ABSTRACT
Virion transmembrane proteins (VTPs) mediate key functions in the herpesvirus infectious cycle. Cyprinid herpesvirus 3 (CyHV-3) is the archetype of fish alloherpesviruses. The present study was devoted to CyHV-3 VTPs. Using mass spectrometry approaches, we identified 16 VTPs of the CyHV-3 FL strain. Mutagenesis experiments demonstrated that eight of these proteins are essential for viral growth in vitro (open reading frame 32 [ORF32], ORF59, ORF81, ORF83, ORF99, ORF106, ORF115, and ORF131), and eight are nonessential (ORF25, ORF64, ORF65, ORF108, ORF132, ORF136, ORF148, and ORF149). Among the nonessential proteins, deletion of ORF25, ORF132, ORF136, ORF148, or ORF149 affects viral replication in vitro, and deletion of ORF25, ORF64, ORF108, ORF132, or ORF149 impacts plaque size. Lack of ORF148 or ORF25 causes attenuation in vivo to a minor or major extent, respectively. The safety and efficacy of a virus lacking ORF25 were compared to those of a previously described vaccine candidate deleted for ORF56 and ORF57 (Δ56-57). Using quantitative PCR, we demonstrated that the ORF25 deleted virus infects fish through skin infection and then spreads to internal organs as reported previously for the wild-type parental virus and the Δ56-57 virus. However, compared to the parental wild-type virus, the replication of the ORF25-deleted virus was reduced in intensity and duration to levels similar to those observed for the Δ56-57 virus. Vaccination of fish with a virus lacking ORF25 was safe but had low efficacy at the doses tested. This characterization of the virion transmembrane proteome of CyHV-3 provides a firm basis for further research on alloherpesvirus VTPs.
IMPORTANCE Virion transmembrane proteins play key roles in the biology of herpesviruses. Cyprinid herpesvirus 3 (CyHV-3) is the archetype of fish alloherpesviruses and the causative agent of major economic losses in common and koi carp worldwide. In this study of the virion transmembrane proteome of CyHV-3, the major findings were: (i) the FL strain encodes 16 virion transmembrane proteins; (ii) eight of these proteins are essential for viral growth in vitro; (iii) seven of the nonessential proteins affect viral growth in vitro, and two affect virulence in vivo; and (iv) a mutant lacking ORF25 is highly attenuated but induces moderate immune protection. This study represents a major breakthrough in understanding the biology of CyHV-3 and will contribute to the development of prophylactic methods. It also provides a firm basis for the further research on alloherpesvirus virion transmembrane proteins.
KEYWORDS: cyprinid herpesvirus 3, alloherpesvirus, herpesvirus, proteome
INTRODUCTION
Cyprinid herpesvirus 3 (CyHV-3; species Cyprinid herpesvirus 3, genus Cyprinivirus, family Alloherpesviridae, order Herpesvirales), also known as koi herpesvirus (KHV), is the etiological agent of a lethal disease in common and koi carp. The common carp (Cyprinus carpio) is one of the main fish grown for human consumption (1). Moreover, its colorful ornamental varieties (koi carp) represent one of the most lucrative markets for individual freshwater fish (2, 3). As a result, CyHV-3 is considered to be the archetypal fish herpesvirus and is the subject of a growing number of studies (4).
The CyHV-3 genome is 295 kbp in size and thus the largest described among all herpesviruses (5). It encodes 155 potential protein-coding open reading frames (ORFs), a large number of which lack similarity to other herpesvirus genes (5–7). Nonetheless, CyHV-3 virions present the characteristic herpesvirus morphology (8), consisting of an icosahedral capsid containing the genome surrounded by an amorphous layer of proteins termed the tegument and enveloped in a lipid membrane bearing virion transmembrane proteins (VTPs) (8).
The protein composition of CyHV-3 virions has been investigated by mass spectrometry-based proteomic approaches for one European and two Chinese strains (9, 10). These analyses led to the identification of 46 virion proteins, of which 34 were detected in all three strains. Among these 34 proteins, 13 were classified as VTPs (9, 10). Since none of these proteins are related to viral proteins with known roles, it is not possible to predict their functions.
VTPs mediate key functions in the herpesvirus infectious cycle, such as binding virions to the cell and mediating entry, providing immune evasion mechanisms, facilitating virion morphogenesis, and promoting virion egress from the host cell. The expression of VTPs on the virion surface may also make them targets for neutralizing antibodies. As a result, it is important to study VTPs in order to understand the biology of infection and to aid in the design of candidate vaccines (attenuated, nonreplicative, or subunit). However, current knowledge of CyHV-3 VTPs is very limited. For example, it is not known whether any of the VTPs identified to date are required for viral replication in vitro.
Here, we performed proteomic and functional analyses of CyHV-3 VTPs. The proteome of CyHV-3 virions was revisited. Based on this new information and that published previously (9, 10), 16 VTPs were identified and selected for further study. Using recombination technologies, VTPs that are essential to viral growth in vitro were identified. Furthermore, the effects of deleting nonessential VTPs on viral growth in vitro and virulence in vivo were investigated. The knowledge gained represents a major breakthrough in understanding the biology of CyHV-3 and provides a firm basis for further research on alloherpesvirus VTPs.
RESULTS AND DISCUSSION
Viral protein composition of CyHV-3 virions.
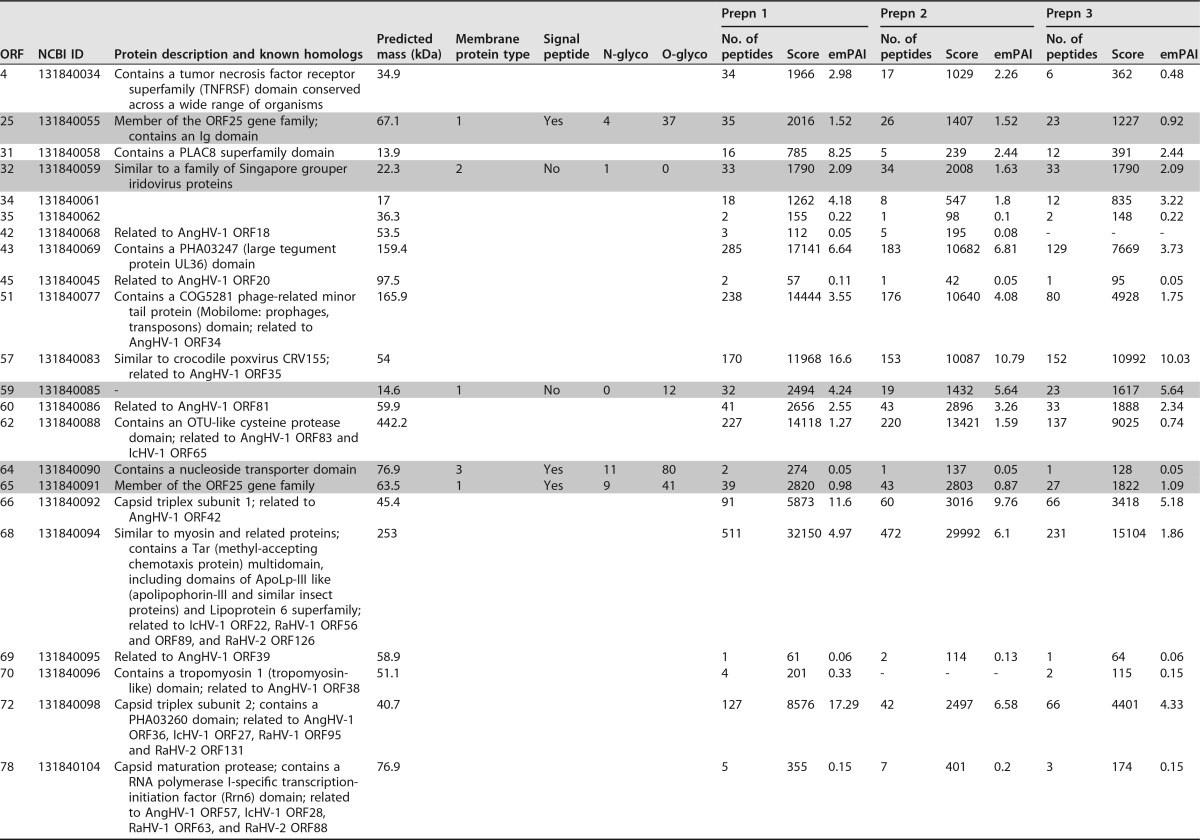
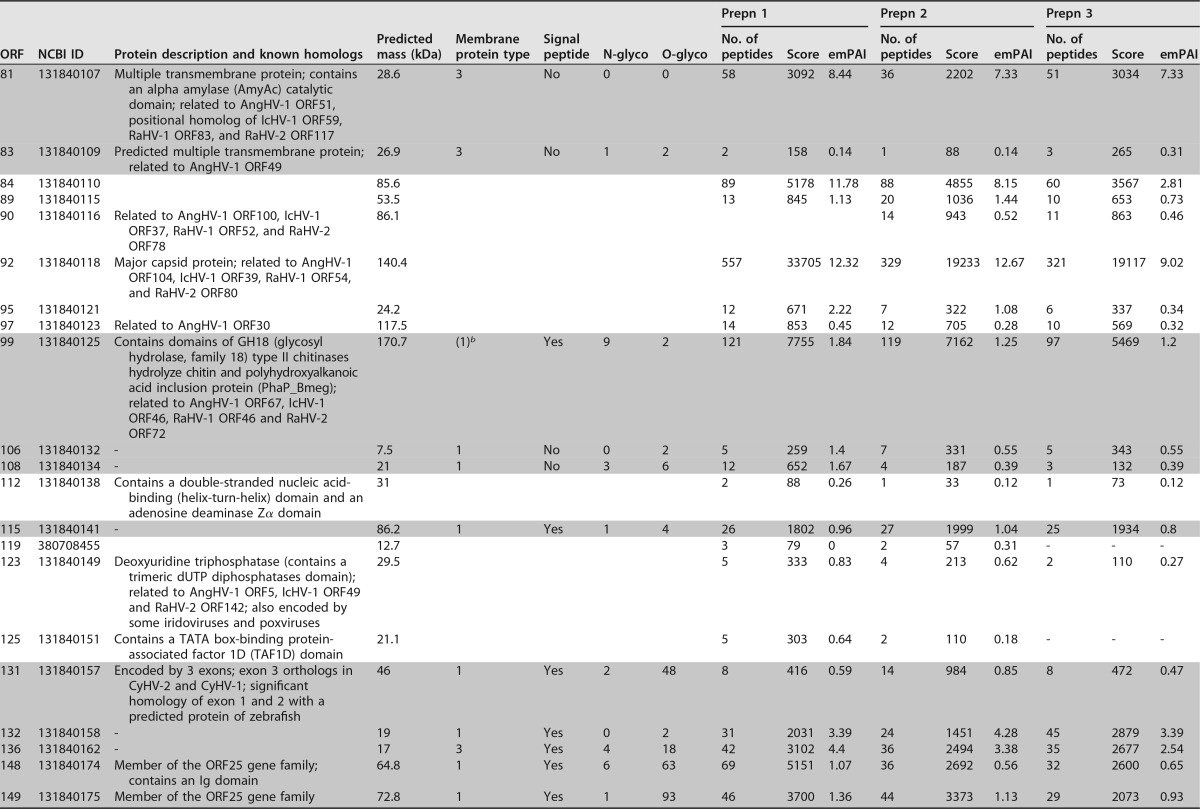
We took advantage of the experience gained during our previous studies of the virion proteomes of herpesviruses (9, 11–13) to revisit the protein composition of extracellular CyHV-3 virions reported by us and others (9, 10). We selected a strategy based on SDS-PAGE separation of proteins, followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) of tryptic peptides, because this approach has been shown to enhance the recovery of peptides derived from proteins that are prone to aggregation or contain hydrophobic domains (9, 11–13). The data listed in Table 1 represent independent analyses of three independent preparations of CyHV-3 virions. A total of 43 viral proteins were identified (based on detection in at least two of the preparations; Table 1 and Fig. 1). This number is identical to that determined by Yi et al. (10). Comparison of the proteomes derived from both studies revealed 39 proteins in common, with four proteins in each study that were not identified in the other (Fig. 1).
