ABSTRACT
In 2014 we observed a noticeable increase in the number of sudden deaths among green tree pythons (Morelia viridis). Pathological examination revealed the accumulation of mucoid material within the airways and lungs in association with enlargement of the entire lung. We performed a full necropsy and histological examination on 12 affected green tree pythons from 7 different breeders to characterize the pathogenesis of this mucinous pneumonia. By histology we could show a marked hyperplasia of the airway epithelium and of faveolar type II pneumocytes. Since routine microbiological tests failed to identify a causative agent, we studied lung tissue samples from a few diseased snakes by next-generation sequencing (NGS). From the NGS data we could assemble a piece of RNA genome whose sequence was <85% identical to that of nidoviruses previously identified in ball pythons and Indian pythons. We then employed reverse transcription-PCR to demonstrate the presence of the novel nidovirus in all diseased snakes. To attempt virus isolation, we established primary cultures of Morelia viridis liver and brain cells, which we inoculated with homogenates of lung tissue from infected individuals. Ultrastructural examination of concentrated cell culture supernatants showed the presence of nidovirus particles, and subsequent NGS analysis yielded the full genome of the novel virus Morelia viridis nidovirus (MVNV). We then generated an antibody against MVNV nucleoprotein, which we used alongside RNA in situ hybridization to demonstrate viral antigen and RNA in the affected lungs. This suggests that in natural infection MVNV damages the respiratory tract epithelium, which then results in epithelial hyperplasia, most likely as an exaggerated regenerative attempt in association with increased epithelial turnover.
IMPORTANCE Novel nidoviruses associated with severe respiratory disease were fairly recently identified in ball pythons and Indian pythons. Herein we report on the isolation and identification of a further nidovirus from green tree pythons (Morelia viridis) with fatal pneumonia. We thoroughly characterized the pathological changes in the infected individuals and show that nidovirus infection is associated with marked epithelial proliferation in the respiratory tract. We speculate that this and the associated excess mucus production can lead to the animals' death by inhibiting normal gas exchange in the lungs. The virus was predominantly detected in the respiratory tract, which renders transmission via the respiratory route likely. Nidoviruses cause sudden outbreaks with high rates of mortality in breeding collections, and most affected snakes die without prior clinical signs. These findings, together with those of other groups, indicate that nidoviruses are a likely cause of severe pneumonia in pythons.
KEYWORDS: Morelia viridis, NGS, epithelial hyperplasia, green tree python, nidovirus, pathogenesis, proliferative pneumonia, type II pneumocyte hyperplasia
INTRODUCTION
The green tree python, or Southern green python, Morelia viridis (Schlegel, 1872), is an oviparous boid constrictor snake with natural habitats in several Indonesian islands, Florida, Papua New Guinea, and northern Australia (1). In recent years, large numbers of M. viridis green tree pythons have been exported from Indonesia to Europe and the United States (2), and they have become increasingly popular in both private and zoological collections (3, 4). So far, knowledge about the infectious diseases that occur in M. viridis is limited (5).
Recently, fatal pneumonias have been reported in two other python species, ball pythons (Python regius) (6) and Indian pythons (Python molurus) (7); infection with novel nidoviruses was found to be the common denominator (8, 9). The described viruses are approximately equidistant from the two current genera, Torovirus and Bafinivirus, of the Torovirinae subfamily in the family Coronaviridae, order Nidovirales (8–15), in which they will likely form a novel genus (9).
The order Nidovirales comprises four families of complex positive-sense single-stranded RNA (ssRNA+) viruses, Arteriviridae, Mesoniviridae, Roniviridae, and Coronaviridae, which are also distinguished by their genome size: roniviruses (∼26 kb) and coronaviruses (26 to 33 kb) are known as “large nidoviruses” (16–18), arteriviruses are known as “small nidoviruses” (13 to 16 kb), and mesoniviruses have genomes with sizes intermediate between those of the large and small nidoviruses (16, 17, 19, 20). While roniviruses and mesoniviruses are known to infect crustaceans and insects (19, 21), arteriviruses have been associated with an acute respiratory syndrome and abortion in pigs (porcine reproductive and respiratory syndrome virus [PRRSV]) or a lethal hemorrhagic disease in nonhuman primates (simian hemorrhagic fever virus [SHFV]) (22). The Coronavirinae subfamily of the Coronaviridae includes several pathogens of mammals (feline coronavirus, transmissible gastroenteritis virus, and equine coronavirus, to mention a few), birds, and fish and has provided a steady supply of emerging threats to human health over the past 15 years, including severe acute respiratory syndrome (SARS) and Middle Eastern respiratory syndrome (MERS) (23). The members of the second subfamily of the Coronaviridae, Torovirinae, have so far not generated similar threats. They include bafiniviruses, which infect ray-finned fish and induce renal tubular necrosis and necrotizing hepatitis (24), and toroviruses, which infect mammals (including humans, cattle, horses, and pigs) (10, 11, 25–27). Toroviruses have a tropism for epithelial cells of both the respiratory and alimentary tracts (28–30); in cattle, an association with pneumonia has been reported (30, 31). Toroviruses exhibit a unique morphology: the viral particles are kidney and/or rod shaped, and toroviruses have a tubular, torus-shaped ribonucleoprotein (RNP) enveloped by a membrane decorated with spikes composed of S protein (32). The RNP comprises the nucleoprotein (N protein) and the ssRNA+ genome (33, 34).
In 2014, a Swiss breeder submitted two adult green tree pythons for diagnostic postmortem examination. Both animals had died with signs of a mucinous pneumonia. In the following year, the same breeder submitted another two individuals. All of these green tree pythons, as well as a further five green tree pythons from three additional breeders, exhibited similar pathological findings. In 2016, additional cases involving two more breeding collections were observed. As all routine diagnostic tests undertaken failed to identify potential causative viruses or specific bacterial agents, we initiated an investigation into the cause of this apparently emerging disease of green tree pythons.
RESULTS
Animals, clinical signs, macroscopic and histological features, and results of screening for infectious agents.
From 2014 to 2016, multiple Morelia viridis green tree pythons affected by a similar disease arrived for postmortem examination: four animals from one breeder (animals A1 and A2 in 2014 and animals A3 and A4 in 2015), five animals from three additional breeders (animals B1, B2, C1, C2, and D1 in 2015), and three animals from three additional breeding collections (animals E1, F1, and G1 in 2016). Of these 12 snakes, 4 had died suddenly without any obvious clinical signs and 7 had exhibited respiratory distress and expulsion of mucus for a period of a few hours to more than 1 week before death; no clinical history was available for 1 animal. All animals were adult and ranged from 1 to 8 years of age (Table 1). Six snakes were female, and six were male. Upon gross postmortem examination, all snakes exhibited a variable amount of mucoid material in the airways, and the mucoid material was most abundant in the faveolar spaces of the lungs. In some animals, the entire trachea, the internal choanae, and the caudal air sacs were obliterated by mucoid material (Fig. 1), and the lung parenchyma appeared thickened. The histological examination revealed a variable degree of epithelial thickening in the trachea and lungs and a mild to moderate interstitial lymphoplasmacellular and heterophilic infiltration of the lung parenchyma (Fig. 2). In one animal (animal E1), a moderate multifocal granulomatous-necrotizing nephritis was additionally observed. The four control snakes (animals G2 to G6), i.e., animals that had been euthanized due to nonrespiratory diseases (Table 1), did not exhibit similar gross and/or histological changes (Fig. 2).
TABLE 1.
Animals tested in this study
| Animala | Age (yr) | Sexb | Clinical signs | Diagnostic testsc | Diagnosis |
|---|---|---|---|---|---|
| MVNV-positive animals | |||||
| A1 | 1 | M | No history | Bacteriology | |
| A2 | 5 | F | Sudden death | NGS | |
| A3 | 8 | M | Sudden death, mucus in oral cavity | Bacteriology | |
| A4 | npd | F | Acute respiratory distress (1 day) | ||
| B1 | 6 | M | Respiratory distress (3–4 days), expulsion of mucus | Virology, virus isolation, NGS | |
| B2 | 7 | F | Respiratory distress (3–4 days), expulsion of mucus | Virology | |
| C1 | 2.5 | M | Chronic anorexia, acute respiratory distress | Virology | |
| C2 | 8 | M | Respiratory distress | Virus isolation | |
| D1 | 6 | F | Respiratory distress (≥1 wk) | NGS | |
| E1 | np | F | Respiratory distress (≥1 wk) | ||
| F1 | 2 | F | Sudden death | ||
| G1 | 6 | F | Sudden death | ||
| Uninfected control animals | |||||
| G2 | 9 | M | Sudden death | Mild colitis | |
| G3 | 6 | F | Emaciation, death | Mild enteritis and nephritis | |
| G4 | 2 | F | Sudden death | BIBD,e severe purulent vasculitis around esophagus and trachea | |
| G5 | 1 | M | Sudden death | Focal suppurative pneumonia in one lobe |
All animals were from breeding collections. MVNV-positive animals tested positive for MVNV by RT-PCR, RNA-ISH, and IH. MVNV-negative animals were negative for MVNV by RT-PCR.
M, male; F, female.
Tests performed prior to or alongside MVNV RT-PCR, IH, and RNA-ISH: bacteriology, routine bacteriological examination performed on lungs; virology, lung tissue screened for viruses (reovirus, paramyxovirus, Sunshine virus, nidovirus) in a commercial laboratory; NGS, next-generation sequencing of lung homogenates.
np, not provided (adult).
BIBD, boid inclusion body disease.
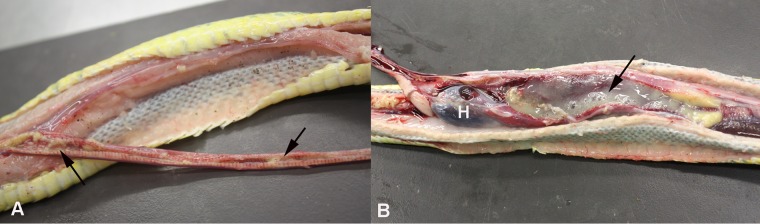
FIG 1.
Gross findings in a green tree python (Morelia viridis) with nidovirus-associated proliferative pneumonia (animal E1). Both trachea (A) and lungs (B) are filled with abundant mucoid material (arrows). H, heart.
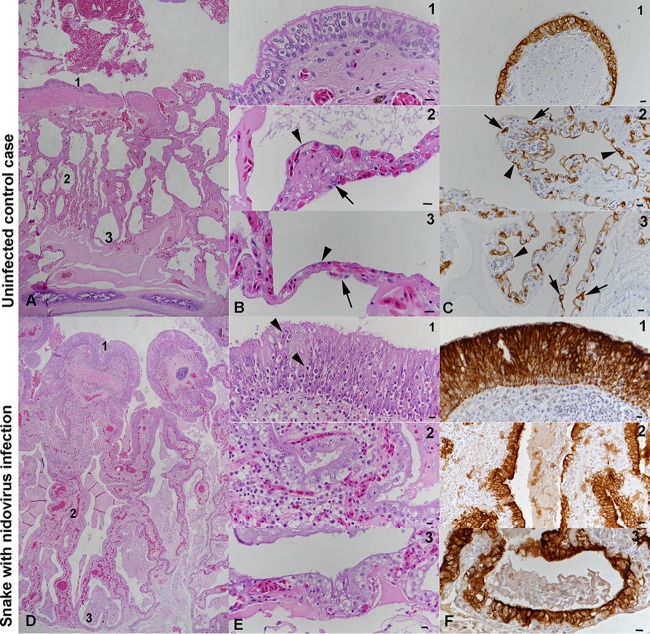
FIG 2.
