Abstract
This study aimed to investigate the association of the GSTP1 gene polymorphism with the outcomes and toxicities of treatments in breast cancer. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated for the association of GSTP1 polymorphism with tumour response and toxicities, and the hazard ratios (HRs) and 95% CIs were calculated for the association between GSTP1 polymorphism and overall survival (OS). The statistical analysis showed that the GSTP1 polymorphism was not associated with tumour response or OS. A significant increase in the incidence of toxicities was observed (GA vs. AA OR = 1.45, 95% CI = 1.04–2.01, P = 0.028; GG vs. AA OR = 1.47, 95% CI = 1.03–2.10, P = 0.036; recessive model OR = 1.54, 95% CI = 1.13–2.09, P = 0.006; and allele model OR = 1.35, 95% CI = 1.07–1.71, P = 0.011), especially in the chemotherapy ± surgery group (GA vs. AA OR = 1.64, 95% CI = 1.05–2.56, P = 0.030; recessive model OR = 1.72, 95% CI = 1.17–2.54, P = 0.006; and allele model OR = 1.57, 95% CI = 1.11–2.21, P = 0.010). Our results indicate that the GSTP1 polymorphism may be associated with increased toxicity, especially in patients treated with chemotherapy ± surgery.
Keywords: GSTP1, breast cancer, treatment outcome, toxicities, meta-analysis
INTRODUCTION
Breast cancer in 2012 was both the most common cancer worldwide, with 1.7 million new cases, and the leading cause of cancer-related deaths in women [1]. Breast cancer is commonly treated with chemotherapy either as an adjuvant systemic treatment after primary surgery or as neoadjuvant therapy before surgery. The neoadjuvant treatment attempts to reduce the tumour stage, with the goal of surgically resecting the mass. Currently, radiotherapy is commonly used after primary surgery to reduce the risk of recurrence [2]. However, patient response to treatment with chemotherapy and radiotherapy is quite variable [3–5]. Inter-patient variations in clinicopathologic characteristics such as clinical disease stage, lymph node status, and hormone receptor expression, could have a large influence on treatment outcomes. Increasing evidence suggests that drug-metabolizing enzymes may play an important role in inter-patient variations, which could affect treatment response and toxicities [6, 7].
Glutathione S-transferases (GSTs) are a superfamily of phase-II metabolic enzymes that play a key role in cellular resistance mechanisms [8]. GSTs detoxify cytotoxic agents by catalysing the reduction of these compounds through their conjugation with glutathione [9]. The GSTP1 gene, a member of the GST family, is located on chromosome 11q13, which contains 7 exons and 6 introns. Genetic polymorphisms involving an adenine to guanine transition (rs1695) at codon 105 in exon 5 of the GSTP1 gene results in amino acid substitution from isoleucine to valine (Ile→Val). This substitution decreases the enzymatic activity of glutathione-S-transferase P1 (GSTP1) and alters the pharmacokinetics of cyclophosphamide, which may influence treatment outcomes and toxicity for breast cancer [10].
Studies have investigated the associations of the GSTP1 (A313G) gene polymorphism with treatment response, prognosis, and toxicities for breast cancer [11–41]. However, these findings failed to reach a consensus owing to a lack of data and inconsistencies in the results between these studies. Therefore, a systematic review and meta-analysis was conducted to evaluate the influence of the GSTP1 (A313G) polymorphism on treatment outcomes and toxicities in patients with breast cancer.
MATERIALS AND METHODS
Literature search and inclusion criteria
PubMed, EMBASE, Cochrane Library, and China National Knowledge Infrastructure were searched for relevant studies up to August 29th, 2016, by using the following terms: “glutathione S-transferase,” “glutathione S-transferase P1,” “GSTP1,” “breast cancer,” “breast carcinoma,” and “breast neoplasm.” Studies were manually filtered without language restrictions. Additional studies were identified by screening references and relevant reviews.
Studies were included if they met the following criteria: (1) inclusion of patients who were treated for breast cancer; (2) evaluation of associations between GSTP1 and treatment outcomes, as well as toxicities after radiotherapy and/or chemotherapy; (3) treatment outcomes including tumour response and overall survival (OS), with toxicities including all adverse effects; and (4) provision of adequate data for calculation of both odds ratios (ORs) and hazard ratios (HRs) with 95% confidence intervals (CIs).
Data extraction
The following items were gathered independently from all eligible studies by two investigators (JM and YL): first author's name, year of publication, country, number of patients, genotyping methods, median follow-up, treatment protocols, treatment outcomes, and toxicities. Any disagreements were resolved through discussion and consensus.
Risk of bias
The risk of bias was assessed by reviewers independently using a modified Ottawa classification for observational studies [42]. Any disagreements were resolved by consensus.
Statistical analysis
The responses were estimated according to Response Evaluation Criteria in Solid Tumors, including complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). Patients with CR and PR were categorized as the responder group, and patients with SD and PD were categorized as the non-responder group. Toxicities were defined as all adverse effects that occurred after treatment with chemotherapy and/or radiotherapy. ORs with 95% CIs were used to evaluate the association between GSTP1 and tumour response and toxicities based on raw data. Cox proportional HRs and 95% CIs for OS were also calculated using the most adjusted HR in each study. In this meta-analysis, we examined the association of variant genotypes of GSTP1 polymorphism with treatment outcome and toxicities.
The heterogeneity was assessed using the Q test with a significance level of P < 0.05. The I2 statistic was used to test the heterogeneity among the included studies [43]. A fixed-effect model (Mantel-Haenszel method) was applied if heterogeneity was not significant [44]. Otherwise, a random effect model (DerSimonian and Laird method) was utilized [45].
Subgroup analysis was carried out based on ethnicity, sample size, and therapeutic method. Ethnic subgroups consisted of three groups: East Asian (Chinese and Japanese), South Asian (Indian and Bangladeshi), and mixed descent (American, Canadian, and Brazilian). Sample size was divided into a large group (≥ 100 cases) and a small group (< 100 cases). Therapeutic methods included chemotherapy ± surgery, radiotherapy ± surgery, and chemotherapy + radiotherapy ± surgery. Potential publication bias was assessed using a Funnel plot [46] and Egger's test [47]. Statistical analyses were conducted with STATA version 11.0 (Stata Corporation, College Station, Texas, USA). All P values were 2-sided and P < 0.05 was considered statistically significant.
