Abstract
Circular RNAs (circRNAs), a novel class of long noncoding RNAs, are characterized by a covalently closed continuous loop without 5′ or 3′ polarities structure and have been widely found in thousands of lives including plants, animals and human beings. Utilizing the high-throughput RNA sequencing (RNA-seq) technology, recent findings have indicated thata great deal of circRNAs, which are endogenous, stable, widely expressed in mammalian cells, often exhibit cell type-specific, tissue-specific or developmental-stage-specific expression. Evidences are arising that some circRNAs might regulate microRNA (miRNA) function as microRNA sponges and play a significant role in transcriptional control. circRNAs associate with related miRNAs and the circRNA-miRNA axes are involved in a serious of disease pathways such as apoptosis, vascularization, invasion and metastasis. In this review, we generalize and analyse the aspects including synthesis, characteristics, classification, and several regulatory functions of circRNAs and highlight the association between circRNAs dysregulation by circRNA-miRNA-mRNA axis and sorts of diseases including cancer- related and non-cancer diseases.”
Keywords: circular RNA, back-splicing, microRNA sponge, tumorgenesis
INTRODUCTION
Circular RNAs (circRNAs) are a class of endogenous non-coding RNAs (ncRNAs) with a structure of covalently closed continuous loops, resulting in a more stable condition than line RNA [1]. In the early 1970s, the first circular RNA was found in some plant viroid and a circular form of RNA in the cytoplasm of monkey renal CV-1 cells was discovered via electro-microscopy by then [2–3]. With advances in high-throughput sequencing technology, recent studies have uncovered that a large number of circRNAs in human beings cells may be involved in human fetal development, myocardial Infarction as well as carcinomas [4–6]. In depth study revealed that some circRNAs might regulate microRNA (miRNA) function as microRNA sponges and the circRNA-miRNA-mRNA axis axes may play a crucial role in cancer-related or non-cancer pathways. For instance, Eur Heart J, et al confirmed that heart-related circRNA HRCR bound to miR-223 directly and suppressed expression of HRCR in human cardiomyocytes enforced attenuated hypertrophic responses [7]. Further studies indicated that circRNA have a potential value to provide novel strategies for clinical diagnostic and treatment of carcinomas [8]. In this review, we generalize synthesis, characteristics, classification, and several regulatory functions of circRNAs and highlight the association between circRNAs dysregulation and sorts of diseases including cancer-related and non-cancer diseases.
Biogenesis of circular RNAs
Numerous studies have disclosed the biogenesis of circRNAs through back-splicing mechanism, which is different from the canonical splicing of long nocoding RNAs. circRNAs including exonic circRNAs, intronic circRNAs, and retained-intron circRNAs can be transcribed from pre-mRNAs sequences by RNA polymerase II (Figure 1) [9]. The distinctive structure of circRNAs contributes to a special form of splicing type: back-splicing that characterizes the synthesis of circular RNAs. During the non-canonical splicing process, RNA exons gain the circular form in a manner of head to tail and formed as exonic circular RNA, while the spliceosomal would cut out intronic genes in the binding part.
Figure 1. Biogenesis of circular RNAs in human diseases.
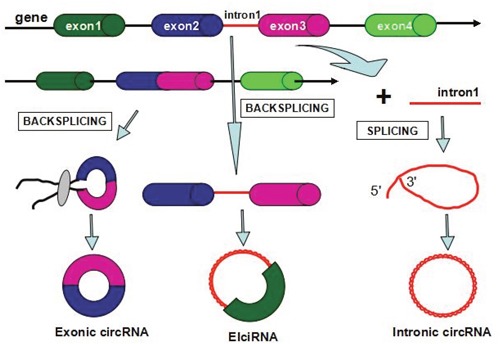
Previous researches have uncovered that exon circularization relies on flanking intronic complementary sequences and alternative formation of inverted repeated Alu pairs that could contribute to alternative circularization as well as resulting in multiple circular RNA transcripts produced from a single sequence [10]. The regulatory rules of circRNA biogenesis are dependent on the cis-regulatory elements and trans-acting factors which control splicing involved in the splicing process [9, 11]. Firstly, the biogenesis of circRNAs is regulated by a variety of RNA-binding proteins [14–15]. Meanwhile, the splicing factor Muscleblind (Mbl) promotes circRNA production from its own pre-mRNA [9]. Quaking (QKI), a type of RBP, are sufficient to bind to two flanking introns and strikingly bring the circularized exons closer together [12]. In addition, adenosine deaminase 1 acting on RNA (ADAR1) also participates in the regulation of synthesis of circRNAs by suppressing circRNA expression independently of its editing activity since it appears to function as an editing enzyme and a double-stranded RNA-binding protein [13]. The process of the back-splicing requires the canonical spliceosomal machinery and is associated with exon circularization efficiency, which is reliable on canonical splice sites bracketing the exons [9, 14–15]. As multiple circRNAs can be produced from the same gene sequences by alternative splicing, a large amount of circRNAs in mammals are processed from internal exons with long flanking introns, containing reverse complementary sequenceswhich are capable of pairing to form RNA duplexes [10].
