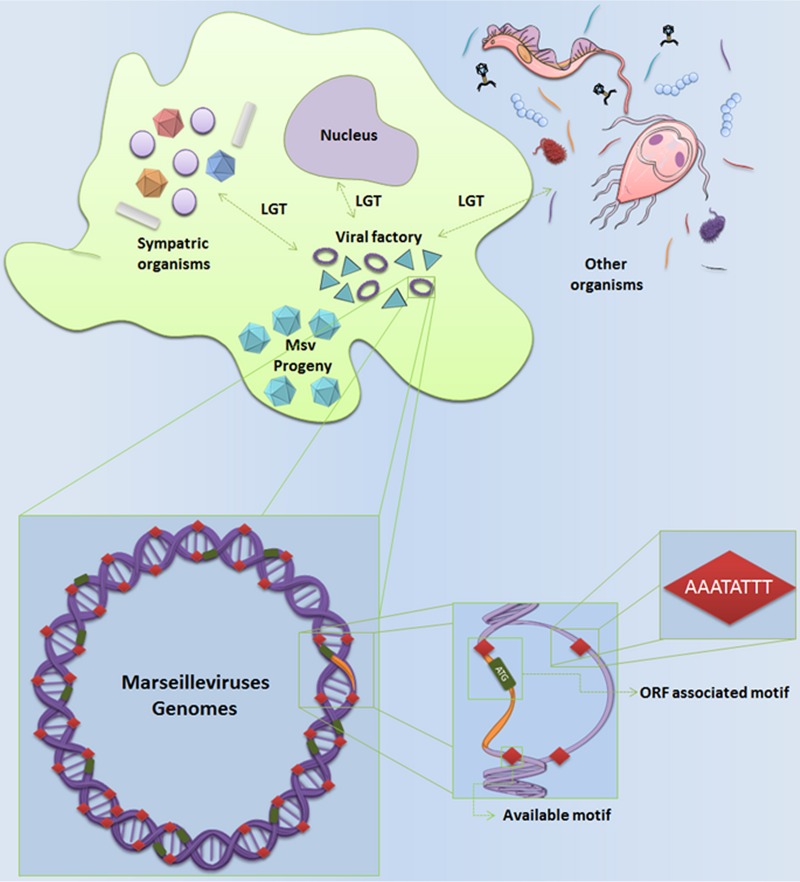
Multiple repetitions of the promoter motif in marseillevirus intergenic regions could help to explain their ability to acquire foreign genes.
Although our results showed that the AAATATTT motif is associated with expression of genes in the marseilleviruses, we also observed the presence of a large percentage of motifs in the IRs that do not appear to be associated with any gene. A total of 52.6% of the motifs found in MsV IR are not associated with genes. This high availability of free motifs was observed in all marseilleviruses lineages: 50.4% in Lausannevirus, 54.5% in Tunisvirus, and 47.7% in BR-MsV (Fig. 4C). In addition, the analysis of the motif distribution within the marseillevirus genome revealed that the motifs are distributed preferentially in the IR and that these motifs are homogeneously distributed in the IR of marseillevirus genomes (Fig. 4D). Interestingly, 62% (148) of the genes that present the AAATATTT motif are core genes and/or genes acquired by LGT (Fig. 4B). Taken together, these data led us to propose a hypothesis supporting that the initial fixation of foreign genes in the marseilleviruses depends not only on stochastic processes, such as genetic drifts and point mutations, but also on an increase in the likelihood of a prompt expression of a given just-acquired gene. This dynamic can speed the fixation of favorable genes and the exclusion of less competitive modified viral genomes (Fig. 5). The homogeneity of the distribution of motifs in genomes reinforces our hypothesis, since LGT can occur in many IRs within the genome. It was hypothesized that the high prevalence of free motifs in IRs reflects the number of potentially available motifs for the regulation of new genes acquired by LGT, revealing the great evolutionary potential of the marseilleviruses. This hypothesis may help to explain the genome plasticity of the marseilleviruses. However, more studies need to be done in order to confirm this hypothesis. To date, there are no differences of GC% content and codon usage among marseilleviruses genes, which suggests an adaptation of a given LGT-acquired gene after the incorporation (data not shown). Moreover, a large number and wide distribution of free promoter motifs may confer a selective adaptive advantage in the sympatric lifestyle of an amoeba.
FIG 5.
AAATATTT motif may contribute to explanation of the plasticity of the marseilleviruses genome. Available genes from amoeba hosts or sympatric organisms would be incorporated into the genome of the marseilleviruses and expressed because of the large number of available promoter motifs in the intergenic regions.
Conclusions.
