Abstract
The cyclin-dependent kinase inhibitor-3 (CDKN3) gene encodes a dual-specificity protein tyrosine phosphatase that dephosphorylates CDK1/CDK2 and other proteins. CDKN3 is often overexpressed in human cancer, and this overexpression correlates with reduced survival in several types of cancer. CDKN3 transcript variants and mutations have also been reported. The mechanism of CDKN3 overexpression and the role of CDKN3 transcript variants in human cancer are not entirely clear. Here, we review the literature and provide additional data to assess the correlation of CDKN3 expression with patient survival. Besides the full-length CDKN3 encoding transcript and a major transcript that skips exon 2 express in normal and cancer cells, minor aberrant transcript variants have been reported. Aberrant CDKN3 transcripts were postulated to encode dominant-negative inhibitors of CDKN3 as an explanation for overexpression of the perceived tumor suppressor gene in human cancer. However, while CDKN3 is often overexpressed in human cancer, aberrant CDKN3 transcripts occur infrequently and at lower levels. CDKN3 mutations and copy number alternation are rare in human cancer, implying that neither loss of CDKN3 activity nor constitutive gain of CDKN3 expression offer an advantage to tumorigenesis. Recently, it was found that CDKN3 transcript and protein levels fluctuate during the cell cycle, peaking in mitosis. Given that rapidly growing tumors have more mitotic cells, the high level of mitotic CDKN3 expression is the most plausible mechanism of frequent CDKN3 overexpression in human cancer. This finding clarifies the mechanism of CDKN3 overexpression in human cancer and questions the view of CDKN3 as a tumor suppressor.
Keywords: CDKN3, CDK1, splicing, mitosis, phosphatase, cancer
1. CDKN3
Cyclin-dependent kinase (CDK) inhibitor-3 (CDKN3) is a dual-specificity protein tyrosine phosphatase of the CDC14 group (Alonso et al., 2004). CDKN3 was originally isolated from a HeLa cDNA library as a CDK2 binding protein via a yeast two-hybrid screen and was termed CDK-Associated Phosphatase (KAP) (Hannon et al., 1994). Although CDKN3 was isolated as a CDK2-binding protein (since experiment was designed to isolate CDK2 partners), it was observed that CDKN3 bound preferentially to CDK1 (CDC2), which functions in the mitotic phase of the cell cycle (Hannon et al., 1994).
As major regulators of cell cycle progression, CDK1 and CDK2 activities are tightly regulated. In addition to the required interaction with cyclins that oscillate through cell cycle, CDK1/CDK2 Ser/Thr kinase activity is further regulated by phosphorylation at three residues; Thr-14 and Tyr-15 of both proteins and Thr-161 of CDK1 and Thr-160 of CDK2 (Gu et al., 1992). Phosphorylation at the dual Thr-14/Tyr-15 sites inhibits kinase activity and is regulated by the PKMYT1 (MYT1)/WEE1 kinase and the dual-specificity protein tyrosine phosphatase CDC25. Thr-161/Thr-160 are activating sites of phosphorylation in the kinase activating loop (Gu et al., 1992) in each protein. CDK7 (CAK) phosphorylates Thr-161/160 thus activating each protein. Phosphorylation of Thr-160 in CDK2 increases its binding affinity for the kinase substrates, but has no effect on cyclin A binding (Brown et al., 1999). CDKN3 is responsible for dephosphorylating Thr-161/Thr-160. Mechanistically, CDKN3 dephosphorylates monomeric CDK2; whereas cyclin A protects CDK2 Thr-160 from being dephosphorylated by CDKN3 (Poon and Hunter, 1995). Thus, the cyclin A-bound CDK2 that drives S phase is resistant to CDKN3. CDKN3 dephosphorylates CDK1 during mitotic exit (Nalepa et al., 2013).