Histological findings in Morelia viridis with nidovirus-associated proliferative pneumonia. (A to C) Uninfected control case (animal G2). Representative photomicrographs of a healthy lung (cross section) are shown. Thin pulmonary septa form the faveolar spaces. At the luminal end, the septa exhibit bundles of smooth muscle cells (myoelastic bundles) that form contractile trabeculae. For detailed assessments, three regions (regions 1, 2, and 3) were identified. The trabeculae are covered by a multilayered pseudostratified bronchus-type epithelium, dominated by ciliated cells (panels B1 and C1). In regions 2 (panels B2 and C2) and 3 (panels B3 and C3), the faveolae are covered by a single-layered gas exchange epithelium, comprised of flat type I pneumocytes (arrowheads); surfactant-producing type II pneumocytes (arrows) are less abundant. (D to F) Snake with nidovirus infection (animal B4). Representative photomicrographs of a diseased lung (cross section) are shown. In the entire lung, the epithelial layer is thickened due to hyperplasia. The faveolar space (region 3) is filled with proteinaceous material, and the interstitium is broadened due to a mixed inflammatory infiltrate (panels E1 and E2). In region 1 (panels E1 and F1), the pseudostratified epithelium covering the trabeculae exhibits increased cellularity with an increase in cell layers and an irregular arrangement (hyperplasia), as well as a loss of cilia and several apoptotic epithelial cells (arrowheads). In regions 2 (panels E2 and F2) and 3 (panels E3 and F3), type I pneumocytes are almost entirely replaced by large, columnar type II pneumocytes. H&E stain (A, B, D, and E); IH for cytokeratin, hematoxylin counterstain (C and F). Bars = 100 μm (A, D) and 10 μm (B, C, E, F).
We suspected an infectious cause and had routine bacteriological examinations performed on the lungs of selected snakes from the 2014 and 2015 cohort (animals A1, A3, and D1). This yielded only a nonspecific bacterial flora (i.e., no primary pathogens, Pseudomonas aeruginosa, a Proteus sp., Citrobacter braakii, Achromobacter xylosoxidans, Stenotrophomonas maltophilia, and Providencia rettgeri). A further three animals, from the 2015 cohort (animals B1, B2, and C1), were then screened in a commercial laboratory for a range of potentially pathogenic viruses (i.e., reovirus, paramyxovirus, Sunshine virus, nidovirus); these tests yielded negative results.
Virus isolation in tissue cultures.
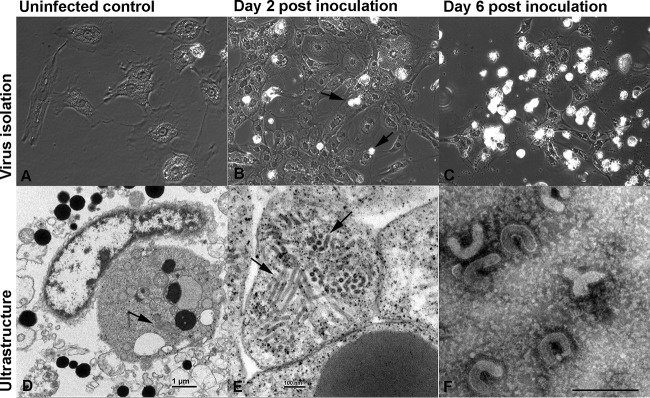
As the routine bacteriological and virological tests failed to identify a common denominator for the diseased snakes, we decided to attempt virus isolation. For this purpose, we prepared primary cultures of green tree python fetal liver and brain cells (Fig. 3A). After four passages, the cells began to proliferate more rapidly and we could expand the cultures enough to attempt isolation of the unknown pathogen. When we used homogenates of lung tissue from the diseased snakes (Table 1) to inoculate both cultures, we observed the first evidence of a cytopathic effect (CPE) (enlargement, rounding, and cytoplasmic vacuolization of cells) and a loss of adherence (Fig. 3B) at approximately 3 days postinfection (dpi). At 6 dpi, almost the entire monolayer was affected and most cells had rounded and/or detached (Fig. 3C). The ultrastructural examination of a cell pellet prepared from the cultures at 1 dpi identified abundant tubular viral structures arranged in stacks within the cytoplasm of infected cells (Fig. 3D and E). We concentrated the supernatants of the infected cells by ultracentrifugation and examined the pelleted material by transmission electron microscopy (TEM) under negative staining. TEM analysis demonstrated rod- and kidney-shaped virions of approximately 120 nm in length (Fig. 3F), consistent with torovirus particles (29, 35).
FIG 3.
Virus isolation in a primary Morelia viridis fetal brain cell culture. (A) Uninfected control cells. (B, C) Cells after inoculation with homogenate of lung tissue from a diseased snake (animal C2). The first evidence of a cytopathic effect is seen after 2 days (B) as enlargement, rounding, and cytoplasmic vacuolization as well as a loss of adherence (arrows). After 6 days (C), only a few intact cells have remained. Most cells are detached and necrotic. (D, E) Ultramicrographs of a cell pellet prepared at 1 dpi. There are abundant tubular structures arranged in stacks within the cytoplasm of infected cells (arrows). (F) Concentrated supernatant of the infected cells, negative staining. Rod- and kidney-shaped virions are approximately 120 nm long. Bars = 1 μm (D) and 100 nm (E).
Identification of nidovirus by NGS and confirmation of the findings by real-time RT-PCR.
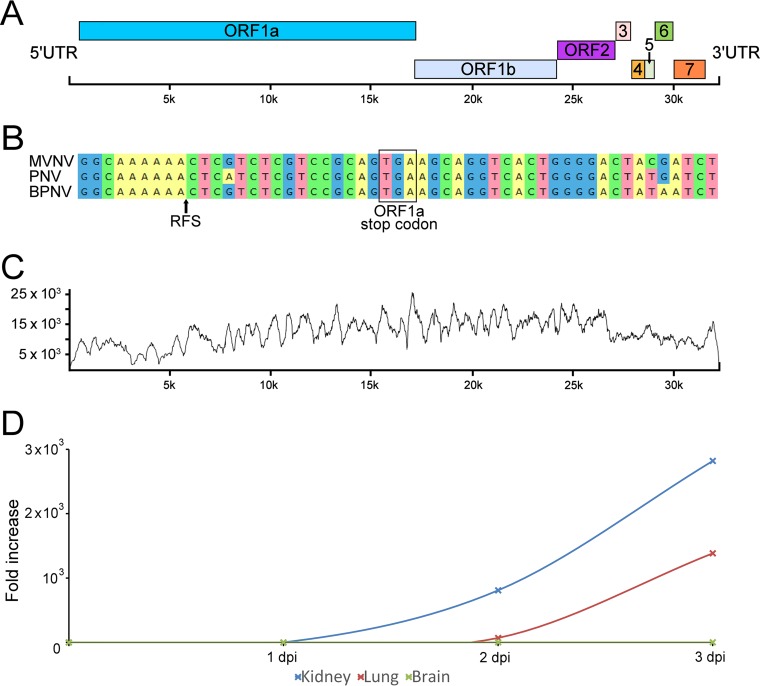
In parallel with our attempts at virus isolation, we decided to employ next-generation sequencing (NGS) for the identification of the causative agent(s) of the pneumonia. We isolated RNA from homogenates of lung tissue from three animals (animals A2, B1, and D1) and performed RNA sequencing. For two snakes (animals B1 and D1), only contiguous sequences (contigs) that matched the sequences of bacterial genomes were identified. We interpreted these bacterial sequences to most likely represent either contamination during postmortem sample collection or, more likely, a secondary bacterial infection, as both had also shown moderate diffuse heterophil infiltration in the lungs. From the third snake (animal A2), however, we obtained a single ∼21,000-nucleotide (nt) contig and several shorter contigs that matched the previously identified sequences of ball python nidovirus (BPNV) and python nidovirus (PNV) (the 21,000-nt contig was ∼85% identical to the sequences of both viruses) (8, 9). In total, we obtained ∼28,000 nt (∼75 to 80%) of the genome from the lung tissue sample. In addition to the novel nidovirus isolate, we identified a contig that matched the sequences of endogenous retrovirus genes with very high coverage; we will describe this contig in a separate report (unpublished data). However, the retrovirus was also present in the supernatants of M. viridis cell cultures inoculated with homogenates of lung tissue. We decided to attempt to produce a nidovirus preparation devoid of the contaminating retrovirus. To do this, we inoculated boid kidney cells with the supernatant collected from the Morelia viridis liver cell culture inoculated with homogenates of lung tissue, since Huder et al. previously reported species-restricted growth of an endogenous python retrovirus (36). Using this approach, we obtained a pure nidovirus isolate, as confirmed by retrovirus-specific reverse transcription-PCR (RT-PCR). We then performed endpoint titration to quantify the number of infectious units in the Morelia viridis nidovirus (MVNV) stock on M. viridis brain cells and could detect 2.25 × 1010 focus-forming units per 1 ml of cell culture supernatant. RNA extracted from the pure nidovirus preparation was subjected to another NGS run, which yielded almost a full-length genome in a single contig. We then performed a reference assembly using BPNV as the template to recover the missing genome ends (some 150 nt in total) for the novel virus, MVNV. The overall genome structure of MVNV (GenBank accession number MF351889) was similar to that of BPNV and PNV (Fig. 4A). The identified contig contains eight partially overlapping open reading frames (ORFs) flanked by the 5′ untranslated region (UTR) (∼650 nt) and the 3′ UTR (∼920 nt). We used the HMMER3 web server to look for functional domains in the detected ORFs, and these are described in Table 2. We also identified a putative ribosomal frameshift signal (RFS) sequence near the end of ORF1a. To further study the RFS, we aligned the sequences of other nidoviruses identified in snakes around the RFS and used the Mfold web server to predict the structure around this region in the MVNV genome (Fig. 4B). The NGS data coverage for the MVNV genome (raw data are available at http://www.ncbi.nlm.nih.gov/biosample/7248312) is shown in Fig. 4C. The identities between the nucleotide and amino acid sequences of the novel nidovirus isolate and those of other python nidoviruses are indicated in Table 3.
FIG 4.
Morelia viridis nidovirus (MVNV) genome organization, NGS coverage, and phylogenetic relationship to related viruses. (A) A schematic representation of the untranslated regions (UTRs) and open reading frames (ORFs) of the 32,414-nt ssRNA+ genome of MVNV. (B) Alignment around the ribosomal frameshift signal (RFS; indicated by an arrow in the alignment and structure prediction) in nidoviruses identified in pythons. The ORF1a stop codon is highlighted. A prediction for the RNA structure around the RFS in MVNV is shown. The arrow highlights a difference from the structure described by Bodewes et al. (8). (C) Contig coverage. The y axis shows the number of reads matching each nucleotide position (x axis) of the contig. The Bowtie2 program was used to map the reads to the contig generated by de novo assembly. (D) Virus growth in different cell lines. The cell culture supernatant of the indicated cell lines inoculated with MVNV was analyzed by qRT-PCR at the indicated time points. The fold increase indicates the increase in the amount of viral RNA in the cell culture supernatant compared to the amount detected at 0 dpi.
TABLE 2.
HMMER3 analysis of functional domains in the ORFs detected in the MVNV genome
| ORF | Family identifier | Pfam accession no.a | Alignment |
Bit score | E value |
|||
|---|---|---|---|---|---|---|---|---|
| Start position | End position | Independent | Conditional | Description | ||||
| ORF1a | Methyltransf_25 | PF13649.5 | 3075 | 3150 | 15.08 | 0.027 | 4.80E−06 | Methyltransferase domain |
| zf-CCCH | PF00642.23 | 1052 | 1072 | 12.09 | 0.13 | 2.40E−05 | Zinc finger C-X8-C-X5-C-X3-H type | |
| zf-CCCH | PF00642.23 | 1098 | 1117 | 9.91 | 0.64 | 0.00011 | Zinc finger C-X8-C-X5-C-X3-H type | |
| ORF1b | NSP13 | PF06460.11 | 2077 | 2239 | 50.2 | 1.80E−13 | 1.20E−16 | Coronavirus NSP13 |
| Viral_helicase1 | PF01443.17 | 1144 | 1380 | 45.91 | 5.20E−12 | 3.40E−15 | Viral (superfamily 1) RNA helicase | |
| AAA_30 | PF13604.5 | 1101 | 1231 | 42.93 | 3.90E−11 | 2.60E−14 | AAA domain | |
| RdRP_1 | PF00680.19 | 479 | 651 | 40.69 | 1.00E−10 | 6.60E−14 | RNA-dependent RNA polymerase | |
| AAA_19 | PF13245.5 | 1100 | 1229 | 33.98 | 3.10E−08 | 2.00E−11 | AAA domain | |
| ORF5 | Arteri_nucleo | PF01481.15 | 32 | 132 | 23.35 | 5.50E−05 | 9.90E−09 | Arterivirus nucleocapsid protein |
TABLE 3.