TABLE 1.
Proteins identified by 2D LC-MS/MS in purified CyHV-3 virionsa
Data are restricted to CyHV-3 proteins detected by SDS-PAGE and LC-MS/MS in at least two of the three independent preparations of purified virions. Predicted transmembrane proteins are shaded in gray. Protein features were based on previous publications (6, 9) or predictions made using TMHMM, SignalP, NetNGlyc 1.0, and the NetOGlyc 3.1 server from the CBS web site. Similarity and domain analyses were performed by BLASTP N-glyco and O-glyco, number of N glycosylations and O glycosylations, respectively.
b No transmembrane domain was detected by software prediction. This sample was classified as a putative type 1 transmembrane protein on the basis of significant similarity to previously studied viral membrane proteins.
FIG 1.
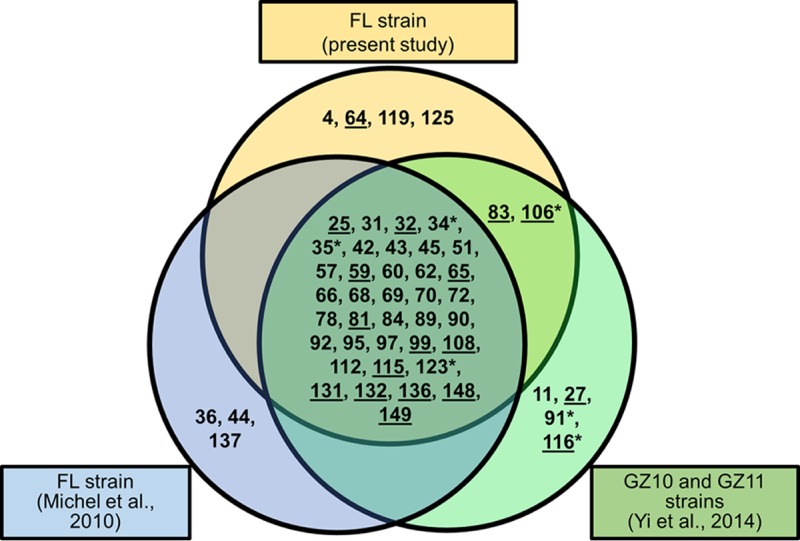
CyHV-3 virion proteome. Schematic representation of CyHV-3 virion-associated proteins identified in independent studies: upper circle, analyses of the European FL strain performed in the present study; lower left circle, analyses of the FL strain performed in a former study (9); and lower right circle, analyses of two Chinese strains (GZ10 and GZ11) (10). Numbers represent CyHV-3 ORFs. Asterisks indicate viral proteins that were detected in only one of the two Chinese isolates. Predicted transmembrane proteins are underlined.
Yi et al. (10) identified ORF11, ORF27, ORF91, and ORF116 as potential virion proteins, whereas we did not. The lack of detection of ORF27 in our study was anticipated because this coding region is disrupted in the FL strain, as has been reported for other CyHV-3 strains (5). ORF91 and ORF116 were not detected in any of the preparations in the present study, and were identified by Yi et al. (10) for only one of the two Chinese strains, with detection being based on a single peptide that was supported by a low Mascot score (10). Similarly, ORF11 was detected on the basis of only one or two peptides (depending on the isolate) with relatively low Mascot scores. Among the four proteins that were detected in the present study and not by Yi et al. (10), two were detected in all three replicates (ORF4 and ORF64), and the two other were detected in only two replicates (ORF119 and ORF125). Finally, three proteins identified in our previous study were detected neither by Yi et al. (10) nor in the present study. Taking into account that these proteins were detected initially at relatively low abundances (9), it is likely that they represented contamination by nonstructural proteins.
In the context of the published bioinformatic analyses of the CyHV-3 genome (5, 6), the results presented above suggest that the FL strain of CyHV-3 encodes 16 predicted VTPs: ORF25, ORF32, ORF59, ORF64, ORF65, ORF81, ORF83, ORF99, ORF106, ORF108, ORF115, ORF131, ORF132, ORF136, ORF148, and ORF149.
CyHV-3 VTPs essential to viral growth in vitro.
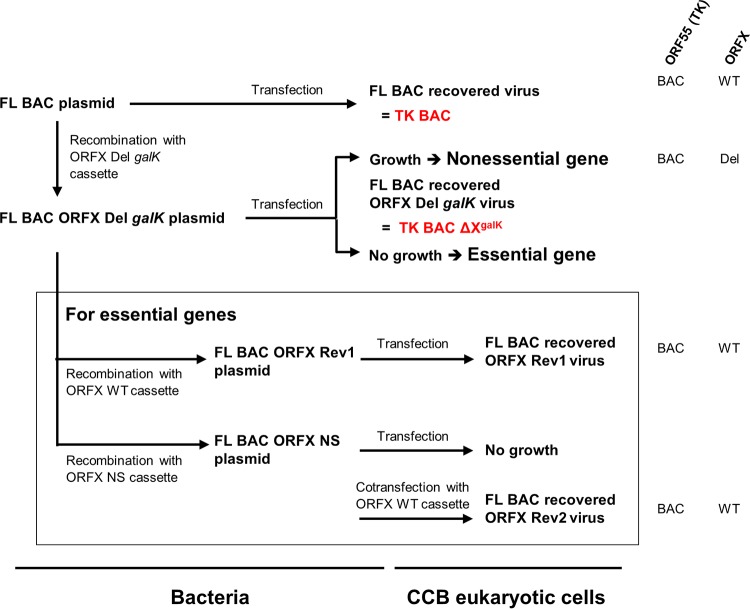
The roles of CyHV-3 VTPs in viral replication in vitro were investigated by using bacterial artificial chromosome (BAC) cloning and prokaryotic recombination technologies to generate viral mutants (Fig. 2). To facilitate the reconstitution of infectious viruses from recombinant plasmids, the BAC cassette was left in the viral genome leading to viruses expressing a truncated form of thymidine kinase (TK; encoded by ORF55) and enhanced green fluorescent protein (EGFP) (the BAC cassette is inserted at the 3′ end of ORF55 and encodes an EGFP expression cassette). The effect of TK truncation on viral replication in vitro and virulence in vivo has been documented previously. TK truncation was shown to have no effect on viral growth in vitro and to reduce virulence slightly in vivo (14). For each ORF predicted to encode a VTP, a single-gene-deletion recombinant plasmid was produced by replacing the ORF by a galK expression cassette. The molecular structures of all recombinant BAC plasmids produced were confirmed by a combined SacI digestion and Southern blotting approach and by sequencing the regions used to target recombination (data not shown). We then investigated the ability of the recombinant plasmids to reconstitute infectious virus after transfection into permissive CCB cells (Fig. 3). Transfection was monitored by detecting EGFP-expressing cells (since the BAC cassette encodes an EGFP reporter gene) at 2 days postinfection (dpi). Examination of cell cultures at 4 dpi revealed the formation of viral plaques for the FL BAC plasmid (used as a positive control) and the BAC plasmids with ORF25, ORF64, ORF65, ORF108, ORF132, ORF136, ORF148, or ORF149 deleted, thereby demonstrating that these ORFs are nonessential for viral growth in vitro. Reconstituted viruses (i.e., viruses mutated in nonessential genes) were amplified, and their genomes were validated by full-length genome sequencing (data not shown) prior to further investigations (see below).
FIG 2.
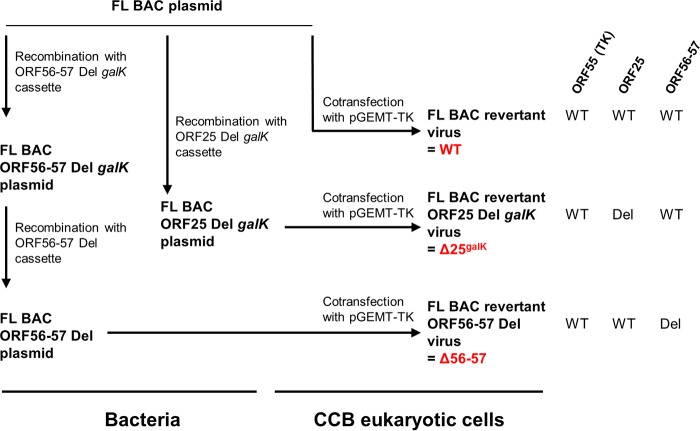
Production of CyHV-3 recombinants to identify essential and nonessential VTPs. Deleted recombinant plasmids were produced for each ORF predicted to encode a VTP (FL BAC ORFX Del galK plasmids, with “X” standing for the number of the ORFs tested; these included ORF25, ORF32, ORF59, ORF64, ORF65, ORF81, ORF83, ORF99, ORF106, ORF108, ORF115, ORF131, ORF132, ORF136, ORF148, and ORF149). The effect of the deletion on the ability of the BAC plasmid to reconstitute infectious virus was tested by transfection into permissive CCB cells. Subsequently, for each gene identified as essential, two additional recombinant plasmids were produced to revert to wild-type ORFX sequence (FL BAC ORFX Rev1) or to insert a nonsense mutation (FL BAC ORFX NS). Plasmids were transfected into CCB cells to determine their ability to induce reconstitution of infectious virus. As an additional revertant control, FL BAC ORFX NS plasmids were cotransfected into CCB cells together with a fragment encoding the WT sequence of ORFX and flanking regions, in order to facilitate reversion to wild-type ORFX sequence by recombination in eukaryotic cells (FL BAC ORFX Rev2). To simplify the reading of the manuscript, some recombinants were given a short name (in red). The right part of the Fig. summarizes the genotype of the strains for ORF55 (TK) and ORFX. Del, deleted; WT, wild type; BAC, presence of the BAC cassette in the 3′ end of ORF55.
FIG 3.