RESULTS
Study characteristics
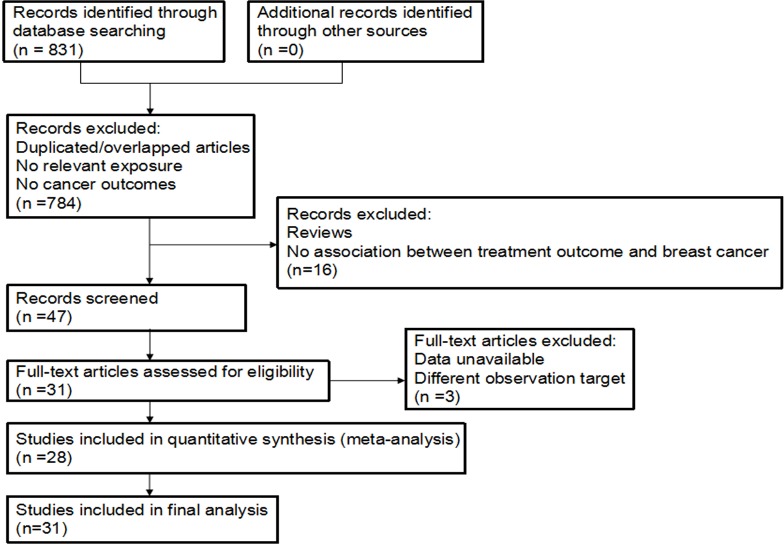
A total of 831 potentially relevant publications were systematically identified. Of them, 784 studies were excluded because they were reviews, letters, comments, or irrelevant studies. An additional study was excluded because data were not provided, and the authors could not be reached [11]. Another study was excluded because it regarded progression-free survival as an observation endpoint [12]. One study was excluded because it regarded CR + PR + SD as the responder group [13]. Another study was excluded owing to inclusion of familial breast cancer and sporadic breast cancer patients [14]. Furthermore, as one study used three different regimens of chemotherapy, it was treated as three articles [15]. After applying the exclusion criteria, 31 studies with a total of 7506 patients were included [14–41]. The study selection flowchart is summarized in Figure 1. The basic characteristics of all included studies are listed in Table 1. Sample sizes ranged from 40 to 1034 patients. Among the studies analysed, 15 reported tumour response events [15, 20–23, 26, 27, 31–35, 37, 38, 41], 13 reported OS [14, 17, 19, 23, 28, 29, 31–34, 38, 41], and 12 reported toxicities [16, 18, 24, 26, 27, 29, 30, 35–37, 39, 40]. Table 2 shows the quality indicators of the included studies.
Figure 1. The flowchart of this meta-analysis.
Table 1. Characteristics of all included studies in this meta-analysis.
| Study | Country | Publication | Genotyping method | Number of patient | Treatment | toxicities | Median follow-up | specimen |
|---|---|---|---|---|---|---|---|---|
| Sweeney, et al. [17] | America | 2000 | PCR-RFLP | 240 | chemotherapy + radiotherapy, chemotherapy, radiotherapy | NP | 58 months | tissue |
| Yang, et al. [29] | China | 2005 | Multiplex PCR | 1034 | chemotherapy | NP | 5.3 years | blood |
| Ambrosone, et al. [16] | German | 2006 | Multiplex PCR | 446 | radiotherapy after surgery | skin toxicities | NP | blood |
| Zárate, et al. [39] | Spain | 2007 | PCR-RFLP | 94 | chemotherapy | haematological and non-haematological | NP | blood |
| Syamala, et al. [14] | India | 2008 | PCR | 347 | NP | NP | NP | blood |
| Kuptsova, et al. [18] | German | 2008 | Multiplex PCR | 390 | radiotherapy after surgery | telangiectasia | 4.1 years | blood |
| Bewick, et al. [19] | Canada | 2008 | PCR | 95 | chemotherapy | NP | 10.4 months | blood or bone marrow |
| Tang, et al. [20] | China | 2009 | PCR | 126 | chemotherapy | NP | 6 weeks | blood |
| Oliveira, et al. [21] | Brazil | 2010 | PCR-RFLP | 40 | chemotherapy | NP | NP | blood |
| Yao, et al. [40] | America | 2010 | PCR-RFLP | 458 | chemotherapy | hematologic | 10.8 years | tumor cell |
| Zhong, et al. [22] | China | 2010 | PCR | 132 | chemotherapy | NP | 9 weeks | blood |
| Zhang (1), et al. [37] | China | 2011 | PCR-RFLP | 120 | chemotherapy | hematologic | NP | blood |
| Bai, et al. [23] | China | 2012 | PCR-RFLP | 159 | chemotherapy | NP | 4 years | blood |
| Terrazzino, et al. [24] | Italy | 2012 | PCR | 237 | radiotherapy | skin fibrosis | 63 days | blood |
| Raabe, et al. [25] | German | 2012 | PCR-RFLP | 83 | radiotherapy | erythema | NP | blood |
| Ji, et al. [26] | China | 2012 | PCR | 153 | chemotherapy | neutropenia | 51 months | blood |
| Tulsyan, et al. [27] | India | 2013 | PCR-RFLP | 100 | chemotherapy | hematologic | NP | blood |
| Duggan, et al. [28] | America | 2013 | PCR | 533 | surgery, surgery and radiotherapy, and chemotherapy | NP | 11.29 years | blood |
| Zhang (2), et al. [38] | China | 2013 | PCR-CTPP | 219 | chemotherapy | NP | 4 years | blood |
| Zhao, et al. [15] | China | 2014 | PCR | 252 | chemotherapy | hematologic | NP | blood |
| Sugishita, et al. [30] | Japan | 2014 | PCR | 102 | chemotherapy | hematologic | 967 days | blood |
| Liu, et al. [34] | China | 2014 | PCR | 382 | chemotherapy after surgery | NP | NP | blood |
| Zhou, et al. [31] | China | 2015 | PCR | 420 | chemotherapy after surgery | NP | 5 years | blood |
| Wang (1), et al. [32] | China | 2015 | PCR-RFLP | 310 | chemotherapy | NP | 5 years | blood |
| Wang (2), et al. [33] | China | 2015 | PCR-RFLP | 262 | chemotherapy | NP | NP | blood |
| Islam, et al. [35] | Bangladesh | 2015 | PCR-RFLP | 256 | chemotherapy | hematologic | NP | blood |
| Eckhoff, et al. [36] | Denmark | 2015 | PCR | 150 | chemotherapy | docetaxel-induced peripheral neuropathy | 7.5 months | blood |
| Yuan, et al. [41] | China | 2015 | PCR-RFLP | 273 | chemotherapy | NP | 5 years | blood |
PCR polymerase chain reaction, NP not provided
Table 2. Quality assessment (risk of bias) of the included studies.