Here, we describe for the first time a conserved eight-nucleotide A/T-rich sequence (AAATATTT) associated with most of the genes of the marseilleviruses and its lineages. The motif AAATATTT is associated with 55% of genes in the marseilleviruses, which is a remarkable level of conservation among giant viruses and eukaryotes. Furthermore, variations in the motif sequence caused a negative effect on gene transcription. LGT events contributed to the diversity of the amoebal virus genomes and are considered to be important for evolutionary processes (2, 31, 32). The homogeneously distributed motifs within the IRs of the marseilleviruses, in addition to the possibility that the IRs were not transferred by LGT, strongly suggest that free motifs can act as major players in the prompt expression of recently acquired genes. In this scenario, within its amoeba host, marseilleviruses are able to find new genes from diverse sources and promptly incorporate those genes. The absence of fragments in MsV IRs that are similar to other organisms suggests that the free AAATATTT motifs did not originate from MsV LGT genes. Instead, the remarkable amount of AAATATTT motifs seems to be an original and ancient characteristic of the Marseilleviridae family, contributing to its genomic mosaicism and plasticity.
MATERIALS AND METHODS
Bioinformatic analyses.
To investigate the promoter motif that regulates the marseilleviruses, the genomic sequences of a representative relative of each lineage of the Marseilleviridae family were analyzed: Marseillevirus marseillevirus strain T19 (representative of lineage A), Lausannevirus (representative of lineage B), Tunisvirus Fontaine 2 strain U484 (representative of lineage C), and Brazilian marseillevirus strain BH2014 (representative of lineage D). These genome sequences are available in GenBank under accession numbers NC_013756.1, NC_015326.1, KF483846.1, and NC_029692.1, respectively.
The IRs of these marseilleviruses species were obtained using the data set that is available under the accession numbers associated with each sequence and by using Artemis software for analysis (33). After extraction of the IRs, we identified the following number of genomic IRs: 402 from the MsV genome (196 were located in the positive strand and 206 were located in the negative strand), 412 from the Lausannevirus genome (198 were in the positive strand and 214 were in the negative strand), 450 from the Tunisvirus genome (225 were in the positive strand and 225 were in the negative strand), and 460 from the BR-MsV genome (221 were in the positive strand and 239 were in the negative strand). The IRs that contained ≥8 bp were not considered, because these IRs were too short to host the motif. The number of IRs that we considered here were 385, 394, 437, and 443 in MsV, Lausannevirus, Tunisvirus, and BR-MsV, respectively.
The search for motif occurrences was performed using the Find Individual Motif Occurrences (FIMO) software and by a manual search to validate the software results (34). The promoter sequence logo (Fig. 1E) was generated using a Berkeley Logo platform (http://weblogo.berkeley.edu). The search for promoter motifs was performed both in marseillevirus IRs and coding regions. The distance between the promoter and the ATG initiation codon was calculated for all genes that contained the motifs of interest. This was undertaken for all viral lineages being investigated in this study.
The LGT analysis was performed on the basis of results described by Boyer et al. (2). Similar searches of the IRs were performed using the Nucleotide Basic Local Alignment Search Tool platform, and regions with more than 40 bp were considered in this analysis. Furthermore, the presence of the AAATATTT motif in the Acanthamoeba castellanii genome was evaluated using a whole-genome shotgun (WGS) project, and sequences were deposited at DDBJ/EMBL/GenBank (accession number AHJI00000000; WGS, AHJI01000001-AHJI01003192; WGS_SCAFLD, KB007791-KB008174). The graphs were completed using GraphPad Prism version 7.00 for Windows (GraphPad Software).
Biological assays: gene expression analyses.
Acanthamoeba castellanii cells (ATCC 30010) were grown in peptose yeast glucose medium that was supplemented with 25 mg/ml of amphotericin B (Fungizone; Cristalia, São Paulo, Brazil), 500 μg/ml of penicillin, and 50 mg/ml gentamicin (Schering-Plough, Brazil) at 32°C. Marseillevirus marseillevirus was replicated in Acanthamoeba castellanii cells at a multiplicity of infection of 0.01. After the appearance of a cytopathic effect, the cells and supernatant were collected, purified by ultracentrifugation with a 25% sucrose cushion at 36,000 × g for 2 h, and then quantified using the 50% tissue culture infectious dose (TCID50) method (35). To understand the biological relevance of the AAATATTT motif in the gene expression pattern of the marseilleviruses, plasmids were drawn that contained the AAATATTT sequence and progressive variations of that sequence (GAATATTT, AGATATTT, AAGTATTT, AAAGATTT, AAATGTTT, AAATAGTT, AAATATGT, and AAATATTG). The sequences were located at position −10 relative to the initiation of the eGFP reporter gene. In addition, a GC-rich sequence (GCGCGCGC), a noninfected/transfected amoeba, and a noninfected/nontransfected amoeba were used as controls. One microgram of each plasmid was used to transfect Acanthamoeba castellanii cells infected with MsV (multiplicity of infection of 10). The transfections were carried out in duplicate using a Lipofectamine reagent according to the manufacturer's instructions.