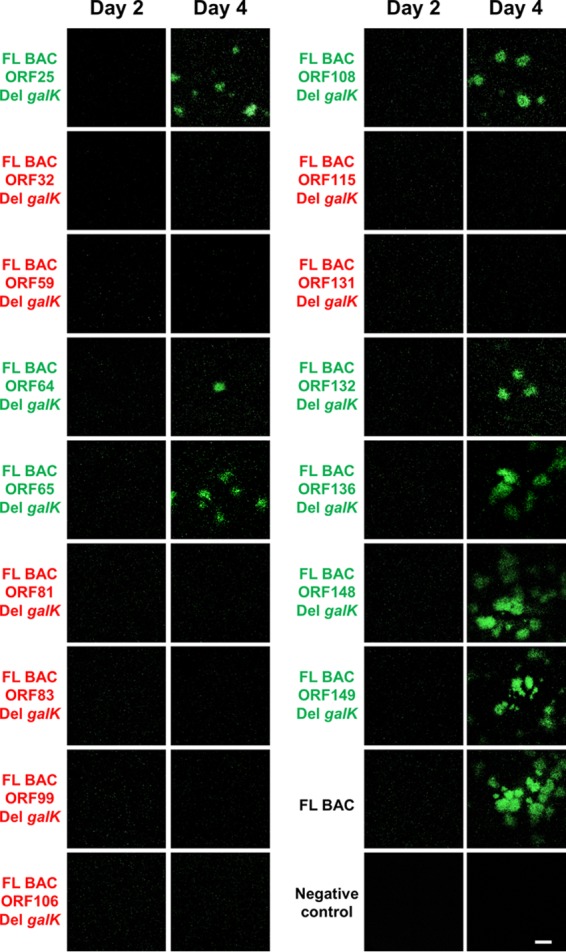
Effect of deleting genes encoding VTPs on the ability of CyHV-3 FL BAC recombinant plasmids to reconstitute infectious virus. CCB cells were transfected with the plasmids indicated. At 2 and 4 dpi, the cells were examined by epifluorescence microscopy (the BAC cassette encodes EGFP). Scale bar, 1 mm. Plasmids able or not able to reconstitute infectious virus are labeled in green or red, respectively.
Transfection of BAC plasmids with ORF32, ORF59, ORF81, ORF83, ORF99, ORF106, ORF115, or ORF131 deleted did not induce the formation of CyHV-3 plaques (Fig. 3), suggesting that these ORFs encode VTPs essential for viral growth in vitro. To test this hypothesis further, additional recombinant BAC plasmids were produced (Fig. 2). The molecular structures of all recombinant BAC plasmids described below were confirmed by a combined SacI digestion and Southern blotting approach and by sequencing the regions used to target recombination (data not shown). To exclude the possibility that the lack of infectivity of the deleted recombinant BAC plasmids resulted from unexpected mutations generated during BAC manipulation, plasmids encoding revertant wild-type (WT) VTP ORF were derived (FL BAC ORFX Rev1 plasmids, Fig. 2) from the deleted BAC plasmids. Transfection of each of the revertant plasmids into CCB cells led to viral growth (Fig. 4, second column). To exclude the possibility that the lack of infectivity observed for the deleted BAC plasmids resulted from a polar effect of the deletion on the expression of a nearby essential gene, a recombinant plasmid in which the ORF was disrupted by a nonsense (NS) mutation was derived for each putative essential VTP gene (FL BAC ORFX NS plasmids, Fig. 2). Transfection of these plasmids into CCB cells did not induce plaque formation (Fig. 4, third column). To exclude the possibility that the lack of infectivity of the NS recombinant BAC plasmids resulted from unexpected mutations generated during BAC manipulation at the stage of NS mutagenesis, the FL BAC ORFX NS plasmids were transfected into CCB cells, together with a WT DNA fragment containing the relevant ORF and flanking regions, in order to induce reversion to a WT sequence after homologous recombination in eukaryotic cells. All plasmids led to viral growth when cotransfected with WT DNA fragments (Fig. 4, fourth column). In summary, the results demonstrated that ORF32, ORF59, ORF81, ORF83, ORF99, ORF106, ORF115, and ORF131 encode VTPs essential to viral growth in vitro.
FIG 4.
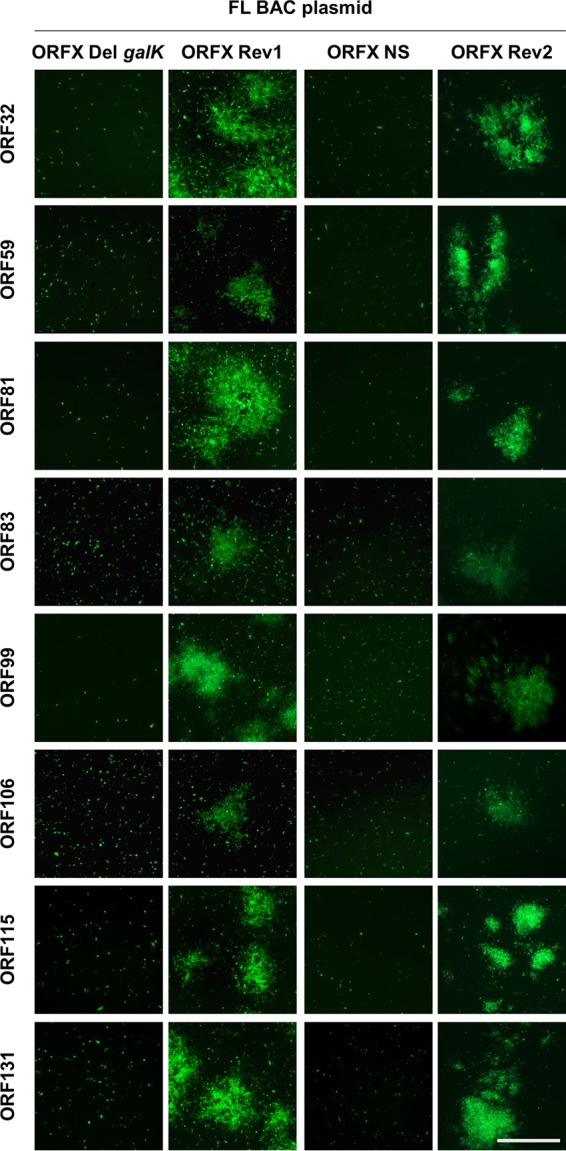
Testing plasmids recombinant for CyHV-3 ORFs predicted to encode essential VTPs. For ORFs identified as essential in Fig. 3, additional recombinant plasmids (ORFX Rev1 and ORFX NS) were produced (see Fig. 2, description in the rectangular frame) and tested for their ability to reconstitute infectious virus after transfection into CCB permissive cells. At 6 days posttransfection, the cells were examined by epifluorescence microscopy for detection of EGFP. Scale bar, 1.5 mm.
These experiments identified essential versus nonessential VTPs of the FL strain of CyHV-3, as summarized in Table 2. To test whether the findings extend across the species Cyprinid herpesvirus 3, the ORFs encoding VTPs were sequenced from nine CyHV-3 strains with different geographical origins and passage histories (Table 2). All of the ORFs encoding essential VTPs were intact and exhibited a limited number of polymorphisms. In contrast, various ORFs encoding nonessential VTPs were truncated by disruptive mutations (highlighted in red in Table 2), the precise pattern depending on the strain.
TABLE 2.
Polymorphisms observed among CyHV-3 strains in ORFs encoding VTPs
| Gene | Requirement in FL strain | Mutationa |
|---|---|---|
| ORF32 | Essential | No mutation |
| ORF59 | Essential | No mutation |
| ORF81 | Essential | No mutation |
| ORF83 | Essential | Several aa inserted in M3, T, and J |
| ORF99 | Essential | Several aa changed in KHV-I, M3, T, and J |
| ORF106 | Essential | No mutation |
| ORF115 | Essential | One aa changed in T |
| ORF131 | Essential | One aa changed in KHV-I, FL, Cavoy, and T |
| ORF25 | Nonessential | Several aa inserted in M3, T, J, and GZ11 |
| ORF27 | Nonessential | Frameshifted in U, KHV-I, T, and GZ11; large regions deleted in FL; stop codon insertion in the beginning of the ORF in Cavoy; several aa changed in M3 and J |
| ORF64 | Nonessential | Frameshifted and several amino acids deleted or inserted in M3, T, and J |
| ORF65 | Nonessential | One aa deleted in KHV-I, FL, Cavoy, I, and E; one aa inserted and several aa deleted in M3, T, and J; one aa changed in GZ11 |
| ORF108 | Nonessential | One aa inserted, one aa changed and a stop codon inserted at the end of the ORF, several aa lost in M3, T, and J |
| ORF132 | Nonessential | One aa changed in KHV-I |
| ORF136 | Nonessential | Several aa inserted and a stop codon inserted in the middle of the ORF in M3, T, and J |
| ORF148 | Nonessential | Several aa inserted in KHV-I, Cavoy, and GZ11; Several aa deleted in M3, T, and J |
| ORF149 | Nonessential | One aa changed in KHV-I, Cavoy, and E, 13 aa inserted in M3 and J |
Boldfacing indicates mutations that create an in-frame stop codon or frameshift. aa, amino acid(s).
Effects of deleting genes encoding nonessential VTPs on CyHV-3 growth in vitro.
Considerations of the CyHV-3 gene arrangement suggested that deletion of ORF25, ORF64, ORF65, ORF108, ORF132, ORF136, or ORF148 would be unlikely to affect the expression of neighboring genes. In contrast, deletion of ORF149 might affect the expression of ORF148, resulting in the phenotype of TK BAC Δ149galK representing the absence of expression of both proteins. To test this hypothesis, TK BAC Δ148galK and TK BAC Δ149galK plaques were stained with monospecific polyclonal antibodies (pAbs) against ORF148 or ORF149. Staining of TK BAC Δ149galK plaques with the anti-ORF148 antibodies demonstrated the expression of the ORF148 protein (data not shown), implying that TK BAC Δ149galK is adequate for investigating the effects of deleting ORF149.
The effect of deleting genes encoding nonessential VTPs on viral growth in vitro was investigated by multistep growth assay (Fig. 5A) and plaque size assay (Fig. 5B). Deletion of ORF25, ORF132, ORF136, ORF148, or ORF149 impaired CyHV-3 replication in the multistep growth assays to various degrees (based on observation of a significant effect for at least two consecutive time points). The more pronounced effect was observed for deletion of ORF25, which was associated with a 10-fold reduction in viral titer. In contrast, deleting ORF64, ORF65, or ORF108 had no major effect on CyHV-3 growth. The plaque size assays (Fig. 5B), based on the results collected at 8 dpi, revealed that deleting ORF25, ORF108, ORF132, or ORF149 resulted in reduced plaque size, whereas deleting ORF64 led to an increased plaque size. Deleting ORF65, ORF136, or ORF148 did not significantly affect plaque size.
FIG 5.
