ABSTRACT
Elicitation of broadly neutralizing antibody (bNAb) responses is a major goal for the development of an HIV-1 vaccine. Current HIV-1 envelope glycoprotein (Env) vaccine candidates elicit predominantly tier 1 and/or autologous tier 2 virus neutralizing antibody (NAb) responses, as well as weak and/or sporadic cross-reactive tier 2 virus NAb responses with unknown specificity. To delineate the specificity of vaccine-elicited cross-reactive tier 2 virus NAb responses, we performed single memory B cell sorting from the peripheral blood of a rhesus macaque immunized with YU2gp140-F trimers in adjuvant, using JR-FL SOSIP.664, a native Env trimer mimetic, as a sorting probe to isolate monoclonal Abs (MAbs). We found striking genetic and functional convergence of the SOSIP-sorted Ig repertoire, with predominant VH4 or VH5 gene family usage and Env V3 specificity. Of these vaccine-elicited V3-specific MAbs, nearly 20% (6/33) displayed cross-reactive tier 2 virus neutralization, which recapitulated the serum neutralization capacity. Substantial similarities in binding specificity, neutralization breadth and potency, and sequence/structural homology were observed between selected macaque cross-reactive V3 NAbs elicited by vaccination and prototypic V3 NAbs derived from natural infections in humans, highlighting the convergence of this subset of primate V3-specific B cell repertories. Our study demonstrated that cross-reactive primary virus neutralizing B cell lineages could be elicited by vaccination as detected using a standardized panel of tier 2 viruses. Whether these lineages could be expanded to acquire increased breadth and potency of neutralization merits further investigation.
IMPORTANCE Elicitation of antibody responses capable of neutralizing diverse HIV-1 primary virus isolates (designated broadly neutralizing antibodies [bNAbs]) remains a high priority for the vaccine field. bNAb responses were so far observed only in response to natural infection within a subset of individuals. To achieve this goal, an improved understanding of vaccine-elicited responses, including at the monoclonal Ab level, is essential. Here, we isolated and characterized a panel of vaccine-elicited cross-reactive neutralizing MAbs targeting the Env V3 loop that moderately neutralized several primary viruses and recapitulated the serum neutralizing antibody response. Striking similarities between the cross-reactive V3 NAbs elicited by vaccination in macaques and natural infections in humans illustrate commonalities between the vaccine- and infection-induced responses to V3 and support the feasibility of exploring the V3 epitope as a HIV-1 vaccine target in nonhuman primates.
KEYWORDS: nonhuman primate, human immunodeficiency virus, neutralizing antibodies, vaccines
INTRODUCTION
Primary circulating HIV-1 variants that dominate viral transmission in humans are typically highly resistant to antibody (Ab)-mediated neutralization and are referred to as tier 2 viruses. A successful HIV-1 vaccine is expected to elicit a broadly neutralizing antibody (bNAb) response that can potently neutralize tier 2 viruses, a goal yet to be met by current vaccine regimens. The HIV-1 envelope glycoproteins (Env) mediate viral entry into the target cell. Env is expressed as a gp160 precursor which, following proteolytic cleavage catalyzed by a cellular protease, is composed of two subunits: the membrane-spanning protein, gp41, and the external subunit, gp120, which are noncovalently linked and form trimers on the surface of the HIV-1 particle (1). As the only target of neutralizing antibodies, the HIV-1 Env functional spike has been the focus of vaccine design for decades.
Various HIV Env-based vaccine candidates have been studied previously. Besides monomeric gp120, the early generation of Env trimer, designated gp140-F or gp140(-/FT) (2), is an uncleaved form of the gp160 ectodomain appended with a heterologous trimerization motif derived from T4 bacteriophage fibritin. The uncleaved gp140-F trimer molecule samples an opened trimer configuration and thus can be recognized by both bNAbs and nonneutralizing Abs, an undesirable feature for selectively eliciting bNAbs in vivo (3). A different Env trimer, designated SOSIP.664, consists of a genetically engineering disulfide bond linkage at the interface of gp120-gp41, an I559P mutation to maintain the gp41 subunits in their prefusion form, and truncation at residue 664 to improve trimer solubility (4–7). SOSIP.664 is a cleaved and well-ordered trimer, mimicking the native form of the Env trimer. Alternatively, a glycine-serine-based (G4S) native flexible peptide linker (NFL) was introduced to replace the furin cleavage site at the gp120-gp41 junction, and it forms a native-like albeit cleavage-independent trimer (8, 9). In general, the well-ordered trimers (e.g., SOSIP or NFL) preferentially present neutralizing epitopes over nonneutralizing ones, in contrast to the uncleaved gp140-F trimers. Despite the remarkable progress in developing trimeric Env immunogens, most current experimental vaccines, including the uncleaved gp140-F, the cleaved and well-ordered SOSIP.664 trimers or NFL trimers in single-chain form, and cleaved trimers in membrane-bound form, so far elicit only tier 1 virus and autologous tier 2 virus NAb responses at moderate or high titers (10–14).
Although bNAbs from chronically infected individuals have been well characterized, isolation of monoclonal antibodies (MAbs) from vaccinated animals was pursued only more recently. These studies revealed the specificities of neutralizing antibody responses in Env-vaccinated animals by isolating MAbs that recapitulate the plasma neutralizing capacity. The determined neutralizing specificities are mostly restricted to tier 1 viruses (10, 15–18) or autologous tier 2 viruses (12, 19, 20). Thus, characterization of vaccine-elicited tier 2 virus NAb responses at a clonal level would provide an improved understanding of Env-specific Ab repertoires induced by vaccination, which is more relevant for future translational vaccine studies, in contrast to the characterization of infection-induced bNAbs pursued extensively in previous studies.
Previously we conducted an HIV-1 Env immunogenicity experiment in rhesus macaques, using gp140-F trimers derived from clade B primary isolate YU2 as a model immunogen to examine the adjuvant effect of saponin-based AbISCO in combination with Toll-like receptor 9 (TLR9) agonist CpG-C ODN (21). The plasma of the immunized animals displayed robust tier 1 virus neutralizing responses but also low titer neutralizing responses against the autologous tier 2 virus (YU2) and several heterologous (cross-reactive) tier 2 viruses (21). In the present study, we set out to define the cross-reactive tier 2 virus NAb response elicited by this vaccination at the MAb level to guide future vaccine design.
To understand the specificities and genetic compositions of the tier 2 virus neutralizing Ab responses elicited in this study, we employed fluorescence-activated cell sorting (FACS)-based single B cell sorting using a SOSIP.664 trimer derived from a heterologous clade B primary isolate JR-FL as the probe. Characterization of a set of tier 2 virus neutralizing MAbs revealed that they targeted predominantly the Env V3 loop, a viral determinant involved in coreceptor binding and critical for HIV-1 virus infectivity (22–24). In addition, selected MAbs shared substantial genetic and structural similarities with the prototypic V3 NAbs derived from natural infections in humans, highlighting the convergence of a subset of primate V3-directed antibodies in infection and vaccination.
RESULTS
Memory B cell sorting using JR-FL SOSIP.664 as a probe.
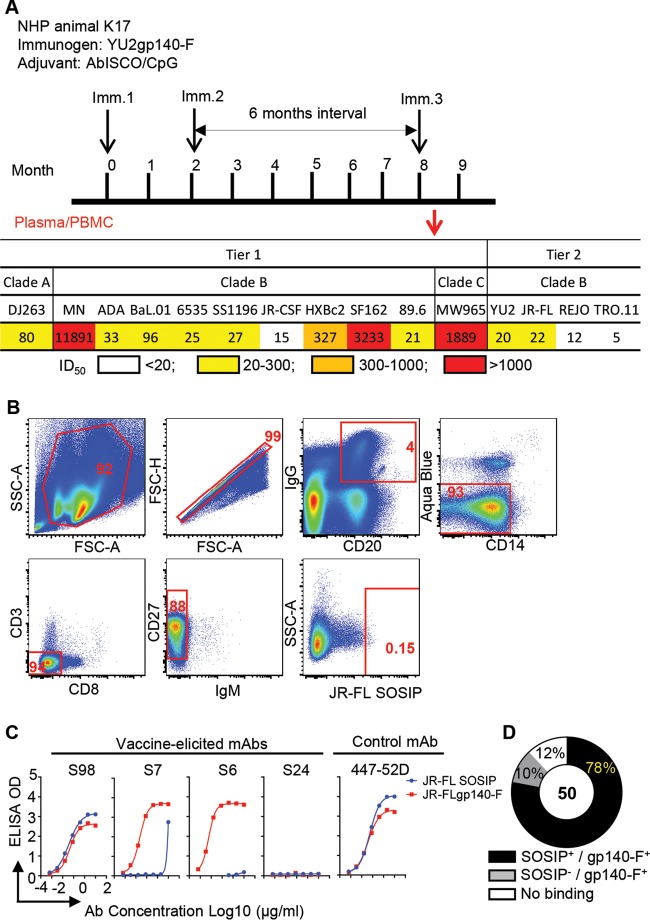
Previously, we performed an immunogenicity study in nonhuman primates (NHPs) using YU2gp140-F, an early-generation HIV-1 Env trimer immunogen, administered three times in adjuvant (21) (Fig. 1A, upper panel). Within the group of 6 animals inoculated with YU2gp140-F and adjuvant AbISCO+CpG, animal K17 showed a representative immunogenicity outcome, which was selected for memory B cell sorting and cloning. Plasma collected from animal K17 2 weeks after the third immunization (Imm 3) displayed robust tier 1 virus neutralizing activity but also moderate neutralization titers against various HIV-1 tier 2 viruses (Fig. 1A, lower panel). To define the cross-reactive tier 2 virus neutralizing B cell response in macaque K17, we sorted antigen-specific memory B cells by fluorescence-activated cell sorting (FACS) and applied Ig cloning methods optimized for NHP B cell repertoire analysis (10, 15). We employed a heterologous Env trimer, JR-FL SOSIP.664, a native clade B Env spike mimetic derived from primary isolate JR-FL (25), as the sorting probe. The JR-FL SOSIP.664 trimers selectively bind bNAbs, which should diminish isolation of non-bNAbs (25).
FIG 1.
JR-FL SOSIP-sorted single memory B cells from K17 and expressed MAb binding profiles. (A) Schematic presentation of the immunization schedule and plasma neutralization activity of the immunized rhesus macaque. Upper panel, immunization/sampling schedule. Rhesus macaques were inoculated with HIV-1 Env YU2gp140-F trimer 3 times (black arrows) using a 6-month interval between the second and the third immunizations (21). Plasma and PBMCs from animal K17 were collected 2 weeks after the third inoculation and subjected to virus neutralization assay and FACS sorting (red arrow). Lower panel, HIV-1 neutralizing activity of K17 plasma (21). Data are shown as the reciprocal plasma dilution giving 50% virus entry inhibition (ID50 titer). A color code is used to illustrate the plasma ID50 neutralization titer. (B) Single-cell sorting for JR-FL SOSIP-specific memory B cells. IgG+ memory B cells were defined as CD3− CD8− Aqua Blue− CD14− CD20+ IgG+ CD27+ IgM−. JR-FL SOSIP-specific memory B cells were then gated by phenotype of JR-FL SOSIPhi. The gate frequency (percent) of the parent population is depicted in red. (C) Specificities of binding of the expressed MAbs to the sorting probe JR-FL SOSIP.664 (blue circles) or uncleaved Env trimer JR-FLgp140-F (red squares) via ELISAs. (D) Binding specificity summary for expressed JR-FL SOSIP-sorted MAbs.