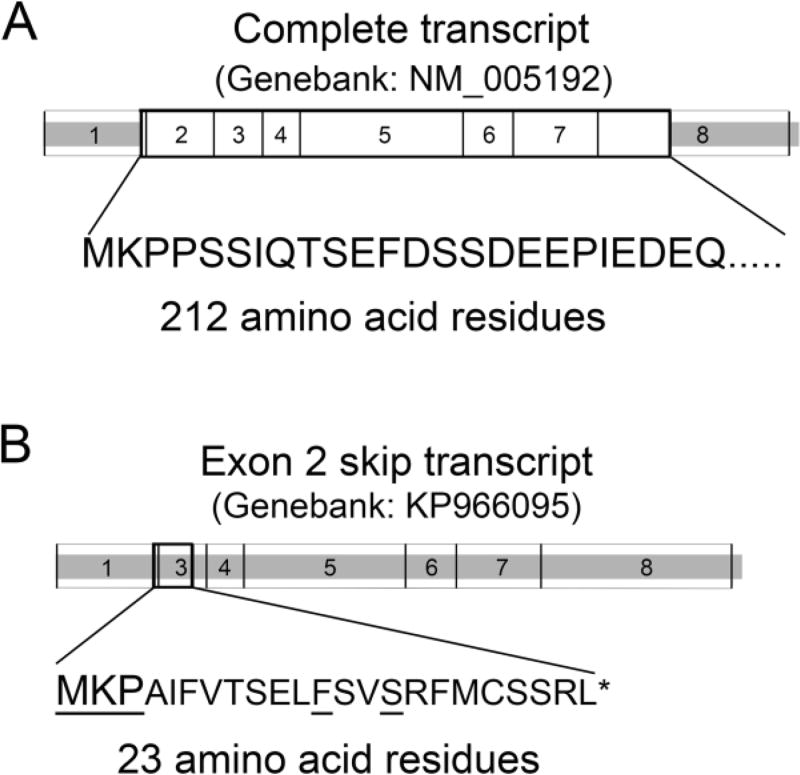
The human CDKN3 gene is located at chromosome 5q13.2 and has 8 exons. The CDKN3 coding mRNA (Genbank accession: NM_005192) has 906 nucleotide residues and encodes a protein of 212 amino acid residues (Fig. 1). In addition to this 212-amino acid residue encoding CDKN3 transcript, several splicing variants have been reported (Barron et al., 2015; Fan et al., 2015; Yeh et al., 2000; Yu et al., 2007). An exon 2-skipping alternative splicing transcript (Fig. 1) was consistently detected in both normal and tumor cells and tissues (Barron et al., 2015; Fan et al., 2015; Yu et al., 2007).
Fig. 1.
Full length and exon 2-skip CDKN3 transcripts. Full length CDKN3 mRNA (Genbank: NM-005192) (A) and exon 2-skip splicing variant (Genbank: KP966095) (B) are shown. Heavily outlined boxes indicate protein coding region. Thin bordered boxes indicate exons. Large letters indicate the N-terminal amino acid sequence of CDKN3. Small letters indicate the frame-shift amino acid sequence in the exon 2-skip transcript. Underlined residues are identical to CDKN3 amino acid residues.
2. CDKN3 overexpression is associated with poor survival in many types of human cancer
Using differential-display PCR, Stuart Aaronson’s laboratory was the first to observe overexpression of CDKN3 mRNA in various human cancer cell lines (Lee et al., 2000). Suppression of CDKN3 expression by antisense CDKN3 inhibited soft-agar colony growth of LNCaP cells and tumor xenograft growth of HeLa cells, suggesting that CDKN3 plays a tumorigenic role in these cells (Lee et al., 2000). Subsequently, increased CDKN3 expression has been observed in many types of cancer (Fan et al., 2015; Yang and Sun, 2015). Moreover, high CDKN3 mRNA levels have been associated with poor survival in glioma (Yu et al., 2007), cervical cancer (Barron et al., 2015; Espinosa et al., 2013), and lung adenocarcinoma (Fan et al., 2015; Zang et al., 2015). Examination of The Cancer Genome Atlas (TCGA) RNA-seq data (via cBioPortal) also indicates that high CDKN3 mRNA levels correlate with poor survival in several types of cancer, including renal clear cell carcinoma, renal papillary cell carcinoma, low grade glioma, and prostate adenocarcinoma (see below).