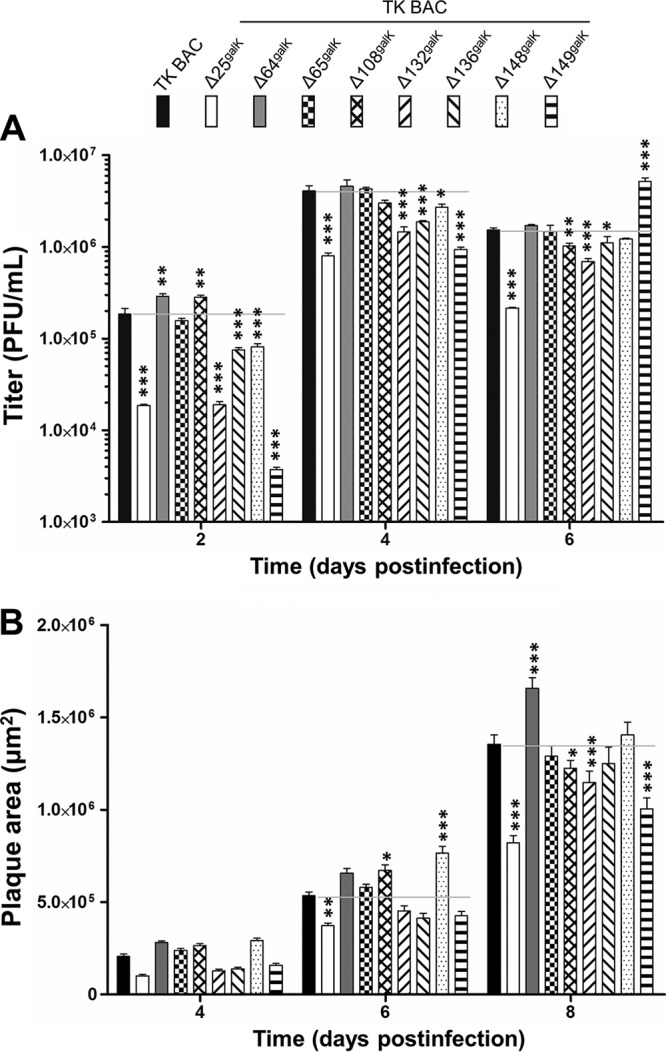
Effects of deleting nonessential VTPs on CyHV-3 growth in vitro. (A) Multistep growth curves. CCB cells were infected with the viruses indicated and the titer (PFU/ml) in the cell supernatant was determined at the indicated time points postinfection. Data presented are the means ± the standard errors of the mean (SEM) of triplicate measurements. Significant differences between the deleted viruses and the TK BAC virus were tested by using two-way ANOVA, taking genotype and time postinfection as variables. (B) Plaque size assay. CCB cells were infected with the viruses indicated, and plaque areas were measured over time. The data presented are means ± the SEM for the measurement of 25 randomly selected plaques. Two-way ANOVA was used to test the significance of the results, taking genotype, time postinfection, and interaction between genotype and time postinfection as variables (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Effect of deleting genes encoding nonessential VTPs on CyHV-3 virulence in vivo.
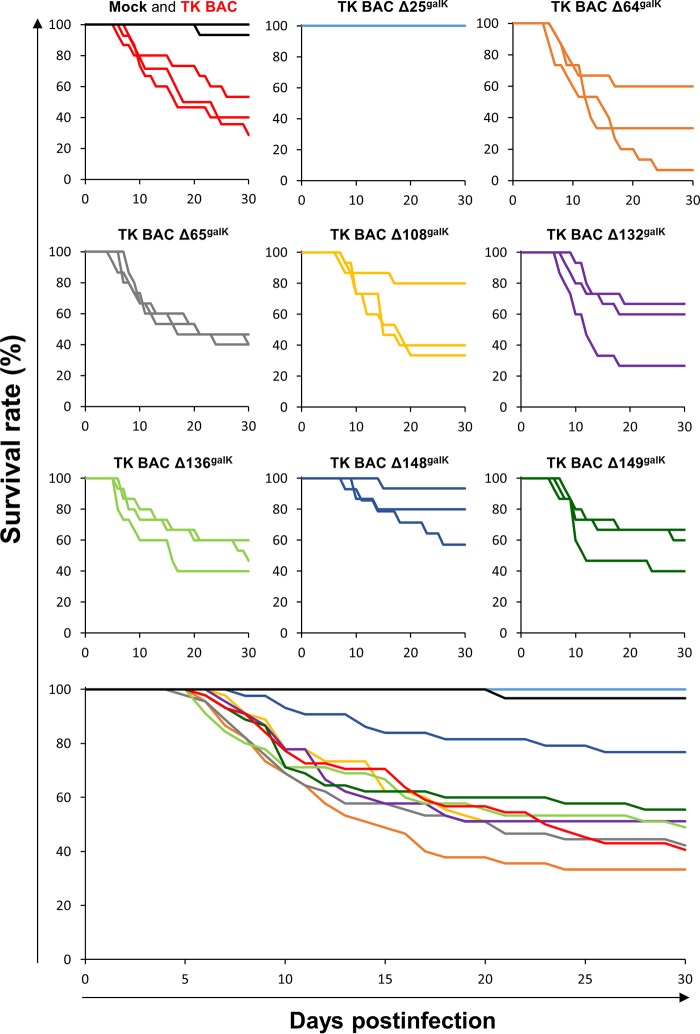
All recombinant viruses deleted for a nonessential ORF (TK BAC ΔXgalK, where X is the number of the ORF) were tested in triplicate for their virulence in carp (Fig. 6). Fish infected with the TK BAC virus (used as a positive control) exhibited all the clinical signs associated with CyHV-3 disease, including apathy, folding of the dorsal fin, hyperemia, increased mucus secretion, skin lesions, suffocation, erratic swimming, and loss of equilibrium. At 30 dpi, the mean survival rate of TK BAC-infected fish was 41%. Fish inoculated with TK BAC Δ64galK, TK BAC Δ65galK, TK BAC Δ108galK, TK BAC Δ132galK, TK BAC Δ136galK, or TK BAC Δ149galK exhibited comparable effects, leading to similar survival rates at 30 dpi (ranging between 33 and 56%). Conversely, fish infected with TK BAC Δ148galK exhibited mild disease and a higher survival rate (77%, P < 0.001). Fish inoculated with TK BAC Δ25galK expressed no symptoms, and all survived the infection.
FIG 6.
Effect of deleting genes encoding nonessential VTPs on CyHV-3 virulence. The virulence of the indicated recombinant viruses was tested in carp (triplicate groups each consisting of 15 subjects; average weight, 1.90 ± 0.68 g, and average age, 3 months old). On day 0, fish were infected for 2 h by immersion in water containing 800 PFU/ml or mock infected. Survival rate was measured over a period of 30 days. The nine upper panels show the survival curves observed for replicates. The lower panel shows the mean survival curves based on the three replicates.
To ensure that each group of fish was infected with the correct virus and to exclude any possibility of viruses spreading among the tanks, PCR assays were performed on three randomly selected dead fish from each infected group (for TK BAC Δ25galK-infected fish, three fish were selected randomly from the living fish at 30 dpi), one fish infected with the TK BAC virus, and one mock-infected fish selected randomly at the end of the experiment (Fig. 7). PCR performed with the ORF112intfw/ORF112intrev primer pair confirmed that samples from all but one group contained the CyHV-3 genome (the exception being the TK BAC Δ25galK-infected group). PCR performed with primers specific for the deleted ORFs confirmed that each group had been infected with the correct virus and that cross-contamination among tanks had not occurred (Fig. 7).
FIG 7.
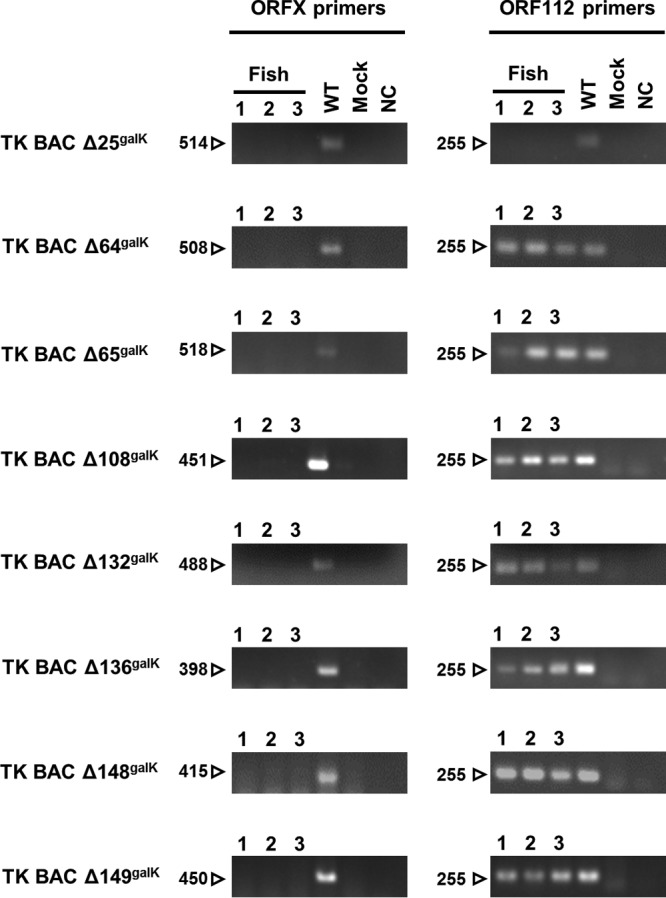
PCR detection and characterization of CyHV-3 genomes recovered from infected dead carp. DNA was extracted from the gills of three dead carp from each of the groups infected with the recombinant viruses deleted for a nonessential gene (TK BAC ΔORFXgalK) during the course of the experiment described in the legend of Fig. 6. In the TK BAC Δ25galK infections, no fish died, and hence three fish were selected randomly from among the living fish at 30 dpi. A carp infected with TK BAC virus and a mock-infected carp were used as positive and negative PCR controls, respectively. PCRs were performed with the appropriate ORFXintfw/ORFXintrev primer pair and with the ORF112intfw/ORF112intrev primer pair. The images are photographs of agarose gels. The numbers on the left of each panel are marker sizes (bp). NC, negative control.
Testing of an ORF25-deleted recombinant as an attenuated vaccine candidate against CyHV-3.
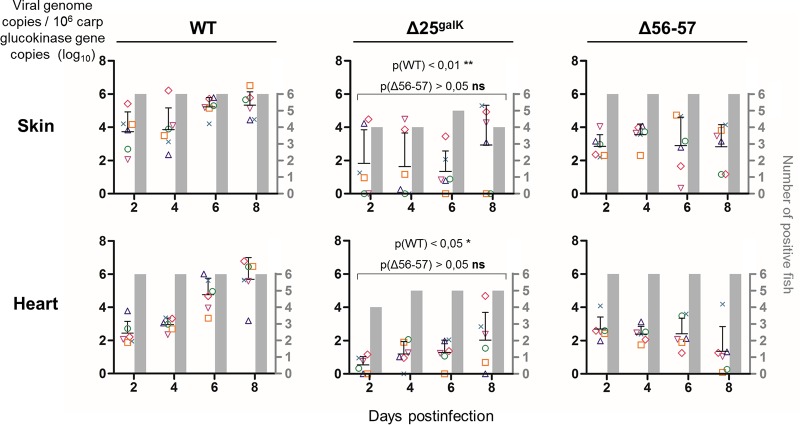
The attenuation described above for TK BAC Δ25galK, from which ORF25 was deleted and encoding a truncated form of TK, suggested that a virus deleted only for ORF25 would have potential as an attenuated recombinant vaccine against CyHV-3. The requisite recombinant (Δ25galK, with ORF25 deleted and encoding a wild-type TK) was produced by following the procedure described in Fig. 8 and in Materials and Methods. The failure to detect virus by PCR in the gills of fish infected with the TK BAC Δ25galK virus (Fig. 7) suggests that deletion of ORF25 could affect the ability of deleted virions to infect fish or to spread within their host. To test this hypothesis and to compare the tropism of a virus lacking ORF25 (Δ25galK) to the tropism of a previously described recombinant vaccine candidate (deleted for ORF56 and ORF57, Δ56-57), the following experiment was performed. Fish were infected with the FL BAC revertant virus (WT, wild-type for TK and for ORF25), FL BAC revertant ORF25 Del galK virus (Δ25galK, wild-type for TK but with ORF25 deleted), or the FL BAC revertant ORF56-57 Del virus (Δ56-57, wild-type for TK but with ORF56-57 deleted) (Fig. 9). The viral charge was measured by qPCR over time in the skin (the portal of entry of CyHV-3) and the heart (selected as an internal organ). The results demonstrated that the ORF25 deleted virus infects fish through skin infection and then spreads to internal organs as reported previously for the WT parental virus and the Δ56-57 virus (15). However, compared to the parental WT virus, the replication of the ORF25 deleted virus was reduced in intensity and duration to levels similar to those observed for the Δ56-57 virus.