We stained peripheral blood mononuclear cells (PBMCs) collected 2 weeks after the third immunization (Imm 3.2) of monkey K17 and sorted cross-reactive Env+ IgG+ memory B cells with the phenotype CD20+ IgG+ Aqua Blue− CD14− CD3− CD8− CD27+ IgM− JR-FL SOSIP.664hi (Fig. 1B). JR-FL-reactive Env+ memory B cells accounted for approximately 0.15% of IgG+ B cells (Fig. 1B), which was lower than the Env+ memory B cell frequency (∼5%) sorted by the autologous antigen, YU2gp140-F, reported previously (15). This suggested that JR-FL SOSIP.664 was a more selective probe than the probes deployed previously.
We recovered the variable domains of the Ig heavy- and light-chain genes of 173 individual JR-FL SOSIP.664-sorted Env+ memory B cells (Table 1) by single-cell PCR. Based on phylogenetic analysis of the isolated antibody Ig heavy- and light-chain sequences (data not shown), we removed the redundant clones with identical sequences and retained 107 unique heavy-chain sequences representing 80 unique clones with matched light chains. We performed clonal lineage assignment for the 80 unique clones and assigned them to 46 clonal lineages, using the criteria that clones with (i) the same VJ segment usage, (ii) the same CDR3 length, and (iii) high nucleotide sequence homology in CDR3 (>75%) are likely derived from the same naive ancestor B cell and thus belong to the same clonal lineage. With each clonal lineage represented with at least one MAb, we expressed 50 MAbs as full-length (FL) IgG1 as described previously for further characterization (10, 15).
TABLE 1.
Properties of the JR-FL SOSIP-specific memory B cell flow cytometric sorts
| Parameter | Value for animal K17 |
|---|---|
| Plasma collection time | 2 wk after immunization 3 |
| Probe | JR-FL SOSIP.664, negatively selected |
| No. of: | |
| Total PBMC | 4 million |
| Memory B cells | |
| Total | 0.11 million |
| Sorted SOSIP+ | 173 |
| Amplified heavy-chain sequences | 118 |
| Productive Ab sequences with matching heavy and light chains | 80 |
| Clonal types based on heavy-chain sequences | 46 |
| Expressed MAbs | |
| Total | 50 |
| Binding to JR-FLgp140-F | 44 |
| Binding to JR-FL SOSIP | 39 |
We used two Env ligands to evaluate the Env binding specificity of the expressed MAbs: the JR-FL SOSIP.664 probe, which preferentially binds bNAbs, and the uncleaved Env trimer mimic JR-FLgp140-F, which binds both bNAbs and non-bNAbs. The binding curves of representative MAbs are shown in Fig. 1C, with the binding specificities of JR-FL SOSIP.664-specific MAbs summarized in Table 2. Most of the MAbs, such as S98, bound both JR-FL SOSIP.664 and JR-FLgp140-F (Fig. 1C), while some MAbs, such as S7, bound JR-FLgp140-F well but JR-FL SOSIP.664 only weakly (Fig. 1C). Of the 50 cloned MAbs, 39 (78%) bound to the sorting probe JR-FL SOSIP.664 as well as to the uncleaved Env trimer JR-FLgp140-F (Fig. 1D). Five of the 50 clones (10%), including S6, bound uncleaved JR-FLgp140-F but not JR-FL SOSIP.664 (Fig. 1C and D), suggesting that a small portion of the JR-FL SOSIP.664 probe possessed less native Env trimer configuration during the cell staining/sorting process. Twelve percent of the MAbs, such as S24, displayed no binding to either of the two Env ligands (Fig. 1C and D). Taken together, these results show that a high proportion of the MAbs (>88%) were Env specific, enabling us to perform further genetic and binding analyses of the JR-FL SOSIP.664-sorted Env+ antibody repertoire.
TABLE 2.
Antigen binding specificities of JR-FL SOSIP-specific MAbs
| MAb | Bindinga |
Specificity | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JR-FL gp140-F | JR-FL SOSIP | gp120 |
V3 peptideb |
||||||||||
| ΔV1V2 | Core | WT | I420R | D368R | ΔV3 | F4 | F5 | F6 | FL | ||||
| S1 | NT | ++ | ++++ | ++++ | ++ | + | ++++ | ++++ | − | +++ | V3 | ||
| S7 | ++++ | + | + | + | + | − | − | − | − | − | V3 dependent | ||
| S12 | ++ | ++ | ++++ | ++++ | + | ++++ | − | − | − | − | CD4bs | ||
| S13 | ++ | ++ | +++ | − | ++++ | ++++ | ++++ | ++++ | − | − | − | − | V1V2 |
| S15 | ++ | ++ | ++++ | ++++ | ++++ | + | + | +++ | − | +++ | V3 | ||
| S18 | ++++ | +++ | ++++ | ++++ | ++++ | − | ++ | +++ | − | +++ | V3 | ||
| S25 | NT | ++++ | NT | NT | NT | NT | +++ | ++++ | + | ++++ | V3 | ||
| S30 | ++ | ++ | ++++ | ++++ | ++++ | − | +++ | ++++ | + | ++++ | V3 | ||
| S33 | ++ | + | ++ | + | ++ | − | − | − | − | − | V3 dependent | ||
| S44 | ++ | ++ | ++++ | ++++ | ++++ | − | ++++ | ++++ | + | ++++ | V3 | ||
| S48 | ++ | ++ | ++++ | ++++ | ++++ | − | ++++ | ++++ | + | ++++ | V3 | ||
| S54 | ++ | ++ | ++++ | ++++ | ++++ | − | +++ | ++++ | + | ++++ | V3 | ||
| S57 | ++++ | + | ++++ | ++++ | ++++ | − | − | +++ | − | +++ | V3 | ||
| S62 | ++ | ++ | ++++ | ++++ | ++++ | − | +++ | +++ | − | +++ | V3 | ||
| S68 | ++ | ++ | ++++ | ++++ | ++++ | − | ++++ | ++++ | − | ++++ | V3 | ||
| S72 | ++++ | +++ | ++++ | ++++ | ++++ | − | − | ++ | − | ++++ | V3 | ||
| S74 | ++ | ++ | ++++ | ++++ | ++++ | + | − | + | − | +++ | V3 | ||
| S75 | ++ | + | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | − | − | − | − | Core |
| S76 | ++ | ++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | − | ++++ | V3 | ||
| S80 | ++++ | +++ | ++++ | ++++ | ++++ | − | − | +++ | +++ | ++++ | V3 | ||
| S81 | NT | ++++ | NT | NT | NT | NT | +++ | ++++ | + | ++++ | V3 | ||
| S87 | ++ | ++ | ++++ | ++++ | ++ | ++++ | − | − | − | − | CD4bs | ||
| S89 | ++++ | +++ | ++ | ++++ | ++++ | − | + | + | − | +++ | V3 | ||
| S96 | ++++ | +++ | ++++ | ++++ | ++++ | − | − | +++ | +++ | ++++ | V3 | ||
| S98 | ++ | ++++ | ++++ | ++++ | ++++ | + | ++++ | +++ | + | ++++ | V3 | ||
| S100 | ++ | ++ | ++++ | ++++ | ++++ | − | ++++ | ++++ | + | ++++ | V3 | ||
| S102 | ++ | ++ | ++++ | ++++ | ++++ | − | +++ | ++++ | + | ++++ | V3 | ||
| S105 | ++ | ++ | ++++ | ++++ | ++ | − | +++ | ++++ | + | +++ | V3 | ||
| S109 | ++ | ++ | ++++ | ++++ | ++++ | − | +++ | ++++ | + | ++++ | V3 | ||
| S110 | + | + | + | + | + | + | NT | NT | NT | NT | unknown | ||
| S118 | ++ | ++ | ++++ | ++++ | ++++ | − | +++ | ++++ | + | ++++ | V3 | ||
| S119 | ++ | ++ | ++++ | ++++ | ++++ | − | ++++ | ++++ | + | +++ | V3 | ||
| S124 | ++++ | +++ | ++ | ++++ | ++++ | − | − | +++ | − | ++++ | V3 | ||
| S128 | ++++ | ++ | ++++ | ++++ | ++++ | − | +++ | +++ | + | +++ | V3 | ||
| S134 | ++ | ++ | ++++ | ++++ | ++++ | − | +++ | ++++ | − | ++++ | V3 | ||
| S138 | ++ | ++ | ++++ | ++++ | ++++ | − | ++++ | +++ | + | +++ | V3 | ||
| S146 | + | + | + | + | + | + | NT | NT | NT | NT | unknown | ||
| S147 | ++ | + | ++++ | ++++ | ++++ | − | + | + | − | +++ | V3 | ||
| S152 | ++ | ++ | ++++ | ++++ | ++++ | − | + | ++++ | + | ++++ | V3 | ||
++++, OD450 ≥ 3 and EC50 ≤ 0.1; +++, OD450 ≥ 3 and EC50 > 0.1; ++, 3 > OD450 ≥ 1; +, 1.0 > OD450 ≥ 0.2; −, OD450 < 0.2; NT, not tested.
FL, full-length V3; F4, F5, and F6, fragments 4 to 6, respectively.
Genetic analysis of JR-FL SOSIP.664-sorted Env+ antibody Ig repertoire.
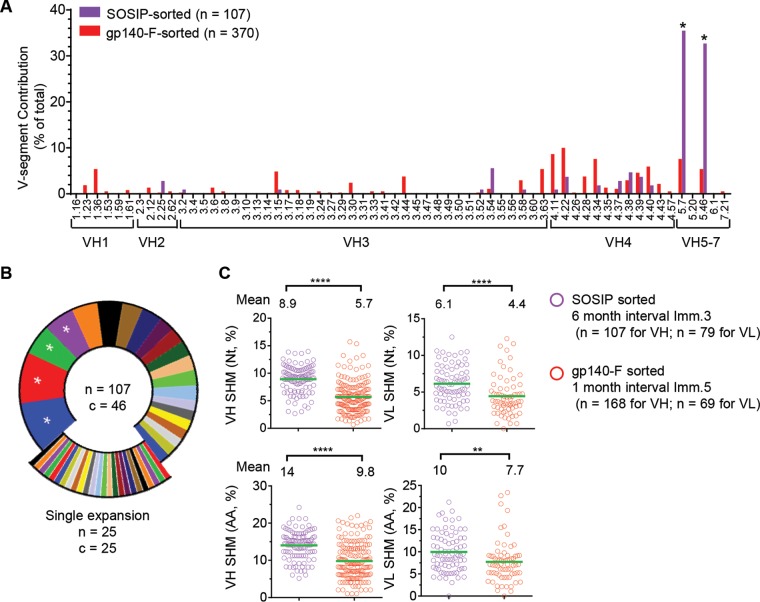
To assess the genetic composition of the JR-FL SOSIP.664-sorted antibody repertoire, we analyzed the heavy-chain V gene usage of the 107 sequences and compared this with the antibody repertoire observed by sorting Env-specific memory B cells from two rhesus macaques (F125 and F128) using a YU2gp140-F probe as previously reported (15). We found that the JR-FL SOSIP-sorted Env+ antibody repertoire was relatively focused, with skewed usage of VH5.7 and VH5.46 gene segments (Fig. 2A) (*, P < 0.05 by two-way analysis of variance [ANOVA]), in contrast to the much-diversified gene segment usage of the YU2gp140-F-sorted Ig repertoire reported in our previous study (15, 26). This was likely the result of the use of a more selective and heterologous JR-FL SOSIP.664 probe in this study.
FIG 2.