Remarkablely, the low back-splicing efficiency is the characteristic of biogenesis of circRNA. This phenomenon possibly results from the presence of canonical splice sites where the exons and spliceosomes are detrimentally assembled at back-splicing sites to catalyze the ligation of upstream 3′ acceptor sites with downstream 5′ donor sites [9, 14–15]. Thus, the biogenesis of circRNAs is still unclear and remains to be further investigated.
Characteristics of circular RNAs
With the development of bioinformatics technology and RNA sequencing, researchers have discovered thousands of circRNAs in most organisms including Archaea, human being and so on. Song X, et al detected >476 distinct circRNAs in human gliomas and Rybak-Wolf A, et al distinguished about 15, 849 distinct circRNAs in mouse and 65, 731 in human being. Moreover, Nair A, et al suggested that circRNA frequency in breast cancer may be useful for the diagnosis and treatment of breast cancer [16–18]. However, some circRNAs expressions are on the contrary, for example, circRNAs including Rims2, Tulp4 and Elf2 were found to be the major isoform of multiple genes in brain tissues [17].
The most important feature of endogenous circRNA is that the closed loop structures of circRNAs with a covalent bond linking the 3 ′ and 5 ′ ends make them avoid exonucleolytic degradation by RNase R [19]. For example, in an enzymatic process, circRNAs, known as Brolling circle amplification (RCA), play a main role in envolution of the development of hepatitis delta virus (HDV) and viroid replication models [20–21]. Noteworthy, some circRNAs are located in the nucleus in organism which harbors miRNA binding miRNA response elements and forming circRNA-miRNA axes are involved in regulation of gene expression at the transcription or post transcription level [10].
Base on the characteristic of circular RNAs, RNA-seq and high-throughput sequencing technology always are used to detect circRNAs. RNA-seq and high-throughput sequencing Technology, as the inevitable outcome of bioinformatics, have different adventage on detecting new circular RNA. Lot of circular RNA databases such as CircBase, CircNet, CircInteractome have been Established and could give more inforation about circRNA.
According to the recent studies, we could find that long noncoding RNAs and circular RNAs have some same aspects. The most meaningful point is that they all could function as a miRNA sponge and regulate downstream mRNA, all of which involved in the pathomechanism of various diseases including cancers. Meanwhile, they have different fields like that circular RNAs have stable structure and have more advantage to be a clinical biomarker than long noncoding RNAs, while long noncoding RNAs are superior to circular RNAs in abundance in human beings cells.
Classification of circular RNAs
The length of CircRNAs differs from each other and can derive from all of region of the genome [22–25]. Classification system of long noncoding RNAs (lncRNAs) could be used as an example to circular RNAs. According to their genomic proximity to the neighboring gene, circRNAs could be classified into five types: a. Sense or exonic, circRNAs may contain just one or multiple exons which come from a linear transcript on the same strand, they show the probability for alternatively spliced isoforms [10, 26]. b. Intronic, they may derive wholly from an intron of the linear transcript depend on a crucial motif which contain a 7-nt GU-rich element close to 11-nt C-rich element and an the 5splice site near the branch point [24]. c. Antisense, if it overlaps one or more exons of the linear sequence on the opposite strand. d. Intragenic or Bidirectional, when circRNAs is transcribed from same gene locus of the linear sequence not classified as ‘sense’ and ‘intronic’ and whereas in close genomic proximity. e. Intergenic, when it is located between the genomic intervals of two genes (Figure 2) [27].
Figure 2. Diagrammatic drawing of circRNA classification.