We found that CDKN3 expression was elevated in non-small cell lung cancer (NSCLC) (Fan et al., 2015). In three cohorts of lung adenocarcinoma, CDKN3 overexpression was prognostic for poor overall survival, while no significant differences in CDKN3 expression were found among different stages of lung adenocarcinoma. Although CDKN3 was overexpressed in lung squamous cell carcinoma, CDKN3 overexpression was not prognostic for patient survival. A likely explanation for the lack of difference between high and low CDKN3 expression to patient survival in lung squamous cell carcinoma is the overall high level of CDKN3 expression within this histological subtype of lung cancer (Fan et al., 2015).
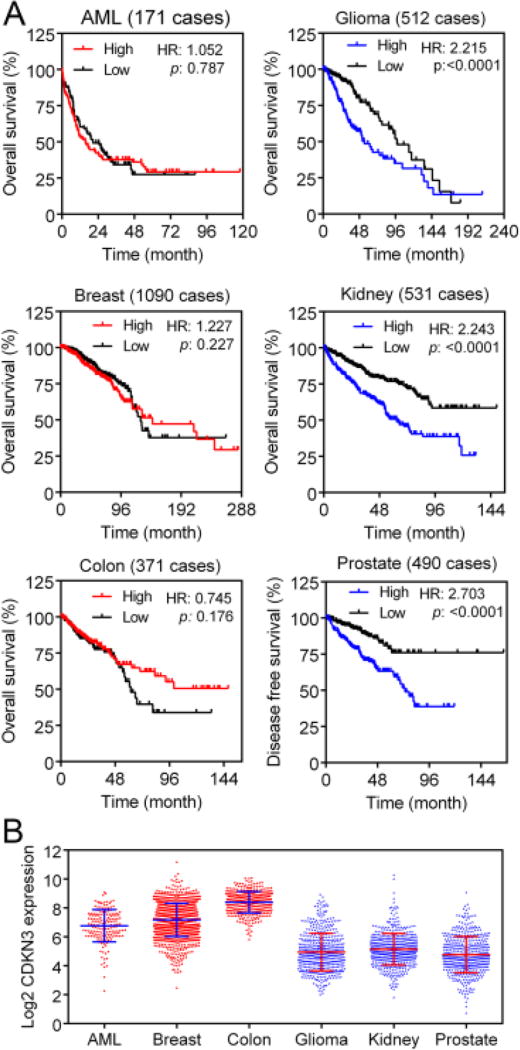
In the TCGA datasets, CDKN3 mRNA levels in acute myeloid leukemia, invasive breast carcinoma, and colorectal adenocarcinoma were not prognostic for overall survival, whereas high CDKN3 mRNA levels were associated with poor overall survival in low grade glioma and renal clear cell carcinoma (Fig. 2A). In prostate adenocarcinoma, while the few deceased cases (2%) limited assessment of a correlation between CDKN3 levels and the overall survival, high CDKN3 mRNA expression was associated with poor disease-free survival (Fig. 2A). Acute myeloid leukemia, invasive breast carcinoma, and colorectal adenocarcinoma had high overall CDKN3 mRNA levels, whereas low grade glioma, renal clear cell carcinoma, and prostate adenocarcinoma had lower overall CDKN3 mRNA levels (Fig. 2B). This difference is similar to what we observed previously between lung adenocarcinoma and lung squamous cell carcinoma.
Fig. 2.
Correlation between CDKN3 levels and cancer patient survival in TCGA datasets. (A) Correlation between CDKN3 mRNA levels and overall or disease-free survival in the indicated types of cancer. High/low CDKN3 mRNA levels were dichotomized on the mean expression value. (B) CDKN3 mRNA levels in six cohorts of cancer from TCGA. TCGA data were obtained via cbioportal.org (Cerami et al., 2012; Gao et al., 2013). AML, acute myeloid leukemia; breast, invasive breast carcinoma; colon, colorectal adenocarcinoma; glioma, low grade glioma; kidney, renal clear cell carcinoma; prostate, prostate adenocarcinoma.