Genetic features of the JR-FL SOSIP-sorted Ig repertoire. (A) V gene segment contribution of the total number of JR-FL SOSIP-sorted sequences (purple) compared to previously reported (15) YU2gp140-F-sorted sequences (red). VH5.7 and VH5.46, overexpressed in the JR-FL SOSIP-specific Ig repertoire, are marked with asterisks. Two-way ANOVA was used for statistical analysis. *, P < 0.05. (B) Clonotype analysis of the JR-FL SOSIP-sorted Ig repertoire defined using the criteria of same V and J gene usage and identical CDR3 length with CDR3 nucleotide homology of >75%. A total of 107 unique VH sequences were recovered from the JR-FL SOSIP-sorted cells (n = 107), classified into 46 clonal lineages (c = 46). Predominant lineages representing >5% of the sequences are marked with asterisks, and lineages represented by a single sequence (n = 25) are illustrated as exploded portions. (C) The JR-FL SOSIP-sorted Ig repertoire (from immunization with a longer immunization interval) displayed a higher level of VH/VL SHM than the YU2gp140-F-specific Ig repertoire (from immunization with a shorter immunization interval). The SHM of the JR-FL SOSIP-sorted Ig repertoire heavy (left, VH)- and light (right, VL)-chain V gene was compared with that of the YU2gp140-F-specific Ig repertoire (15) at the nucleotide (Nt) (upper panel) or amino acid (AA) (lower panel) level. Statistical differences in SHM were evaluated using the t test. **, P < 0.01; ****, P < 0.0001.
We further analyzed the clonality of the JR-FL SOSIP-sorted VH sequences and found that 25 out of 46 lineages (54%) were represented by single sequences (Fig. 2B), which is comparable to the gp140-F- or CD4bs-specific repertoire in our previous report (15). Interestingly, the majority of the JR-FL SOSIP-specific clones (90%) used λ chains (Table 3).
TABLE 3.
Genetic properties of JR-FL SOSIP-specific MAbsa
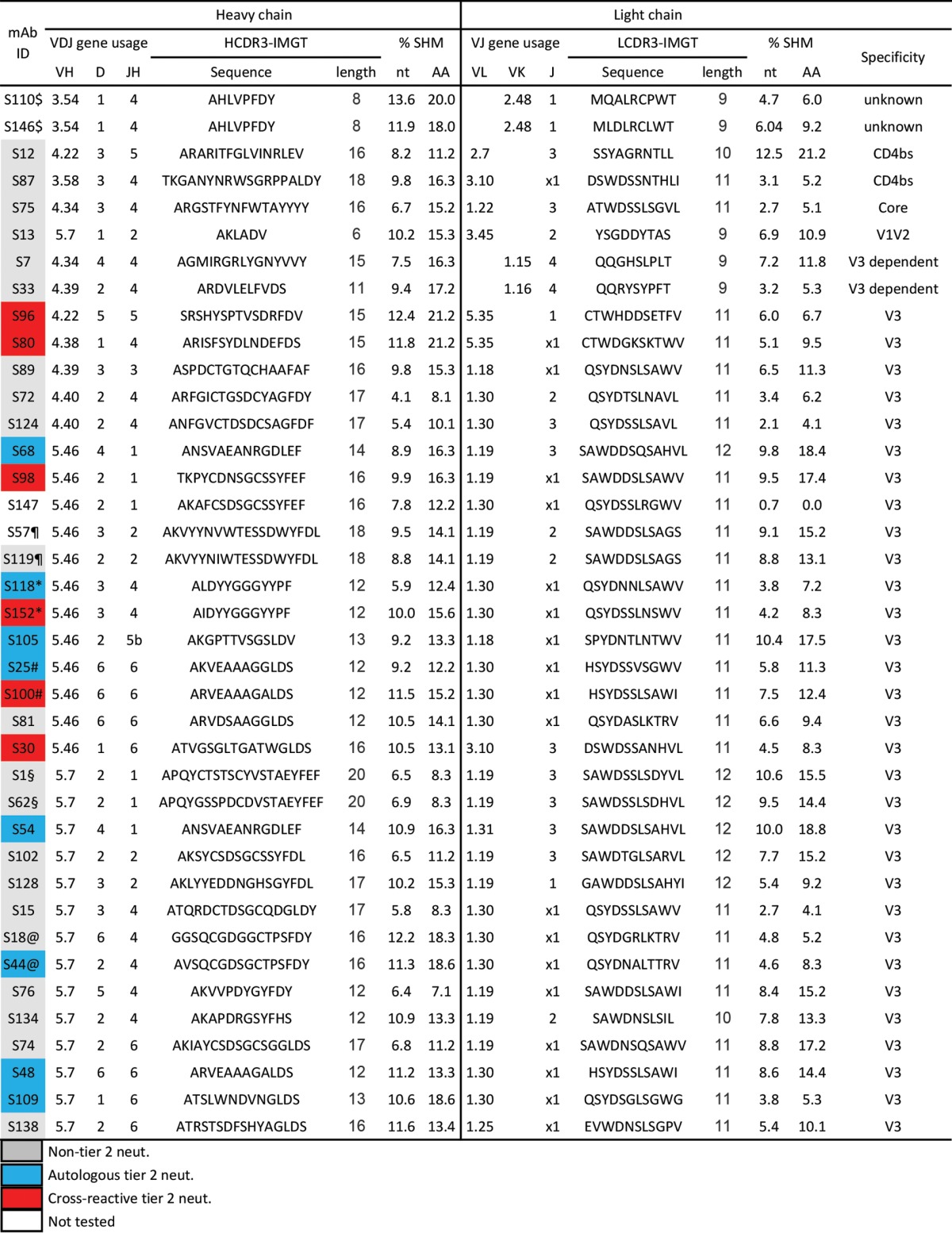
Ab heavy- and light-chain sequences with same V and J gene usage and identical CDR3 length with >75% CDR3 identity at the nucleotide level were identified as variants from the same clonal lineage and are indicated as follows: $, S110 and S146; ¶, S57 and S119; §, S1 and S62; @, S18 and S44; #, S25 and S100; *, S118 and S152. Gray, non-tier 2 neutralizer; blue, autologous tier 2 neutralizer; red, cross-reactive tier 2 neutralizer; white, not tested.
The accumulation of somatic hypermutation (SHM) is important for antibody affinity maturation and virus neutralization (15, 16, 27–29). The SHM levels of the JR-FL SOSIP-sorted Env+ antibody Ig repertoire heavy chain and light chain were approximately 8.9% at the nucleotide level and 14% at the amino acid residue level for the heavy chain and 6.1% at the nucleotide level and 10% at the amino acid residue level for the light chain (Fig. 2C). The SHM of the JR-FL SOSIP-sorted clones was significantly higher than that of the gp140-F-sorted Env+ Ig repertoire reported previously (Fig. 2C) (****, P < 0.0001; **, P < 0.01 [t test]). This may relate to the fact that the JR-FL SOSIP-sorted Env+ memory B cells were isolated from a macaque that was immunized 3 times (months 0, 2, and 8) with a 6-month interval between the second and third immunizations, while the gp140-F-sorted Env+ Ig repertoire was from two macaques (F125 and F128) immunized 5 times with a 1- or 1.5-month interval between each immunization (15). Our data using SHM as the indicator of antibody affinity maturation suggested that a longer interval between immunizations, similar to the successful hepatitis B vaccination schedule, may elicit an antibody response of superior quality compared to that when shorter intervals are used (e.g., 1 month).
MAbs isolated from JR-FL SOSIP-sorted B cells predominantly target the V3 loop.
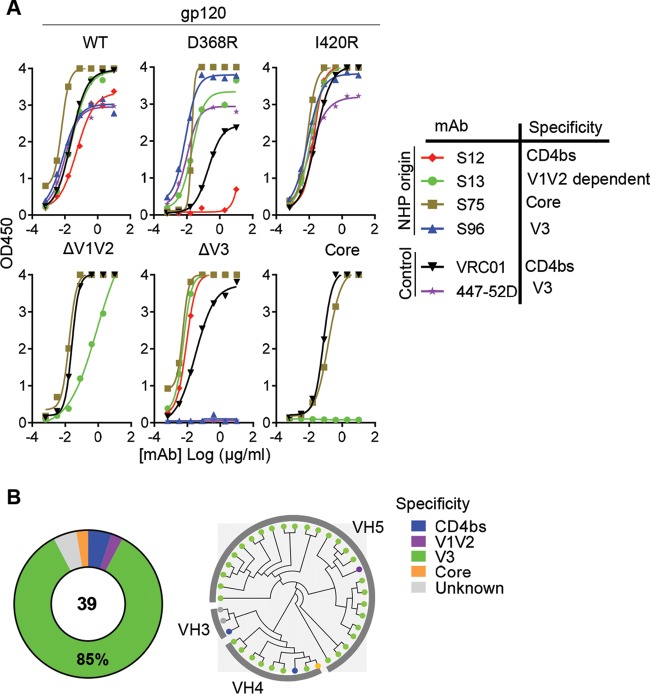
We took advantage of a panel of established gp120 variants derived from virus isolate Bal.01 (30) to further delineate the antigenic specificities of the JR-FL SOSIP-specific MAbs. After verifying the binding affinity of JR-FL SOSIP-specific MAbs to the wild type (WT) of Bal.01 gp120, we compared the binding between the WT and mutants of different Env antigenic domains to assess antibody binding specificity. The mutants included the CD4bs knockout mutant D368R, the CD4i (coreceptor binding site) knockout mutant I420R, the V1 and V2 loop-deleted mutant ΔV1V2, the V3 loop-deleted mutant ΔV3, and a mutant with all loops deleted (core). Based on the binding profile of each MAb (representative curves are shown in Fig. 3A), we identified the epitope specificities on Env as follows: (i) CD4bs MAbs with substantially weaker binding to the CD4bs mutant D368R than the WT gp120, similar to prototypic CD4bs MAb VRC01 (e.g., S12 in Fig. 3A); (ii) V1V2-dependent MAbs with diminished binding to the ΔV1V2 mutant compared to the WT gp120 (e.g., S13 in Fig. 3A); (iii) V3-specific MAbs with abolished binding to gp120 mutant ΔV3, similar to V3 MAb 447-52D (e.g., S96 in Fig. 3A); and (iv) core-specific MAbs with unchanged binding to mutants D368R, I420R, ΔV1V2, ΔV3, and core (e.g., S75 in Fig. 3A). The majority of the JR-FL SOSIP-isolated MAbs failed to bind to gp120 mutant ΔV3 (Table 2) and thus were defined as V3 specific. Overall, 33 out of 39 (85%) JR-FL SOSIP-binding MAbs were V3 specific, while CD4bs-specific, V1V2-dependent, and core-specific MAbs were infrequent (Fig. 3B, left panel). Phylogenetic analysis indicated that the V3-specific MAbs had skewed usage of VH4 or VH5 gene segments (Fig. 3B, right panel), indicating a genetic bias for this class of antibodies.
FIG 3.
JR-FL SOSIP-specific MAbs epitope mapping. (A) Representative JR-FL SOSIP-specific MAb binding profiles with Bal.01 gp120 WT and mutant variants via ELISAs. VRC01 (CD4bs specific) and 447-52D (V3 specific) were used as control MAbs. (B) Summary of binding specificities of the expressed JR-FL SOSIP-specific MAbs (left) and phylogenetic analysis of the Ig molecules (right). The heavy-chain VDJ sequences of JR-FL SOSIP-specific MAbs were used to generate the phylogenetic tree. The dot at the end of each edge is colored based on the corresponding antibody binding specificity.
V3 MAb binding specificity associated with tier 2 virus neutralization capacity.