Nucleotide and amino acid sequence identities between MVNV and the nidoviruses identified in Python molurus (Indian python) and Python regius (ball python)
| ORF | % identity |
|||
|---|---|---|---|---|
| Nucleotide sequence |
Amino acid sequence |
|||
| Python nidovirus S1536/13 (KJ935003)a | Ball python nidovirus 07-53 (KJ541759) | Python nidovirus S1536/13 (KJ935003) | Ball python nidovirus 07-53 (KJ541759) | |
| 5′ UTR | 85.1 | 87.0 | ||
| ORF1a | 72.8 | 72.8 | 69.5 | 69.6 |
| ORF1b | 86.5 | 86.3 | 91.0 | 90.5 |
| ORF2 | 80.9 | 80.0 | 85.0 | 84.8 |
| ORF3 | 69.5 | 67.0 | 64.7 | 65.4 |
| ORF4 | 76.5 | 79.2 | 83.3 | 82.9 |
| ORF5 | 75.1 | 74.0 | 74.2 | 74.8 |
| ORF6 | 65.6 | 66.9 | 59.7 | 59.2 |
| ORF7 | 77.1 | 78.6 | 68.8 | 71.9 |
| 3′ UTR | 95.2 | 93.9 | ||
| Complete genome/CDSb | 77.3 | 77.3 | 77.2 | 77.1 |
GenBank accession numbers are given in parentheses.
CDS, coding sequence.
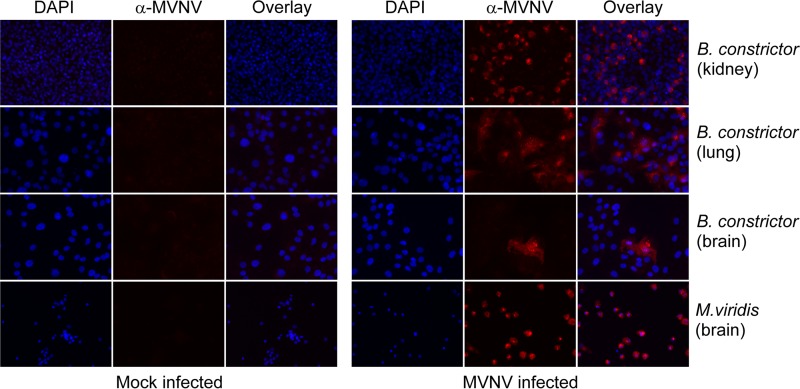
We then tested whether the isolated MVNV could infect other cell lines. For this purpose, we selected brain, kidney, and lung cell lines of Boa constrictor and used M. viridis brain cells as the positive control. We also designed a TaqMan real-time RT-PCR to be able to quantify and monitor the nidovirus RNA in the cell culture supernatants and lung tissue samples from the diseased snakes. Analysis of the cell culture supernatants collected from different cell lines inoculated with MVNV showed clear amplification in B. constrictor kidney and lung cells (Fig. 4D). The cytopathic effect on M. viridis brain cells was extremely severe, and therefore, a similar quantification could not be performed. The infected cells were stained using anti-MVNV N protein antiserum at 3 dpi, and all cell lines, except B. constrictor brain cells (only a few infected cells were detected), were shown to be extremely permissive for MVNV (Fig. 5).
FIG 5.
Immunofluorescence staining of M. viridis and B. constrictor cell lines with anti-MVNV N protein antiserum at a 1:2,000 dilution. MVNV- or mock-infected cells were stained at 3 dpi, and images were recorded at a magnification of ×40 under an inverted fluorescence microscope. (Right) Mock-infected cells; (left) MVNV-infected cells. Blue, DAPI; red, MVNV-specific staining.
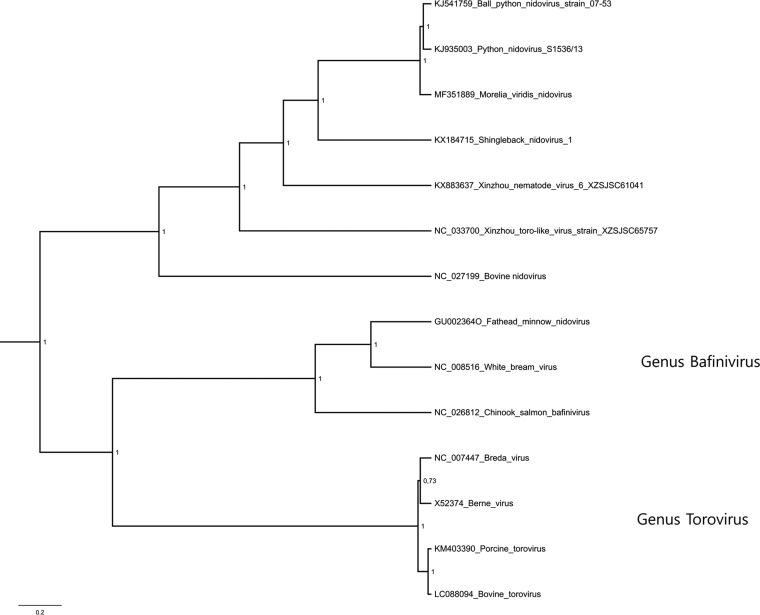
As ORF1b is the most conserved ORF among nidoviruses (17), we selected this region for the phylogenetic analysis. The phylogenetic tree suggests that MVNV forms an outgroup to BPNV and PNV (Fig. 6). Further, the python nidoviruses clustered together with shingleback nidovirus (GenBank accession number KX184715) (37), Xinzhou nematode virus 6 (GenBank accession number KX883637), Xinzhou toro-like virus (NCBI reference sequence accession number NC_033700), and bovine nidovirus (NCBI reference sequence accession number NC_027199) (31). Consistent with the previous reports, these form a clade that is separate from both the genus Bafinivirus and the genus Torovirus (Fig. 6).
FIG 6.
Maximum clade credibility tree constructed from the ORF1b amino acid sequences of representatives of the subfamily Torovirinae. The phylogenetic tree was constructed using the Bayesian MCMC method with the LG+G+I model of substitution. Posterior probabilities are shown at each node.
Finally, we used RNA isolated from cell cultures inoculated with an homogenate of lung tissue from an infected snake as the positive control and RNA isolated from a Boa constrictor snake as the negative control in the real-time RT-PCR. All diseased snakes tested positive for nidovirus RNA in the lungs. In contrast, the four snakes without gross and/or histological evidence of pneumonia were negative.
Nidoviruses are associated with pneumonia in M. viridis.
The histological examination confirmed that all diseased snakes had suffered from a chronic pneumonia with epithelial thickening in the trachea and lungs (Fig. 2) and excess mucus in the lumen of the lungs and airway. The inflammatory component was represented by mild to moderate multifocal interstitial infiltration of lymphocytes, plasma cells, and/or heterophils (Fig. 2E). The mucus filling the faveolar space often contained heterophils and cell debris. In three of the nine tracheas examined, we also observed a variable degree of infiltration with a mixture of inflammatory cells (heterophils, macrophages, lymphocytes). Over its entire length, the lung epithelium exhibited numerous epithelial cells that contained mucus, as indicated by periodic acid-Schiff (PAS)–alcian blue staining (Fig. 7A, C, and E). In region 1, at the trabeculae, the mucus-containing cells had the ultrastructural features of secretory cells (38) (Fig. 7B). In regions 2 and 3 (faveolar epithelium), where the increase in the amounts of these cells was the most striking, they exhibited the morphology of type II pneumocytes (Fig. 7D and F) (39); their numbers varied in affected snakes and ranged from occasional patchy aggregates to a diffuse lining of the entire faveolae in association with an almost complete absence of type I pneumocytes (Fig. 2E). Ultrastructurally, these cells exhibited serous/mucous granules instead of the lamellar bodies that are characteristic of type II pneumocytes (Fig. 7B, D, and F); they have recently been described to be transformed type II pneumocytes (40).
FIG 7.
Changes in the lung epithelium of Morelia viridis green tree pythons with nidovirus-associated proliferative pneumonia. Representative photomicrographs of cross sections of the diseased lungs of animal B4 (A, C, E) and ultramicrographs of cross sections of the diseased lungs of animal A1 (B, D, and F) are shown. (A, B) Region 1. (A) The PAS-alcian blue stain highlights positive secretory epithelial cells (arrowheads) in all cell layers of the hyperplastic multilayered trabecular epithelium. Abundant mucus fills the lumen. (B) The ultrastructural features of most cells (cytoplasmic mucous granules and short microvilli) show that they are secretory cells (38). Only a few ciliated cells are seen (arrow). (C to F) In regions 2 (C, D) and 3 (E, F), the faveolae are diffusely lined by cuboidal cells containing serous/mucous granules (arrowheads, C and E), consistent with type II pneumocytes. Ultrastructurally, these cells resemble so-called transformed type II pneumocytes, as they contain serous/mucous granules (arrows) and lack the lamellar bodies characteristic for normal type II pneumocytes (39). C, capillary.
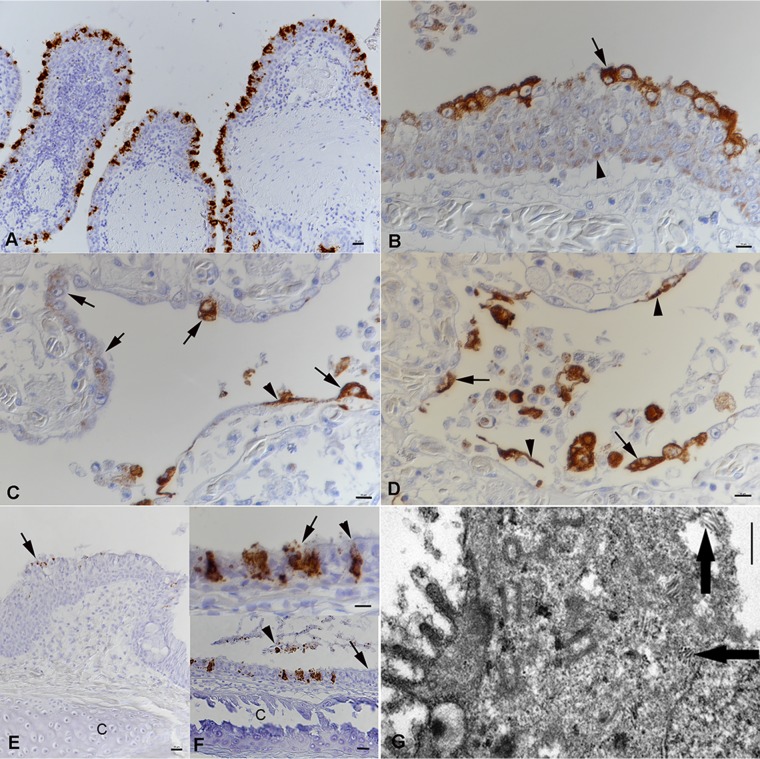
Having detected nidoviral RNA in the affected lungs, we aimed to identify the viral target cells. For this purpose, we employed both RNA in situ hybridization (RNA-ISH) and immunohistology (IH). Viral RNA and antigen (N protein) were detected in the cytoplasm of the trabecular pseudostratified epithelium (region 1) and the cytoplasm of both type I and type II pneumocytes lining the faveolar space (regions 2 and 3) in all affected snakes (Fig. 8A to D). Intact and degenerated cells shed into the mucus were found to be infected, and occasionally, we also detected cell-free viral RNA and antigen (Fig. 8B). Infected cells also varied in number between animals and in different areas of the lung in individual snakes. In the hyperplastic epithelium of region 1, most cells exhibited a weak, focal cytoplasmic reaction, and only the superficial cell layer contained abundant viral antigen (Fig. 8B). The new type II pneumocytes that seemed to progressively replace the type I cells exhibited a similarly weak focal reaction, whereas the fully differentiated pneumocytes were strongly positive for viral antigen (Fig. 8C and D), suggesting more efficient virus replication. When examined, the trachea (n = 9) and the nasopharyngeal epithelium (n = 1) also exhibited infected, degenerating epithelial cells (Fig. 8E and F). Viral antigen was detected in few epithelial cells of the esophagus in one animal with acute pneumonia (animal G1). Interestingly, viral antigen was also found cell free and within a few macrophages in the focal granulomatous-necrotizing nephritis in another affected animal (animal E1). We did not detect viral RNA or antigen in other tissues of the infected snakes, including the stomach and intestines, or in the lungs of the control snakes (animals G2 to G6).