| Study | Ascertainment of outcome | Adjusting for confounders | Attrition bias | Patient selection |
|---|---|---|---|---|
| Sweeney, et al. [17] | Main confounders and any additional confounders | Yes | No risk | Consecutive |
| Yang, et al. [29] | Main confounders and any additional confounders | Yes | No risk | Consecutive |
| Ambrosone, et al. [16] | Main confounders and any additional confounders | No | No risk | Consecutive |
| Zárate, et al. [39] | Main confounders and any additional confounders | No | No risk | Selected/non-consecutive patients |
| Syamala, et al. [14] | Main confounders and any additional confounders | No | Unclear reporting | Selected/non-consecutive patients |
| Kuptsova, et al. [18] | Main confounders and any additional confounders | No | No risk | Consecutive |
| Bewick, et al. [19] | Main confounders and any additional confounders | Yes | No risk | Selected/non-consecutive patients |
| Tang, et al. [20] | Main confounders and any additional confounders | No | Unclear reporting | Consecutive |
| Oliveira, et al. [21] | Main confounders and any additional confounders | No | Unclear reporting | Consecutive |
| Yao, et al. [40] | Main confounders and any additional confounders | No | No risk | Consecutive |
| Zhong, et al. [22] | Main confounders and any additional confounders | No | Unclear reporting | Consecutive |
| Zhang (1), et al. [37] | Main confounders and any additional confounders | No | Unclear reporting | Selected/non-consecutive patients |
| Bai, et al. [23] | Main confounders and any additional confounders | Yes | No risk | Consecutive |
| Terrazzino, et al. [24] | Main confounders and any additional confounders | No | No risk | Consecutive |
| Raabe, et al. [25] | Main confounders and any additional confounders | No | Unclear reporting | Consecutive |
| Ji, et al. [26] | Main confounders and any additional confounders | No | Unclear reporting | Selected/non-consecutive patients |
| Tulsyan, et al. [27] | Main confounders and any additional confounders | No | Unclear reporting | Consecutive |
| Duggan, et al. [28] | Main confounders and any additional confounders | Yes | No risk | Consecutive |
| Zhang (2), et al. [38] | Main confounders and any additional confounders | Yes | No risk | Consecutive |
| Zhao, et al. [15] | Main confounders and any additional confounders | No | Unclear reporting | Consecutive |
| Sugishita, et al. [30] | Main confounders and any additional confounders | No | Unclear reporting | Selected/non-consecutive patients |
| Liu, et al. [34] | Main confounders and any additional confounders | Yes | No risk | Consecutive |
| Zhou, et al. [31] | Main confounders and any additional confounders | Yes | No risk | Consecutive |
| Wang (1), et al. [32] | Main confounders and any additional confounders | Yes | No risk | Consecutive |
| Wang (2), et al. [33] | Main confounders and any additional confounders | No | No risk | Selected/non-consecutive patients |
| Islam, et al. [35] | Main confounders and any additional confounders | No | Unclear reporting | Consecutive |
| Eckhoff, et al. [36] | Main confounders and any additional confounders | No | Unclear reporting | Consecutive |
| Yuan, et al. [41] | Main confounders and any additional confounders | Yes | No risk | Consecutive |
Quantitative synthesis
Tables 3 and 4 summarize the meta-analyses of the association of the GSTP1 polymorphism with tumour response, OS, and toxicities, respectively. The meta-analysis was conducted using a fixed-effect model when P > 0.05 for the Q test, which indicated a lack of heterogeneity among studies; otherwise, a random-effect model was used.
Table 3. Summary of results in the association of GSTP1 polymorphism with tumor response and overall survival.
| GSTP1 Genotype | No. of studies | Tumor response | No. of studies | Overall survival | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of patients | OR (95 % CI) | P value | P of heterogeneity | I2 (%) | No. of patients | HR (95 % CI) | P value | P of heterogeneity | I2 (%) | |||||
| GA vs. AA | Overall | 15 | 2941 | 1.32 (0.97–1.80) | 0.073 | 0.001 | 63.2 | 13 | 4274 | 1.14 (0.97–1.33) | 0.106 | 0.328 | 11.7 | |
| Ethnicity | East Asian | 12 | 2684 | 1.38 (0.99–1.92) | 0.058 | 0.001 | 64.6 | 8 | 3059 | 1.10 (0.90–1.33) | 0.347 | 0.289 | 17.9 | |
| South Asian | 2 | 217 | 1.43 (0.46–4.43) | 0.533 | 0.054 | 73.1 | 2 | 347 | 1.53 (0.78–2.99) | 0.218 | 0.739 | 0.0 | ||
| Mixed descent | 1 | 40 | 0.38 (0.10–1.15) | 0.158 | NA | NA | 3 | 868 | 1.19 (0.78–1.81) | 0.423 | 0.132 | 50.6 | ||
| Sample size | Large | 14 | 2901 | 1.38 (1.02–1.88) | 0.038 | 0.001 | 63.1 | 12 | 4179 | 1.11 (0.94–1.31) | 0.231 | 0.306 | 14.1 | |
| Small | 1 | 40 | 0.38 (0.10–1.45) | 0.143 | NA | NA | 1 | 95 | 1.37 (0.88–2.13) | 0.163 | NA | NA | ||
| Overall | 14 | 2901 | 1.29 (0.79–2.13) | 0.312 | 0.000 | 71.6 | 12 | 4149 | 0.94 (0.56–1.57) | 0.814 | 0.000 | 80.3 | ||
| Ethnicity | East Asian | 12 | 2684 | 1.19 (0.69–2.03) | 0.531 | 0.000 | 73.7 | 8 | 3059 | 1.08 (0.57–2.06) | 0.806 | 0.000 | 85.7 | |