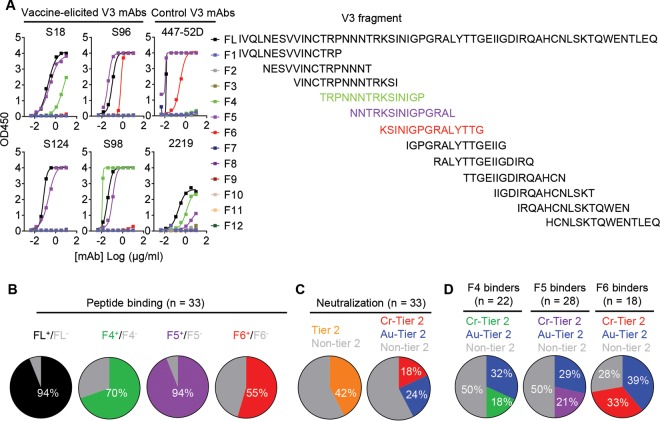
To further define the epitopes of the V3-specific MAbs, we synthesized 12 overlapping 15-mer V3 peptide fragments (Fig. 4A, right panel) based on the YU2 Env sequence (16) and tested their binding to the V3 MAbs by enzyme-linked immunosorbent assays (ELISAs) (Table 2). We found that 94% (31/33) of V3 MAbs bound full-length (FL) V3 peptide, which confirmed that most of the MAbs directly recognize the V3 region (Fig. 4B). The affinities of the MAbs binding to the FL V3 peptide were within the nanomolar range, with an average KD (equilibrium dissociation constant) value of around 0.7 nM (data not shown), as measured by biolayer light interferometry (BLI).
FIG 4.
JR-FL SOSIP-sorted V3 MAb binding specificity is associated with tier 2 virus neutralization capacity. (A) JR-FL SOSIP-specific V3-directed MAb binding profile with peptides within the V3 loop region. Left, binding curves of the representative JR-FL SOSIP-specific V3-MAbs to the peptides, with the prototypic human V3-directed MAbs 447-52D and 2219 as controls. Right, peptides with the sequences of the full length (FL) of YU2 V3 peptide and 12 overlapping 15-mer peptide fragments. Three overlapping peptide fragments (F4, F5, and F6) targeted by different V3 MAbs are highlighted in different colors. (B) Summary of V3 MAb binding specificity for the full-length (FL) peptide and the F4, F5, and F6 peptide fragments. The color of each slice represents the binding specificity for each fragment, while the area of each slice is proportional to the respective frequency of MAbs. (C) Pie charts of the distribution of JR-FL SOSIP V3 MAb HIV-1 pseudovirus neutralization capacity. MAbs are categorized as cross-reactive tier 2 virus neutralizing MAbs (Cr-tier 2 Neut.), autologous tier 2 virus neutralizing MAbs (Au-tier 2 Neut.), or non-tier 2 virus neutralizing MAbs (Non-tier 2 Neut.). Left, the frequencies of MAbs capable of neutralizing tier 2 viruses (orange) and those are not (gray). Right, MAbs neutralizing heterogeneous tier 2 viruses (red) and autologous tier 2 virus (YU2) (blue). (D) V3 MAb binding specificities to peptides and tier 2 virus neutralization capacities. MAbs that bind V3 fragment 6 have higher tier 2 virus neutralizing frequency than those that bind only fragment 4 or 5. Left, MAbs binding to fragment 4; middle, MAbs binding to fragment 5; right, MAbs binding to fragment 6.
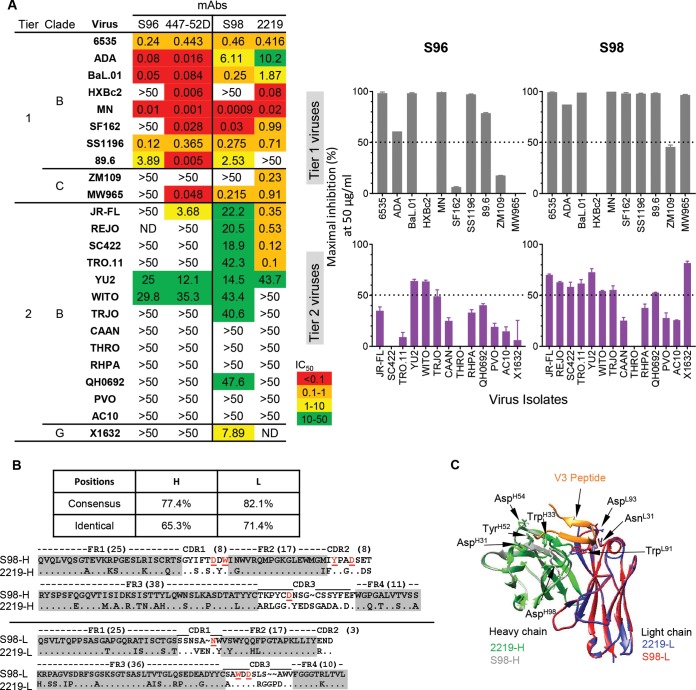
Besides the FL V3 peptide, the V3 MAbs recognized V3 fragments F4, F5, and F6, at different frequencies, with representative binding curves shown in Fig. 4A (left panel). Notably, similar to the frequency of binding to the FL V3 peptide, 94% of the V3 MAbs recognized the V3 F5 fragment (Fig. 4A and B), with fragments F4 and F6 recognized at lower frequencies (Fig. 4A and B). None of the V3 MAbs recognized the other V3 peptide fragments (Fig. 4A). The data suggest that the V3 F5 fragment encompasses the majority of the epitopes recognized by these V3 MAbs, which frequently reach toward either side of the F5 fragment region overlapping with F4 or F6 fragments. Interestingly, we found that some of the V3 MAbs displayed a binding preference for these V3 peptides that was similar to that of two prototypic cross-reactive tier 2 virus neutralizing V3 MAbs derived from natural infections, 447-52D and 2219 (Fig. 4A). For instance, MAb S96 bound V3 peptides FL, F5, and F6, reminiscent of 447-52D, while MAb S98 bound FL, F4, and F5, similar to 2219 (Fig. 4A).
We next evaluated the neutralizing activity of these JR-FL SOSIP-binding MAbs against a panel of HIV-1 virus isolates (10 tier 1 and 9 tier 2 isolates) from clades A, B, and C, using the TZM-bl assay (Table 4; see Fig. S1 in the supplemental material). We initially tested tier 1 virus neutralization capacity using MAbs with diverse binding specificities (Table 4; Fig. S1). We found that with the exception of the V1V2-dependent MAb S13, all the other CD4bs, core, and V3 MAbs displayed various degrees of tier 1 virus neutralization capacity (Table 4; Fig. S1). We then assessed the tier 2 virus neutralization profiles of the MAbs. We found that none of the CD4bs-, core-, and V1V2-specific MAbs displayed any neutralization against tier 2 viruses, while several V3 MAbs did (Table 4; Fig. S1), suggesting that the moderate tier 2 virus neutralizing capacity of the concurrent animal plasma is mediated by V3-specific Abs (Fig. 1A).
TABLE 4.
HIV-1 neutralizing activities of JR-FL SOSIP-specific MAbsa
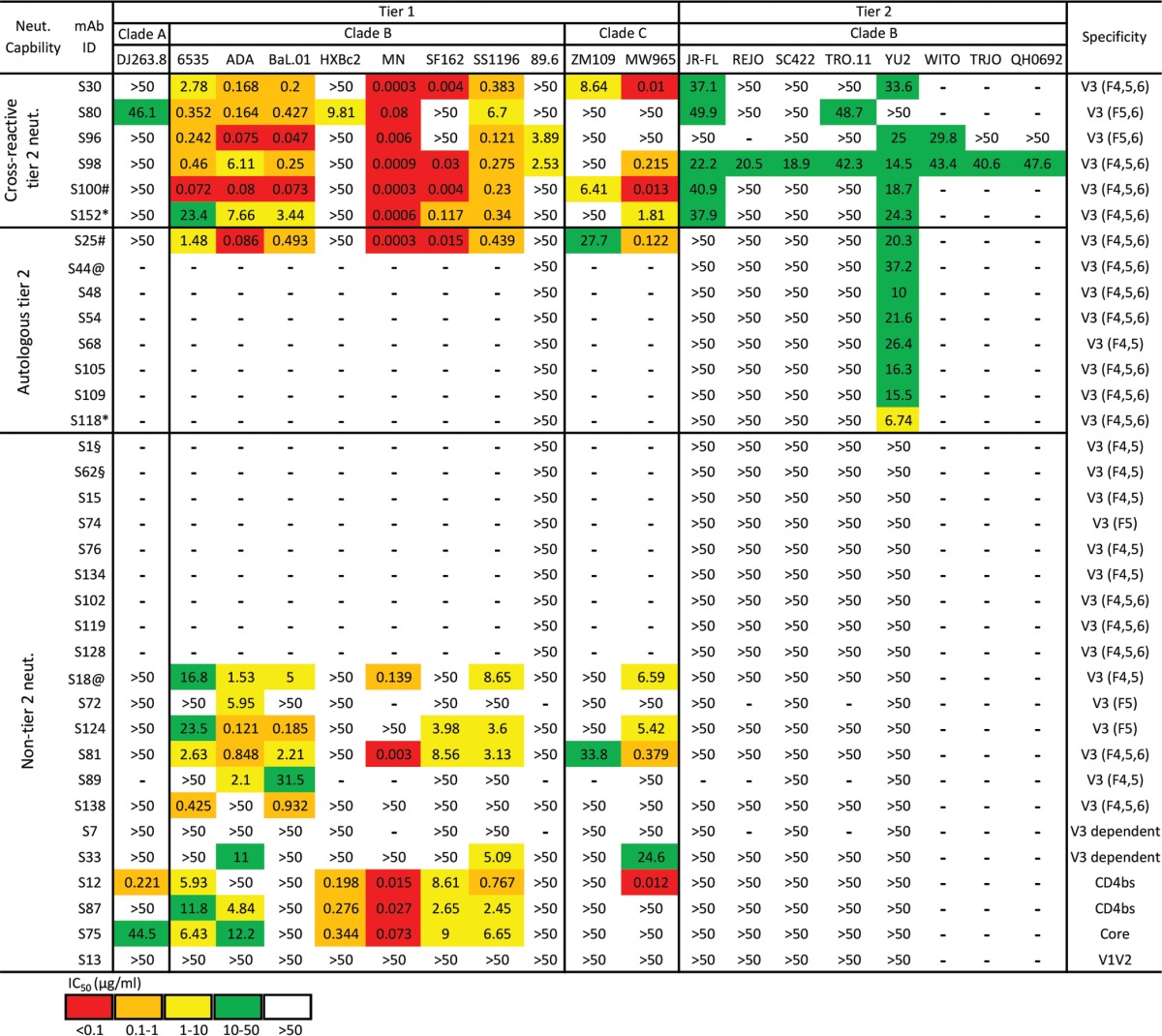
IC50 values: red, <0.1 μg/ml; orange, 0.1 to 1 μg/ml; yellow, 1 to 10 μg/ml; green, 10 to 50 μg/ml; white, >50 μg/ml; —, not tested. Tested MAb sequences with same V and J gene usage and identical CDR3 length with >75% CDR3 identity at the nucleotide level were identified as clonal variants and are indicated as follows: §, S1 and S62; @, S18 and S44; #, S25 and S100; *, S118 and S152.
We then tested the tier 2 virus neutralization capacity of the remaining V3 MAbs (Table 4; Fig. S1). Surprisingly, we observed that 42% of V3 MAbs neutralized tier 2 viruses (Fig. 4C [left panel], Table 4, and Fig. S1), with 24% exclusively neutralizing autologous tier 2 virus YU2 (Au-tier 2 neut.) and 18% displaying cross-reactive neutralization against YU2 and/or other heterologous tier 2 viruses (Cr-tier 2 neut.) (Fig. 4C [right panel], Table 4, and Fig. S1). Furthermore, we sought to examine the relationship between the binding phenotype and neutralization capacity of these V3 MAbs. As shown in Fig. 4D and Table 4, V3 fragment 6 was frequently recognized by tier 2 virus neutralizing MAbs (72%, both Au- and Cr-tier 2 virus neutralizing MAbs), especially by cross-reactive tier 2 virus neutralizing MAbs (33%).
Vaccine-elicited NHP V3 MAbs share genetic and functional homology with V3 antibodies derived from natural infections in humans.