FIG 8.
Viral target cells in lungs and airways of Morelia viridis green tree pythons with nidovirus-associated proliferative pneumonia. (A to D) The lungs of animals A1 (A) and A2 (B to D). (A) The pseudostratified trabelular epithelium (region 1) exhibits abundant viral RNA mainly in superficial epithelial cell layers. (B) Viral antigen expression is seen in all cell layers. In the basal and middle layers, it is represented by a focal cytoplasmic reaction (arrowhead). The fully differentiated superficial epithelial cells exhibit abundant viral antigen (arrow) and a loss of cilia. Infected cells are also found to be shed into the lumen. (C) Region 2. In areas with intact faveolar epithelium, both type I pneumocytes (arrowhead) and differentiated type II pneumocytes (arrows) exhibit strong viral antigen expression. The new, hyperplastic type II pneumocytes in other areas display a weaker focal cytoplasmic reaction (arrows). (D) At the base of the faveolae, type I pneumocytes (arrowheads) and type II pneumocytes (arrows) are also found to be infected. They are also shed into the faveolar lumen. (E) Nasal cavity of animal A1. The epithelium is thickened due to hyperplasia and exhibits nidovirus infection mainly of the fully differentiated superficial epithelial cells (arrow). C, cartilage. (F) Trachea of animal A1. (Bottom) The ciliated epithelium is largely intact (arrow), but there are scattered individual cells or small groups of infected cells. Sloughed, degenerate epithelial cells are also found to carry viral RNA (arrowhead). (Top) Infected cells are intact (arrowhead) or degenerate, shedding virus (arrow). C, tracheal ring cartilage. RNA-ISH (RNAscope technology) (A, E, F) and IH (HRP method) for the detection of MVNV N protein (B to D) and hematoxylin counterstain were used. (G) Ultramicrograph of a type II pneumocyte with staples of tubular structures of 100 to 150 nm in length (arrows). Bars = 20 μm (A, E, and F [bottom]), 10 μm (B to D and F [top]), and 1 μm (G).
We could demonstrate cytoplasmic accumulations of tubular structures that ranged from 150 nm to 250 nm in length in rare epithelial cells that had the morphology of type II pneumocytes by ultrastructural examination (Fig. 8G).
The epithelial thickening was the most striking feature in the lungs of the nidovirus-positive snakes. We therefore performed a more detailed histological examination in the attempt to identify the processes underlying this phenomenon. The multilayered epithelium covering the trabeculae (region 1) displayed increased cellularity with an increase in the number of cell layers and an irregular arrangement, indicating hyperplasia (Fig. 2E). The epithelial hyperplasia extended to the upper respiratory tract and was also observed in the trachea, larynx, and nasal cavity. The middle and basal areas of the faveolae (regions 2 and 3), where the epithelium is unilayered, showed nuclear crowding, which also suggested hyperplasia (Fig. 2E). The thickening appeared to result from an increase in individual cell height, associated with a more columnar appearance of the cells (Fig. 2E and F). Additionally, we noted an increase in septal connective tissue (interstitial fibrosis). The described changes resulted in thickening of the septa and narrowing of the faveolar lumen (Fig. 2D).
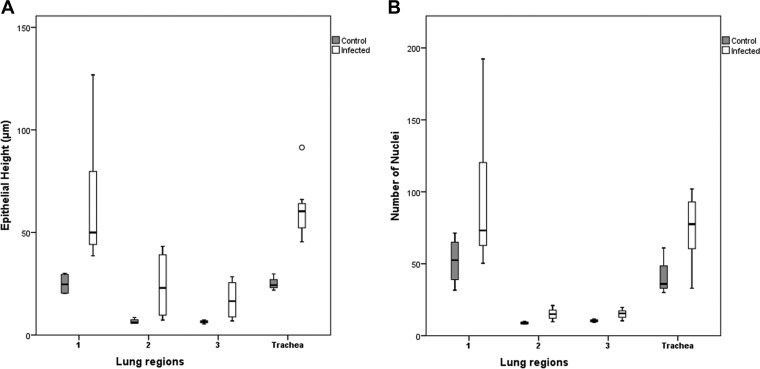
In the attempt to quantify the epithelial hyperplasia, we measured the average total epithelial height in the three defined regions (Fig. 2 and 9) in all diseased, i.e., RT-PCR-positive, animals and compared it to that in the RT-PCR-negative animals without pneumonia (controls). In all locations, the epithelium was significantly (P < 0.05) higher in the diseased animals than in the control snakes (Fig. 10A; Table 4). Hyperplasia was confirmed in both the lungs (regions 1, 2, and 3) and the trachea, where the average number of nuclei in the epithelial layer was significantly higher in the diseased snakes (Fig. 10B; Table 4).
FIG 9.
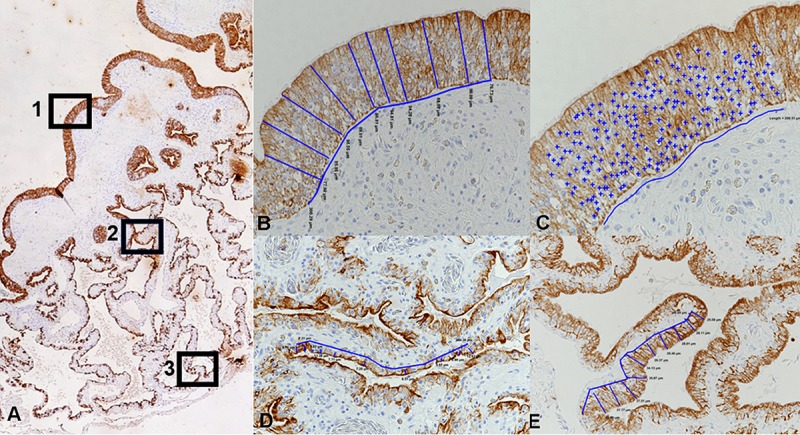
Illustration of the morphometric approach taken to measure the epithelial height and number of epithelial cell nuclei in cytokeratin-stained cross sections of the lungs. (A) Three regions were defined in the proximal, bronchiolar lung: (i) the luminal part of the primary trabeculae, covered by a multilayered pseudostratified ciliated epithelium (region 1), and (ii) the faveolae at the middle level (region 2) and at the base (region 3) covered by a single-layered epithelium. (B) Measurement of epithelial height in region 1. The blue lines identify the locations of the 10 measurements taken over a 300-μm-wide epithelial segment, determining the distance between the apical epithelial cell border and the basal membrane. Magnification, ×200. (C) Determination of cell number in region 1. The total amount of epithelial nuclei (+) counted in a 200-μm-wide epithelial segment. Magnification, ×400. (D, E) Measurement of epithelial height in regions 2 (D) and 3 (E). The blue lines identify the locations of the 10 measurements taken over a 300-μm-wide epithelial segment, determining the distance between the apical epithelial cell border and the basal membrane. Magnification, ×200.
FIG 10.
The average epithelial height measured in three lung regions and the trachea of infected animals was significantly higher than that in noninfected (control) animals. Box plots illustrate the third quartile (Q3) and first quartile (Q1) range of the data, and the open circle represents a data outlier (>1.5 times the interquartile range). Box plots were created with SPSS Statistics software. A Mann-Whitney U test for independent samples (t test) was used for statistical analysis, and a P value of less than 0.05 was considered significant.
TABLE 4.
Results of quantitative assessment of epithelial height and cellularity in lungs and tracheas of MVNV-positive animals in comparison to uninfected control animalsa
| Tissue and regionb | MVNV-positive animals |
Uninfected control animals |
||
|---|---|---|---|---|
| Epithelial ht (μm) | No. of nuclei | Epithelial ht (μm) | No. of nuclei | |
| Lung | ||||
| Region 1 | 62.97 ± 25.39 | 62.97 ± 25.39 | 24.96 ± 26.13 | 6.54 ± 0.81 |
| Region 2 | 24.4 ± 14.16 | 14.97 ± 3.57 | 6.61 ± 14.59 | 8.83 ± 0.76 |
| Region 3 | 17.43 ± 8.1 | 15.22 ± 2.66 | 6.46 ± 8.41 | 10.33 ± 0.85 |
| Trachea | 61.5 ± 14.06 | 82.33 ± 39.15 | 28.21 ± 5.74 | 44.25 ± 12.09 |
MVNV-positive animals (n = 12) tested positive by RT-PCR, RNA-ISH, and IH, and uninfected control animals (n = 4) were negative for MVNV by RT-PCR and IH. Data represent means ± standard deviations. For all regions and the trachea, the values were significantly different between infected and control animals (P < 0.05).
Region 1, multilayered pseudostratified epithelium covering the luminal part of the primary trabeculae; region 2, single layer of epithelium at the middle level of the faveolae; region 3, single layer of epithelium at the base of the faveolae.
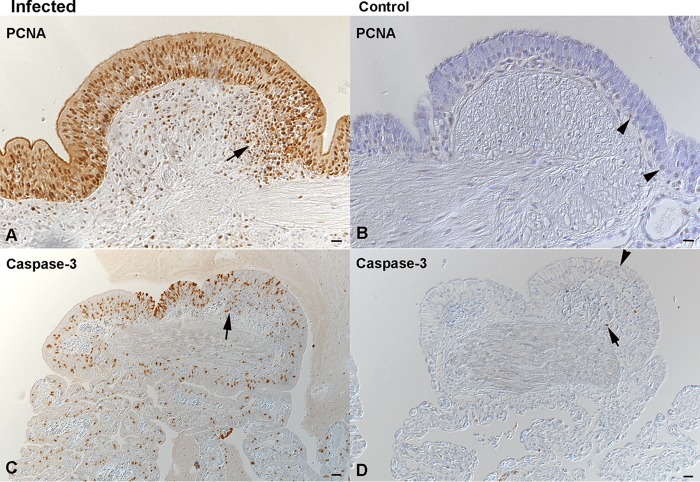
Epithelial hyperplasia was associated with increased proliferative activity, as up to 90% of epithelial cells in all lung regions and in the trachea of the diseased animals were found to express PCNA (Fig. 11A). Alongside this, a moderate number of epithelial cells were found to undergo apoptosis, based on cleaved caspase-3 staining (Fig. 11C). In comparison, control animals exhibited rare PCNA-positive type II pneumocytes in all layers (Fig. 11B) as well as scattered apoptotic (cleaved caspase-3-positive) type I and type II pneumocytes (Fig. 11D), likely representing the physiological turnover of the epithelium.
FIG 11.
Increased epithelial turnover in the lungs of Morelia viridis green tree pythons with nidovirus-associated proliferative pneumonia. (A, C) Trabeculae (region 1) of diseased animal C1. The hyperplastic pseudostratified epithelium is proliferating, as confirmed by the expression of PCNA in almost all epithelial cells. Infiltrating lymphocytes (arrow, A) are also positive. There are also abundant, cleaved caspase 3-positive apoptotic epithelial cells (C). A proportion of infiltrating leukocytes is also found to undergo apoptosis (arrow, C). (B, D) Trabeculae (region 1) of control animal G3. In the control animal, only scattered individual basal epithelial cells are found to express PCNA (arrowhead, B). Similarly, only rare epithelial cells are found to undergo apoptosis (arrowhead, D). Apoptosis (i.e., cleaved caspase-3 expression) is also seen in occasional lymphocytes (arrow) of the lymphoid aggregates. IH (HRP method) was used to detect PCNA (A, B) and cleaved caspase-3 (C, D). Hematoxylin counterstain was used. Bars = 20 μm.