| GG vs. AA | South Asian | 2 | 217 | 2.41 (0.88–6.58) | 0.086 | 0.467 | 0.0 | 1 | 222 | 0.30 (0.03–2.99) | 0.305 | NA | NA | |
| Mixed descent | NA | NA | NA | NA | NA | NA | 3 | 868 | 0.72 (0.26–2.00) | 0.525 | 0.052 | 66.2 | ||
| GA + GG vs. AA (dominant model) | Sample size | Large | 14 | 2901 | 1.29 (0.79–2.13) | 0.312 | 0.000 | 71.6 | 11 | 4054 | 0.89 (0.51–1.57) | 0.692 | 0.000 | 81.6 |
| Small | NA | NA | NA | NA | NA | NA | 1 | 95 | 1.51 (0.75–3.02) | 0.246 | NA | NA | ||
| Overall | 15 | 2941 | 1.37 (0.97–1.94) | 0.074 | 0.000 | 76.3 | 3 | 1048 | 1.74 (1.32–2.30) | < 0.001 | 0.140 | 49.2 | ||
| Ethnicity | East Asian | 12 | 2684 | 1.43 (0.97–2.10) | 0.068 | 0.000 | 78.6 | 1 | 420 | 2.53 (1.60–4.03) | NA | NA | NA | |
| South Asian | 2 | 217 | 1.58 (0.55–4.56) | 0.399 | 0.056 | 72.6 | NA | NA | NA | NA | NA | NA | ||
| Mixed descent | 1 | 40 | 0.38 (0.10–1.15) | 0.158 | NA | NA | 2 | 628 | 1.41 (0.99–2.00) | 0.053 | 0.956 | 0.0 | ||
| Sample size | Large | 14 | 2901 | 1.45 (1.02–2.06) | 0.040 | 0.000 | 77.0 | 2 | 953 | 1.98 (1.14–3.45) | 0.015 | 0.152 | 51.2 | |
| Small | 1 | 40 | 0.38 (0.10–1.45) | 0.143 | NA | NA | 1 | 95 | 1.40 (0.92–2.12) | 0.100 | NA | NA | ||
| GG vs. AA+GA (recessive model) | Overall | 14 | 2901 | 1.05 (0.70–1.57) | 0.829 | 0.000 | 66.2 | NA | NA | NA | NA | NA | NA | |
| Ethnicity | East Asian | 12 | 2684 | 0.97 (0.63–1.48) | 0.872 | 0.000 | 68.7 | NA | NA | NA | NA | NA | NA | |
| South Asian | 2 | 217 | 2.07 (0.78–5.47) | 0.143 | 0.797 | 0 | NA | NA | NA | NA | NA | NA | ||
| Mixed descent | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| Sample size | Large | 14 | 2901 | 1.05 (0.70–1.57) | 0.829 | 0.000 | 66.2 | NA | NA | NA | NA | NA | NA | |
| Small | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| Allele model (G vs. A) | Overall | 15 | 2941 | 1.26 (0.93–1.70) | 0.134 | 0.000 | 82.2 | 2 | 601 | 1.32 (0.47–3.74) | 0.601 | 0.004 | 87.7 | |
| Ethnicity | East Asian | 12 | 2684 | 1.28 (0.91–1.79) | 0.156 | 0.000 | 84.6 | 2 | 601 | 1.32 (0.47–3.74) | 0.601 | 0.004 | 87.7 | |
| South Asian | 2 | 217 | 1.54 (0.76–3.10) | 0.231 | 0.101 | 62.9 | NA | NA | NA | NA | NA | NA | ||
| Mixed descent | 1 | 40 | 0.38 (0.10–1.45) | 0.143 | NA | NA | NA | NA | NA | NA | NA | NA | ||
| Sample size | Large | 14 | 2901 | 1.31 (0.96–1.78) | 0.089 | 0.000 | 83.1 | 2 | 601 | 1.32 (0.47–3.74) | 0.601 | 0.004 | 87.7 | |
| Small | 1 | 40 | 0.38 (0.10–1.45) | 0.143 | NA | NA | NA | NA | NA | NA | NA | NA | ||
GSTP1 glutathione S-transferase P1, OR odds ratio, CI confidence interval, HR hazard ratio, NA not available.
Table 4. Summary of results in the association of GSTP1 polymorphism with toxicities.
| GSTP1 Genotype | No. of studies | toxicities | ||||||
|---|---|---|---|---|---|---|---|---|
| No. of patients | OR (95 % CI) | P value | P of heterogeneity | I2 (%) | ||||
| GA vs. AA | Overall | 11 | 1950 | 1.45 (1.04–2.01) | 0.028 | 0.024 | 51.5 | |
| Therapeutic method | Chemotherapy ± surgery | 8 | 1031 | 1.64 (1.05–2.56) | 0.030 | 0.032 | 54.2 | |
| Radiotherapy ± surgery | 3 | 919 | 1.10 (0.79–1.52) | 0.579 | 0.144 | 48.5 | ||
| GG vs. AA | Overall | 11 | 1950 | 1.47 (1.03–2.10) | 0.036 | 0.709 | 0.0 | |
| Therapeutic method | Chemotherapy ± surgery | 8 | 1031 | 1.58 (0.98–2.55) | 0.059 | 0.776 | 0.0 | |
| Radiotherapy ± surgery | 3 | 919 | 1.33 (0.77–2.28) | 0.306 | 0.207 | 36.4 | ||
| GA + GG vs. AA (dominant model) | Overall | 14 | 2747 | 1.35 (0.99–1.83) | 0.058 | 0.001 | 61.3 | |
| Therapeutic method | Chemotherapy ± surgery | 10 | 1591 | 1.40 (0.90–2.18) | 0.133 | 0.001 | 68.8 | |
| Radiotherapy ± surgery | 4 | 1156 | 1.24 (0.93–1.65) | 0.143 | 0.213 | 33.3 | ||
| GG vs. AA+GA (recessive model) | Overall | 12 | 2044 | 1.54 (1.13–2.09) | 0.006 | 0.330 | 11.8 | |
| Therapeutic method | Chemotherapy ± surgery | 9 | 1125 | 1.72 (1.17–2.54) | 0.006 | 0.526 | 0.0 | |
| Radiotherapy ± surgery | 3 | 919 | 1.12 (0.47–2.67) | 0.792 | 0.092 | 58.2 | ||
| G vs. A (Allele model) | Overall | 11 | 1950 | 1.35 (1.07–1.71) | 0.011 | 0.023 | 51.6 | |
| Therapeutic method | Chemotherapy ± surgery | 8 | 1031 | 1.57 (1.11–2.21) | 0.010 | 0.013 | 60.7 | |
| Radiotherapy ± surgery | 3 | 919 | 1.12 (0.89–1.40) | 0.346 | 0.568 | 0.0 | ||
GSTP1 glutathione S-transferase P1, OR odds ratio, CI confidence interval, NA not available.
Tumor response
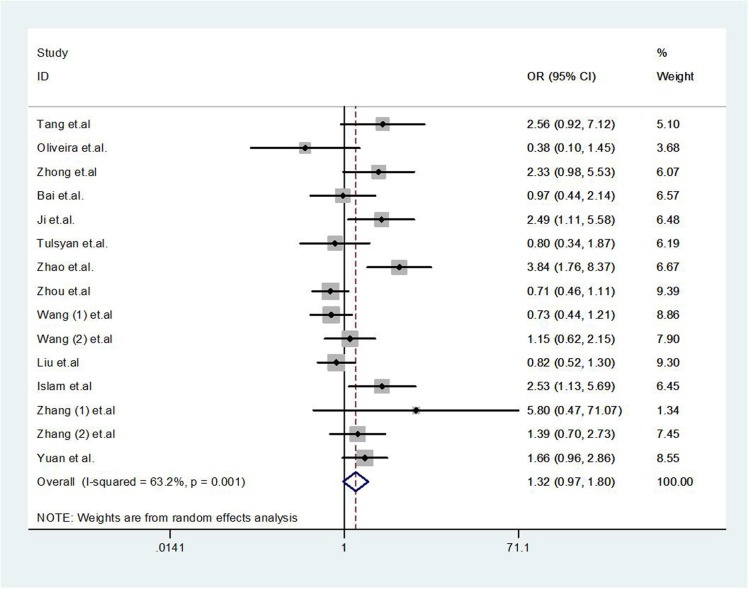
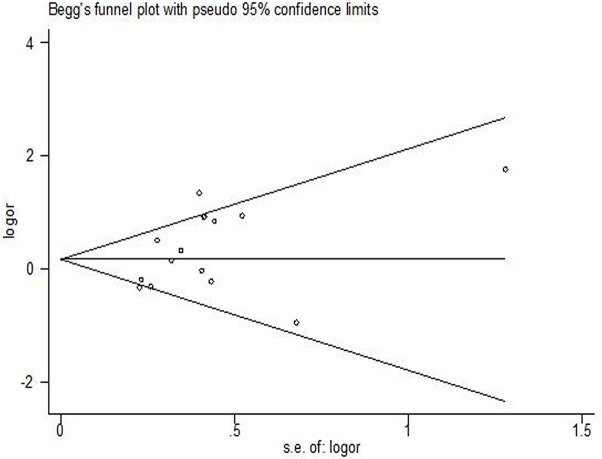
There was no significant association between the GSTP1 polymorphism and tumour response (GA vs. AA OR = 1.32, 95% CI 0.97–1.80, P = 0.073, Figure 2; GG vs. AA OR = 1.29, 95% CI 0.79–2.13, P = 0.312; dominant model OR = 1.37, 95% CI 0.97–1.94, P = 0.074; recessive model OR = 1.05, 95% CI 0.70–1.57, P = 0.829; or allele model OR = 1.26, 95% CI 0.93–1.70, P = 0.134). Publication bias was observed in the Funnel plot and Egger's test (GG vs. AA, P = 0.023; and recessive model, P = 0.034), but not for other models (GA vs. AA, P = 0.066, Figure 3; dominant model P = 0.052, or allele model P = 0.054).