It is intriguing that two V3 MAbs, S96 and S98, with binding specificity profiles similar to those of prototypic cross-reactive tier 2 virus neutralizing MAbs 447-52D and 2219, respectively (Fig. 4A), demonstrated neutralization breadths similar to or slightly better than those of 447-52D and 2219 (Fig. 5A, left panel). The most broadly neutralizing MAb, S98, neutralized 9/14 of the tested tier 2 viruses, including JR-FL, SC422, REJO, TRO.11, YU2, WITO, TRJO, QH0692, and X1632, with all virus neutralization slightly higher than 50% inhibition at 50 μg/ml of MAb (Fig. 5A and Table 4). Although the neutralization potency of these V3 MAbs was relatively moderate as judged by 50% inhibitory concentrations (IC50s) (Fig. 5A and Table 4), with virus entry inhibition seldom above 80% (Fig. S1) and mostly restricted to clade B viruses, the cloned MAb panel largely recapitulated the plasma neutralization breadth (Fig. 1A, lower panel).
FIG 5.
Similarities between selected cross-reactive V3 NAbs elicited by vaccination in macaque and prototypic V3 NAbs derived from natural infections in humans. (A) Vaccine-elicited S96 and S98 display neutralization profiles similar to those of the control MAbs 447-52D and 2219, respectively. Left, the neutralization potency (MAb concentration to achieve 50% virus entry inhibition, IC50 in μg/ml) is color coded as indicated. Right, the maximal virus entry inhibition (%) mediated by MAbs at 50 μg/ml is displayed by bar graphs with standard errors. (B) Macaque V3 MAb S98 displays sequence homology with human V3 MAb 2219. The consensus and identical positions of heavy- and light-chain sequences between MAbs S98 and 2219 are indicated. The critical residues for V3 MAb 2219 contacting the V3 region are indicated in red and underlined. (C) MAbs S98 and 2219 share a similar binding mode for V3 peptide as illustrated by homology modeling. S98 binding to V3 peptide was modeled using human MAb 2219 as the template (PDB no. 2B0S). The structural elements of MAb 2219 heavy chain (green) and light chain (blue), MAb S98 heavy chain (gray) and light chain (red), and V3 peptide (orange) are shown. The side chains of the critical residues for V3 MAb 2219 contacting the V3 region, as well as their counterparts in S98, are indicated by arrows.
In addition, we found that the VH/VL genes of NHP-derived MAb S98 shared high degrees of sequence homology with those used by human V3 MAb 2219, with consensus positions of 77.4% and 82.1% for heavy and light chains, respectively (Fig. 5B). We identified the most critical residues for MAb 2219 contacting the V3 region (31), including AsnL31, TrpL91, AspL93, AspH31, TrpH33, TyrH52, AspH54, and AspH98 present in the VH/VL genes of S98 (Fig. 5B), based on Kabat numbering system. Furthermore, we performed a homology modeling of S98 using the structure of the 2219-V3 peptide complex (PDB no. 2B0S) as the template based on the substantial VH/VL sequence homology of these two MAbs. In the resulting structural model, we found that both the frameworks and CDR loops of these two MAbs, including the critical contacting residues, were positioned in similar manners (Fig. 5C), suggesting that these two MAbs share similar modes of antigen interaction. The structural similarity between S98 and 2219 is consistent with their analogous virus neutralization breadth (Fig. 5A). The striking genetic, structural, and functional similarities between the NHP V3 MAb S98 elicited by immunization and the V3 MAb 2219 derived from natural infection in humans imply the genetic and structural convergence of this subset of primate V3-specific antibodies.
The epitopes of the V3 MAbs are partially/transiently accessible on the Env trimer.
The epitopes and neutralization mechanisms of V3 MAbs derived from natural infections have been well studied. This reveals that the V3 crown is largely occluded by structural elements, including the V1V2 loops and N-linked glycans on the static Env functional trimer, as demonstrated by the structures of trimeric Env SOSIP molecules in either the unliganded (32) or bNAb-bound (in complex with PGT122 and VRC01) form (33) (Fig. 6A, left). However, the V3 crown can also sample an accessible state (Fig. 6A, right) when the Env trimer engages CD4, causing relocation of the V1V2 loops, or spontaneously flickers to an open configuration due to intrinsic flexibility. Therefore, the cryptic nature of V3 neutralizing epitopes on the virus Env functional trimer restricts the neutralization potency and breadth of V3 MAbs derived from natural infection, raising the question how vaccine-elicited tier 2 virus neutralizing V3 MAbs interact with their epitopes.
FIG 6.
The epitopes of the V3 MAbs are partially/transiently accessible on the Env trimer. (A) Crystal structures of the cleaved, soluble BG505 SOSIP.664 in the native state (left) (PDB no. 4NCO) and the Env-CD4-complexed JR-FLgp120 core with the V3 region (right) (PDB no. 2B4C) indicating structural rearrangement of the V3 loop in the pre-CD4 triggered (gp120 of BG505 SOSIP) (middle) and post-CD4 binding (JR-FLgp120 core) (right) conformations. The three protomers of the BG505 SOSIP.664 trimer are denoted in gray, green, and gold, respectively. V3 fragment 6 region is shown in red and the rest of the V3 region in blue. (B) Representative negative-stain electron microscopy 2D class averages of V3 MAb Fab in complex with JR-FL SOSIP.664 trimer. Fabs of vaccine-elicited MAbs S30, S96, and S100 as well as the control MAb 447-52D were incubated with JR-FL SOSIP.664 trimer individually to form complexes. The regions corresponding to the trimer and Fab are colored in white and cyan, respectively. For more details, see Fig. S2 in the supplemental material and Materials and Methods. (C) V3 MAbs display enhanced binding to sCD4-triggered JR-FL SOSIP.664 trimer. Left, binding curves of V3 MAb 447-52D and V2 glycan-directed MAb PG16 to sCD4-triggered or nontriggered JR-FL SOSIP. Right, binding curves of selected vaccine-elicited V3 MAbs S25, S80, S96, and S98. (D) V3 MAbs S96 and S98 show elevated neutralizing activity against sCD4-treated viruses YU2 and 89.6. S96 and S98 show increasing neutralization activity in the presence of sCD4 at 0.4 μg/ml. Neutralization with sCD4 alone is marked with a box. (E) Glycans at the Env V3 base and V2 loop modulate virus sensitivity to V3 MAbs. A comparison of V3 MAb neutralizing activity against WT JR-CSF pseudovirus and variants with glycans knocked out at residues 160, 295, 301, and 332 is shown. IC50 is color coded as indicated.
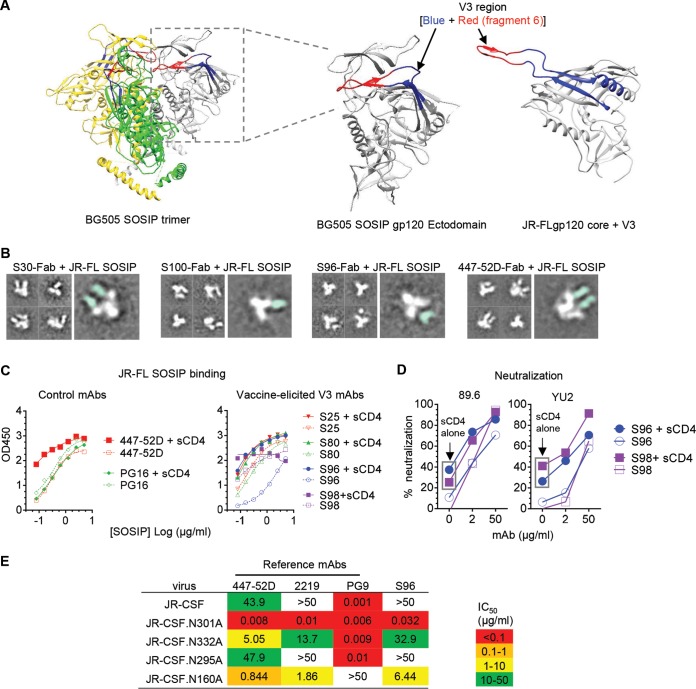
To examine the interaction of the vaccine-elicited V3 MAbs and Env trimers, we performed negative-stain electron microscopy of complexes of JR-FL SOSIP.664 with antigen-binding fragments (Fabs) of selected V3 MAbs. We found that each JR-FL SOSIP.664 trimer bound either one or two Fab molecules of immunization-elicited V3 MAbs S30 and S100 (Fig. 6B; see Fig. S2 in the supplemental material). For V3 MAb S96, we observed a maximum of only one Fab bound to a SOSIP trimer (Fig. 6B; Fig. S2), while one to three Fab molecules of the prototypic V3 MAb 447-52D (Fig. 6B) bound to a SOSIP trimer (Fig. 6B; Fig. S2), consistent with the partially/transiently exposed property of the V3 loop on the Env functional trimer. In all cases, we found unbound Fabs (Fig. S2), likely due to the excess amount added in the complex formation process, as well as unbound SOSIP trimers (Fig. 6B; Fig. S2), which may sample a more closed configuration.
It has been reported that the presence of soluble CD4 (sCD4) could elevate the exposure of V3 epitopes on the well-ordered SOSIP molecule (32, 34). Therefore, we speculated that sCD4 binding might enhance vaccine-elicited V3 MAb binding to the SOSIP trimers. As expected, the selected V3 MAbs (S25, S80, S96, and S98) showed increased binding to sCD4-bound JR-FL SOSIP trimers compared to binding in the absence of sCD4 (Fig. 6C, right panel), similar to the control V3 MAb 447-52D but not the V2 glycan-binding MAb PG16 (Fig. 6B, left panel). Enhanced neutralizing activity of S96 and S98 against viruses 89.6 and YU2, preincubated with sCD4, further confirmed this notion (Fig. 6D). Our data suggested that sCD4 induced a substantial configuration change of the Env functional trimer, resulting in elevated V3 epitope exposure and susceptibility to neutralization by immunization-elicited V3 MAbs, similar to the observation with V3 MAbs derived from natural infections (34, 35). This implies that in addition to directly binding to the static Env trimer with a cryptic cognate epitope, V3 MAbs may capture the CD4-engaged Env functional trimer with augmented epitope access to accomplish virus neutralization.
To investigate the effect of Env trimer glycans on virus neutralization sensitivity to V3 MAbs, we tested the alternation of antibody neutralizing activity against WT and glycan knockout mutants of virus isolate JR-CSF. We found that while WT JR-CSF virus was resistant to V3 MAbs S96 and 2219, mutant JR-CSF viruses N160A, N301A, and N332A, which lack N-glycans at Env residues 160, 301, and 332, respectively, were sensitive to neutralization of these two MAbs (Fig. 6E). Previous studies found that N-glycans on residues 160, 301, and 332, located in the V2 loop (N160) and the base of V3 loop (N301 and N332), respectively, play critical roles in stabilizing the Env trimer configuration while restricting the accessibility of the V3 loop and subsequently reducing virus sensitivity to V3 MAb neutralization (36). Similarly, WT JR-CSF virus was moderately neutralized by V3 MAb 447-52D (IC50 = 43.9 μg/ml), whereas mutant viruses with N-glycan deletions at these residues were 8- to 500-fold more sensitive to 447-52D neutralization (Fig. 6E). These results demonstrated that glycans in the V2 loop and the base of V3 loop impeded Env sensitivity to V3 MAbs, including S96, presumably due to the reduced accessibility of the V3 loop in the presence of these glycans. Thus, our data reinforce the notion that steric hindrance imposed by Env structural elements such as glycans impedes V3 MAb neutralization capability, while CD4 engagement of the Env trimer may enhance the exposure of the V3 neutralizing epitope.
Affinity maturation is critical for immunization-elicited V3 MAb to neutralize primary virus isolates.