DISCUSSION
We initiated the present study by the urge to identify the causative agent of a fatal pneumonia observed in green tree pythons (Morelia viridis), characterized by the accumulation of mucoid material in the airways and a histologically notable thickening of the lung epithelium. NGS revealed the presence of a novel nidovirus, MVNV, which by phylogenetic analysis groups with toroviruses. RT-PCR, immunohistology, and RNA-ISH served to confirm its association with the disease and to identify the viral target cells, i.e., epithelial cells in the airways and the luminal trabeculae, as well as faveolar type I and II pneumocytes. We were able to isolate and grow MVNV in both M. viridis and B. constrictor cell cultures. MVNV induced a cytopathic effect both in vitro and in vivo. However, we also found nidovirus infection to associate with generalized hyperplasia of the airway and lung epithelium, which exhibited a distinct proliferative activity and a degree of apoptotic cell death. Together these findings would suggest that nidovirus infection increases the turnover of the epithelium. The mucus accumulation in the air-conducting space was accompanied by a significant increase in secretory epithelial cells at the trabeculae and in (transformed) type II pneumocytes in the faveolae, indicating increased mucus and/or surfactant production.
These results indicate that MVNV infects and damages the differentiated respiratory and faveolar epithelium but then persists and induces increased turnover of the infected epithelial cells. The observed type II pneumocyte hyperplasia is obviously not specific to nidovirus infection in the python, since it has been described in snakes as a consequence of pneumocyte injury in a range of infectious diseases of viral, bacterial, and mycotic origin (39, 40). It might therefore represent an exaggerated regenerative attempt. In mammals, type II pneumocyte hyperplasia is also seen to be part of a lung defense mechanism against various insults (41–43). In our cases, the hyperplastic epithelial cells exhibited abundant cytoplasmic serous/mucous granules instead of the lamellar bodies that represent surfactant (38). This suggests excess mucus production and release and reduced surfactant production and release and would explain the clinical findings (38, 40). Previous studies have shown that the surfactant of snakes, due to its phospholipid composition, which is different from that in other animals, is likely less important for airway stabilization but, rather, functions as an antiadherent (also known as an “antiglue”) factor and an antiedemic factor in the faveolar space (44); the reduction in the amount of surfactant could therefore have added to the respiratory distress observed in the affected snakes.
Increased turnover and/or hyperplasia of the epithelium has been described for several coronaviral diseases, such as infectious bronchitis in chickens, Breda virus infection in calves, and coronavirus infection in rats (45–47). So far, however, the mechanisms underlying this process have not been explained. Members of the subfamily Torovirinae are also known to induce epithelial cell apoptosis (35, 48, 49); however, though our findings point toward this, further studies are required to elucidate whether this also applies to MVNV and other snake nidoviruses.
In our study, only fatal cases were examined. The pathological findings suggest that death was mainly due to impaired gas exchange as a consequence of type I pneumocyte loss. In the healthy snake lung, thin cytoplasmic extensions of type I pneumocytes cover the capillary walls, forming the gas-blood barrier (38). Their replacement by mucus/surfactant-secreting type II pneumocytes with their excessive height due to cell crowding is unlikely to allow effective gas exchange. With an average thickness of 24.40 μm (region 2) and 17.43 μm (region 3), the barrier was more than 1.7 × 104 times thicker than the normal blood-gas barrier in the reptilian lung, which ranges from 0.4 to 1 nm (38).
Further anatomical peculiarities of the Morelia viridis lung (or the boid snake lung in general) could have contributed to the fatal outcome of the disease (39, 50–52). Boidae have a well-developed right lung and a rudimental left lung. The right lung displays two anatomically distinct regions: the anterior region, which contains the profusely compartmented gas exchange tissue, and the posterior saccular region, which is devoid of respiratory tissue and has therefore been referred to as the “air sac” (53, 54). The combination of elongated lungs and caudal air sacs may contribute considerably to the outcome of the disease, as they create a cul-de-sac that impairs removal of the mucus and thereby significantly reduces the amount of air-filled space and the gas exchange capacity.
Though toroviruses mainly associate with enteric diseases, recent studies have shown that they can be both entero- and pneumotropic (31, 55, 56). New nidoviruses were recently identified in the lungs of cattle and wild shingleback lizards with pneumonia, though their direct association with disease has so far not been examined (31, 37). We found MVNV-associated lesions almost exclusively in the airways and lungs, similar to previous reports on nidovirus infections in other python species (8, 9, 15). The detection of viral RNA by PCR in other tissues, such as the liver, spleen, kidney, and intestine, however, indicated that the viruses spread systemically. We found further evidence of that and of its pathogenicity, as we detected nidovirus N protein within a focal granulomatous-necrotizing nephritis in one animal. We also detected viral antigen in epithelial cells of the cranial esophagus in one affected animal; however, in M. viridis the esophagus carries a ciliated epithelium (data not shown), a feature also known for other snake species (57, 58); infection could therefore be due to an overspill from the trachea and nasal cavity. We did not detect viral antigen in any cells of the stomach or intestine, and a previous study also did not find viral RNA by RNA-ISH, suggesting that the python nidoviruses are primarily respiratory (8). Viral spread via the expelling of mucus from the nasal cavity would be a likely route of transmission.
MATERIALS AND METHODS
Animals.
The study was performed on 16 green tree pythons (Morelia viridis) from six breeding collections in Switzerland and one collection in Germany (Table 1). The collections varied in both size (i.e., the number of breeding animals) and species range, from a single snake up to a collection of 50 snakes of various species.
All animals were submitted to the Institute of Veterinary Pathology, Vetsuisse Faculty, University of Zurich, for diagnostic purposes. Fifteen snakes had died spontaneously, and one was euthanized following an Animals Scientific Procedures Act 1986 (ASPA) schedule 1 procedure (appropriate methods of humane killing [http://www.legislation.gov.uk/ukpga/1986/14/schedule/1]). In each case, a full diagnostic postmortem examination was performed with the owner's consent. For these diagnosis-motivated necropsies, no ethical permission is required at the University of Zurich.
The initial study population was represented by nine snakes, of which four (animals A1 to A4) were from one breeder (breeder A), two each were from a second breeder (breeder B; animals B1 and B2) and a third breeder (breeder C; animals C1 and C2), and one (animal D1) was from a fourth breeder (breeder D) (Table 1). These animals were submitted between September 2014 and November 2015. After completion of the next-generation sequencing (NGS) study, another seven snakes were submitted (animals E1, F1, and G1 to G5) by another three breeders (breeders E, F, and G). All initial snakes and another three snakes of the second cohort exhibited one common gross feature: the airways contained a variable amount of mucoid material which was most abundant in the faveolar lumen (Table 1). The remaining four snakes (animals G2 to G5) did not show any changes in the airways or lungs (Table 1) and later served as controls.
Sample collection and screening for infectious agents.
During postmortem examinations, samples from all organs were collected and fixed in 10% buffered formalin for histological examination. Additional samples from the brain, lung, liver, and kidney were stored at −80°C for further analysis. Also, lung tissue samples from the freshly euthanized snake (animal A2) and from two snakes that were necropsied within a few hours after death (animals B1 and D1) were fixed in glutaraldehyde-paraformaldehyde and processed for transmission electron microscopy (TEM) as described previously (59).
Prior to the performance of NGS, six of the initial cases were screened for infectious agents: the cultivation of bacteria from the lungs of three snakes (animals A1, A3, and D1) was attempted at the Institute of Veterinary Bacteriology, Vetsuisse Faculty, University of Zurich, and lung tissue samples from another three snakes (animals B1, B2, and C1) were submitted to a commercial lab (Laboklin, Basel, Switzerland) for virus detection (reovirus, paramyxovirus, Sunshine virus, nidovirus) (Table 1).
Virus isolation and ultrastructural characterization.
Cultures of primary tissues from the Morelia viridis snakes were established as described previously (59), using brain and liver material from a fetus (from a clutch of animals B1 and B2). The tissue was trimmed into blocks (1 mm), suspended in 5 ml of minimal essential medium (MEM; Thermo Fisher Scientific, Gibco) supplemented with HEPES (25 mM), 10% fetal bovine serum (FBS; Biochrom), gentamicin (0.05 mg/ml), l-glutamine (2 mM, Biochrom), 10% tryptose phosphate broth (Difco), and 20 μl α-d-glucose (in 90 g/liter phosphate-buffered saline [PBS]) in sterile cell culture dishes (diameter, 5 cm), and incubated at 30°C in 5% CO2. Primary Boa constrictor snake lung (V/4Lu) and brain (V/4Br) cell lines were established as described above.
Cell cultures were used for inoculations at passages 8 to 15 for the M. viridis cell lines and passages 30 to 35 for the V/4Lu and V/4Br cell lines. Briefly, after initial trimming of the lung tissue into blocks (>1 mm), the pieces were mechanically homogenized in 1 ml of trypsin-EDTA solution (0.25%; Thermo Fisher Scientific, Gibco), the cell debris was pelleted by centrifugation (5 min at 1,000 × g), and the remaining supernatant was diluted in 5 ml of MEM supplemented with 25 mM HEPES (Thermo Fisher Scientific) and 15% FBS (Biochrom) and filtered through a 0.45-μm-pore-size filter. One milliliter of the filtered homogenate of lung tissue was further diluted 1:10 in growth medium and was used to inoculate 75-cm2 flasks of both brain and liver cells. The medium was changed at 1- to 2-day intervals until most cells detached or died. The supernatants were frozen at −20°C, pooled, and filtered through a 0.45-μm-pore-size filter. The cleared supernatant was loaded onto a cushion of 30% (wt/vol) sucrose in PBS, concentrated by centrifugation at 100,000 × g for 2 h at 4°C, and solubilized in PBS. For protection, protease inhibitor (protease inhibitor cocktail tablets [cOmplete; mini; EDTA free; Roche Diagnostics, Mannheim, Germany]) was added to the supernatant. For negative staining, samples were adsorbed onto carbon-coated Parlodion films mounted on 300-mesh/inch copper grids (EMS, Fort Washington, PA, USA) for 10 min, washed once with H2O, and stained with 2% phosphotungstic acid (PTA; pH 7.0; Aldrich, Steinheim, Germany) for 1 min. Specimens were analyzed in a transmission electron microscope (CM12; Philips, Eindhoven, The Netherlands) equipped with a charge-coupled-device camera (Ultrascan 1000; Gatan, Pleasanton, CA, USA) at an acceleration voltage of 100 kV.
M. viridis brain and liver cells were harvested at 3, 4, and 5 days postinfection (dpi) and pelleted by centrifugation at 5,000 rpm for 5 min at room temperature (Eppendorf centrifuge 5415C; NIST). The pellets were fixed with 2.5% glutaraldehyde in PBS (pH 7.4) and embedded in resin by routine procedures for TEM (59). To obtain a retrovirus-free virus preparation, the supernatant collected from M. viridis liver cell cultures inoculated with an homogenate of lung tissue was used to inoculate a B. constrictor kidney cell line, I/1Ki (59). The supernatant was collected from the infected I/1Ki cells at 2-day intervals until 8 dpi. The supernatants were filtered through a 0.45-μm-pore-size filter, pooled, aliquoted, and stored at −80°C.
Next-generation sequencing.