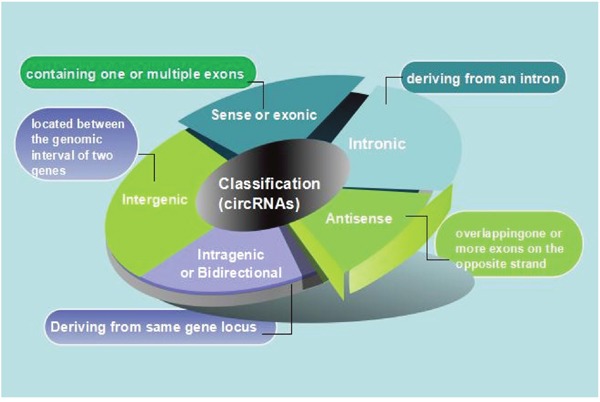
(a) Sense or exonic, CircRNAs may contain just one or multiple exons which come from a linear transcript on the same strand, they show the probability for alternatively spliced isoforms. (b) Intronic, they may derive wholly from an intron of the linear transcript depend on a crucial motif which contain a 7-nt GU-rich element close to 11-nt C-rich element and an the 5splice site near the branch point. (c) Antisense, if it overlaps one or more exons of the linear sequence on the opposite strand. (d) Intragenic or Bidirectional, when circRNAs is transcribed from same gene locus of the linear sequence not classified as ‘sense’ and ‘intronic’ and whereas in close genomic proximity. (e) Intergenic, when it is located between the genomic interval of two genes.
Function of circular RNAs
CircRNAs play a crucial role in gene regulation at the post-transcription or transcription level and then have an impact at the level of gene expression. In this review, several functions of circular RNAs were listed as follows.
MiRNA sponges
MiRNAs, an abundant class of small noncoding RNAs (∼22 nt), posttranscriptionally modulate the translation of target mRNAs via corresponding miRNA response elements [28]. The current studies have provided evidences that some of circRNAs harbor MREs, suggesting a potential role as competitive endogenous RNAs (ceRNAs) to compete for miRNA-binding sites, thus affecting miRNA activities [23, 29–30]. For example, CiRS-7, as a Human circRNA cerebellar degeneration-related protein 1 transcript (CDR1), acts as a miR-7 sponge by binding with related miRNA and influencing the availability of miR-7 to bind to its target mRNAs [30]. Besides ciRS-7, testis-specific circRNA, sex-determining region Y (circSRY), serves as the miR-138 sponge and functions as a platform for binding miR-138 and regulates linked mRNA translation [25, 30–32]. Additionally, Zheng Q, et al revealed an abundant circHIPK3 was observed to sponge to 9 miRNAs with 18 potential binding sites and in particular regulates cell growth by sponging multiple miR-124 and inhibiting miR-124 activity in malignant tumors [6]. Overall, the circ-miRNA axis, regardless of promotion or suppression, have significant effects on various pathways in human diseases and is worthy more thorough study (Figure 3) [33].
Figure 3. Schematic diagram of circRNAs functioning as miRNA sponges or competing endogenous RNAs.
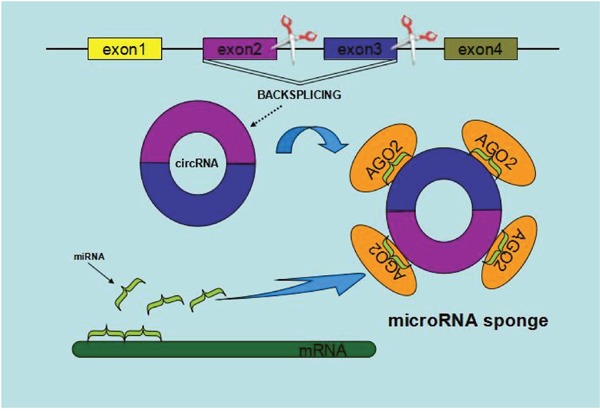
Transcription regulation
Although most circRNAs regulate miRNAs as the role of miRNA sponge, some circular intronic RNAs (ciRNAs) or exon-intron circRNAs (EIciRNAs) are predicted to function as post-transcriptional regulators of gene expression and regulate linear RNA transcription and protein expression [22, 34–35]. Some ciRNAs such as c-sirt7, derived from lariats and interacting with the Pol II complex make the transcription levels of the relevant ankyrin repeat domain 52 (ANKRD52) or sirtuin 7 (SIRT7) genes decline. All together, abundant ciRNAs promote transcription of their parental genes with the unknown mechanism [24]. Meanwhile, EIciRNAs mainly localized in the nucleus, connect with U1 snRNP and ascend transcription of related parental genes [36].