FIG 8.
Flow chart of the production of CyHV-3 recombinants deleted for ORF25 or ORF56-57. Recombinants were given a short name (in red) for use in the text. Viral genotypes for ORF55 (TK), ORF25, and ORF56-57 are given on the right. Del, deleted; WT, wild type.
FIG 9.
Effect of ORF25 deletion on viral tropism according to qPCR analysis. At time zero, carp (average weight, 6.10 g ± 1.39 g; average age, 6 months old) were infected for 2 h by immersion in water containing the virus indicated at 800 PFU/ml and then returned to larger tanks. At the indicated time points, fish were sampled and submitted to qPCR analysis. The data obtained for each fish (according to viral genotype and time postinfection) are represented by the same symbol to allow correlation of the data obtained for the two tested organs. The number of viral genome copies is expressed as the log10 per 106 carp glucokinase gene copies. Individual values represent the mean of duplicate measurements. Mock-infected fish were used as a negative control and no viral genome copies were detected in these fish. The number of positive fish among six analyzed fish is represented by gray bars. Viral charges [viral genome copies/106 carp glucokinase gene copies (log10)] were compared using one-way ANOVA taking the viral genotype as a variable.
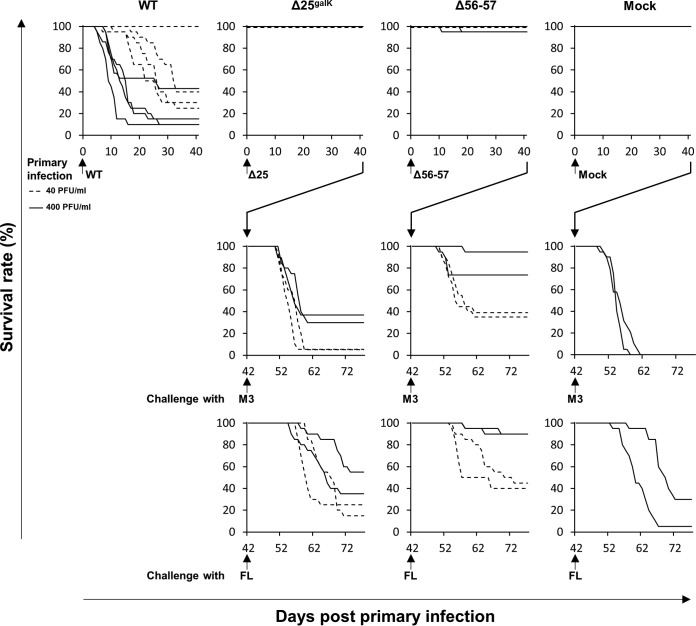
Next, the virulence of the Δ25galK virus was compared to that of the WT strain and the previously described Δ56-57 attenuated virus (Fig. 10) (15). A high degree of attenuation of Δ25galK was indicated by the finding that infected fish exhibited no mortality or symptoms (Fig. 10). The level of immune protection conferred by primary infection with Δ25galK or Δ56-57 virus was tested by challenging infected fish with the M3 or FL strains of CyHV-3 (Fig. 10, middle and lower lines of the graph, respectively). The fish vaccinated with Δ25galK exhibited a poor level of protection, independent of the dose used for primary infection and the challenge strain used, with the relative percentage survival at a maximum of 34 when the fish were vaccinated at a dose of 400 PFU/ml and challenged with the M3 strain. In comparison, vaccination with the Δ56-57 attenuated virus resulted in a relative percentage survival of 84 under the same conditions (Fig. 10).
FIG 10.
Testing of Δ25galK and Δ56-57 deletion recombinants as attenuated recombinant vaccine candidates. The virulence of the recombinant viruses indicated was first tested in carp (first line of graph: four replicates per condition; n = 20 fish; average weight, 2.42 ± 0.95 g; average age, 6 months old). On day 0, fish were mock infected or infected for 2 h by immersion in water containing 40 (dotted line) or 400 (continuous line) PFU of virus/ml. The survival rate was measured over a period of 42 days. The immune protection conferred by primary infection was then tested for Δ25galK and Δ56-57-infected fish at 42 dpi by distributing mock-infected fish and fish that survived the primary infection into duplicate tanks (n = 20) and challenging them by cohabitation with fish infected with the M3 strain (middle line of graphs) or the FL strain of CyHV-3 (lower line of graphs). The survival rate was measured according to time postchallenge.
The data above demonstrated that the VTP encoded by ORF25 is nonessential for viral replication in vitro but acts as a key virulence factor in vivo. ORF25 is the prototypical gene of the ORF25 family, which contains six paralogs (ORF25, ORF26, ORF27, ORF65, ORF148, and ORF149) encoding type I transmembrane proteins (5). Although ORF26 is a pseudogene, all other paralogs encode CyHV-3 VTPs. The FL strain used in this study as the parental strain encodes a disrupted ORF27, as has been reported for other strains of CyHV-3 (5). Individual deletions of each member of the ORF25 family in this ORF27-negative background revealed that none of them is essential for viral replication in vitro, although it is possible that this is the consequence of redundant functions expressed by the paralogs. The attenuation observed for a mutant lacking ORF25 suggested that this locus might serve as a target for generating recombinant attenuated vaccines. However, there are two points of caution in relation to this hypothesis: the ORF25 deletion virus induced weak immune protection, and it is possible that reversion to virulence could occur by mutation of the paralogous genes.
Concluding remarks.
Herpesvirus VTPs play crucial roles in the viral life cycle. Although the functions of VTPs in members of the family Herpesviridae have been studied extensively, very little information is available on VTPs in members of the family Alloherpesviridae. The very distant evolutionary origin of these families has resulted in a lack of detectable genetic similarity between VTPs in the two groups. This situation makes it impossible to advance hypotheses on the roles of alloherpesvirus VTPs on the basis of the much more extensive knowledge available for other herpesviruses. The present study of the archetypal fish alloherpesvirus has enabled major steps to be taken toward characterizing alloherpesvirus VTPs: (i) the number of CyHV-3 VTPs was expanded from 14 to 16; (ii) these were characterized as consisting of eight essential and nine nonessential proteins; (iii) individual deletion of seven nonessential VTPs affected viral growth in vitro, and individual deletion of two affected virulence in vivo; and (iv) a mutant lacking ORF25 proved to be highly attenuated in vivo but induced a poor immune protection against a lethal challenge. This study provides a firm basis for the further study of alloherpesvirus VTPs.
MATERIALS AND METHODS
Cells and viruses.
Cyprinus carpio brain (CCB) cells (16) were cultured as described previously (14). The CyHV-3 FL strain was isolated in Belgium from a fish that had died from CyHV-3 infection and had been used to produce the FL BAC plasmid (14). Other CyHV-3 strains from different geographical origins were also used: the M3 strain from Belgium, the U strain from the United States (5); the J strain from Japan (5), the E strain from England (kindly provided by K. Way), the T strain from Taiwan (17), the GZ11 strain from China (18), and the KHV-I and I strains from Israel (5). The attenuated Cavoy strain from Israel was produced by passaging in cell culture (19).
Production and purification of CyHV-3 virions.
CCB cells were infected with the CyHV-3 FL strain at a multiplicity of infection (MOI) of 0.02 PFU/cell (20). The supernatant was harvested at the beginning of viral release, when cell lysis was minimal (4 dpi). Virions were purified from the cell supernatant and monitored for purity as described previously (9).
SDS-PAGE and LC-MS/MS proteomic analysis.
Virion proteins were separated by SDS-PAGE and detected by Coomassie blue staining. The entire lanes of the gel were cut into 30 equally sized slices, which were subjected to in-gel trypsin digestion (21). Peptides were separated by reversed-phase chromatography using a 40-min CAN gradient (4 to 45%) and analyzed by using an HCT ultra-ion trap (Bruker) in data-dependent AutoMS (2) mode with a scan range of 100 to 2,800 m/z and three averages, and five precursor ions at 300 to 1,500 m/z were selected from the MS scan. Precursors were actively excluded within a 0.5 min window, and all singly charged ions were excluded. The MS data were processed by using Mascot distiller, and all single-slice analyses were merged into a single data set. The data were searched for matches to CyHV-3 protein sequences (160 entries) by using an in-house Mascot 2.2 server (Matrix Science). The default search parameters used were as follows: enzyme = trypsin, maximum missed cleavages = 1, fixed modifications = carbamidomethyl (C), variable modifications = oxidation (M), peptide tolerance ± 1.3 Da; MS/MS tolerance = ± 0.5 Da, and instrument = ESI-TRAP. Only sequences identified by a Mascot score of >30 were considered, and single peptide identification was systematically evaluated manually. In addition, the emPAI (22) value was calculated to estimate relative protein abundance in the GC extract only.
Antibodies.
Mouse pAbs against CyHV-3 ORF148 or ORF149 were produced by customized DNA immunization (DelphiGenetics).
Cloning and site-directed mutagenesis of CyHV-3 ORFs.
DNA fragments containing CyHV-3 ORFs of interest plus at least 200 bp upstream and downstream were amplified by PCR using CyHV-3 FL BAC DNA as a template (see Table 3 for primer details). The amplification products were TA cloned into the pGEM-T-Easy vector (Promega), and the resulting clones (pGEM-T-ORFX WT, X standing for the number of the ORF tested) with the correct sequence were used as a recombination cassette or as a template for mutagenesis. For ORFs encoding VTPs, site-directed mutagenesis was used to create 1- or 2-bp substitutions changing a tyrosine or a tryptophan codon into a stop codon, resulting in pGEM-T-ORFX NS. Oligonucleotide primers were designed by using the QuikChange primer design tool (http://www.genomics.agilent.com/primerDesignProgram.jsp) (Table 3).
TABLE 3.