RNA was isolated from the lungs of three diseased snakes (animals A2, B1, and D1, Table 1) with the TRIzol reagent (Life Technologies) according to the manufacturer's protocol, using 5 μg of RNA-grade glycogen (Thermo Fisher Scientific) as the carrier. The RNA samples were initially treated with DNase I (Fermentas), followed by repurification with a GeneJET RNA purification kit (Thermo Fisher Scientific). A Ribo-Zero Gold rRNA removal kit for epidemiology (Illumina) was used according to the manufacturer's protocol to further clean the RNA. In the case of sequencing of the virus from the cell culture supernatants, RNA isolation was done using a QIAamp viral RNA minikit (Qiagen) following the manufacturer's protocol without the addition of carrier RNA. For RNA isolated from cell culture supernatants, the rRNA was removed by use of a NEBNext rRNA depletion kit (New England BioLabs). Indexing and NGS library preparation were accomplished with a NEBNext Ultra RNA library preparation kit (New England BioLabs) according to the manufacturer's protocol. The libraries were quantified using a NEBNext Library Quant kit for Illumina (New England BioLabs). Pooled libraries were sequenced on an Illumina MiSeq sequencer (Illumina) using an MiSeq reagent kit (version 3; Illumina) with 291-bp reads (for lung tissue samples) and 300-bp reads (for RNA isolated from the cell culture supernatant) from both ends (paired-end reads). After the removal of reads matching the host genome, de novo sequence assembly of both reads was performed with the MIRA sequence assembler (version 4.9.5; http://mira-assembler.sourceforge.net/) on a CSC Taito supercluster server (IT Center for Science Ltd., Espoo, Finland). The generated contiguous sequences (contigs) from the lung tissue sample run were initially screened by nucleotide sequence analysis with the BLAST program (blastn; https://blast.ncbi.nlm.nih.gov/Blast.cgi), and several contigs matching the sequences of ball python nidovirus (BPNV; NCBI reference sequence accession number NC_024709.1) and python nidovirus (PNV; GenBank accession number KJ935003.1) were identified. In attempt to obtain a full-length genome, the contigs were mapped to BPNV and PNV genomes using the BWA-SW tool (60) in the Unipro UGENE bioinformatics tool kit (61). However, a contig with an almost full-length genome for a novel nidovirus subsequently named Morelia viridis nidovirus (MVNV) was obtained via de novo assembly from RNA isolated from the cell culture supernatant. The genome open reading frames (ORFs) were detected using the Unipro UGENE bioinformatics tool kit. The contig coverage was determined using the reference alignment of the entire NGS data determined by use of the Bowtie2 program in the Unipro UGENE bioinformatics tool kit.
Phylogenetic analysis and bioinformatics.
Since the BLASTN search of the contig obtained by de novo assembly from purified virus material suggested that python nidoviruses (BPNV, PNV, a proposed new genus in the Torovirinae subfamily of nidoviruses) were the group most homologous to MVNV, representative sequences of this subfamily (i.e., genus Torovirus, genus Bafinivirus, and python nidoviruses) were downloaded from GenBank. The amino acid sequences of conserved ORF1b were aligned using the ClustalW algorithm implemented in the MEGA (version 6.06) program (62), followed by manual refinement. The best-fit substitution model was sought using the maximum likelihood method implemented in MEGA (version 6.06). Phylogenetic trees were constructed using the Bayesian Monte Carlo Markov chain (MCMC) method implemented in the BEAST (version 1.8.0) program (63). The analyses were performed with the LG+G+I model of substitution, a strict clock, and a constant-size demographic model. The Bayesian analyses were run for 10 million states and sampled every 1,000 states. The analyses were carried out on the CSC server (IT Center for Science Ltd., Espoo, Finland). Posterior probabilities were calculated with a burn-in of 1 million states and checked for convergence using the Tracer (version 1.6) program (64). The Mfold web server (available at http://unafold.rna.albany.edu/?q=mfold) was utilized for determining the RNA folding (65) around the ribosomal frameshift signal (RFS). The Unipro UGENE bioinformatics tool kit (61) was utilized for alignment of the nucleotides (by use of the MUSCLE program [66]) around the RFS. Biosequence analysis with the HMMER web server by the use of profile hidden Markov models (available at http://hmmer.org/) (67) was used to identify conserved domains in the identified ORFs.
Quantitative reverse transcription-PCR (qRT-PCR).
RNA was extracted from lung tissue samples with the TRIzol reagent (Life Technologies) and mechanical homogenization with a MagNA Lyser homogenizer (Roche). After addition of chloroform and separation of the RNA-containing phase by centrifugation (15 min, 12,000 × g, 4°C), the RNA was purified with a Qiagen RNeasy minikit (Qiagen) following the manufacturer's protocol for RNA cleanup.
The amount of RNA in the samples was measured with a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific), and the samples were subsequently diluted with RNase-free water to a concentration of 150 ng/μl to allow comparative assessment.
A TaqMan qRT-PCR assay was set up using the following primers (Microsynth AG, Switzerland): Nido-fwd (5′-AGTCATCTGTCTCGACCACCT-3′) and Nido-rev (5′-ACATGTAGAGCACTTTGACTGGTT-3′). The sequence for the Nido-probe probe (Microsynth AG, Switzerland) was FAM-CGACAACTGGGTCATCAGACGC-TAMRA, where FAM represents 6-carboxyfluorescein and TAMRA represents 6-carboxytetramethylrhodamine.
The qRT-PCR was performed on an Applied Biosystems 7500 fast real-time PCR system using 96-well plates. The reaction volume (25 μl) consisted of 12.5 μl of One-Step qRT-PCR master mix (Eurogentec), 1 μl each of the forward and reverse primers (10 μM), 1 μl of probe (10 μM), 0.125 μl of reverse transcriptase (Euroscript RT and RNase inhibitor mix; Eurogentec), 5 μl (750 ng) of template RNA, and 4.5 μl of diethyl pyrocarbonate-treated H2O.
The qRT-PCR was set up with the following cycling conditions: (i) 30 min at 48°C, (ii) 10 min at 95°C, (iii) 15 s at 95°C, and (iv) 1 min at 60°C, with 40 cycles being performed between the third and fourth steps. The data were collected during the fourth step of the RT-PCR program.
Recombinant protein expression and generation of anti-MVNV N protein antiserum.
To identify the ORF for the nucleoprotein (N protein) gene in the genome of the novel virus isolate, the de novo assembled genome was aligned with the BPNV (NCBI reference sequence NC_024709.1) and PNV (GenBank accession number KJ935003.1) genomes using the MUSCLE program (66) in the Unipro UGENE bioinformatics tool kit (61). The N protein-coding region was obtained from cDNA transcribed with random primers using RNA isolated from the supernatants of cultured cells inoculated with lung tissue homogenate (see above). cDNA transcription was done using RevertAid transcriptase (Thermo Fisher Scientific) following the protocol for random hexamer amplification. PCR amplification of the whole N protein-coding region was done with primers designed according to the Champion pET directional TOPO expression kit's manual (Thermo Fisher Scientific) using Phusion Flash high-fidelity PCR master mix (Thermo Fisher Scientific). The PCR product was purified from the agarose gel using a GeneJET gel extraction kit (Thermo Fisher Scientific) and ligated to the pET101/D-TOPO vector with an N-terminal V5 epitope and His tags (Thermo Fisher Scientific). The ligated vector was transformed into One Shot TOP10 competent cells (Thermo Fisher Scientific), and clones with the desired insert were screened by plating on Luria broth (LB) agar plates with 100 μg/ml ampicillin. Individual colonies were grown in 5 ml of LB supplemented with 100 μg/ml ampicillin overnight, and the plasmids were purified using a GeneJET plasmid miniprep kit (Thermo Fisher Scientific) following the manufacturer's recommendation. The plasmids were sequenced (Microsynth AG, Switzerland) using T7 forward and reverse primers. The N protein-containing plasmid was transformed into BL21 Star(DE3) One Shot chemically competent Escherichia coli cells (Thermo Fisher Scientific), and protein expression was done in LB supplemented with 100 μg/ml carbenicillin (Sigma-Aldrich) and 1% glucose (Sigma-Aldrich) according to the manufacturer's recommendation. The recombinant N protein expressed in inclusion bodies was purified using chelating Sepharose fast-flow medium (GE Healthcare Life Sciences) with cobalt as the immobilized metal ion and 8 M urea in all buffers. The protein in the elution buffer (50 mM Tris, 500 mM NaCl, 300 mM imidazole, 8 M urea, pH 7.5) was concentrated using a 10-kDa-cutoff Amicon Ultra-15 centrifugal filter unit (EMD Millipore), and buffer exchange (to 25 mM Tris, 75 mM NaCl, pH 7.5) was achieved by decreasing the urea concentration slowly using a Slide-A-Lyzer dialysis cassette (molecular weight cutoff, 3.5; Thermo Fisher Scientific). The purified recombinant protein was used to immunize a rabbit by applying the following scheme: initial immunization on day 0, first booster on day 7, second booster on day 14, third booster on day 42, and a final bleeding on day 49 (BioGenes GmbH, Berlin, Germany; BioGenes adheres to EU and global animal welfare regulations). The generated antiserum was cleaned by affinity purification by a method similar to the methods described previously (68, 69). Briefly, 500 μg of purified recombinant N protein was dialyzed in PBS, followed by coupling to CNBr-activated Sepharose 4B (GE Healthcare Life Sciences) following the manufacturer's protocol. The affinity matrix was packed into Econo-Pac chromatography columns (Bio-Rad), and antiserum was passed through the column by gravity flow. After the antibodies were washed with several column volumes of PBS, the bound antibodies were eluted with 0.1 M glycine (pH 2.5), and the fractions were neutralized by addition of 1 M Tris, pH 8.5. The affinity-purified antibody was dialyzed against PBS and concentrated using a 10-kDa-cutoff Amicon Ultra-15 centrifugal filter unit (EMD Millipore). The purified antibody was mixed with glycerol (50%) and stored in aliquots at −20°C.
Immunofluorescence staining, qRT-PCR of cell culture supernatants, and estimation of virus titer.
For immunofluorescence staining, M. viridis cells (brain) and B. constrictor cells (kidney, lung, brain) were detached by the use of trypsin, washed with growth medium, and seeded on glass-bottom 24-well plates (Cellvis, USA). The attached cells were inoculated by incubation with 250 μl of 1/10-diluted (in growth medium) MVNV isolated on I/1Ki cells. After 1 h of incubation at 30°C, the cells were washed twice with supplemented growth medium, and finally, 1 ml of supplemented growth medium was added to each well. For each cell line, three wells were inoculated with MVNV and three wells were mock infected with supplemented growth medium. At 3 dpi, the cells were fixed by replacing the supplemented growth medium with 4% electron microscopy-grade paraformaldehyde in PBS, followed by 15 min of incubation at room temperature. After fixation, the cells were washed once with PBS, permeabilized (0.25% Triton X-100 and 3% bovine serum albumin in PBS), and left in PBS until they were stained. The replicate wells (three replicate wells with MVNV-infected cells and three replicate wells with mock-infected cells for each cell line) were incubated with anti-MVNV N protein antibody at a 1:1,000, 1:2,000, or 1:4,000 dilution (in PBS) for 1 h at room temperature; washed 5 times with PBS; incubated for 45 min at room temperature with 1:1,000-diluted (in PBS) Alexa Fluor 594-labeled goat anti-rabbit immunoglobulin secondary antibody (Invitrogen); washed 4 times with PBS; incubated for 1 min with DAPI (4′,6-diamidino-2-phenylindole) in PBS; washed twice with distilled H2O; and air dried; and a coverslip was placed with FluoreGuard mounting medium (Biosystems, Switzerland). Images of MVNV- and mock-infected cells were taken at a ×400 magnification with a Nikon Eclipse Ti-U inverted microscope with NIS Advanced Research software. Anti-MVNV N protein antiserum diluted 1:2,000 yielded the best signal-to-noise ratio.
For the monitoring of virus production, the cell culture supernatant was collected at 0, 1, 2, and 3 dpi. At each time point, 47 μl was collected from each well, and samples from replicate wells were pooled to make ∼140 μl. The samples were stored frozen at −20°C prior to RNA isolation with a QIAamp viral RNA minikit (Qiagen). The cell culture supernatants were analyzed by qRT-PCR in duplicate, and the relative amount of virus RNA (compared to that at 0 dpi) at each time point was determined using the 2−ΔCT threshold cycle (CT) method.