Competition with linear splicing
Back-splicing as circRNA biogenesis itself has been suggested that the usage of 5′ and 3′splice sites can compete with pre-mRNA splicing contribute to lower levels of linear mRNAs that include the exons circularRNAs [9–10]. An circularized exon means that it will not be involved in the processed mRNA and occurs in expense of linear splicing to produce mRNAs [37]. therefore, the synthesis of back-spliced circRNAs lead to the consequence of the downregulation of protein coding mRNAs. Future work revealed a large proportion of genes could take part in generating circRNAs and the process of circRNA biogenesis is happening along with the whole life. alternative splicing mechanisms like a switch who make the decision to back-splice or splice linearly. Consequently, a crucial gene regulatory mechanism involving this competition could be established and get more attention [22, 35].
Other functions
Increasing evidences from the studies of circ-Foxo3 uncover that circ-Foxo3 can sequester proteins in the cytoplasm and prevent their nuclear entry by binding these proteins in the cytoplasm. On the contrary, knockdown of circ-Foxo3 which interacting with stress-related proteins FAK and HIF-1α may enhance nuclear expression of these proteins [22, 38–39]. Moreover, circRNAs with open reading frames also may be translated into peptides. For example, Haidar et al. found a circRNA of the virusoid in rice yellow mottle virus can code for a protein [40].
Association between circRNA-miRNA axis and progression of human diseases
CircRNAs have been reported to be involved in many human diseases especially for carcinomas. To date, several studies suggest that the circRNA-miRNA-mRNA axis axis is probably involved in signaling pathway of human diseases by regulating pathogenicity-related gene expression.
Circular RNAs in non-cancer diseases
In this review, we listed the expression of circRNAs and circRNA-miRNA axis in signaling pathway of various types of non-cancer diseases such as major depressive disorder, myocardial infarction and provide potential implications in targeted therapy (Table 1).
Table 1. Summary of the expression of circRNAs and circRNA-miRNA axis in signaling pathway of non-cancer diseases.
| Type of non-tumor diseases | Sequence name or more content | Expression | Intersection molecules and/or pathway | First author |
|---|---|---|---|---|
| TLR4/LPS | circ-RasGEF1B | ↑ | circ-RasGEF1B↓-ICAM-1↓-protecting cells against microbial infection | Ng, W. L [41]. |
| Myocardial Infarction | CircRNA Cdr1 | ↑ | CircRNA Cdr1↑-miR-7a↓-PARP, SP1-Myocardial Infarction | Geng, H.H. [42]. |
| osteoarthritis | Circ-CER | ↓ | CircRNA-CER/MMP13-miR-136- chondrocyte ECM degradation | Liu, Q. [43]. |
| cardiac senescence | circ-Foxo3 | ↑ | circ-Foxo3↑-ID-1↓, E2F1↓, FAK↓, HIF1alpha↓-cardiac senescence | Du, W. W [38]. |
| heart failure | circ-HRCR | ↓ | circ-HRCR↑- miR-223-ARC-reduced hypertrophic responses | Wang, K [7]. |
| immunosenescence | circRNA_100783 | ↑ | circRNA100783-miR-mRNA-CD28-related CD8(+) T cell ageing | Wang, Y. H [44]. |
| diabetes | Cdr1as | ↑ | long-term forskolin/PMA-Cdr1as↑-miR7-cAMP-PKC-Pax6/Myrip | Xu, H. [45]. |
| brain tissue | ciRS-7 | ↑ | ciRS-7 - miR-7(Ago) | Hansen, T. B [30]. |
| Circ-Sry | ↑ | Sry-miR-138 | ||
| lead-induced neuronal cell apoptosis | circ-Rar1 | unknown | circRar1-miR-671- apoptosis-associated caspase8/p38 | Nan, A. [46]. |
| nonalcoholic steatohepatitis | circRNA_002581 | unknown | circRNA_002581-miR-122-Slc1a5/Plp2/Cpeb1 | Jin, X. [47]. |
| circRNA_007585 | unknown | circRNA_007585-miR-326- UCP2 | ||
| Hirschsprung's disease | Circ-ZNF609 | ↓ | ZNF609- miR-150-5p - AKT3-proliferation, migration | Peng, L. [48]. |
| cardiac fibroblasts | CircRNA_000203 | ↑ | CircRNA_000203↑-miR-26b-5p↓- Col1a2 and CTGF | Tang, C. M. [49]. |
↓ means down-regulated.