Oligonucleotide primersa
| Analysis | Primer | Sequence (5′–3′)b | Coordinates or accession no. |
|---|---|---|---|
| Synthesis of inserts | |||
| CyHV-3 ORF25 | ORF25outF | CCTCCGACTCTGAAGACGAT | 45389–45408 |
| ORF25outR | GGTACGTGATGCTGTAGAAG | 47569–47588 | |
| CyHV-3 ORF32 | ORF32outF | CCAGGTCCAGGCAGTCTC | 54216–54233 |
| ORF32outR | ATGCGCGTCTACAAGATCG | 55619–55601 | |
| CyHV-3 ORF59 | ORF59outF | GACGCCTGTCTCTGGTAGA | 101541–101559 |
| ORF59outR | GCGTCTCCAAAAAGAGCGG | 102941–102923 | |
| CyHV-3 ORF64 | ORF64outF | GTCGAGATATCCGAGGCAGA | 119716–119735 |
| ORF64outR | TTCATAGCCACAACCGGAGT | 122203–122184 | |
| CyHV-3 ORF65 | ORF65outF | TGGAGAACATCAAGCAGCAC | 121920–121939 |
| ORF65outR | GAAGTCTGAAACTGTCTGAATG | 124106–124127 | |
| CyHV-3 ORF81 | ORF81outF | GGTTTAGTCCAAATCCGACCT | 150519–150539 |
| ORF81outR | GCAGCTTCTGCTTCAGAGTG | 152023–152004 | |
| CyHV-3 ORF83 | ORF83outF | ACACGCTGATGGTCACGAG | 152549–152567 |
| ORF83outR | ACACCAACCAGTTCGTGCAG | 153638–153619 | |
| CyHV-3 ORF99 | ORF99intF | GCTTAGCCTGTTCGGCAC | 184445–184462 |
| ORF99intR | AAGATCTGGGACACGGACTG | 185515–185496 | |
| CyHV-3 ORF106 | ORF106outF | GCTGACACCTGTCACAACCA | 195629–195648 |
| ORF106outR | TCGTGAGGACAAACCGTCTC | 196189–196170 | |
| CyHV-3 ORF108 | ORF108outF | GATGATGAAGGGTGTTCATG | 201067–201086 |
| ORF108outR | CATCTACAAGTCGGACAACC | 201937–201956 | |
| CyHV-3 ORF115 | ORF115outF | CACGTAAACGAAGCCCCATA | 207951–207970 |
| ORF115outR | ATTCGTGGCGGCTGTTATAC | 210734–210715 | |
| CyHV-3 ORF131 | ORF131outF | GCCCTGGTCCTCGTACTTTT | 224971–224990 |
| ORF131outR | CTGATCAGATTCTCAGGAGCAG | 227271–227250 | |
| CyHV-3 ORF132 | ORF132outF | GCTCCTGAGAATCTGATCAG | 227252–227271 |
| ORF132outR | GTTAATGGTCACAGAAGCGC | 228155–228174 | |
| CyHV-3 ORF136 | ORF136outF | GTGTCAAGTACGTGGAGCGT | 231157–231176 |
| ORF136outR | GGCCTCTGAATGTGTTATTGC | 231991–232011 | |
| CyHV-3 ORF148 | ORF148outF | CATGGTTCGAGGTTGTGAAG | 253808–253827 |
| ORF148outR | CAGCATTCGTCTCTTAGTGC | 255933–255952 | |
| CyHV-3 ORF149 | ORF149outF | ACCCTATCATGATTGACGGC | 255770–255789 |
| ORF149outR | CTTTCGTTCTACTGTTCCTC | 258172–258191 | |
| Synthesis of galK recombination cassettes | |||
| ORF 25 Del galK | ORF25 galK F | GTCATTTTTCCTCGTGGTGTGCAGCTTTTTCTAACGTGTGGTAGGACGCTAACCATTACCAAGTAAACCATTACGCCTGTTGACAATTAATCATCGGCA | 45495–45569 |
| ORF25 galK R | TGACCAGGGTAGAATGATAGACCCGCCGCTAAGAAAAATGACTCTCCGTCCGCTACCGTCGCCCGCTATAGTGTTTCAGCACTGTCCTGCTCCTT | 47376–47450 | |
| ORF 32 Del galK | ORF32 galK F | ATAATACGCACCCTGGTCACTTTCTTTAACGTGTCCGTGTCGGGCTCCCTACACGTTTCCTCCGAGTCGCCGATCCCTGTTGACAATTAATCATCGGCA | 54417–54491 |
| ORF32 galK R | CTGTATCCCATCAGGGGCTGATCTGCTGACGGCCTCGACGTGGTGGTGATGGTGGTGGTGGTGGTGATGATGATGTCAGCACTGTCCTGCTCCTT | 54491–54417 | |
| ORF 59 Del galK | ORF59 galK F | CCCGCACGACGTCGGACGCTCCCTACCGAGTGAGCCGTCTCTCTGACCCGGGTCGTGCGGGCGCCACCAGCCCTTCCTGTTGACAATTAATCATCGGCA | 101961–102035 |
| ORF59 galK R | AGCAGTGGTAGTCCTCTCACCCCCCCCCCCCCCCCCTTGACCTTCACCCTTCACCCCTAACCCCAACAACCAACCTCAGCACTGTCCTGCTCCTT | 102521–102447 | |
| ORF 64 Del galK | ORF64 galK F | TGGTCCTCCCATGCTTTTTATTCAATCCTGCTCCTCTTGAAAAGCTTAAGCCTGTTGACAATTAATCATCGGCA | 119814–119863 |
| ORF64 galK R | GTACACTATATTGCGTTTATTTGTTTTTCATAAAAAGCACATACATAATATCAGCACTGTCCTGCTCCTT | 122082–122033 | |
| ORF 65 Del galK | ORF65 galK F | CAATATAGTGTACACCGTGGTCTTTATTAATTTGATAACATAATAATGTACCTGTTGACAATTAATCATCGGCA | 122070–122119 |
| ORF65 galK R | CAAGACGCCTCACACGACGGAAACCCGCAACACCACCCCACAAACAAACGTCAGCACTGTCCTGCTCCTT | 123911–123960 | |
| ORF 81 Del galK | ORF81 galK F | GCACTTGAGAGCAGCGGAGGAGAAGACCGGTGCATCCATCTCGACGCTCGTCAGGTTGTGACTGTCCTCGTCCTCCCTGTTGACAATTAATCATCGGCA | 150722–150796 |
| ORF81 galK R | CGGCTGCTGTGTTTTGTGGTGCTGGCCTCGACTGTGGGGTGGTGGTGGCGGCGACTGCGAGAGGCTTCGGAATCAGCACTGTCCTGCTCCTT | 150796–150722 | |
| ORF 83 Del galK | ORF83 galK F | CACAATAAAAAGCTCCCCTCGGGGATCACCAAGACAACCTTTCAGTAGCCCCTGTTGACAATTAATCATCGGCA | 153481–153530 |
| ORF83 galK R | TCTTCGCCCTCGCCCTCGTCCCTCAACCTGTTGTACCTATACGCGCTCATTCAGCACTGTCCTGCTCCTT | 152758–152807 | |
| ORF 99 Del galK | ORF99 galK F | TGCAGGCCACCGCCGCCAACGCCGAGATGTCCAACAACCTCCAGACGCAGCCTGTTGACAATTAATCATCGGCA | 184714–184763 |
| ORF99 galK R | AGGGCTTGCTCTTGAGGAAGGGCTGGTTGACGGGGCAGCCCTGCAGGTAATCAGCACTGTCCTGCTCCTT | 185214–185165 | |
| ORF 106 Del galK | ORF106 galK F | ACTACTCGCCCACCGCCTTATACAACTTCACCGCGTATCCGCCGTACATCCCTGTTGACAATTAATCATCGGCA | 195727–195776 |
| ORF106 galK R | GGGTTTGGTAGAGGGCGCTGCGGGGGATGAGCCCGACGATACCGCTGCGGTCAGCACTGTCCTGCTCCTT | 196063–196014 | |
| ORF 108 Del galK | ORF108 galK F | CACTTAGTCTACCACTCACTTTACCTTACAACCACCCAGACACCAACACACCACACCACCCACACTACGAGCGAGCCTGTTGACAATTAATCATCGGCA | 201156–201230 |
| ORF108 galK R | ACCAAGCAACACGTTCATGTGATTTTTTTTTATTGTTGATGTTGTTTGATGTATTACAGTTACAATAAACGTAATTCAGCACTGTCCTGCTCCTT | 201819–201893 | |
| ORF 115 Del galK | ORF115 galK F | ATCGTGTCCTCCGTCTCGGTCATCTCTGTGGTTTTTTCCAGCTTTGCGCCCCTGTTGACAATTAATCATCGGCA | 208152–208226 |
| ORF115 galK R | GCTCAAAATAAAAAAAACATGTCTGGACCTTTACATCTTGTTTCAATAATTCAGCACTGTCCTGCTCCTT | 210641–210567 | |
| ORF 131 Del galK | ORF131 galK F | GTGAGGGAGTGATATGGAGTGAACGTAAATGGAGGGGCGCTGCGGAGGTTCCTGTTGACAATTAATCATCGGCA | 225410–225484 |
| ORf131 galK R | TCGAGACGCCCGAACTGGTCGAGGCCTACGTGAACGACGTCAAGGTCCGCTCAGCACTGTCCTGCTCCTT | 226426–226352 | |
| ORF 132 Del galK | ORF132 galK F | TCAAGTCTTTATTATTACTTTATTTAATGGGGTTCGGTTGGAGAAGAGCAGTCAAGTGAGCAGAAGAGAAGATCGCCTGTTGACAATTAATCATCGGCA | 227369–227443 |
| ORF132 galK R | AACTGAGGCCTCTCCGGCTATCCGCACCGAGTGATATACCGCCCGCCACTACCCCAGAACCCCCACACAGCCGCATCAGCACTGTCCTGCTCCTT | 227957–228031 | |
| ORF 136 Del galK | ORF136 galK F | AAGCTTGCCTTTGAAACATGGAAAAGAGCTCTCGTGTGTTTCTTTAGAATAAGACGACGTCGAGCGACGACCATCCCTGTTGACAATTAATCATCGGCA | 231264–231337 |
| ORF136 galK R | TGTTGGTTTGCTTTGGCTTGATAGCACACAAAAATCACATTTGTAAGAGTTTTATTGTTTGACAATGATACATCATCAGCACTGTCCTGCTCCTT | 231801–231875 | |
| ORF 148 Del galK | ORF148 galK F | GAGGGTAGAGAGGGCTGGAGAGTAGGAGAGGAGAGAGGGGAAAGTTTTGAAGTTCTTGCACATGTTGAGGGGAAGCCTGTTGACAATTAATCATCGGCA | 253881–253955 |
| ORF148 galK R | AAAACCAAAATTTTTCTGTTATCGTATCGTGATGAGCGCGACCGAGGCGACGGTCCAGTAACTCCGCCGTCAATCTCAGCACTGTCCTGCTCCTT | 255780–255854 | |
| ORF 149 Del galK | ORF149 galK F | TTTGCTTTTTTTATTATGTGATGCTGTAGTGTACACACGTATTAGAAACAATAGTAGAGCAAGCAAGCAAAAGCCCTGTTGACAATTAATCATCGGCA | 255861–255935 |
| ORF149 galK R | TTTTCTTTTGACGCGCAATGACCGACGGCTCAAGTGACGGCTACCTAGACGATACGTCACAGACGGCTGAGGACCTCAGCACTGTCCTGCTCCTT | 257997–258071 | |
| Nonsense mutagenesis of essential genesc | |||
| ORF32 | ORF32.T237G-sense | GACAACGGCTGGGGCTAGACCTTTCTGATCCAATC | 54711–54745 |
| ORF32.A237C-antisense | GATTGGATCAGAAAGGTCTAGCCCCAGCCGTTGTC | 54745–54711 | |
| ORF59 | ORF59.C144G-sense | CATGCAGCGCTAGCAACTGTGCGAG | 102315–102291 |
| ORF59.G144C-antisense | CTCGCACAGTTGCTAGCGCTGCATG | 102291–102315 | |
| ORF81 | ORF81.C207G-sense | TTGCGCACGCCATGTAGTCCAACGATCCC | 150987–151015 |
| ORF81.G207C-antisense | GGGATCGTTGGACTACATGGCGTGCGCAA | 151015–150987 | |
| ORF83 | ORF83.G225A-sense | GTTCATGTACTGAGTCACCCTACGC | 153268–153244 |
| ORF83.C225T-antisense | GCGTAGGGTGACTCAGTACATGAAC | 153244–153268 | |
| ORF99 | ORF99.C3039G-sense | CAACGACAGGTTCTCGTAGGTGGCGAGCC | 184946–184974 |
| ORF99.G3039C-antisense | GGCTCGCCACCTACGAGAACCTGTCGTTG | 184974–184946 | |
| ORF106 | ORF106.CA108AT-sense | CTTCGTCGTCTAATAGGCCCAGCAC | 195872–195896 |
| ORF106GT108TA-antisense | GTGCTGGGCCTATTAGACGACGAAG | 195896–195872 | |
| ORF115 | ORF115.C744A-sense | GGACCTGGTCCTGTAATCCCAGACGAACGTG | 208955–208985 |
| ORF115.G744T-antisense | CACGTTCGTCTGGGATTACAGGACCAGGTCC | 208985–208955 | |
| ORF131 | ORF131.C735G-sense | TCCAGGGCCGTGTAGGAGGCCGC | 226054–226032 |