To estimate the virus titer in the MVNV stock isolated on the B. constrictor kidney cell line (I/1Ki), M. viridis brain cells were trypsinized, washed with supplemented growth medium, and seeded onto a 96-well tissue culture plate (TPP, Switzerland) in 135 μl of supplemented growth medium per well. A serial 10-fold dilution of the MVNV stock was prepared to reach a 1:106 dilution, and 15 μl of each dilution was pipetted into 12 parallel wells. The plate was incubated for 5 days at 30°C (until a clear cytopathic effect [CPE] was seen in all wells inoculated with MVNV dilutions).
Histology, IH, and RNA-ISH.
Formalin-fixed tissue samples were trimmed and embedded in paraffin wax by routine procedures. Sections (4 to 5 μm) were prepared and stained with hematoxylin and eosin (H&E) and the PAS-alcian blue stain for the evaluation of mucus-producing cells or were used for immunohistology (IH) and RNA in situ hybridization (RNA-ISH) (performed on the lungs of all affected snakes plus all major organs or tissues from five affected animals [animals B1, B2, A4, G1, and E1] and one healthy individual [animal G5]).
IH was performed using the custom-made rabbit polyclonal antibody (see above) to demonstrate nidovirus N protein in tissue sections. The EnVision horseradish peroxidase (HRP) detection system (Dako, Baar, Switzerland) was applied. After deparaffinization, sections were incubated in peroxidase-blocking solution (Dako) for 10 min at room temperature to block any endogenous peroxidase activity, followed by incubation with the primary antibody (anti-MVNV N protein diluted 1:1,000 in Dako dilution buffer) for 12 to 15 h at 4°C. This was followed by incubation with the EnVision+ system HRP-labeled rabbit antibody (Dako) according to the manufacturer's protocol. The reaction was visualized with diaminobenzidine tetrahydrochloride (DAB), followed by counterstaining with hematoxylin. Between each incubation step, the sections underwent a wash in Tris-buffered saline solution containing Tween 20 (TBS-Tween; pH 7.6) for 10 min. A formalin-fixed, paraffin-embedded cell pellet prepared from the infected cell cultures served as a positive control. Consecutive sections incubated with the preimmune serum instead of the specific primary antibody served as negative controls.
Sections from the lungs and trachea were also stained by IH for cytokeratins (clone PCK-26; Novus Biologicals) to highlight respiratory and epithelial cells, for cleaved caspase-3 (rabbit anti-human cleaved caspase-3 monoclonal antibody; Asp175; clone 5A1E; Cell Signaling Technology) to demonstrate apoptotic cells, and for proliferating cell nuclear antigen (mouse anti-rat PCNA; clone PC10; Dako) to detect proliferating cells, using routine protocols established for other species (70–72). Intestinal epithelium was used as a positive control for all three detection systems after the cross-reactivity of the antibodies was confirmed by a comparison of the expression pattern in the python with those seen in the respective mammalian tissues.
For RNA-ISH, the RNAscope technology (Advanced Cell Diagnostics Inc., USA) was employed by using a set of 30 Z oligonucleotide probes (based on the MVNV genome) and following the manufacturer's instructions (73). DAB was used as the chromogen, and hematoxylin was used for counterstaining. A formalin-fixed, paraffin-embedded pellet of virus-infected cells served as a positive control. dapB (the bacterial gene coding for dihydrodipicolinate reductase) was used as a negative control. RNA preservation was confirmed by the detection of a housekeeping gene encoding ubiquitin C (ubc).
Morphometry.
Quantitative assessment of the changes observed in the (respiratory) epithelium was undertaken in the proximal, bronchial lung, as the caudal part is nonrespiratory and taken up by the air sacs. Three regions in the proximal, bronchial lung were defined on the basis of their epithelial cover and functions (Fig. 2A and 9): (i) region 1, which is the luminal part of the primary trabeculae and which is covered by multilayered pseudostratified bronchus-type epithelium, dominated by ciliated cells with few secretory and goblet cells, and supported by a thick smooth muscle layer (i.e., myoelastic bundles) (38, 39); (ii) region 2, which is the middle level of the faveolae; and (iii) region 3, which is the base of the faveolae. The faveolae are covered by a single-layered epithelium similar to that in the mammalian lung (gas exchange epithelium), are dominated by flat type I pneumocytes with thin, cytoplasmic extensions that cover the pulmonary capillaries, and contain micropinocytotic vesicles (74). Between the capillaries lie the surfactant-producing type II pneumocytes, which are cuboidal cells with short microvilli that contain lamellar bodies and numerous micropinocytotic vesicles (75). In addition, so-called secretory cells containing secretory granules have been described (38).
H&E- and cytokeratin-stained cross sections of the lungs were examined and photographed using a light microscope (Eclipse Ni-U; Nikon Corp.), and the images were analyzed with the associated imaging software (NIS-Elements AR; Nikon Corp.).
The average total height of the epithelial layer in each of the three regions was determined. For this purpose, the distance between the apical epithelial cell border and the basal membrane was measured at each of 10 points over three 300-μm-wide epithelial segments in each region in the cytokeratin-stained section at a ×200 magnification (Fig. 9B, D, and E). For each location, the average value obtained for each of the three segments was used to determine the average height.
As we also observed an apparent increase in the cell layers of the generally multilayered epithelium in region 1 in several diseased animals and saw evidence of increased cellularity in the other two regions, we counted the total number of epithelial cell nuclei in each of three 200-μm segments in all three regions in the cytokeratin-stained section at a ×400 magnification (Fig. 9C).
Statistical analysis.
Statistical analyses were performed using SPSS Statistics (version 17.0.0) software (IBM) to assess the epithelial thickness and cellularity. A P value of 0.05 for significance was utilized for all calculations. A Mann-Whitney U test for independent samples (t test) was applied to compare lung parameters between noninfected and infected snakes.
Accession number(s).
The MVNV genome is available in GenBank under accession number MF351889, and the raw NGS data are available under BioSample accession number SAMN07248312 (http://www.ncbi.nlm.nih.gov/biosample/7248312).
ACKNOWLEDGMENTS
We are grateful to the technical staff of the Histology Laboratory and the Electron Microscopy Unit, Institute of Veterinary Pathology, and to Elisabeth M. Schraner, Institute of Virology and Institute of Veterinary Anatomy, Vetsuisse Faculty, University of Zurich, for excellent technical support. We also thank our colleagues at the Institute of Veterinary Bacteriology, Vetsuisse Faculty, University of Zurich, for performing the bacteriological examinations. Particular thanks are due to the breeders who submitted their snakes for diagnostic purposes.
REFERENCES
- 1.McDiarmid RW, Campbell JA, Touré T. 1999. Snake species of the world: a taxonomic and geographic reference, vol.1 Herpetologists' League, Washington, DC. [Google Scholar]
- 2.More G, Pantchev N, Herrmann DC, Vrhovec MG, Ofner S, Conraths FJ, Schares G. 2014. Molecular identification of Sarcocystis spp. helped to define the origin of green pythons (Morelia viridis) confiscated in Germany. Parasitology 141:646–651. doi: 10.1017/S0031182013001960. [DOI] [PubMed] [Google Scholar]
- 3.Nijman V, Shepherd CR. 2009. Wildlife trade from ASEAN to the EU: issues with the trade in captive-bred reptiles from Indonesia. TRAFFIC Europe Report for the European Commission. European Commission, Brussels, Belgium. [Google Scholar]
- 4.Rawlings LH, Donnellan SC. 2003. Phylogeographic analysis of the green python, Morelia viridis, reveals cryptic diversity. Mol Phylogenet Evol 27:36–44. doi: 10.1016/S1055-7903(02)00396-2. [DOI] [PubMed] [Google Scholar]
- 5.Aqrawi T, Stöhr AC, Knauf-Witzens T, Krengel A, Heckers KO, Marschang RE. 2015. Identification of snake arenaviruses in live boas and pythons in a zoo in Germany. Tierarztl Prax Ausg K Kleintiere Heimtiere 43:239–247. doi: 10.15654/TPK-140743. [DOI] [PubMed] [Google Scholar]
- 6.Shaw G. (ed). 1802. General zoology. vol III G. Kearsley, London, United Kingdom. [Google Scholar]
- 7.Linné CV, Salvius L. 1758. Systema naturae, vol. 1 Holmiae:Impensis Direct. Laurentii Salvii. [Google Scholar]
- 8.Bodewes R, Lempp C, Schurch AC, Habierski A, Hahn K, Lamers M, von Dornberg K, Wohlsein P, Drexler JF, Haagmans BL, Smits SL, Baumgartner W, Osterhaus AD. 2014. Novel divergent nidovirus in a python with pneumonia. J Gen Virol 95:2480–2485. doi: 10.1099/vir.0.068700-0. [DOI] [PubMed] [Google Scholar]
- 9.Stenglein MD, Jacobson ER, Wozniak EJ, Wellehan JFX, Kincaid A, Gordon M, Porter BF, Baumgartner W, Stahl S, Kelley K, Towner JS, DeRisi JL. 2014. Ball python nidovirus. A candidate etiologic agent for severe respiratory disease in Python regius. mBio 5:e01484-14. doi: 10.1128/mBio.01484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun H, Lan D, Lu L, Chen M, Wang C, Hua X. 2014. Molecular characterization and phylogenetic analysis of the genome of porcine torovirus. Arch Virol 159:773–778. doi: 10.1007/s00705-013-1861-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draker R, Roper RL, Petric M, Tellier R. 2006. The complete sequence of the bovine torovirus genome. Virus Res 115:56–68. doi: 10.1016/j.virusres.2005.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagerland JA, Pohlenz JFL, Woode GN. 1986. A morphological study of the replication of Breda virus (proposed family Toroviridae) in bovine intestinal cells. J Gen Virol 67:1293–1304. doi: 10.1099/0022-1317-67-7-1293. [DOI] [PubMed] [Google Scholar]
- 13.Hoet AE, Cho K-O, Chang K-O, Loerch SC, Wittum TE, Saif LJ. 2002. Enteric and nasal shedding of bovine torovirus (Breda virus) in feedlot cattle. Am J Vet Res 63:342–348. doi: 10.2460/ajvr.2002.63.342. [DOI] [PubMed] [Google Scholar]
- 14.Woode GN, Reed DE, Runnels PL, Herrig MA, Hill HT. 1982. Studies with an unclassified virus isolated from diarrheic calves. Vet Microbiol 7:221–240. doi: 10.1016/0378-1135(82)90036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uccellini L, Ossiboff RJ, de Matos REC, Morrisey JK, Petrosov A, Navarrete-Macias I, Jain K, Hicks AL, Buckles EL, Tokarz R, McAloose D, Lipkin W. 2014. Identification of a novel nidovirus in an outbreak of fatal respiratory disease in ball pythons (Python regius). Virol J 11:144. doi: 10.1186/1743-422X-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Groot RJ, Cowley JA, Enjuanes L, Faaberg KS, Perlman S, Rottier PJ, Snijder EJ, Ziebuhr J, Gorbalenya AE. 2011. Order Nidovirales, p 785–795. Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 17.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales. Evolving the largest RNA virus genome. Virus Res 117:17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nga PT, del Carmen Parquet M, Lauber C, Parida M, Nabeshima T, Yu F, Thuy NT, Inoue S, Ito T, Okamoto K. 2011. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog 7:e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauber C, Ziebuhr J, Junglen S, Drosten C, Zirkel F, Nga PT, Morita K, Snijder EJ, Gorbalenya AE. 2012. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch Virol 157:1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauber C, Gorbalenya AE. 2012. Toward genetics-based virus taxonomy: comparative analysis of a genetics-based classification and the taxonomy of picornaviruses. J Virol 86:3905–3915. doi: 10.1128/JVI.07174-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker PJ, Winton JR. 2010. Emerging viral diseases of fish and shrimp. Vet Res 41:51. doi: 10.1051/vetres/2010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snijder EJ, Kikkert M, Fang Y. 2013. Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 23.To KKW, Hung IFN, Chan JFW, Yuen K-Y. 2013. From SARS coronavirus to novel animal and human coronaviruses. J Thorac Dis 5(Suppl 2):S103–S108. doi: 10.3978/j.issn.2072-1439.2013.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baird A, Faisal M. 2016. Fathead minnow nidovirus infects spotfin shiner Cyprinella spiloptera and golden shiner Notemigonus crysoleucas. Dis Aquat Org 119:37–44. doi: 10.3354/dao02970. [DOI] [PubMed] [Google Scholar]