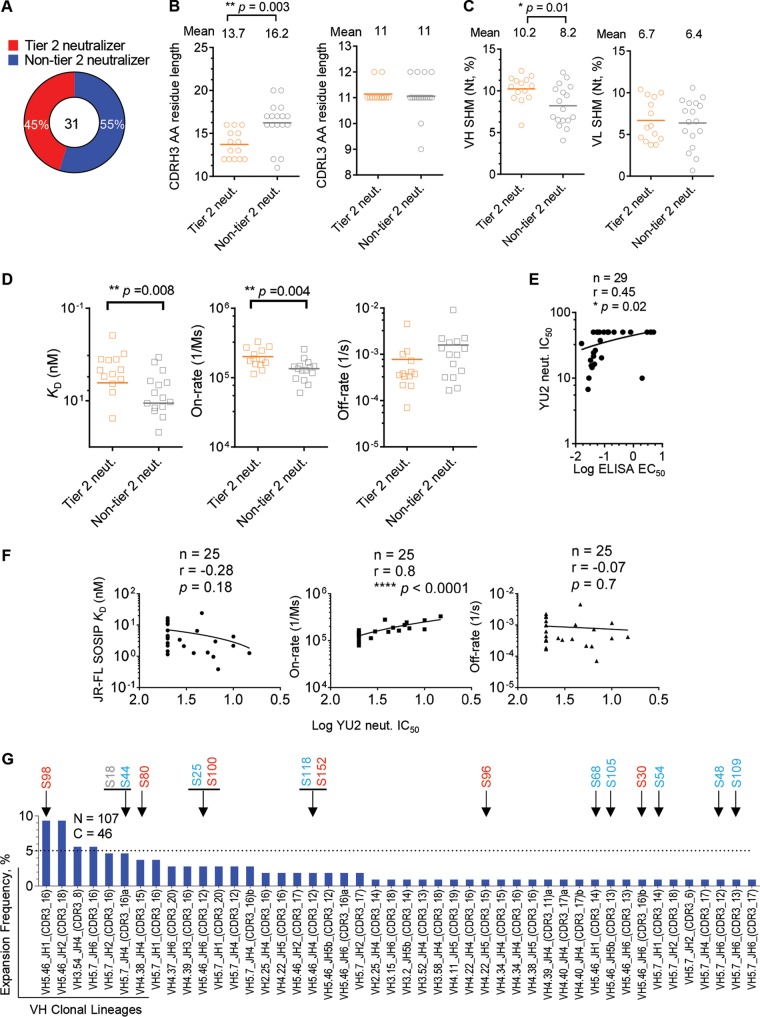
Since the elicitation of an antibody response capable of neutralizing primary (tier 2) virus isolates at substantial titers is critical for HIV-1 vaccine development, we examined factors that may contribute to the tier 2 virus neutralization capacities of the immunization-elicited V3 MAbs. We chose 31 V3 MAbs that bound full-length V3 peptide to perform the analysis by dividing these V3 MAbs into two categories: (i) tier 2 neutralizers that can neutralize at least one primary virus isolate and (ii) non-tier 2 neutralizers, accounting for 45% and 55% of the V3 MAbs, respectively (Fig. 7A).
FIG 7.
Affinity maturation is critical for V3 MAb neutralization capacity. (A) A total of 45% of immunization-elicited V3 MAbs binding full-length V3 peptide were able to neutralize tier 2 viruses. (B) Comparison of heavy-chain (left) or light-chain (right) CDR3 lengths of tier 2 virus neutralizing (orange) or non-tier 2 virus neutralizing (gray) MAbs, with the same color code as in panel A. Statistical differences were evaluated using the t test. (C) Comparison of heavy-chain (left) or light-chain (right) somatic hypermutation (SHM) of V regions of V3 MAbs, with the same color code as in panel A. (D) Comparison of JR-FL SOSIP.664 trimer binding affinity (dissociation constants [KD] and association [on-rate] and dissociation [off-rate] rates) of V3 MAbs, with the same color code as in panel A. (E) Antibody autologous neutralization (strain YU2) potency (IC50) correlates with binding avidity (log ELISA EC50) for the JR-FL SOSIP.664 trimer. Correlation was evaluated using the nonparametric Spearman test. The total MAb number (n), correlation coefficient (r), and P value are shown. (F) Autologous neutralization potency (log YU2 neutralization IC50) correlates with V3 MAb-trimer binding association rate (on-rate). The correlation between binding affinity (KD), on-rate or off-rate, and autologous neutralization IC50 (log transformed) of each antibody was determined as for panel E. (G) Expansion frequencies of tier 2 neutralizer lineages. The JR-FL SOSIP-sorted Ig repertoire possessed expansions (n = 107) (Fig. 2B) that were classified into 46 clonal lineages based on heavy-chain sequences. Arrows indicate lineages of tier 2 virus neutralizing MAbs, with cross-reactive (Cr-tier 2) neutralizing MAbs colored in red, autologous (Au-tier 2) neutralizing MAbs in blue, and non-tier 2 neutralizing MAbs in gray. Note that certain lineages consist of Cr-tier 2, Au-tier 2, and/or non-tier 2 neutralizing MAbs simultaneously.
Previous studies suggest that a long CDRH3 might contribute to virus neutralization of natural-infection-derived V3 MAbs (37). As such, we first compared the CDR3 residue length between tier 2 and non-tier 2 virus neutralizing V3 MAbs. Surprisingly, tier 2 neutralizing V3 MAbs possessed, on average, a shorter CDRH3 (**, P = 0.003 by t test) and similar CDRL3 length (Fig. 7B) compared to non-tier 2 neutralizers, suggesting that long CDR3 is not required for tier 2 virus neutralization. However, the vaccine-elicited tier 2 neutralizers possessed significantly higher SHMs in VH (Fig. 7C, left panel) (*P = 0.01 by t test) but not in the VL domain of light chain (Fig. 7C, right panel).
The level of SHM usually indicates the affinity maturation extent of antibody. Therefore, we further tested the binding affinity of these V3 MAbs to the sorting probe, JR-FL SOSIP.664 Env trimer. We observed that tier 2 neutralizers had significantly higher affinity (KD) than non-tier 2 neutralizers (Fig. 7D, left panel) (**, P = 0.008 by t test), conferred by the association rate (on-rate) of the MAb-Env interaction (Fig. 7D, middle panel) (**, P = 0.004 by t test) but not by the off-rate (Fig. 7D, right panel). Furthermore, we plotted the autologous YU2 neutralization IC50 titers of V3 MAbs versus their JR-FL SOSIP.664 trimer binding ELISA 50% effective concentration (EC50) titers for correlation analysis. We found that the neutralization potency correlated with trimer binding affinity (Fig. 7E) (*, P = 0.02 by nonparametric Spearman correlation). Consistently, we found that this correlation was dependent on the MAb-trimer association rate (on-rate) (Fig. 7F, middle panel) (****, P < 0.0001 by nonparametric Spearman correlation), suggesting that V3 MAbs capable of efficiently overcoming the steric hindrance to approach the virus Env functional spikes have higher neutralization potency against tier 2 virus. We observed no significant difference in CDR3 length, SHM level, and binding affinity for JR-FL SOSIP.664 trimer between the cross-reactive and autologous tier 2 virus neutralizing MAbs (data not shown), presumably due to the limited MAb sample sizes.
Finally, to examine the overall expansion level of the V3-specific tier 2 neutralizer clonal lineages in the JR-FL SOSIP.664 sorted B cell repertoire, we assessed the expansion frequency of each tier 2 neutralizer lineage, based on the amplified VH sequences of each SOSIP-sorted single B cell. Tier 2 neutralizer clonal lineages demonstrated expansion frequencies with various expansion levels (Fig. 7G). More than half of the tier 2 neutralizer lineages had single expansion, while others had multiple expansions. There was no obvious difference in the clonal lineage expansion level between cross-reactive and autologous tier 2 neutralizers (Fig. 7G), suggesting that the concurrent immunogen, YU2gp140-F, imposed virtually equal selection pressures on B cells encoding antibodies with different neutralizing breadths and potencies. The S98 clonal lineage demonstrated the highest expansion frequency, accounting for ∼9% of the total SOSIP-sorted memory B cells (Fig. 7G). However, since we were able to amplify both the heavy- and light-chain genes of only clone S98 to reconstitute full-length IgG for functional analysis within this lineage, the neutralization capacities of the other clones in this lineage and their contributions to the polyclonal serum neutralization breadth remain unknown. Furthermore, we found that there were clonal lineages consisting of MAbs with different levels of neutralization capacity (e.g., S18 and S44, S25 and S100, and S118 and S152) (Fig. 7G). This observation suggests that it is possible for a B cell clone to evolve to possess an elevated level of neutralization capacity by affinity maturation, e.g., from non-tier 2 neutralization to autologous and even cross-reactive tier 2 neutralization. To pin down this point, future works with larger MAb sampling size and MAbs isolated from multiple animals are required.
DISCUSSION
In this study, we delineated the neutralization specificity of cross-reactive tier 2 virus neutralizing antibody responses elicited by vaccination in nonhuman primates. We performed single memory B cell sorting from the peripheral blood of a rhesus macaque immunized with YU2gp140-F trimers in adjuvant, using JR-FL SOSIP.664, a native Env trimer mimetic, as a sorting probe. With the selective SOSIP sorting probe, we isolated 39 SOSIP-binding MAbs, with 85% targeting the HIV-1 gp120 V3 loop (n = 33), of which nearly 20% (6/33) displayed cross-reactive tier 2 virus neutralization that recapitulated the serum neutralization capacity. In addition, we observed that there were substantial similarities in binding specificity and neutralization breadth and potency between selected cross-reactive V3 NAbs elicited by vaccination in macaques and prototypic V3 NAbs derived from natural infections in humans. Furthermore, we found striking sequence and structural homology between some of the V3 NAbs with macaque (S98) and human (e.g., 2219) origins. It has been reported that some V3 MAbs, including 2219, derived from natural infections in human share VH5-51/VL lambda gene usages as well as a conserved antigenic structure (38). All of these observations underscore the genetic and functional convergence of this subset of the primate V3-specific B cell repertoire. Thus, our study demonstrates that it is possible to elicit neutralizing V3 NAbs with some tier 2 cross-reactivity, similar to the human MAbs 447-52D and 2219 in humans, by immunization.
It is well known that the V3 loop, particularly the V3 crown, is immunogenic (39). However, on the static unliganded Env functional spike, the V3 region is occluded by adjacent structural elements, including the V1V2 loop and stem (32, 33, 36, 40), dampening enthusiasm to target this region by vaccination. However, recent studies reveal that the unliganded Env trimer spikes have various degrees of transient forms that differ between different virus isolates (41–43). This intrinsic Env trimer spike conformational dynamics confers to certain tier 2 and 3 viruses susceptibility to V3 Abs. This is consistent with our observation that selected vaccine-induced V3-specific tier 2 virus NAbs display substantial binding affinity for the JR-FL SOSIP.664 trimer, despite steric hindrance masking their cognate epitopes (Fig. 6E). In fact, recent studies show that SOSIP.664 or NFL trimer immunogens elicit a V3-specific neutralizing antibody response quite frequently in immunized animals (44, 45), suggesting that V3 on SOSIP or NFL trimeric Env can be well exposed under physiological conditions and highly immunogenic in vivo. In addition, our data (Fig. 6D) along with those of others, showed that sCD4 could enhance selected V3 MAb neutralizing capacity toward tier 2 viruses (34), suggesting a virus neutralization mechanism with which a subset of V3 MAbs may capture the Env trimer after the CD4 receptor engagement, resulting in augmented V3 epitope exposure (46, 47). Furthermore, recent emerging in vivo data demonstrating that the V3 Ab response correlates with reduced infection of infants born to HIV-infected mothers (48) and a reduced rate of infection in human vaccinees (49) suggest that responses against this region may be effective in some settings despite the modest neutralization potency.