| SDM131.G735C-antisense | GCGGCCTCCTACACGGCCCTGGA | 226032–226054 | |
| Synthesis of probes for Southern analysis (probe name) | |||
| GalK | GalK.Int2-F | AGGTGAGGAACTAAACCCAG | |
| GalK.Int2-R | CGTATTGCAGCAGCTTTATC | ||
| ORF32 | ORF32.SB-F | CAGATGTGGCTGGACATGA | 54567–54585 |
| ORF32.SB-R | GCTACCCAGGTGGTGTTGTT | 55006–54987 | |
| ORF59 | ORF59.SB-F | GTTCTGCGTCGAGGATGATG | 102042–102061 |
| ORF59.SB-R | CGTTCAGCACCTCGGTCTAC | 102427–102408 | |
| ORF81 | ORF81.SB-F | AAAGCTCAACTGGCCAAGAG | 150809–150828 |
| ORF81.SB-R | ACACCGCCGTCTTCGAGTAG | 151259–151240 | |
| ORF83 | ORF83.SB-F | TGCTCTCCACGTCCTTGTA | 152838–152856 |
| ORF83.SB-R | ATGTCTCCTTTGTGCGGTC | 153480–153462 | |
| ORF99 | ORF99.SB-F | CCTCCAGACGCAGATCAACT | 184751–184770 |
| ORF99.SB-R | CTTGAGGAAGGGCTGGTTG | 185204–185186 | |
| ORF106 | ORF106.SB-F | GCAGCAGCAGCAGAAGTG | 195783–195800 |
| ORF106.SB-R | TCAGTTGGTCTTGGGGCC | 196013–195996 | |
| ORF115 | ORF115.SB-F | CTACGCCAACGACGAACC | 209426–209443 |
| ORF115.SB-R | AGCACCACGAACCACACTC | 209776–209758 | |
| ORF131 | ORF131.SB-F | ACTGGAGCGGGTGATAGTTG | 225487–225506 |
| ORF131.SB-R | GGACTCGTCGTGCCTCTC | 225937–225920 | |
| PCR | |||
| ORF25 | ORF25int_F | ATCAAGCGCTACGACGACTT | 45948–45967 |
| ORF25int_R | TGTTGCAGGAGGTGTAGACG | 46442–46461 | |
| ORF64 | ORF64int_F | CCATAGTCCAGGACGACGAT | 121431–121450 |
| ORF64int_R | TGCTGCTTGATGTTCTCCAC | 121919–121938 | |
| ORF65 | ORF65int_F | GATGGTCATGTTGGTGTTGC | 122996–123015 |
| ORF65int_R | CCAAGAACGAGCTCTTCACC | 123494–123513 | |
| ORF108 | ORF108int_F | ACCAACTACACGACCGTCTC | 201246–201265 |
| ORF108int_R | CCGGGTTGGTGTAGGTAGAA | 201677–201696 | |
| ORF132 | ORF132int_F | TTGGTTTTTGTTGGTGACGA | 227468–227487 |
| ORF132int_R | TGACGGGTTCCAAGATTAGC | 227936–227955 | |
| ORF136 | ORF136int_F | CTGGTTACATGGGTGGCTTT | 231360–231379 |
| ORF136int_R | TAGCCCCTGTTGTAGGATGC | 231738–231757 | |
| ORF148 | Pr148fw2 | GGTTGTTGGAGTAGTGGTGC | 254196–254215 |
| Pr148rev2 | CTCATTCAGGCTCGGAGAC | 254592–254610 | |
| ORF149 | Pr149fw1 | GTAGTCGCTGGATGTGACG | 256662–256680 |
| Pr149rev1 | GTCAACACGGACTGCTCCG | 257093–257111 | |
| ORF112 | ORF112intFW | CCAGTGTCATCACCACAAGC | 205336–205355 |
| ORF112intREV | ACGGACATCCTGGGTATCAA | 205571–205590 | |
| qPCR | |||
| CyHV-3 ORF89 | KHV-86F | GACGCCGGAGACCTTGTG | AF411803 |
| KHV-163R | CGGGTTCTTATTTTTGTCCTTGTT | ||
| KHV-109P | (6FAM) CTTCCTCTGCTCGGCGAGCACG (BHQ1) | ||
| Carp glucokinase | CgGluc-162F | ACTGCGAGTGGAGACACATGAT | AF053332 |
| CgGluc-230R | TCAGGTGTGGAGCGGACAT | ||
| CgGluc-185P | (6FAM) AAGCCAGTGTCAAAATGCTGCCCACT (BHQ1) |
Coordinates are listed in relation to GenBank accession number NC_009127.1.
Underlined: segments correspond to the CyHV-3 sequence; italicization indicates sequences corresponding to galK sequence. Mutated nucleotides are both underlined and italicized.
Production of CyHV-3 FL BAC recombinant plasmids and CyHV-3 recombinant viruses using BAC cloning and prokaryotic recombination technologies.
CyHV-3 FL BAC recombinant plasmids and CyHV-3 recombinant viruses were produced on the basis of the strategy outlined in Fig. 2 and 8, by using a two-step galactokinase gene (galK) positive/negative selection in bacteria (15, 23). Single-gene deleted recombinant plasmids were produced for various ORFs by replacing the sequence (initiation codon to stop codon) by a galK expression cassette (galK positive selection). Recombination cassettes encoding galK were produced by PCR using the primers listed in Table 3 and the pgalK vector as the template. The derived ORFX Del galK cassette consisted of the galK gene flanked by 75-bp sequences homologous to the regions of the CyHV-3 genome immediately upstream and downstream of ORFX. The flanking sequences served as targets to induce homologous recombination with the FL BAC parental plasmid in competent E. coli cells, resulting in replacement of the entire ORF by the galK expression cassette (FL BAC ORFX Del galK plasmid).
The recombinant FL BAC plasmids were transfected into permissive CCB cells in the presence of polyethylenimine (PEI). Transfection of the BAC plasmids and eventual reconstitution of infectious virus were monitored by epifluorescence microscopy, since the FL BAC plasmids and derived viruses contained an EGFP expression cassette. For mutations that did not prevent reconstitution of infectious virus (i.e., in nonessential genes), cell supernatants were collected and the viruses were amplified, leading to FL BAC recovered ORFX Del galK viruses. For mutations that prevented reconstitution of infectious virus (i.e., in essential genes), further investigations were performed. For each ORF identified as being essential for viral growth, WT revertant (Rev1) and nonsense (NS) mutation clones were produced by using a second recombination process (galK negative selection). ORFX Rev1 and ORFX NS recombination cassettes were produced by PCR using WT FL BAC DNA and pGEMT-ORFX NS as the templates, respectively (see Table 3 for primers). The resulting FL BAC ORFX Rev1 and FL BAC ORF X NS plasmids were transfected into CCB cells as described above. To exclude the possibility that the lack of infectivity of the NS-null recombinant BAC plasmids was due to unexpected mutations during BAC manipulation, FL BAC ORFX NS plasmids were also transfected into CCB cells together with a WT DNA fragment containing the appropriate ORF and upstream and downstream flanking regions, in order to induce reversion to a WT sequence after homologous recombination in eukaryotic cells.
Genetic characterization of CyHV-3 recombinant BAC plasmids and derived recombinant viruses.
CyHV-3 FL BAC recombinant plasmids and derived recombinant viruses were characterized by sequencing regions of interest and by assessing the sizes of SacI fragments by agarose gel electrophoresis and Southern blotting (24). Probes for Southern blotting were produced by PCR using the primers listed in Table 3. CyHV-3 recombinant viruses deleted for nonessential genes (the FL BAC recovered ORFX Del galK viruses and the FL BAC revertant ORF25 Del galK virus) were characterized further by full-length genome sequencing as described previously (15).
Indirect immunofluorescence staining.
Cells grown on glass coverslips were fixed in phosphate-buffered saline (PBS) containing 4% (wt/vol) paraformaldehyde at 4°C for 15 min and then 20°C for 10 min. After a washing step with PBS, samples were permeabilized in PBS containing 0.1% (vol/vol) NP-40 at 37°C for 15 min (15). Immunofluorescence staining (incubation and washes) was performed in PBS containing 10% (vol/vol) fetal calf serum (FCS). Monospecific mouse pAbs (dilution 1:100) against CyHV-3 ORF148 or ORF149 were used as the primary antibody, followed by Alexa Fluor 568 goat anti-mouse immunoglobulin G(H+L) (Invitrogen) as the secondary antibody (1:1,000 dilution). After a washing step, the cells were mounted by using Prolong Gold antifade reagent with DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen). Samples were analyzed by confocal microscopy using a Leica SP5 confocal microscope (14).
Multistep growth curves.
Triplicate cultures of CCB cells were infected with CyHV-3 viruses at an MOI of 0.05 PFU/cell. After incubation for 2 h, the cells were washed with PBS and overlaid with Dulbecco modified Eagle medium (DMEM) containing 4.5 g/liter glucose and 10% (vol/vol) FCS. The supernatant was removed from the infected cultures at successive intervals (2, 4, and 6 dpi) and stored at −80°C. Titers of infectious viral particles were determined by duplicate plaque assays in CCB cells (24).
Plaque size assay.