↑ means up-regulated.
Some circRNAs present as an upward trend to regulate the pathways. Circ-Foxo3, originated from one of the forkhead family of transcription factors Foxo3, could both act as miRNA sponges for miR-136, miR-138, miR-433, miR-762, miR-3614-5p and influencing their function [50]. Du WW, et al tested the roles of a circular RNA circ-Foxo3 in senescence both in vitro and in vivo and found that circ-Foxo3 was highly expressed in heart of aged patients and mice and it binded to the anti-senescent protein ID-1 and the transcription factor E2F1 and the anti-stress proteins FAK and HIF1α. On the contrary, they also found that silencing endogenous circ-Foxo3 suppressed senescence of mouse embryonic fibroblasts and ectopic expression of circ-Foxo3 relieved senescence [38].
Meanwhile, some other circRNAs play a downward regulation in non-cancer related diseases. For instance, Liu Q. et al discovered that circRNA-CER decreased expression in osteoarthritis (OA) and regulated MMP13 expression as a competing endogenous RNA (ceRNA) and involving in the processof chondrocyte ECM degradation, suggesting that circRNA-CER could be used as a potential target in OA therapy [43]. Peng L, et al reported that the cir-ZNF609 was down-regulated in Hirschsprung disease (HSCR) compared with normal adjacent tissues and may also act as a sponge for miR-150-5p to regulate the expression of AKT3 which is linked with the proliferation and migration of cells. Furthermore, silencing cir-ZNF609 inhibited the migration and proliferation of cells. In general, these findings expounded that cir-ZNF609 participate in the onset of HSCR through the cir-ZNF609-miR-150-5p-AKT3 axis [48].
Circular RNAs in carcinomas
We also offered the table about the expression of circRNAs and circRNA-miRNA axis in signaling pathway of some sorts of human cancers such as neuroglioma, colorectal cancer (Table 2).
Table 2. Summary of the expression of circRNAs and circRNA-miRNA axis in signaling pathway of cancers.
| Type of cancer | Sequence name or more content | Expression | Intersection molecules and/or pathway | First author |
|---|---|---|---|---|
| bladder carcinoma | circ-TCF25 | ↑ | circTCF25↑-miR-103a-3p↓/miR-107↓-CDK6↑-proliferation, migration↑ | Zhong, Z. [50]. |
| esophageal cancer | CircRNA_001059 | ↑ | CircRNA_001059-miRNA (Wnt pathway) | Su, H. [51]. |
| CircRNA_000167 | ↓ | CircRNA_000167-miRNA (Wnt pathway) | ||
| hepatoma carcinoma | CircRNA Cdr1 | ↑ | CircRNA Cdr1↑-miR-7↓-CCNE1, PIK3CD↓-proliferation, invision↑ | Yu, L. [52]. |
| bladder carcinoma | Circ-MYLK | unknown | CircRNA MYLK-miR-29a-3p-DNMT3B | Huang, M. [53]. |
| hepatoma carcinoma | hsa_circ_0005075 | unknown | hsa_circ_0005075-miR-23B-5P/93-3P/581/23A-5P-HCC tumor size | Shang, X. [54]. |
| Colorectal Cancer | hsa_circ_001569 | ↑ | circRNA_001569-miR-145↓-E2F5, BAG4, FMNL2↑-proliferation, invision | Xie, H. [55]. |
| seven cancers | circ-HIPK3 | ↓ | circHIPK3-miR-124-cell growth | Zheng, Q. [6]. |
| neuroglioma | circRNA | unknown | miR-671-5p↑/CDR1-AS↓/CDR1↓/VSNL1↓-migration↑, migration↑ | Barbagallo, D [56]. |
| carcinoma | circ-Foxo3 | ↓ | circ-Foxo3↑--p21-CDK2↓-cell cycle progression↓ | Du, W. W [57]. |
| Colorectal Cancer | cir-ITCH | ↓ | cir-ITCH↓-ITCH↑-Wnt/beta-catenin↓ | Huang, G [58]. |
| esophageal carcinoma | cir-ITCH | ↓ | cir-ITCH↓-miR7/17/214-ITCH↑-phosphorylated DV12-Wnt/beta-catenin↓ | Li, F [59]. |
| breast cancer | circ-Foxo3 | ↑ | Foxo3P↑-Foxo3 circ/Foxo3 mRNA-proliferation↓ | Yang, W. [8]. |
| carcinoma | circ-CDR1 | unknown | CDR1↓-miR-671-CDR1 mRNA ↓ | Hansen, T. B [60]. |
| neuroglioma | cZNF292 | ↓ | cZNF292↓-Wnt/beta-catenin signaling pathway-PRR11, Cyclin A, p-CDK2, VEGFR-1/2, p-VEGFR-1/2, EGFR-proliferation, cell cycle progression↓ | Yang, P. [61]. |
| hepatoma carcinoma | ciRS-7 (Cdr1as) | ↓ | ciRS-7-miR-7-PIK3CD/p70S6K | Xu, L. [62]. |