Figure 2. Forest plot of tumor response for GSTP1 gene polymorphism in breast cancer patients (GA vs. AA).
Figure 3. Funnel plot of GSTP1 gene polymorphism for assessment of publication bias: tumor response (GA vs. AA).
Overall survival
The GSTP1 polymorphism was associated with OS in the dominant genetic model (HR = 1.74, 95% CI 1.32–2.30, P < 0.001), but not other genetic models (GA vs. AA, HR = 1.14, 95% CI 0.97–1.33, P = 0.106; GG vs. AA, HR = 0.94, 95% CI 0.56–1.57, P = 0.814; or allele model HR = 1.32, 95 % CI 0.47–3.74, P = 0.601), as shown in Table 3. Publication bias was not observed in the Funnel plot or Egger's test (GA vs. AA, P = 0.365; and GG vs. AA, P = 0.719).
Toxicities
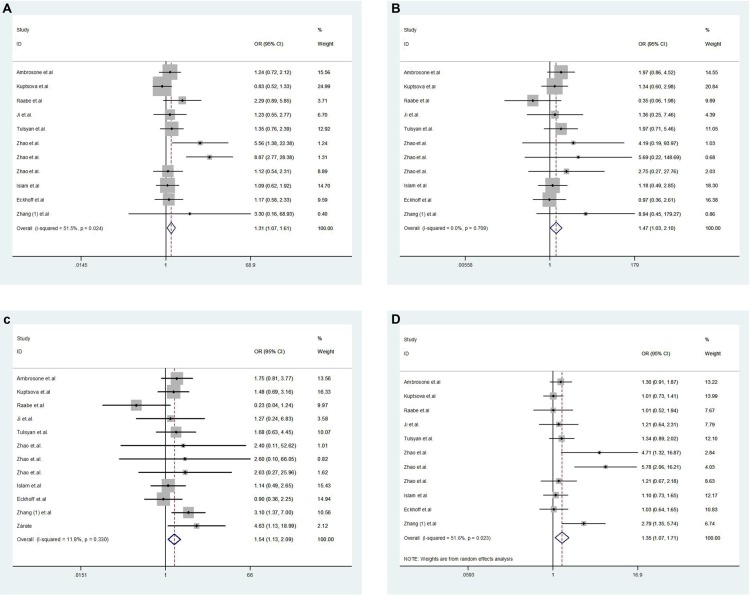
Pooled results showed that there was a significant increase of toxicities (GA vs. AA OR = 1.45, 95% CI 1.04–2.01, P = 0.028, Figure 4A; GG vs. AA OR = 1.47, 95% CI 1.03–2.10, P = 0.036, Figure 4B; recessive model OR = 1.54, 95% CI 1.13–2.09, P = 0.006, Figure 4C; and allele model OR = 1.35, 95% CI 1.07–1.71, P = 0.011, Figure 4D; Table 4). The dominant model was not found to be significantly associated with toxicity (OR = 1.35, 95% CI 0.99–1.83, P = 0.058). Publication bias was observed in the Funnel plot and Egger's test for the genetic models (GA vs. AA, P = 0.008; dominant model, P = 0.011; and allele model, P = 0.008) There was no publication bias in other models (GG vs. AA, P = 0.271; and the recessive model, P = 0.957).
Figure 4.
Forest plot of toxicities for GSTP1 gene polymorphism in breast cancer patients ((A): GA vs. AA; (B): GG vs. AA; (C): recessive model; (D): allele model).
Subgroup analysis
In subgroup analyses, the GSTP1 polymorphism was associated with increased tumour response when the sample size was large (GA vs. AA OR = 1.38, 95% CI 1.02–1.88, P = 0.038; and dominant model OR = 1.45, 95% CI 1.02–2.06, P = 0.829), but this association was not found when the sample size was small, or with any ethnicity subgroup. In addition, no associations between GSTP1 polymorphism and OS were found in either large or small sample sizes. The estimated results showed that there was an increased incidence of toxicities after chemotherapy ± surgery in three genetic models: GA vs. AA (OR = 1.64, 95% CI 1.05–2.56, P = 0.030), recessive model (OR = 1.72, 95% CI 1.17–2.54, P = 0.006), and allele model (OR = 1.57, 95% CI 1.11–2.21, P = 0.010; Table 4). However, we failed to find such an association in the radiotherapy ± surgery group.
DISCUSSION
While the breast cancer treatment response of chemotherapy and/or radiotherapy cannot be optimally predicted, the consequence of gene polymorphism affecting drug efficacy, through encoding metabolizing enzymes and drug transporters, has been confirmed [48]. For breast cancer, anthracycline/paclitaxel-based agents are often effective, which is due to DNA damage, as well as mitochondrial membrane disruption, triggering the apoptotic mechanism and contributing to tumour cell death by the generation of reactive oxygen species (ROS). GSTs (particularly GSTP1) are multifunctional enzymes involved in the protection of cellular components targeted by anticancer drugs. GSTs detoxify chemotherapeutic drugs, or their metabolites, by catalysing the reduction of these compounds through conjugation with glutathione [49]. Therefore, this function of GSTs may result in tumour resistance to cytotoxic agents during chemotherapy [50]. However, the substitution of Ile to Val at codon 105 would result in the decrease of this function, and thus potentially cause an increase in the efficacy of chemotherapy [10]. Previous studies have investigated the association between the GSTP1 polymorphism and treatment outcomes in other cancers [52–53]. In these studies, no significant association between the GSTP1 polymorphism and tumour response from platinum-based chemotherapy was found in either colorectal cancer or gastric cancer [51, 52]. On the contrary, the variant G allele was significantly associated with positive response to platinum-based chemotherapy in East-Asian patients with non-small cell lung cancer [53]. A significantly longer OS was observed in GG + GA genotypes than in AA genotypes in gastric cancer [52]. In the current meta-analysis, there was no significant association between the GSTP1 polymorphism and tumour response. With these inconsistent results, additional studies using uniform evaluation standards are needed in future to sufficiently determine the association between the GSTP1 polymorphism and tumour response.