For the gene expression analysis, total RNA was purified from cells at 0.5 h (T0.5), 1 h (T1), 2 h (T2), and 3 h (T3) postinfection using a TRIzol reagent (Invitrogen) that was treated with DNase (Invitrogen) and reverse transcribed using Moloney murine leukemia virus reverse transcriptase (200 U/liter; Thermo Fisher Scientific) according to the manufacturer's instructions. The resulting cDNAs were used as a template in a StepOne thermocycler (Applied Biosystems) quantitative PCR assay to target the eGFP gene. Thermal cycling conditions were the following: one cycle at 95°C for 10 min, 40 cycles at 95°C for 10 s and 60°C for 40 s, a melting curve analysis at 95°C for 15 s and 58°C for 15 s, a temperature increase of 1°C per 2 s until a temperature of 95°C was reached, and a final cycle at 95°C for 15 s. The results were analyzed by StepOne software and were analyzed with the 2−ΔΔCT method. The amoebas were observed using a Zeiss (LSM 510 META) microscope at 3 hpi.
ACKNOWLEDGMENTS
We are grateful to our colleagues from Laboratório de Vírus of Universidade Federal de Minas Gerais. In addition, we thank CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), and FAPEMIG (Fundação de Amparo à Pesquisa do estado de Minas Gerais).
E.G.K., C.A.B., F.G.F., and J.S.A. are CNPq researchers. B.L.S., J.S.A., and E.G.K. are members of a CAPES-COFECUB project.
We have no conflicts of interest to declare.
REFERENCES
- 1.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie J-M, Raoult D. 2003. A giant virus in amoebae. Science 299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A 106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabre E, Jeudy S, Legendre M, Trauchessec M, Claverie J, Abergel C. 2017. Noumeavirus replication relies on a transient remote control of the host nucleus. Nat Commun 8:15087. doi: 10.1038/ncomms15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aherfi S, Pagnier I, Fournous G, Raoult D, La Scola B, Colson P. 2013. Complete genome sequence of Cannes 8 virus, a new member of the proposed family “marseilleviridae.” Virus Genes 47:550–555. [DOI] [PubMed] [Google Scholar]
- 5.Popgeorgiev N, Boyer M, Fancello L, Monteil S, Robert C, Rivet R, Nappez C, Azza S, Chiaroni J, Raoult D, Desnues C. 2013. Marseillevirus-like virus recovered from blood donated by asymptomatic humans. J Infect Dis 208:1042–1050. doi: 10.1093/infdis/jit292. [DOI] [PubMed] [Google Scholar]
- 6.Popgeorgiev N, Michel G, Lepidi H, Raoult D, Desnues C. 2013. Marseillevirus adenitis in an 11-month-old child. J Clin Microbiol 51:4102–4105. doi: 10.1128/JCM.01918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dornas FP, Assis FL, Aherfi S, Arantes T, Abrahão JS, Colson P, La Scola B. 2016. A Brazilian marseillevirus is the founding member of a lineage in family Marseilleviridae. Viruses 8:76–85. doi: 10.3390/v8030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aherfi S, Boughalmi M, Pagnier I, Fournous G, La Scola B, Raoult D, Colson P. 2014. Complete genome sequence of Tunisvirus, a new member of the proposed family Marseilleviridae. Arch Virol 159:2349–2358. doi: 10.1007/s00705-014-2023-5. [DOI] [PubMed] [Google Scholar]
- 9.Doutre G, Philippe N, Abergel C, Claverie JM. 2014. Genome analysis of the first Marseilleviridae representative from Australia indicates that most of its genes contribute to virus fitness. J Virol 88:14340–14349. doi: 10.1128/JVI.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aherfi S, Colson P, Raoult D. 2016. Marseillevirus in the pharynx of a patient with neurologic disorders. Emerg Infect Dis 22:2008–2010. doi: 10.3201/eid2211.160189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aherfi S, Colson P, Audoly G, Nappez C, Xerri L, Valensi A, Million M, Lepidi H, Costello R, Raoult D. 2016. Marseillevirus in lymphoma: a giant in the lymph node. Lancet Infect Dis 10:225–234. [DOI] [PubMed] [Google Scholar]
- 12.Takemura M. 2016. Morphological and taxonomic properties of Tokyovirus, the first Marseilleviridae member isolated from Japan. Microbes Environ 31:442–448. doi: 10.1264/jsme2.ME16107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas V, Bertelli C, Collyn F, Casson N, Telenti A, Goesmann A, Croxatto A, Greub G. 2011. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ Microbiol 13:1454–1466. doi: 10.1111/j.1462-2920.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- 14.Boughalmi M, Pagnier I, Aherfi S, Colson P, Raoult D, La Scola B. 2013. First isolation of a marseillevirus in the diptera Syrphidae eristalis tenax. Intervirology 56:386–394. doi: 10.1159/000354560. [DOI] [PubMed] [Google Scholar]