- 25.McVey DS, Kennedy M, Chengappa MM (ed). 2013. Veterinary microbiology, 3rd ed Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 26.Beards GM, Campbell AD, Cottrell NR, Peiris JS, Rees N, Sanders RC, Shirley JA, Wood HC, Flewett TH. 1984. Enzyme-linked immunosorbent assays based on polyclonal and monoclonal antibodies for rotavirus detection. J Clin Microbiol 19:248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss M, Steck F, Horzinek MC. 1983. Purification and partial characterization of a new enveloped RNA virus (Berne virus). J Gen Virol 64:1849–1858. doi: 10.1099/0022-1317-64-9-1849. [DOI] [PubMed] [Google Scholar]
- 28.Cann AJ. 2001. Particles. The function and formation of virus particles, p 25–27. In Cann AJ. (ed), Principles of molecular virology (standard edition). Elsevier, Academic Press, London, United Kingdom. [Google Scholar]
- 29.Cornelissen LA, van Woensel PA, de Groot RJ, Horzinek MC, Visser N, Egberink HF. 1998. Cell culture-grown putative bovine respiratory torovirus identified as a coronavirus. Vet Rec 142:683–686. doi: 10.1136/vr.142.25.683. [DOI] [PubMed] [Google Scholar]
- 30.Vanopdenbosch E, Wellemans G, Petroff K. 1991. Breda virus associated with respiratory disease in calves. Vet Rec 129:203. [DOI] [PubMed] [Google Scholar]
- 31.Tokarz R, Sameroff S, Hesse RA, Hause BM, Desai A, Jain K, Lipkin WI. 2015. Discovery of a novel nidovirus in cattle with respiratory disease. J Gen Virol 96:2188–2193. doi: 10.1099/vir.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smits SL, Lavazza A, Matiz K, Horzinek MC, Koopmans MP, de Groot RJ. 2003. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J Virol 77:9567–9577. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petric M. 2003. Other viruses causing gastroenteritis. Epidemiology of toroviruses. In Desselberger U, Gray J (ed), Viral gastroenteritis, 9th ed Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 34.Snijder EJ, Ederveen J, Spaan WJ, Weiss M, Horzinek MC. 1988. Characterization of Berne virus genomic and messenger RNAs. J Gen Virol 69:2135–2144. doi: 10.1099/0022-1317-69-9-2135. [DOI] [PubMed] [Google Scholar]
- 35.Kuwabara M, Wada K, Maeda Y, Miyazaki A, Tsunemitsu H. 2007. First isolation of cytopathogenic bovine torovirus in cell culture from a calf with diarrhea. Clin Vaccine Immunol 14:998–1004. doi: 10.1128/CVI.00475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huder JB, Boni J, Hatt J-M, Soldati G, Lutz H, Schupbach J. 2002. Identification and characterization of two closely related unclassifiable endogenous retroviruses in pythons (Python molurus and Python curtus). J Virol 76:7607–7615. doi: 10.1128/JVI.76.15.7607-7615.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Dea MA, Jackson B, Jackson C, Xavier P, Warren K. 2016. Discovery and partial genomic characterisation of a novel nidovirus associated with respiratory disease in wild shingleback lizards (Tiliqua rugosa). PLoS One 11:e0165209. doi: 10.1371/journal.pone.0165209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastor LM. 1995. The histology of the reptilian lung, p 127–155. In Pastor LM. (ed), Histology, ultrastructure and immunohistochemistry of the respiratory organs in non mammalian vertebrates, 1st ed Secretariado de Publicaciones de la Universidad de Murcia, Murcia, Spain. [Google Scholar]
- 39.Starck JM, Weimer I, Aupperle H, Muller K, Marschang RE, Kiefer I, Pees M. 2015. Morphological pulmonary diffusion capacity for oxygen of burmese pythons (Python molurus): a comparison of animals in healthy condition and with different pulmonary infections. J Comp Pathol 153:333–351. doi: 10.1016/j.jcpa.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Jacobson ER, Adams HP, Geisbert TW, Tucker SJ, Hall BJ, Homer BL. 1997. Pulmonary lesions in experimental ophidian paramyxovirus pneumonia of Aruba Island rattlesnakes, Crotalus unicolor. Vet Pathol 34:450–459. doi: 10.1177/030098589703400509. [DOI] [PubMed] [Google Scholar]
- 41.Ward HE, Nicholas TE. 1984. Alveolar type I and type II cells. Aust N Z J Med 14:731–734. [PubMed] [Google Scholar]
- 42.Fehrenbach H, Kasper M, Tschernig T, Pan T, Schuh D, Shannon JM, Muller M, Mason RJ. 1999. Keratinocyte growth factor-induced hyperplasia of rat alveolar type II cells in vivo is resolved by differentiation into type I cells and by apoptosis. Eur Respir J 14:534–544. doi: 10.1034/j.1399-3003.1999.14c10.x. [DOI] [PubMed] [Google Scholar]
- 43.Caswell JL, Williams KJ. 2016. Respiratory system, p 465–591. In Maxie MG. (ed), Jubb, Kennedy & Palmer's pathology of domestic animals, 6th ed, vol 2 Elsevier, St. Louis, MO. [Google Scholar]
- 44.Veldhuizen R, Nag K, Orgeig S, Possmayer F. 1998. The role of lipids in pulmonary surfactant. Biochim Biophys Acta 1408:90–108. doi: 10.1016/S0925-4439(98)00061-1. [DOI] [PubMed] [Google Scholar]
- 45.Grgiæ H, Hunter DB, Hunton P, Nagy É. 2008. Pathogenicity of infectious bronchitis virus isolates from Ontario chickens. Can J Vet Res 72:403–410. [PMC free article] [PubMed] [Google Scholar]
- 46.Schunk MK, Percy DH, Rosendal S. 1995. Effect of time of exposure to rat coronavirus and Mycoplasma pulmonis on respiratory tract lesions in the Wistar rat. Can J Vet Res 59:60–66. [PMC free article] [PubMed] [Google Scholar]
- 47.Koopmans M, Horzinek MC. 1995. The pathogenesis of torovirus infections in animals and humans, p 403–413. In Siddell SG. (ed), The Coronaviridae. Springer US, Boston, MA. [Google Scholar]
- 48.Maestre AM, Garzon A, Rodriguez D. 2011. Equine torovirus (BEV) induces caspase-mediated apoptosis in infected cells. PLoS One 6:e20972. doi: 10.1371/journal.pone.0020972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding X, Xu F, Chen H, Tesh RB, Xiao S-Y. 2005. Apoptosis of hepatocytes caused by Punta Toro virus (Bunyaviridae. Phlebovirus) and its implication for phlebovirus pathogenesis. Am J Pathol 167:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stinner JN. 1987. Gas exchange and air flow in the lung of the snake, Pituophis melanoleucus. J Comp Physiol B 157:307–314. doi: 10.1007/BF00693357. [DOI] [Google Scholar]
- 51.Wallach V. 1998. The lungs of snakes. Biol Reptilia 19:93–295. [Google Scholar]
- 52.Perry SF, Bauer AM, Russell AP, Alston JT, Maloney JE. 1989. Lungs of the gecko Rhacodactylus leachianus (reptilia: Gekkonidae): a correlative gross anatomical and light and electron microscopic study. J Morphol 199:23–40. doi: 10.1002/jmor.1051990104. [DOI] [PubMed] [Google Scholar]
- 53.McDonald HS. 1959. Respiratory functions of the ophidian air sac. Herpetologica 15:193–198. [Google Scholar]
- 54.Stinner HN. 1982. Ventillation, gas exchange and blood gases in the snake, Pituophis melanoleucus. Respir Physiol 47:279–298. doi: 10.1016/0034-5687(82)90058-5. [DOI] [PubMed] [Google Scholar]
- 55.Saif LJ, Bohl EH. 1986. Transmissible gastroenteritis, p 255–274. In Leman AD, Straw B, Glock RD (ed), Diseases of swine, 6th ed Iowa State University Press, Ames, IA. [Google Scholar]
- 56.Koopmans M, Horzinek MC. 1994. Toroviruses of animals and humans. A review. Adv Virus Res 43:233–273. doi: 10.1016/S0065-3527(08)60050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imai M, Shibata T, Moriguchi K. 1991. Pepsinogen granules in the esophageal epithelium of the rock snake. Okajimas Folia Anat Jpn 68:231–234. doi: 10.2535/ofaj1936.68.4_231. [DOI] [PubMed] [Google Scholar]
- 58.Cundall D, Tuttman C, Close M. 2014. A model of the anterior esophagus in snakes, with functional and developmental implications. Anat Rec (Hoboken) 297:586–598. doi: 10.1002/ar.22860. [DOI] [PubMed] [Google Scholar]
- 59.Hetzel U, Sironen T, Laurinmaki P, Liljeroos L, Patjas A, Henttonen H, Vaheri A, Artelt A, Kipar A, Butcher SJ, Vapalahti O, Hepojoki J. 2013. Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J Virol 87:10918–10935. doi: 10.1128/JVI.01123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 62.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6. Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. http://tree.bio.ed.ac.uk/software/tracer/.
- 65.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Finn RD, Clements J, Eddy SR. 2011. HMMER web server. Interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hepojoki J, Kipar A, Korzyukov Y, Bell-Sakyi L, Vapalahti O, Hetzel U. 2015. Replication of boid inclusion body disease-associated arenaviruses is temperature sensitive in both boid and mammalian cells. J Virol 89:1119–1128. doi: 10.1128/JVI.03119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korzyukov Y, Hetzel U, Kipar A, Vapalahti O, Hepojoki J. 2016. Generation of anti-boa immunoglobulin antibodies for serodiagnostic applications, and their use to detect anti-reptarenavirus antibodies in Boa constrictor. PLoS One 11:e0158417. doi: 10.1371/journal.pone.0158417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ressel L, Ward S, Kipar A. 2015. Equine cutaneous mast cell tumours exhibit variable differentiation, proliferation activity and KIT expression. J Comp Pathol 153:236–243. doi: 10.1016/j.jcpa.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 71.Antoine DJ, Williams DP, Kipar A, Laverty H, Park BK. 2010. Diet restriction inhibits apoptosis and HMGB1 oxidation and promotes inflammatory cell recruitment during acetaminophen hepatotoxicity. Mol Med 16:479–490. doi: 10.2119/molmed.2010.00126. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Swadzba E, Rupik W. 2010. Ultrastructural studies of epidermis keratinization in grass snake embryos Natrix natrix L. (Lepidosauria, Serpentes) during late embryogenesis. Zoology (Jena) 113:339–360. doi: 10.1016/j.zool.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 73.Wang F, Flanagan J, Su N, Wang L-C, Bui S, Nielson A, Wu X, Vo H-T, Ma X-J, Luo Y. 2012. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maina JN. 1989. The morphology of the lung of the black mamba Dendroaspis polylepis (Reptilia: Ophidia: Elapidae). A scanning and transmission electron microscopic study. J Anat 167:31–46. [PMC free article] [PubMed] [Google Scholar]
- 75.Perry SF. 1983. 1. Introduction. Descriptive classification of lung types. Histological structure, p 1–3. In Perry SF. (ed), Reptilian lungs: functional anatomy and evolution. Springer Verlag, Berlin, Germany. [PubMed] [Google Scholar]