CCB cells grown in 6-well plates were infected with viruses at an MOI of 200 PFU/well. After incubation for 2 h, the cells were overlaid with DMEM containing 10% (vol/vol) FCS and 0.6% (wt/vol) carboxymethyl cellulose (medium viscosity; Sigma) in order to obtain isolated plaques (25). At successive intervals after infection, the plaques were fixed in PBS containing 4% (wt/vol) paraformaldehyde at 4°C for 15 min and then 20°C for 10 min. Images were captured with a confocal microscope (Nikon A1R). For each virus, 25 plaques were measured in duplicate wells. The plaque area was measured by using ImageJ 1.46 software (26).
Fish.
Common carp (Cyprinus carpio) were kept in 60-liter tanks at 24°C. Water parameters were checked twice per week. The health of the fish was confirmed by microbiological, parasitic, and clinical examination immediately prior to the experiments, all of which were preceded by an acclimation period of at least 2 weeks.
Inoculation of fish with CyHV-3.
Two modes of inoculation were used: immersion in infectious water and cohabitation with newly infected fish. For inoculation by immersion, the fish were immersed for 2 h in constantly aerated water containing virus, the volume of water being adjusted to a biomass of around 10% on the basis of fish size and number. At the end of the incubation period, the fish were returned to 60-liter tanks. For inoculation by cohabitation, naive fish were immersed for 2 h in water containing 400 PFU/ml of the FL or M3 strain of CyHV-3. The newly infected fish were then released into tanks at a ratio of 2 newly infected fish per 20 fish to be infected.
Ethics statement.
The maintenance and care of fish and the experiments conducted complied with the guidelines of the European Convention for the Protection of Vertebrate Animals Used for Experimental and other Scientific Purposes (CETS 123). The animal studies were approved by the local ethics committee of the University of Liège, Belgium (laboratory accreditation 1610008, protocol 1414). All efforts were made to minimize suffering.
Quantification of virus genome copies in organs by real-time TaqMan PCR.
The virus genome was quantified by real-time TaqMan PCR as described previously (24), by amplifying fragments of the CyHV-3 ORF89 and carp glucokinase genes. The primers and probes are listed in Table 3.
Statistical analysis.
Viral growth (Fig. 5A) and plaque size (Fig. 5B) results were compared by using two-way analysis of variance (ANOVA) with interactions, followed by a post hoc Student t test. qPCR results (Fig. 9) were compared using one-way ANOVA, followed by a post hoc Student t test. Survival curves (Fig. 6 and 10) were compared by using log-rank tests. These analyses were done by using GraphPad Prism 5. The variables used and comparisons retained for statistical illustrations are presented in the figure legends. Statistical significance is represented in the figures by asterisks as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
ACKNOWLEDGMENTS
C.V. is a research fellow of the Fonds National Belge de la Recherche Scientifique (FNRS). This study was supported by the University of Liège (ARC15/19-12), the Belgian Science Policy (Belspo) (BELVIR IAP7/45), the FNRS (grant 2877824), and the Medical Research Council (MC_UU_12014/3).
We are grateful to Cédric Delforge, Emeline Deglaire, Aurélie Vanderlinden, Justine Javaux, and Lorène Dams for excellent technical assistance. We are grateful to S. Bergman, M. Kotler, F. Lieffrig, and K. Way for providing CyHV-3 strains.
REFERENCES
- 1.Food and Agriculture Organization of the United Nations. 2015. Global aquaculture production, 1950–2013. FAO, Rome, Italy: http://www.fao.org/fishery/statistics/global-aquaculture-production/query/fr/en/. [Google Scholar]
- 2.Ilouze M, Dishon A, Kotler M. 2006. Characterization of a novel virus causing a lethal disease in carp and koi. Microbiol Mol Biol Rev 70:147–156. doi: 10.1128/MMBR.70.1.147-156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakus KL, Ouyang P, Boutier M, Ronsmans M, Reschner A, Vancsok C, Jazowiecka-Rakus J, Vanderplasschen A. 2013. Cyprinid herpesvirus 3: an interesting virus for applied and fundamental research. Vet Res 44:85. doi: 10.1186/1297-9716-44-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutier M, Ronsmans M, Rakus K, Jazowiecka-Rakus J, Vancsok C, Morvan L, Peñaranda MMD, Stone DM, Way K, van Beurden SJ, Davison AJ, Vanderplasschen A. 2015. Cyprinid herpesvirus 3: an archetype of fish alloherpesviruses. Adv Virus Res 93:161–256. doi: 10.1016/bs.aivir.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Aoki T, Hirono I, Kurokawa K, Fukuda H, Nahary R, Eldar A, Davison AJ, Waltzek TB, Bercovier H, Hedrick RP. 2007. Genome sequences of three koi herpesvirus isolates representing the expanding distribution of an emerging disease threatening koi and common carp worldwide. J Virol 81:5058–5065. doi: 10.1128/JVI.00146-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison AJ, Kurobe T, Gatherer D, Cunningham C, Korf I, Fukuda H, Hedrick RP, Waltzek TB. 2013. Comparative genomics of carp herpesviruses. J Virol 87:2908–2922. doi: 10.1128/JVI.03206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waltzek TB, Kelley GO, Stone DM, Way K, Hanson L, Fukuda H, Hirono I, Aoki T, Davison AJ, Hedrick RP. 2005. Koi herpesvirus represents a third cyprinid herpesvirus (CyHV-3) in the family Herpesviridae. J Gen Virol 86:1659–1667. doi: 10.1099/vir.0.80982-0. [DOI] [PubMed] [Google Scholar]
- 8.Miyazaki T, Kuzuya Y, Yasumoto S, Yasuda M, Kobayashi T. 2008. Histopathological and ultrastructural features of Koi herpesvirus (KHV)-infected carp Cyprinus carpio, and the morphology and morphogenesis of KHV. Dis Aquat Organ 80:1–11. doi: 10.3354/dao01929. [DOI] [PubMed] [Google Scholar]
- 9.Michel B, Leroy B, Stalin Raj V, Lieffrig F, Mast J, Wattiez R, Vanderplasschen AF, Costes B. 2010. The genome of cyprinid herpesvirus 3 encodes 40 proteins incorporated in mature virions. J Gen Virol 91:452–462. doi: 10.1099/vir.0.015198-0. [DOI] [PubMed] [Google Scholar]
- 10.Yi Y, Zhang H, Lee X, Weng S, He J, Dong C. 2014. Extracellular virion proteins of two Chinese CyHV-3/KHV isolates, and identification of two novel envelope proteins. Virus Res 191:108–116. doi: 10.1016/j.virusres.2014.07.034. [DOI] [PubMed] [Google Scholar]
- 11.Lete C, Palmeira L, Leroy B, Mast J, Machiels B, Wattiez R, Vanderplasschen A, Gillet L. 2012. Proteomic characterization of bovine herpesvirus 4 extracellular virions. J Virol 86:11567–11580. doi: 10.1128/JVI.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Beurden SJ, Leroy B, Wattiez R, Haenen OL, Boeren S, Vervoort JJ, Peeters BP, Rottier PJ, Engelsma MY, Vanderplasschen AF. 2011. Identification and localization of the structural proteins of anguillid herpesvirus 1. Vet Res 42:105. doi: 10.1186/1297-9716-42-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidick S, Leroy B, Palmeira L, Machiels B, Mast J, François S, Wattiez R, Vanderplasschen A, Gillet L. 2013. Proteomic characterization of murid herpesvirus 4 extracellular virions. PLoS One 8:e83842. doi: 10.1371/journal.pone.0083842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costes B, Fournier G, Michel B, Delforge C, Raj VS, Dewals B, Gillet L, Drion P, Body A, Schynts F, Lieffrig F, Vanderplasschen A. 2008. Cloning of the koi herpesvirus genome as an infectious bacterial artificial chromosome demonstrates that disruption of the thymidine kinase locus induces partial attenuation in Cyprinus carpio koi. J Virol 82:4955–4964. doi: 10.1128/JVI.00211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boutier M, Ronsmans M, Ouyang P, Fournier G, Reschner A, Rakus K, Wilkie GS, Farnir F, Bayrou C, Lieffrig F, Li H, Desmecht D, Davison AJ, Vanderplasschen A. 2015. Rational development of an attenuated recombinant cyprinid herpesvirus 3 vaccine using prokaryotic mutagenesis and in vivo bioluminescent imaging. PLoS Pathog 11:e1004690. doi: 10.1371/journal.ppat.1004690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neukirch M, Böttcher K, Bunnajrakul S. 1999. Isolation of a virus from koi with altered gills. Bull Eur Assoc Fish Pathol 19:221–224. [Google Scholar]
- 17.Torrent F, Villena A, Lee PA, Fuchs W, Bergmann SM, Coll JM. 2016. The amino-terminal domain of ORF149 of koi herpesvirus is preferentially targeted by IgM from carp populations surviving infection. Arch Virol 161:2653–2665. doi: 10.1007/s00705-016-2934-4. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Lee X, Weng S, He J, Dong C. 2015. Whole-genome sequence of a novel Chinese cyprinid herpesvirus 3 isolate reveals the existence of a distinct European genotype in East Asia. Vet Microbiol 175:185–194. doi: 10.1016/j.vetmic.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 19.Ronen A, Perelberg A, Abramowitz J, Hutoran M, Tinman S, Bejerano I, Steinitz M, Kotler M. 2003. Efficient vaccine against the virus causing a lethal disease in cultured Cyprinus carpio. Vaccine 21:4677–4684. doi: 10.1016/S0264-410X(03)00523-1. [DOI] [PubMed] [Google Scholar]
- 20.Costes B, Raj VS, Michel B, Fournier G, Thirion M, Gillet L, Mast J, Lieffrig F, Bremont M, Vanderplasschen A. 2009. The major portal of entry of koi herpesvirus in Cyprinus carpio is the skin. J Virol 83:2819–2830. doi: 10.1128/JVI.02305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leroy B, Roupie V, Noel-Georis I, Rosseels V, Walravens K, Govaerts M, Huygen K, Wattiez R. 2007. Antigen discovery: a postgenomic approach to paratuberculosis diagnosis. Proteomics 7:1164–1176. doi: 10.1002/pmic.200600988. [DOI] [PubMed] [Google Scholar]
- 22.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Rakus K, Ronsmans M, Forlenza M, Boutier M, Piazzon MC, Jazowiecka-Rakus J, Gatherer D, Athanasiadis A, Farnir F, Davison AJ, Boudinot P, Michiels T, Wiegertjes GF, Vanderplasschen A. 2017. Conserved fever pathways across vertebrates: a herpesvirus expressed decoy TNF-α receptor delays behavioral fever in fish. Cell Host Microbe 21:244–253. doi: 10.1016/j.chom.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouyang P, Rakus K, Boutier M, Reschner A, Leroy B, Ronsmans M, Fournier G, Scohy S, Costes B, Wattiez R, Vanderplasschen A. 2013. The IL-10 homologue encoded by cyprinid herpesvirus 3 is essential neither for viral replication in vitro nor for virulence in vivo. Vet Res 44:53. doi: 10.1186/1297-9716-44-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanderplasschen A, Bublot M, Dubuisson J, Pastoret P-P, Thiry E. 1993. Attachment of the gammaherpesvirus bovine herpesvirus 4 is mediated by the interaction of gp8 glycoprotein with heparinlike moieties on the cell surface. Virology 196:232–240. doi: 10.1006/viro.1993.1471. [DOI] [PubMed] [Google Scholar]
- 26.Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int 11:36–42. [Google Scholar]