Chemotherapy and radiotherapy can potentially be interrupted by treatment toxicities. Radiotherapy often has short-term toxicities such as skin erythema and irritation, as well as medium- and long-term toxicities such as breast oedema, pain, fibrosis, and/or telangiectasia. Chemotherapy is usually accompanied by toxicities, such as hematologic, cardiac, and hepatic dysfunction, and vomiting. Severe toxicities might result in treatment interruption and thereby affect treatment efficacy. The GSTP1 Ile to Val substitution has been associated with reduced enzyme activity in the removal of chemotherapy agents [3]. Therefore, it may lead to several toxicities during chemotherapy. Two previous studies have evaluated the association between the GSTP1 polymorphism and toxicities. Both studies found that patients with the AA genotype were at significantly higher risk of haematological and neurological toxicities, compared with patients expressing the AG or GG genotypes [54, 55]. In the current study, we found that the GSTP1 polymorphism was associated with increased toxicities, especially in patients treated with chemotherapy ± surgery. However, this function is advantageous for decreasing toxicities of patients receiving radiotherapy ± surgery. As radiation results in the generation of ROS and lipid peroxidation, nuclear GSTP1 plays a direct role in the cellular sensitivity to oxidative stress. This oxidative stress is caused by hydrogen peroxide through the formation of lipid-peroxide modified DNA. In this meta-analysis, no association was observed between the GSTP1 polymorphism and toxicities due to radiotherapy ± surgery.
Although an effort was made to conduct an accurate and comprehensive analysis, this study has several limitations. First, some factors that may lead to heterogeneity and thus have an influence on treatment outcome—such as treatment options (adjuvant and/or neoadjuvant chemotherapy), chemotherapeutic agents, breast cancer subtypes (hormone receptor-positive, Her2-positive, and triple-negative), and TNM staging status—were not strictly described in some studies. Therefore, these factors could not be stratified into subgroups with great detail. Furthermore, the major sources of heterogeneity where not detected, except for subgroup analysis by ethnicity, sample size, and therapeutic method. Second, publication bias existed in this meta-analysis. This may be due to the absence of some negative trials, which may lead to overestimate the treatment effects.
In conclusion, our meta-analysis suggests that GSTP1 polymorphism may be associated with increased incidence of toxicities, especially in patients treated with chemotherapy ± surgery. Nevertheless, no significant associations were found between the GSTP1 polymorphism and tumour response or OS.
Acknowledgments
We thank all the people who provided technical support and useful discussion of the article.
Footnotes
CONFLICTS OF INTEREST
There is no conflicts of interest.
FUNDING
The study was supported by grants from the Guangxi Science and Technology Development Program (Gui Ke Gong14124004-1-11) and Guangxi Self-financing Scientific Research Subject (Z2013418). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.van der Hage JA, van de Velde CJ, Julien JP, Tubiana-Hulin M, Vandervelden C, Duchateau L. Preoperative chemotherapy in primary operable breast cancer: results from the European Organization for Research and Treatment of Cancer trial 10902. J Clin Oncol. 2001;9:4224–4237. doi: 10.1200/JCO.2001.19.22.4224. [DOI] [PubMed] [Google Scholar]
- 3.Rodrigues FF, Santos RE, Melo MB, Silva MA, Oliveira AL, Rozenowicz RL, Ulson LB, Aoki T. Correlation of polymorphism C3435T of the MDR-1 gene and the response of primary chemotherapy in women with locally advanced breast cancer. Genet Mol Res. 2008;7:177–183. doi: 10.4238/vol7-1gmr400. [DOI] [PubMed] [Google Scholar]
- 4.Arun BK, Granville LA, Yin G, Middleton LP, Dawood S, Kau SW, Kamal A, Hsu L, Hortobagyi GN, Sahin AA. Glutathione-s-transferase-pi expression in early breast cancer: association with outcome and response to chemotherapy. Cancer Invest. 2010;28:554–559. doi: 10.3109/07357900903286925. [DOI] [PubMed] [Google Scholar]
- 5.Peters LJ. Radiation therapy tolerance limits: For one or for all?--Janeway lecture. Cancer. 1996;77:2379–2385. doi: 10.1002/(SICI)1097-0142(19960601)77:11<2379::AID-CNCR29>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 6.Huang MY, Wang YH, Chen FM, Lee SC, Fang WY, Cheng TL, Hou MF, Wang JY, Lin SR. Multiple genetic polymorphisms of GSTP1 313AG, MDR1 3435CC, and MTHFR 677CC highly correlated with early relapse of breast cancer patients in Taiwan. Ann Surg Oncol. 2008;15:872–880. doi: 10.1245/s10434-007-9719-7. [DOI] [PubMed] [Google Scholar]
- 7.Franco RL, Schenka NG, Schenka AA, Rezende LF, Gurgel MS. Glutathione S-transferase Pi expression in invasive breast cancer and its relation with the clinical outcome. J BUON. 2012;17:259–264. [PubMed] [Google Scholar]
- 8.Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482:21–26. doi: 10.1016/s0027-5107(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 9.Townsend D, Tew K. Cancer drugs, genetic variation and the glutathione-S-transferase gene family. Am J Pharmacogenomics. 2003;3:157–172. doi: 10.2165/00129785-200303030-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Board PG, Webb GC, Coggan M. Isolation of a cDNA clone and localization of the human glutathione S-transferase 3 genes to chromosome bands 11q13 and 12q13-14. Ann Hum Genet. 1989;53:205–213. doi: 10.1111/j.1469-1809.1989.tb01786.x. [DOI] [PubMed] [Google Scholar]
- 11.Oliveira AL, Oliveira Rodrigues FF, Dos Santos RE, Rozenowicz RL, Barbosa de Melo M. GSTT1, GSTM1, and GSTP1 polymorphisms as a prognostic factor in women with breast cancer. Genet Mol Res. 2014;13:2521–2530. doi: 10.4238/2014.January.22.9. [DOI] [PubMed] [Google Scholar]
- 12.Ge J, Tian AX, Wang QS, Kong PZ, Yu Y, Li XQ, Cao XC, Feng YM. The GSTP1 105Val allele increases breast cancer risk and aggressiveness but enhances response to cyclophosphamide chemotherapy in North China. PloS one. 2013;8:e67589. doi: 10.1371/journal.pone.0067589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou XN, Dong NN, Yu J, Wang XL, Ren J, Shao H. Study on the Polymorphisms of CYP3A5 and GSTP1 Genes in the Prediction of Short-Term Effcacy of Docetaxel Plus Thiotepa For Patients With Metastatic Breast Cancer. Chin Pharm J. 2012;47:127–131. [Google Scholar]
- 14.Syamala VS, Sreeja L, Syamala V, Raveendran PB, Balakrishnan R, Kuttan R, Ankathil R. Influence of germline polymorphisms of GSTT1, GSTM1, and GSTP1 in familial versus sporadic breast cancer susceptibility and survival. Fam cancer. 2008;7:213–220. doi: 10.1007/s10689-007-9177-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhao YJ, Liu XL. The research of the relationship between the polymorphisms of GSTM1, GSTT1 and GSTP1 with clinicopathology features, chemotherapy response and toxicities in breast cancer. 2014.