| lung cancer | circ-ITCH | ↓ | ITCH-miR-7/miR-214 (Wnt/beta-catenin signaling pathway) -A549/NIC-H460 | Wan, L. [63]. |
| carcinoma | circ-Foxo3 | ↓ | circ-Foxo3↑- Foxo3↑, p53↓- tumor apoptosis | Du, W. W. [64]. |
| KRAS mutant colon cancer | circRNA | ↓ | mutant KRAS- circRNA abundance↓ | Dou, Y. [65]. |
| clear cell renal cell carcinoma | circ-HIAT1 | ↓ | AR - circHIAT1↓-miR-195-5P/29a-3p/29c-3p↓-CDC42↑-CCRCC migration and invasion | Wang, Kefeng [66]. |
↓ means down-regulated.
↑ means up-regulated.
Some circRNAs, participating in the interaction with miRNAs, play a role as a upward trend to regulate the pathways significantly in human cancers. Zhong et, al reported that circTCF25 binded to miR-103a-3p/miR-107 and potentially contributed to the up-regulation of thirteen targets about cell proliferation, migration and invasion. Subsequently, the phenomena that down-regulating miR-103a-3p and miR-107, increasing CDK6 expression, and promoting proliferation and migration were observed by over-expression of circTCF25. All of these suggested that circTCF25 could be involved in pathway of bladder carcinoma via circTCF25-miR-103a-3p/miR-107-CDK6 axis and be a new potential marker for this cancer [50].
We also pay attention to some circRNAs that playing a downward regulation in carcinomas. Up to date, several evidence has demonstrated that androgen receptor (AR) is related with promoting the mechanisms about metastasis of clear cell renalcell carcinoma (ccRCC). Wang K et al identified a new circRNA (named as circHIAT1) with lower expression in ccRCCs, compareing with adjacent nomal tissues. AR-suppressed circHIAT1 suppress ccRCC cell progression by rising circ HIAT1 expression. In this newly signal process, circHIAT1 could deregulate miR-195-5p /29a-3p/29c-3p axis and increase CDC42 expression to fulfil the goal of enhancing ccRCC cell migration and invasion. Conclusively, they suggested that AR-circHIAT1 -mediated miR-195-5p/29a-3p/29c-3p/CDC42 axis may give us novel insights into develop new therapies to cure ccRCC [66]. Yang P et al reported cZNF292 is an important circular oncogenic RNA and plays a critical role in tube formation. According to the information that the Wnt/β-catenin signaling pathway and related genes such as Cyclin A, p-CDK2, VEGFR-1/2 and EGFR were involved in the process of halting the Cell cycle progression in humanglioma U87MG and U251 cells. they found that cZNF292 silencing plays an critical role in suppressing tube formation by inhibiting glioma cell proliferation and cell cycle progression and may serve as new potential targets and biomarkers for glioma therapy [61].
Conclusion and perspective
CircRNAs, a recently discovered RNA type, were thought to be the products of transcription errors. With the development of high-throughput sequencing technologies and bioinformatics, they have become a popular research subject and gain more attention currently. circRNAs as miRNA sponge specifically bind to microRNAs and then modulate gene expression by competing with competing endogenous RNA, providing a more novel understanding of the RNA language and their role in signaling pathway of human diseases. CircRNAs are expressed widely in tissues, saliva, blood, exosomes and much more stable owning to their circular structure. According to these characteristics, circRNAs could be as biological markers of human diseases and thus improve the accuracy and specificity of diagnosis and therapies.
In addition, the understanding and achievements in the study of circRNAs are still very limited at present. Numerous problems about circular RNA are unknown. For example, with the natural stable structure, how circRNAs are degraded rightly in final? Are there any other molecular mechanism of synthesis method of circRNAs? The meaning and value of circRNA is obvious and it is crucial to do more research on circRNAs, especially the circRNA biogenesis.