3. Normal and aberrant CDKN3 transcripts
As mentioned above, the full-length 212-amino acid residue CDKN3 is encoded by 906-nucleotide mRNA derived from 8 exons. In addition, an exon 2-skip alternative splicing isoform is present in all human cell lines and tissues that have been analyzed (Barron et al., 2015; Fan et al., 2015; Yu et al., 2007). The exon 2-skip transcript results in a frameshift and encodes a short 23-amino acid residue peptide non-homologous to CDKN3. Since this 23-amino acid residue peptide lacks homology to CDKN3 (Fan et al., 2015), it is unlikely that it has any function related to CDKN3. Because these two transcripts are present in all cell lines and tissues that we have examined, we consider these two alternative splicing isoforms to be the normal human CDKN3 transcripts.
Yeh et al. (Yeh et al., 2000) were the first to report 15 aberrant transcripts in 27 hepatocellular carcinoma tissue samples and six aberrant transcripts in 12 noncancerous liver tissues. That is, ~50% of cancerous and noncancerous liver tissues in this study had abnormal CDKN3 transcripts (although these alterations were not validated in the corresponding genomic DNA). These aberrant CDKN3 transcripts include nonsense mutations and deletions that resulted in truncated CDKN3 and CDKN3 with missense or internal deletion mutations. Potentially, some of these aberrant transcripts may result in dominant-negative CDKN3 mutants that could interfere with normal CDKN3 function. We found no CDKN3 mutations among 30 cases of hepatocellular carcinoma from Total Cancer Care™ (TCC) data (Fan et al., 2015). Among 442 cases of hepatocellular carcinoma in TCGA and 231 cases of hepatocellular carcinoma reported by Ahn et al. (Ahn et al., 2014), there were two cases with missense mutations (I72T, E17K) (cbioportal.org). Therefore, the high incidence of CDKN3 missense and nonsense mutations reported by Yeh et al. (Yeh et al., 2000) appeared inconsistent with genomic data in hepatocellular carcinoma from other laboratories, and some of the deletion mutations were probably generated post-transcriptionally via alternative splicing. Subsequently, Yu et al. (Yu et al., 2007) reported two aberrant splicing variants of CDKN3 “b” and “d” involving exons 2 and 3, respectively, in glioblastoma. The b variant resulted in an 8-amino acid peptide. The d variant was derived from using an alternative splice site in exon 2, resulting in a 179-amino acid protein skipping part of exon 2 and the entire exon 3. This d variant is the same as the BX-09 transcript previously detected by Yeh et al. (Yeh et al., 2000) in hepatocellular carcinoma and it could interfere with CDKN3 activity, since it lacks a functional catalytic domain, but may compete for CDK binding (Yu et al., 2007).
Barron et al. (Barron et al., 2015) reported five minor aberrant transcripts in cervical cancer and control tissues. These transcripts were generated from alternative splicing lacking internal exons or distal exons, but retaining exons 4, 6, 7, and/or 8 that encode CDK2 interacting regions of CDKN3 (Song et al., 2001). Thus, proteins encoded by these aberrant transcripts are predicted to be able to interfere with the wildtype CDKN3 function. These transcripts were detected in normal and cervical cancer samples with similar frequencies (Barron et al., 2015). However, the qPCR measurement of CDKN3 levels in control and tumor samples mainly represented the wildtype CDKN3 transcript (Barron et al., 2015). In all cases, the CDKN3 transcript encoding the wildtype CDKN3 is the predominant transcript.
We examined CDKN3 transcripts in six non-small cell lung cancer tissues, four non-cancerous cell culture or cell lines, and 14 cancer cell lines that include lung, breast, cervical, colorectal, liver, pancreatic, and prostate cancer and osteosarcoma (Fan et al., 2015). Although we used RT-PCR primer pairs that were designed to detect potential alternative splicing in every exon of CDKN3, no aberrant alternative splicing or mutation was found in these 24 samples.