- 16.Ambrosone CB, Tian C, Ahn J, Kropp S, Helmbold I, von Fournier D, Haase W, Sautter-Bihl ML, Wenz F, Chang-Claude J. Genetic predictors of acute toxicities related to radiation therapy following lumpectomy for breast cancer: a case-series study. Breast Cancer Res. 2006;8:R40. doi: 10.1186/bcr1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sweeney C, McClure GY, Fares MY, Stone A, Coles BF, Thompson PA, Korourian S, Hutchins LF, Kadlubar FF, Ambrosone CB. Association between survival after treatment for breast cancer and glutathione S-transferase P1 Ile105Val polymorphism. Cancer Res. 2000;60:5621–5624. [PubMed] [Google Scholar]
- 18.Kuptsova N, Chang-Claude J, Kropp S, Helmbold I, Schmezer P, von Fournier D, Haase W, Sautter-Bihl ML, Wenz F, Onel K, Ambrosone CB. Genetic predictors of long-term toxicities after radiation therapy for breast cancer. Int J Cancer. 2008;122:1333–1339. doi: 10.1002/ijc.23138. [DOI] [PubMed] [Google Scholar]
- 19.Bewick MA, Conlon MS, Lafrenie RM. Polymorphisms in manganese superoxide dismutase, myeloperoxidase and glutathione-S-transferase and survival after treatment for metastatic breast cancer. Breast Cancer Res Treat. 2008;111:93–101. doi: 10.1007/s10549-007-9764-8. [DOI] [PubMed] [Google Scholar]
- 20.Tang JH, Zhao JH, Wu JZ, Lu JW, Pan LQ, Xu ZY. Establishment of a multiplex ligationdependent SNP genotyping method and its application in the detection of genes related to chemotherapeutic drugs in breast cancer. Chin J Oncol. 2009;31:108–113. [PubMed] [Google Scholar]
- 21.Oliveira AL, Rodrigues FF, Santos RE, Aoki T, Rocha MN, Longui CA, Melo MB. GSTT1, GSTM1, and GSTP1 polymorphisms and chemotherapy response in locally advanced breast cancer. Genet Mol Res. 2010;9:1045–1053. doi: 10.4238/vol9-2gmr726. [DOI] [PubMed] [Google Scholar]
- 22.Zhong SL, Jiang SG, Tang JH, Li J, Li WJ, Zhao JH. Association of GSTs gene polymorphism with response to chemotherapy in breast cancer. Chin J Clin Lab Sci. 2010;28:438–440. [Google Scholar]
- 23.Bai YL, Zhou B, Jing XY, Zhang B, Huo XQ, Ma C, He JM. Predictive role of GSTs on the prognosis of breast cancer patients with neoadjuvant chemotherapy. Asian Pac J Cancer Prev. 2012;13:5019–5022. doi: 10.7314/apjcp.2012.13.10.5019. [DOI] [PubMed] [Google Scholar]
- 24.Terrazzino S, La Mattina P, Gambaro G, Masini L, Franco P, Canonico PL, Genazzani AA, Krengli M. Common variants of GSTP1, GSTA1, and TGFβ1 are associated with the risk of radiation-induced fbrosis in breast cancer patients. Int J Radiat Oncol Biol Phys. 2012;83:504–511. doi: 10.1016/j.ijrobp.2011.06.2012. [DOI] [PubMed] [Google Scholar]
- 25.Raabe A, Derda K, Reuther S, Szymczak S, Borgmann K, Hoeller U, Ziegler A, Petersen C, Dikomey E. Association of single nucleotide polymorphisms in the genes ATM, GSTP1, SOD2, TGFB1, XPD and XRCC1 with risk of severe erythema after breast conserving radiotherapy. Radiat Oncol. 2012;7:65. doi: 10.1186/1748-717X-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji M, Tang J, Zhao J, Xu B, Qin J, Lu J. Polymorphisms in genes involved in drug detoxifcation and clinical outcomes of anthracycline-based neoadjuvant chemotherapy in Chinese Han breast cancer patients. Cancer Biol Ther. 2012;13:264–271. doi: 10.4161/cbt.13.5.18920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tulsyan S, Chaturvedi P, Agarwal G, Lal P, Agrawal S, Mittal RD, Mittal B. Pharmacogenetic influence of GST polymorphisms on anthracycline-based chemotherapy responses and toxicities in breast cancer patients: a multianalytical approach. Mol Diagn Ther. 2013;17:371–379. doi: 10.1007/s40291-013-0045-4. [DOI] [PubMed] [Google Scholar]
- 28.Duggan C, Ballard-Barbash R, Baumgartner RN, Baumgartner KB, Bernstein L, McTiernan A. Associations between null mutations in GSTT1 and GSTM1, the GSTP1 Ile (105) Val polymorphism, and mortality in breast cancer survivors. SpringerPlus. 2013;2:450. doi: 10.1186/2193-1801-2-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang G, Shu XO, Ruan ZX, Cai QY, Jin F, Gao YT, Zheng W. Genetic polymorphisms in glutathione-Stransferase genes (GSTM1, GSTT1, GSTP1) and survival after chemotherapy for invasive breast carcinoma. Cancer. 2005;103:52–58. doi: 10.1002/cncr.20729. [DOI] [PubMed] [Google Scholar]
- 30.Sugishita M, Imai T, Kikumori T, Mitsuma A, Shimokata T, Shibata T, Morita S, Inada-Inoue M, Sawaki M, Hasegawa Y, Ando Y. Pharmacogenetic association between GSTP1 genetic polymorphism and febrile neutropenia in Japanese patients with early breast cancer. Breast Cancer. 2016;23:195–201. doi: 10.1007/s12282-014-0547-x. [DOI] [PubMed] [Google Scholar]
- 31.Zhou L, Huang A, Zhang D, Yao J, Zhang Y, Li X. Genetic variability of glutathione S-transferases influences treatment outcome of breast cancer. Tumour Biol. 2015;36:5925–5929. doi: 10.1007/s13277-015-3266-9. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Huang ZH. Predictive potential role of glutathione S-transferase polymorphisms in the prognosis of breast cancer. Genet Mol Res. 2015;14:10236–10241. doi: 10.4238/2015.August.28.7. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Wang T, Yin GY, Yang L, Wang ZG, Bu XB. Glutathione S-transferase polymorphisms influence chemotherapy response and treatment outcome in breast cancer. Genet Mol Res. 2015;14:11126–11132. doi: 10.4238/2015.September.22.6. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Luo J, Wang Y, Li L, Yang S. Predictive potential role of glutathione S-transferases polymorphisms on prognosis of breast cancer. Int J Clin Exp Pathol. 2014;7:8935. [PMC free article] [PubMed] [Google Scholar]