In summary, we dissected the cross-reactive tier 2 virus neutralizing antibody response elicited by the early generation of HIV Env trimer gp140-F in rhesus macaques at the clonal level by antigen-specific single B cell sorting and cloning, using the heterologous native Env trimer mimetic SOSIP.664 as selective probe. We found that most of the trimer SOSIP.664-sorted MAbs were directed toward the gp120 V3 loop region, particularly the V3 crown, which recapitulated the serum neutralization capacity. Our study revealed striking similarities in binding specificity, neutralization breadth and potency, sequence, and structural homology between selected cross-reactive V3 NAbs elicited by vaccination in macaques and prototypic V3 NAbs derived from natural infections in humans, strengthening the notion that certain subset of V3-specific B cell repertories share a high level of convergence in primates and even other species (39, 50). Our data demonstrate that unusually high levels of SHM and long CDRH3 regions are not required for the cross-reactive V3-directed neutralizing antibody activities described here, suggesting that these types of responses merit further investigation.
MATERIALS AND METHODS
Macaque peripheral blood sample.
In the immunization regimen with long intervals, rhesus macaques of Chinese origin, approximately 5 to 6 years old, were immunized three times at weeks 0, 8, and 32 by intramuscular injection (given in a total volume of 1 ml, divided equally between the left and right hind legs). Soluble YU2gp140-F Env trimers in combination with 75 μg AbISCO (Isconova AB [now Novavax]) and 500 μg CpG ODN2395 (InvivoGen) adjuvant (Fig. 1A) (21) were used in the inoculation. Two hundred micrograms of Env protein/animal was given for the first inoculation, and 100 μg was given for the following injections. Peripheral blood was collected 2 weeks after each immunization, followed by PBMC preparation as described previously (15). In this study, frozen PBMCs from rhesus macaque K17 at 2 weeks after the third immunization were used (Fig. 1A) (21). As a comparison, an immunization regimen with short intervals was used previously, with immunizations occurring at weeks 0, 4, 8, 12, and 18 (51). The animal studies were conducted with the approval of the regional Committee for Animal Ethics, and all methods were carried out in accordance with the approved guidelines.
Cell staining and single-cell flow cytometric sorting.
PBMCs were stained by a cocktail of antibodies for identifying memory B cells as described previously (15). To sort JR-FL SOSIP-specific memory B cells, JR-FL SOSIP.664 containing an Avi tag was expressed, purified, biotinylated (15), and negatively selected by non-bNAb F105 (25), followed by conjugation with streptavidin-allophycocyanin (APC [Life Technologies]) before incorporation into the above-stated antibody cocktail (15). Following staining, the cells were sorted into 96-well plates at single-cell density as described previously (15).
Single-cell reverse transcription-PCR (RT-PCR), antibody cloning, and expression.
The sorted cells were lysed with the lysis buffer, followed by single-cell reverse transcription and PCRs to amplify Ig sequences as described previously (15, 27, 52). After sequencing verification of correct V(D)J sequences, the Ig variable domains of selected B cell clones were cloned into Ab IgG heavy- and light-chain expression vectors, respectively, as described previously (15). The antibody expression vectors were cotransfected into 293F cells, and the cell culture supernatants containing secreted IgG antibody were purified by columns packed in-house containing protein A-Sepharose beads (GE Healthcare). To express Fab antibody, the heavy-chain variable domain was inserted into a Fab expression vector containing a His tag as previously described (3), followed by cotransfection with light-chain expression vector. Fab was purified from cell culture supernatant with cOmplete His tag purification resin (Roche).
Analysis of Env- or peptide-binding Abs.
The purified antibodies were tested for specificity by enzyme-linked immunosorbent assays (ELISAs). For SOSIP-Ab binding analysis, anti-His tag Ab (R&D Systems, clone AD1.1.10) was applied to MaxiSorp plates (Nunc, Thermo Scientific) at 2 μg/ml in phosphate-buffered saline (PBS) overnight at 4°C. After the plates were blocked with PBS–2% dry milk–5% fetal bovine serum (FBS) for 1 h at 37°C, JR-FL SOSIP.664 with a His6 tag was added to each well at 2 μg/ml and incubated for 30 min at room temperature, followed by the addition of antibodies in a 5-fold dilution series starting from 10 μg/ml for 45 min at room temperature. Goat anti-human IgG-horseradish peroxidase (HRP) conjugate (Jackson ImmunoResearch) was used at a 1:10,000 dilution in PBS–0.05% Tween 20 for 1 h at room temperature. The bound MAb was detected with 100 μl/well of tetramethylbenzidine (TMB) substrate (Life Technologies) for 5 min before the addition of 100 μl of 3% H2SO4 to stop the reaction. The optical density at 450 nm (OD450) was measured. After each step of the ELISA, plates were washed four times with PBS supplemented with 0.05% Tween 20.
For soluble CD4 (sCD4)-induced JR-FL SOSIP-Ab binding analysis, MAbs were applied to plates at 2 μg/ml in PBS and left overnight at 4°C. Biotin-labeled JR-FL SOSIP.664 was preincubated with sCD4 (Progenics Pharmaceuticals) at a constant molar concentration of 0 or 1 μM for 1 h at room temperature. After plates were blocked, sCD4-induced (1 μM sCD4) or noninduced (0 μM sCD4) JR-FL SOSIP was added to wells coated with MAbs in a 2-fold dilution series starting from 5 μg/ml for 2 h at room temperature. Poly-HRP-conjugated streptavidin (Pierce) was then used at a 1:5,000 dilution in 10-fold-diluted blocking buffer in PBS–0.05% Tween 20 for 1 h at room temperature. Binding signals were developed and detected with TMB substrate as described above.
For MAb binding ELISA with peptides, gp120, or gp140-F, peptides or gp120/gp140-F was applied to plates at 2 μg/ml in PBS and left overnight at 4°C. After blocking the wells in the plate, MAbs were added into wells in a 5-fold dilution series starting from 10 μg/ml and incubated for 1 h at 37°C, followed by the same procedures stated above to detect bound MAbs.
JR-FL SOSIP-sorted Ig sequence genetic analysis.
The antibody V(D)J gene segments generated by single-cell sorting and PCRs were analyzed as described previously (15). The rhesus macaque Ig germ line databases published previously were used to assign the germ line gene usage and analyze somatic hypermutation (SHM). SHM at the level of amino acid and clonal lineage definition were analyzed as described previously (15).
Electron microscopy and image processing.
Negatively selected JR-FL SOSIP.664 trimers were incubated with a 6 M excess of selected antibody Fab overnight at room temperature in Tris-buffered saline (TBS). A 3-μl aliquot containing ∼0.05 mg/ml of Fab plus JR-FL SOSIP complex was applied for 15 s on a carbon-coated 400 Cu mesh grid that had been glow discharged at 20 mA for 30 s, followed by negative staining with 2% uranyl formate for 30 s. Data were collected using an FEI Tecnai Spirit electron microscope operating at 120 keV, with an electron dose of ∼36 e-/Å2 and a magnification of ×52,000, which resulted in a pixel size of 2.05 Å at the specimen plane. Images were acquired with a Tietz 4 k × 4 k TemCam-F416 CMOS camera using a nominal defocus of 1,000 nm and the Leginon package at 10° tilt increments, up to 50°. The tilts provided additional particle orientations to improve the image reconstructions. Particles were picked automatically using DoG Picker and put into a particle stack using the Appion software package (53, 54). Two-dimensional (2D) class averages were calculated using particles binned by two via Xmipp clustering 2D alignment (55) and sorted into classes. Particles corresponding to complexes were selected into a substack and binned by two before another round of reference-free alignment was carried out using the Xmipp clustering and 2D alignment. For sample S30-Fab plus JRFL SOSIP, 9,996 particles were selected from 100 images (see Fig. S2 in the supplemental material), with 0 to 2 Fabs binding to SOSIP trimer. Unbound Fab also appeared in the class averages. For sample S96-Fab plus JRFL SOSIP, 11,368 particles were selected from 107 images. Complexes with a single Fab bound and unliganded trimer appeared in the class averages. For sample S100-Fab plus JRFL SOSIP, 12,492 particles were selected from 112 images. Trimers with 0 to 2 Fabs and unbound Fab appeared in the class averages. For sample 447-52D Fab plus JRFL SOSIP, 16,855 particles were selected from 130 images. Zero to 3 Fabs were visible on trimers, as well as unbound Fab. In all cases, the unbound Fab was likely due to excess added during the complex formation process.
Antibody binding affinity.
Biolayer light interferometry (BLI) was performed using an Octet RED96 instrument (ForteBio, Pall Life Sciences) as described previously (15). Biotin-labeled full-length V3 peptide or JR-FL SOSIP was captured onto streptavidin biosensors at concentration of 10 μg/ml as ligand and the tested MAb was diluted in a 2-fold series starting from 50 nM to 0.78 nM for peptide and 250 nM to 7.8 nM for SOSIP as the analyte in solution. Briefly, biosensors, prehydrated in binding buffer (PBS with 0.2% Tween 20) for 10 min, were first immersed in binding buffer for 60 s to establish a baseline, followed by submerging in a solution containing biotin-labeled V3 peptide or JR-FL SOSIP for 60 s to capture the ligand. The biosensors were then submerged in binding buffer for a wash for 60 s. The biosensors were then immersed in a solution containing various concentrations of analyte MAb to detect analyte/ligand association, followed by 120 s in binding buffer to assess analyte/ligand dissociation. Binding affinity constants (dissociation constant, KD; on-rate, Kon; off-rate, Koff) were determined using Octet Analysis version 7 software.
HIV-1 neutralization assays.
The neutralization assays of antibody (IgG or Fab) were performed in the format of a single round of infection using HIV-1 Env pseudoviruses and TZM-bl target cells, as described previously (56–58). Neutralization curves were fit by nonlinear regression using a 5-parameter hill slope equation as previously described (57). The 50% inhibitory concentrations (IC50) of antibody were reported as the concentration of antibody required to inhibit infection by 50%. For sCD4-treated virus neutralization, sCD4 at a concentration of 0 or 0.4 μg/ml was added to wells in a plate containing viruses and left for 30 min at 37°C in a cell culture incubator, followed by the addition of purified MAb at a concentration of 0, 2, or 50 μg/ml to neutralize virus.
Structure modeling.
Comparative modeling was performed with MODELLER 9.14 (59) using Chimera interface and the crystal structure of anti-HIV-1 V3 Fab 2219 in complex with MN peptide as a template (PDB no. 2B0S) (31).
Statistical analysis.
Comparisons of individual V gene segment compositions in the JR-FL SOSIP- and gp140-F-specific Ig repertoires were carried out with two-way ANOVA. Comparison of three or more groups was conducted with one-way ANOVA. Statistical evaluation of the difference between two groups was performed with the t test. The correlation was determined with the nonparametric Spearman correlation test, with a two-tailed P value calculated for significance. Statistical significance was expressed as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. All statistical analysis was performed with GraphPad Prism version 6.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01 AI102766 to Y.L., P01 AI104722 to R.T.W., Y.L., and G.B.K.H., and UM1 AI100663 to A.B.W. and R.T.W.), the Bill and Melinda Gates Foundation CAVD (OPP1115782 and OPP1084519 to A.B.W.), and the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (to J.R.M.). This work was also partially funded by the International AIDS Vaccine Initiative (IAVI) (to A.B.W. and R.T.W.) with the generous support of USAID, the Ministry of Foreign Affairs of the Netherlands, and the Bill & Melinda Gates Foundation; a full list of IAVI donors is available at www.iavi.org.
The contents of this article are the responsibility of the authors and do not necessarily reflect the views of USAID or the U.S. Government.
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: human soluble CD4 recombinant protein (sCD4) from Progenics and anti-HIV-1 gp120 monoclonal antibody (2219) from Susan Zolla-Pazner. We are grateful to Dennis Burton (The Scripps Research Institute) for providing JR-CSF gp120 alanine-scanning mutant panel plasmids. We thank Andrey Galkin (Institute for Bioscience and Biotechnology Research) for structural modeling.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00910-17.