Acknowledgments
This study was supported by the Development of Medical Science and Technology Foundation of Nanjing (grant No.ZKX14035) to Professor Hongyong Cao.
Footnotes
CONFLICTS OF INTEREST
The authors report no conflicts of interest in this work.
REFERENCES
- 1.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 2.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547–55. doi: 10.1016/0092-8674(76)90223-3. [DOI] [PubMed] [Google Scholar]
- 4.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vausort M, Salgado-Somoza A, Zhang L, Leszek P, Scholz M, Teren A, Burkhardt R, Thiery J, Wagner DR, Devaux Y. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol. 2016;68:1247–1248. doi: 10.1016/j.jacc.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 8.Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2015;35:3919–3931. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- 9.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Chen T, Xiang JF, Zhu S, Chen S, Yin QF, Zhang XO, Zhang J, Feng H, Dong R, Li XJ, Yang L, Chen LL. ADAR1 is required for differentiation and neural induction by regulating microRNA processing in a catalytically independent manner. Cell Res. 2015;25:459–476. doi: 10.1038/cr.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 18.Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher JP, Subramanian S, Kalari KR. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7:80967–80979. doi: 10.18632/oncotarget.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores R, Grubb D, Elleuch A, Nohales MÁ, Delgado S, Gago S. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis δ virus: variations on a theme. RNA Biol. 2011;8:200–206. doi: 10.4161/rna.8.2.14238. [DOI] [PubMed] [Google Scholar]
- 21.Abe N, Hiroshima M, Maruyama H, Nakashima Y, Nakano Y, Matsuda A, Sako Y, Ito Y, Abe H. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem Int Ed Engl. 2011;52:7004–7008. doi: 10.1002/anie.201302044. [DOI] [PubMed] [Google Scholar]
- 22.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu S, Zhong Y, Shang R. The emerging landscape of circular RNA in life processes. RNA Biol. 2016;11:1–8. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas LF, Saetrom P. Circular RNAs are depleted of poly-morphisms at microRNA binding sites. Bioinformatics. 2014;30:2243–2246. doi: 10.1093/bioinformatics/btu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 31.Andreeva K, Cooper NG. Circular RNAs: new players in gene regulation. Adv Biosci Biotechnol. 2015;06:433–441. [Google Scholar]
- 32.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLos One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 37.Kelly S, Greenman C, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 38.Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 39.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocation. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 40.Abouhaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci USA. 2014;111:14542–14547. doi: 10.1073/pnas.1402814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, Ea CK. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13:861–871. doi: 10.1080/15476286.2016.1207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, Ao Y. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a mir-136 'sponge' in human cartilage degradation. Sci Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing. 2015;12:17. doi: 10.1186/s12979-015-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nan A, Chen L, Zhang N, Liu Z, Yang T, Wang Z, Yang C, Jiang Y. A novel regulatory network among LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch Toxicol. 2017;91:1671–1684. doi: 10.1007/s00204-016-1837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin X, Feng CY, Xiang Z, Chen YP, Li YM. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. 2016;7:66455–66467. doi: 10.18632/oncotarget.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng L, Chen G, Zhu Z, Shen Z, Du C, Zang R, Su Y, Xie H, Li H, Xu X, Xia Y, Tang W. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget. 2016;8:808–818. doi: 10.18632/oncotarget.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, Wu SL, Cheng JD, Shan ZX. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, Zhao L, Zhang X, Pan H, Xie D, Jin X, Xie C. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Translat Med. 2016;14:225. doi: 10.1186/s12967-016-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11:e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. J Urol. 2017;197:51. doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shang X1, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine. 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barbagallo D, Condorelli A, Ragusa M, Salito L, Sammito M, Banelli B, Caltabiano R, Barbagallo G, Zappalà A, Battaglia R, Cirnigliaro M, Lanzafame S, Vasquez E, et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7:4746–4759. doi: 10.18632/oncotarget.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu C, Li G, Zhu Y. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7:63449–63455. doi: 10.18632/oncotarget.11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PubMed] [Google Scholar]
- 63.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG, Zhang B. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang K, Sun Y, Tao W, Fei X, Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12. doi: 10.1016/j.canlet.2016.12.036. [DOI] [PubMed] [Google Scholar]