- 35.Islam MS, Islam MS, Parvin S, Ahmed MU, Bin Sayeed MS, Uddin MM, Hussain SM, Hasnat A. Effect of GSTP1 and ABCC4 gene polymorphisms on response and toxicities of cyclophosphamideepirubicin-5-fluorouracil-based chemotherapy in Bangladeshi breast cancer patients. Tumor Biol. 2015;36:5451–5457. doi: 10.1007/s13277-015-3211-y. [DOI] [PubMed] [Google Scholar]
- 36.Eckhoff L, Feddersen S, Knoop AS, Ewertz M, Bergmann TK. Docetaxel-induced neuropathy: A pharmacogenetic casecontrol study of 150 women with early-stage breast cancer. Acta Oncologica. 2015;54:530–537. doi: 10.3109/0284186X.2014.969846. [DOI] [PubMed] [Google Scholar]
- 37.Zhang BL, Sun T, Zhang BN, Zheng S, Lv N, Xu BH, Wang X, Chen GJ, Yu DK, Lin DX. Polymorphisms of GSTP1 is associated with differences of chemotherapy response and toxicities in breast cancer. Chin Med J. 2011;124:199–204. [PubMed] [Google Scholar]
- 38.Zhang XY, Li MY, Wang HL, Niu WY, Wu ZX, Shi GC, He JM, Wu JQ. Polymorphism in glutathione S-transferase P1 (GSTP1) on the effect of epirubicin or doxorubicin chemotherapy among breast cancer. African Journal of Pharmacy and Pharmacology. 2013;7:54–57. [Google Scholar]
- 39.Zárate R, González-Santigo S, de la Haba J, Bandres E, Morales R, Salgado J, Gómez A, Aranda E, García-Foncillas J. GSTP1 and MTHFR polymorphisms are related with toxicities in breast cancer adjuvant anthracycline-based treatment. Curr Drug Metab. 2007;8:481–486. doi: 10.2174/138920007780866780. [DOI] [PubMed] [Google Scholar]
- 40.Yao S, Barlow WE, Albain KS, Choi JY, Zhao H, Livingston RB, Davis W, Rae JM, Yeh IT, Hutchins LF, Ravdin PM, Martino S, Lyss AP, et al. Gene polymorphisms in cyclophosphamide metabolism pathway, treatment-related toxicities, and disease-free survival in SWOG 8897 clinical trial for breast cancer. Clin Cancer Res. 2010;16:6169–6176. doi: 10.1158/1078-0432.CCR-10-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan P, Yuan L, Xu BL, Wang CZ, Yang HZ, Li Y. Predictive potential role of glutathione S-transferases polymorphisms in response to chemotherapy and breast cancer prognosis. Genet Mol Res. 2015;14:16675–16681. doi: 10.4238/2015.December.11.15. [DOI] [PubMed] [Google Scholar]
- 42.Wells GA, Shea B, O'Connell D, Peterson J, Welch V, Losos M, Tugwell. P. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in metaanalyses. 2013. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp
- 43.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexander J, Sutton KR, Jones DR, Sheldon TA, Song F. Methods for meta-analysis in medical research. Chichester: Wiley; 2000. [Google Scholar]
- 45.DerSimonian R, Laird N. Meta-analysis in clinical trials. Contemp Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 46.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 47.Egger M, Smith GD, Schneider M, Minder C. Bias in metaanalysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosch TM, Meijerman I, Beijnen JH, Schellens JH. Genetic polymorphisms of drug-metabolizing enzymes and drug transporters in the chemotherapeutic treatment of cancer. Clin Pharmacokinet. 2006;45:253–285. doi: 10.2165/00003088-200645030-00003. [DOI] [PubMed] [Google Scholar]
- 49.Townsend DM, Tew KD. The role of glutathioneStransferase in anti-cancer drug resistance. Oncogene. 2003;22:7369–7375. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salinas AE, Wong MG. Glutathione S-transferases—a review. Curr Med Chem. 1999;6:279–309. [PubMed] [Google Scholar]
- 51.Ye F, Liu Z, Tan A, Liao M, Mo Z, Yang X. XRCC1 and GSTP1 polymorphisms and prognosis of oxaliplatin-based chemotherapy in colorectal cancer: a meta-analysis. Cancer Chemother Pharmacol. 2013;71:733–740. doi: 10.1007/s00280-012-2067-8. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Chen JQ, Liu JL, Qin XG, Huang Y. Polymorphisms in ERCC1, GSTs, TS and MTHFR predict clinical outcomes of gastric cancer patients treated with platinum/5-Fu-based chemotherapy: a systematic review. BMC Gastroenterol. 2012;12:137. doi: 10.1186/1471-230X-12-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang YL, Xian L. The association between the GSTP1 A313G and GSTM1 null/present polymorphisms and the treatment response of the platinum-based chemotherapy in non-small cell lung cancer (NSCLC) patients: a metaanalysis. Tumor Biol. 2014;35:6791–6799. doi: 10.1007/s13277-014-1866-4. [DOI] [PubMed] [Google Scholar]
- 54.Li QF, Yao RY, Liu KW, Lv HY, Jiang T, Liang J. Genetic polymorphism of GSTP1: prediction of clinical outcome to oxaliplatin/5-FU-based chemotherapy in advanced gastric cancer. J Korean Med Sci. 2010;25:846–852. doi: 10.3346/jkms.2010.25.6.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goekkurt E, Al-Batran SE, Hartmann JT, Mogck U, Schuch G, Kramer M, Jaeger E, Bokemeyer C, Ehninger G, Stoehlmacher J. Pharmacogenetic analyses of a phase III trial in metastatic gastroesophageal adenocarcinoma with fluorouracil and leucovorin plus either oxaliplatin or cisplatin: A study of the Arbeitsgemeinschaft Internistische Onkologie. J Clin Oncol. 2009;27:2863–2873. doi: 10.1200/JCO.2008.19.1718. [DOI] [PubMed] [Google Scholar]