REFERENCES
- 1.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 2.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol 76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran K, Poulsen C, Guenaga J, de Val N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, Karlsson Hedestam GB, Wyatt RT. 2014. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc Natl Acad Sci U S A 111:E738–E747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley JM, Sanders RW, Master A, Cayanan CS, Wiley CL, Schiffner L, Travis B, Kuhmann S, Burton DR, Hu SL, Olson WC, Moore JP. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol 76:2606–2616. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders RW, Schiffner L, Master A, Kajumo F, Guo Y, Dragic T, Moore JP, Binley JM. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J Virol 74:5091–5100. doi: 10.1128/JVI.74.11.5091-5100.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. 2015. Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J Virol 90:2806–2817. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. 2015. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep 11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O'Dell S, Feng Y, Mascola JR, Wyatt RT, Karlsson Hedestam GB. 2012. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Transl Med 4:142ra196. doi: 10.1126/scitranslmed.3003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakrabarti BK, Feng Y, Sharma SK, McKee K, Karlsson Hedestam GB, Labranche CC, Montefiori DC, Mascola JR, Wyatt RT. 2013. Robust neutralizing antibodies elicited by HIV-1 JRFL envelope glycoprotein trimers in nonhuman primates. J Virol 87:13239–13251. doi: 10.1128/JVI.01247-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, Kulp DW, Macauley MS, Sok D, Pauthner M, Menis S, Cottrell CA, Torres JL, Hsueh J, Schief WR, Wilson IA, Ward AB, Sanders RW, Burton DR. 2016. Holes in the glycan shield of the native HIV envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep 16:2327–2338. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 vaccines. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, Stewart-Jones G, Destefano J, O'Dell S, LaBranche C, Robinson JE, Montefiori DC, McKee K, Du SX, Doria-Rose N, Kwong PD, Mascola JR, Zhu P, Schief WR, Wyatt RT, Whalen RG, Binley JM. 2015. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLoS Pathog 11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Sundling C, Wilson R, O'Dell S, Chen Y, Dai K, Phad GE, Zhu J, Xiao Y, Mascola JR, Karlsson Hedestam GB, Wyatt RT, Li Y. 2016. High-resolution longitudinal study of HIV-1 Env vaccine-elicited B cell responses to the virus primary receptor binding site reveals affinity maturation and clonal persistence. J Immunol 196:3729–3743. doi: 10.4049/jimmunol.1502543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phad GE, Vazquez Bernat N, Feng Y, Ingale J, Martinez Murillo PA, O'Dell S, Li Y, Mascola JR, Sundling C, Wyatt RT, Karlsson Hedestam GB. 2015. Diverse antibody genetic and recognition properties revealed following HIV-1 envelope glycoprotein immunization. J Immunol 194:5903–5914. doi: 10.4049/jimmunol.1500122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Wilson R, O'Dell S, Guenaga J, Feng Y, Tran K, Chiang CI, Arendt HE, DeStefano J, Mascola JR, Wyatt RT, Li Y. 2016. An HIV-1 Env-antibody complex focuses antibody responses to conserved neutralizing epitopes. J Immunol 197:3982–3998. doi: 10.4049/jimmunol.1601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Vaine M, Wallace A, Han D, Wan S, Seaman MS, Montefiori D, Wang S, Lu S. 2013. A novel rabbit monoclonal antibody platform to dissect the diverse repertoire of antibody epitopes for HIV-1 Env immunogen design. J Virol 87:10232–10243. doi: 10.1128/JVI.00837-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Murillo P, Tran K, Guenaga J, Lindgren G, Adori M, Feng Y, Phad GE, Vazquez Bernat N, Bale S, Ingale J, Dubrovskaya V, O'Dell S, Pramanik L, Spangberg M, Corcoran M, Lore K, Mascola JR, Wyatt RT, Karlsson Hedestam GB. 2017. Particulate array of well-ordered HIV clade C Env trimers elicits neutralizing antibodies that display a unique V2 cap approach. Immunity 46:804–817 e807. doi: 10.1016/j.immuni.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradley T, Fera D, Bhiman J, Eslamizar L, Lu X, Anasti K, Zhang R, Sutherland LL, Scearce RM, Bowman CM, Stolarchuk C, Lloyd KE, Parks R, Eaton A, Foulger A, Nie X, Karim SS, Barnett S, Kelsoe G, Kepler TB, Alam SM, Montefiori DC, Moody MA, Liao HX, Morris L, Santra S, Harrison SC, Haynes BF. 2016. Structural constraints of vaccine-induced tier-2 autologous HIV neutralizing antibodies targeting the receptor-binding site. Cell Rep 14:43–54. doi: 10.1016/j.celrep.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez P, Sundling C, O'Dell S, Mascola JR, Wyatt RT, Karlsson Hedestam GB. 2015. Primate immune responses to HIV-1 Env formulated in the saponin-based adjuvant AbISCO-100 in the presence or absence of TLR9 co-stimulation. Sci Rep 5:8925. doi: 10.1038/srep08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung CS, Vander Heyden N, Ratner L. 1999. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J Virol 73:8216–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cormier EG, Dragic T. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol 76:8953–8957. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148. doi: 10.1016/S0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 25.Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. 2015. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog 11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundling C, Zhang Z, Phad GE, Sheng Z, Wang Y, Mascola JR, Li Y, Wyatt RT, Shapiro L, Karlsson Hedestam GB. 2014. Single-cell and deep sequencing of IgG-switched macaque B cells reveal a diverse Ig repertoire following immunization. J Immunol 192:3637–3644. doi: 10.4049/jimmunol.1303334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereno-Orbea J, Kalyuzhniy O, Deresa I, Hu X, Spencer S, Jones M, Georgeson E, Adachi Y, Kubitz M, deCamp AC, Julien JP, Wilson IA, Burton DR, Crotty S, Schief WR. 2016. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Svehla K, Louder MK, Wycuff D, Phogat S, Tang M, Migueles SA, Wu X, Phogat A, Shaw GM, Connors M, Hoxie J, Mascola JR, Wyatt R. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol 83:1045–1059. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. 2006. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol 80:6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang GY, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GB, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schon A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, et al. 2015. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, Sambor A, Nason MC, Yang ZY, Wu L, Zolla-Pazner S, Nabel GJ, Mascola JR. 2008. Soluble CD4 broadens neutralization of V3-directed monoclonal antibodies and guinea pig vaccine sera against HIV-1 subtype B and C reference viruses. Virology 380:285–295. doi: 10.1016/j.virol.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Upadhyay C, Mayr LM, Zhang J, Kumar R, Gorny MK, Nadas A, Zolla-Pazner S, Hioe CE. 2014. Distinct mechanisms regulate exposure of neutralizing epitopes in the V2 and V3 loops of HIV-1 envelope. J Virol 88:12853–12865. doi: 10.1128/JVI.02125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zolla-Pazner S, Cohen SS, Boyd D, Kong XP, Seaman M, Nussenzweig M, Klein F, Overbaugh J, Totrov M. 2015. Structure/function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J Virol 90:636–649. doi: 10.1128/JVI.01645-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burke V, Williams C, Sukumaran M, Kim SS, Li H, Wang XH, Gorny MK, Zolla-Pazner S, Kong XP. 2009. Structural basis of the cross-reactivity of genetically related human anti-HIV-1 mAbs: implications for design of V3-based immunogens. Structure 17:1538–1546. doi: 10.1016/j.str.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorny MK, Sampson J, Li H, Jiang X, Totrov M, Wang XH, Williams C, O'Neal T, Volsky B, Li L, Cardozo T, Nyambi P, Zolla-Pazner S, Kong XP. 2011. Human anti-V3 HIV-1 monoclonal antibodies encoded by the VH5-51/VL lambda genes define a conserved antigenic structure. PLoS One 6:e27780. doi: 10.1371/journal.pone.0027780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balasubramanian P, Kumar R, Williams C, Itri V, Wang S, Lu S, Hessell AJ, Haigwood NL, Sinangil F, Higgins KW, Liu L, Li L, Nyambi P, Gorny MK, Totrov M, Nadas A, Kong XP, Zolla-Pazner S, Hioe CE. 2017. Differential induction of anti-V3 crown antibodies with cradle- and ladle-binding modes in response to HIV-1 envelope vaccination. Vaccine 35:1464–1473. doi: 10.1016/j.vaccine.2016.11.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB 3rd, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munro JB, Mothes W. 2015. Structure and dynamics of the native HIV-1 Env trimer. J Virol 89:5752–5755. doi: 10.1128/JVI.03187-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guttman M, Cupo A, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. 2015. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun 6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Y, Tran K, Bale S, Kumar S, Guenaga J, Wilson R, de Val N, Arendt H, DeStefano J, Ward AB, Wyatt RT. 2016. Thermostability of well-ordered HIV spikes correlates with the elicitation of autologous tier 2 neutralizing antibodies. PLoS Pathog 12:e1005767. doi: 10.1371/journal.ppat.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng C, Pancera M, Bossert A, Schmidt SD, Chen RE, Chen X, Druz A, Narpala S, Doria-Rose NA, McDermott AB, Kwong PD, Mascola JR. 2015. Immunogenicity of a prefusion HIV-1 envelope trimer in complex with a quaternary-structure-specific antibody. J Virol 90:2740–2755. doi: 10.1128/JVI.02380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scharf L, Wang H, Gao H, Chen S, McDowall AW, Bjorkman PJ. 2015. Broadly neutralizing antibody 8ANC195 recognizes closed and open states of HIV-1 Env. Cell 162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. 2016. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci U S A 113:E7151–E7158. doi: 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Permar SR, Fong Y, Vandergrift N, Fouda GG, Gilbert P, Parks R, Jaeger FH, Pollara J, Martelli A, Liebl BE, Lloyd K, Yates NL, Overman RG, Shen X, Whitaker K, Chen H, Pritchett J, Solomon E, Friberg E, Marshall DJ, Whitesides JF, Gurley TC, Von Holle T, Martinez DR, Cai F, Kumar A, Xia SM, Lu X, Louzao R, Wilkes S, Datta S, Sarzotti-Kelsoe M, Liao HX, Ferrari G, Alam SM, Montefiori DC, Denny TN, Moody MA, Tomaras GD, Gao F, Haynes BF. 2015. Maternal HIV-1 envelope-specific antibody responses and reduced risk of perinatal transmission. J Clin Invest 125:2702–2706. doi: 10.1172/JCI81593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorny MK, Wang XH, Williams C, Volsky B, Revesz K, Witover B, Burda S, Urbanski M, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S, Nadas A. 2009. Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol Immunol 46:917–926. doi: 10.1016/j.molimm.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sundling C, Forsell MN, O'Dell S, Feng Y, Chakrabarti B, Rao SS, Lore K, Mascola JR, Wyatt RT, Douagi I, Karlsson Hedestam GB. 2010. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J Exp Med 207:2003–2017. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sundling C, Phad G, Douagi I, Navis M, Karlsson Hedestam GB. 2012. Isolation of antibody V(D)J sequences from single cell sorted rhesus macaque B cells. J Immunol Methods 386:85–93. doi: 10.1016/j.jim.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B. 2009. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. 2009. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol 166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorzano CO, Bilbao-Castro JR, Shkolnisky Y, Alcorlo M, Melero R, Caffarena-Fernandez G, Li M, Xu G, Marabini R, Carazo JM. 2010. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol 171:197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu XL, Zhou TQ, O'Dell S, Wyatt RT, Kwong PD, Mascola JR. 2009. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody b12 that effectively targets the site of CD4 attachment. J Virol 83:10892–10907. doi: 10.1128/JVI.01142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. 2000. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.