- 15.García-Escudero R, Viñuela E. 2000. Structure of African swine fever virus late promoters: requirement of a TATA sequence at the initiation region. J Virol 74:8176–8182. doi: 10.1128/JVI.74.17.8176-8182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nalçacioǧlu R, Ince IA, Vlak JM, Demirbǎ Z, van Oers MM. 2007. The Chilo iridescent virus DNA polymerase promoter contains an essential AAAAT motif. J Gen Virol 88:2488–2494. doi: 10.1099/vir.0.82947-0. [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald LA, Boucher PT, Yanai-Balser GM, Suhre K, Graves MV, Van Etten JL. 2008. Putative gene promoter sequences in the chlorella viruses. Virology 380:388–393. doi: 10.1016/j.virol.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salem TZ, Turney CM, Wang L, Xue J, Wan XF, Cheng XW. 2008. Transcriptional analysis of a major capsid protein gene from Spodoptera exigua ascovirus 5a. Arch Virol 153:149–162. doi: 10.1007/s00705-007-1081-3. [DOI] [PubMed] [Google Scholar]
- 19.Legendre M, Audic S, Poirot O, Hingamp P, Seltzer V, Byrne D, Lartigue A, Lescot M, Bernadac A, Poulain J, Abergel C, Claverie JM. 2010. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in mimivirus. Genome Res 20:664–674. doi: 10.1101/gr.102582.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Reynolds SE, Martens CA, Bruno DP, Porcella SF, Moss B. 2011. Expression profiling of the intermediate and late stages of poxvirus replication. J Virol 85:9899–9908. doi: 10.1128/JVI.05446-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliveira G, Andrade A, Rodrigues R, Arantes T, Boratto P, Silva L, Dornas F, Trindade G, Drumond B, La Scola B, Kroon E, Abrahão J. 2017. Promoter motifs in NCLDVs: an evolutionary perspective. Viruses 9:16–32. doi: 10.3390/v9010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suhre K, Audic S, Claverie J-M. 2005. Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes. Proc Natl Acad Sci U S A 102:14689–14693. doi: 10.1073/pnas.0506465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Etten JL, Graves MV, Müller DG, Boland W, Delaroque N. 2002. Phycodnaviridae-large DNA algal viruses. Arch Virol 147:1479–1516. doi: 10.1007/s00705-002-0822-6. [DOI] [PubMed] [Google Scholar]
- 24.Jakob NJ, Müller K, Bahr U, Darai G. 2001. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology 286:182–196. doi: 10.1006/viro.2001.0963. [DOI] [PubMed] [Google Scholar]
- 25.Fischer MG, Allen MJ, Wilson WH, Suttle CA. 2010. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci U S A 107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haberle V, Lenhard B. 2016. Promoter architectures and developmental gene regulation. Semin Cell Dev Biol 57:11–23. doi: 10.1016/j.semcdb.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat Rev Microbiol 2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 28.Davison AJ, Moss B. 1989. Structure of vaccinia virus early promoters. J Mol Biol 210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 29.Yates PR, Dixon LK, Turner PC. 1995. Promoter analysis of an African swine fever virus gene encoding a putative elongation factor. Biochem Soc Trans 23:139S. doi: 10.1042/bst023139s. [DOI] [PubMed] [Google Scholar]
- 30.Raoult D, Boyer M. 2010. Amoebae as genitors and reservoirs of giant viruses. Intervirology 53:321–329. doi: 10.1159/000312917. [DOI] [PubMed] [Google Scholar]
- 31.Andersson JO. 2009. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol 63:177–193. doi: 10.1146/annurev.micro.091208.073203. [DOI] [PubMed] [Google Scholar]
- 32.Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, Bertelli C, Schilde C, Kianianmomeni A, Bürglin TR, Frech C, Turcotte B, Kopec KO, Synnott JM, Choo C, Paponov I, Finkler A, Heng Tan CS, Hutchins AP, Weinmeier T, Rattei T, Chu JSC, Gimenez G, Irimia M, Rigden DJ, Fitzpatrick DA, Lorenzo-Morales J, Bateman A, Chiu C-H, Tang P, Hegemann P, Fromm H, Raoult D, Greub G, Miranda-Saavedra D, Chen N, Nash P, Ginger ML, Horn M, Schaap P, Caler L, Loftus BJ. 2013. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol 14:R11. doi: 10.1186/gb-2013-14-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Genomics 16:944–945. [DOI] [PubMed] [Google Scholar]
- 34.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc Second Int Conf Intell Syst Mol Biol 28–36. [PubMed] [Google Scholar]
- 35.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. [Google Scholar]