The reasons that others detected minor aberrant CDKN3 transcripts in their samples and that we did not remain to be determined. One possibility is the difference in tissue types and samples. Another possibility is that others used more sensitive methods. For instance, Barron et al. (Barron et al., 2015) used nested primers to re-amplify RT-PCR products. Yeh et al. (Yeh et al., 2000) cloned PCR products and sequenced cloned cDNA. We used the same primers to amplify the PT-PCR products and sequenced these PCR products directly. Thus, we did not exclude the possibility that minor aberrant transcripts might exist in our samples, but focused on major transcripts.
4. CDKN3 expression is elevated in the mitotic phase and controls cell division
While CDKN3 is often overexpressed in human cancer, CDKN3 gene amplification occurs infrequently. For instance, among the 516 cases of TCGA lung adenocarcinoma for which copy number alteration data were available, 10 cases (2%) had CDKN3 gene amplification. Thus, the frequent overexpression of CDKN3 in human cancer is unlikely to be caused by gene amplification.
In a panel of synchronized cell lines, we found that the CDKN3 protein level fluctuated during the cell cycle (Fan et al., 2015). CDKN3 protein was expressed at low levels in G0/1 and S phase and was increased in M phase in parallel with phosphorylation of histone H3 Ser-10, which is a marker of M phase. RT-qPCR analysis indicated that M phase cells consistently had high levels of CDKN3 mRNA and G0/1-arrested cells had low levels of CDKN3 mRNA (Fan et al., 2015). In some cell lines (HCC827, H2228, H1299), high levels of CDKN3 mRNA were also detected in S phase although we did not observe a corresponding increase at the protein level (Fan et al., 2015). These data suggest that the frequent CDKN3 overexpression in human cancer is primarily due to the increased number of mitotic cells.
In a spindle assembly checkpoint (SAC) assay, in which SAC failure produced a multinucleated phenotype when HeLa cells were treated with taxol, CDKN3 siRNAs yielded the stronger SAC failure phenotype among 801 siRNAs targeting human phosphatases (Nalepa et al., 2013). Further analysis indicated that the centrosome-located CDK1 was phosphorylated at Thr-161 during mitotic entry. CDK1 Thr-161 was dephosphorylated in anaphase until completion of mitosis, and CDKN3 was responsible for dephosphorylation of CDK1 Thr-161 in late mitosis (Nalepa et al., 2013). Mitotic exit is mediated by inactivation of CDK1/cyclin B. Inhibition of residual CDK1 activity by dephosphorylating CDK1 Thr-161 may be a mechanism to ensure normal mitotic exit (Nalepa et al., 2013).
During S phase, CDK2/cyclin A phosphorylates and activates monopolar spindle 1 (MPS1), which is a dual-specificity kinase that promotes centrosome duplication. Elevated MPS1 results in overduplication of centrosomes that causes abnormal spindle assembly and interferes with cell division. It was found that CDK2/cyclin A phosphorylated MPS1 at Thr-468 and possibly at Thr453, which promotes its centrosomal stability, whereas CDKN3 dephosphorylated MPS1 to induce proteasome-mediated degradation of MPS1 at centrosomes (Srinivas et al., 2015). Recall that CDK2 Thr-160 phosphorylation state does not affect cyclin A binding (Brown et al., 1999), and that only after cyclin A is degraded post S phase, can CDK2 Thr-160 be dephosphorylated by CDKN3 (Poon and Hunter, 1995). Thus, it has been postulated that, after cyclin A degradation, CDKN3 dephosphorylates CDK2 and MPS1 to prevent centrosome overduplication (Srinivas et al., 2015). Taken together, these results indicate that CDKN3 has positive roles in regulating cell division by controlling mitotic exit and centrosome duplication.
5. Conclusion and perspective
CDKN3 was perceived as a tumor suppressor gene analogous to CDKN2A/CDKN2B. Counter- intuitively, CDKN3 is often overexpressed in human cancer, whereas CDKN2A/CDKN2B are often deleted in certain types of human cancer, such as non-small cell lung cancer. Early studies attributed CDKN3 overexpression to dominant-negative protein, caused by either mutations or aberrant splicing transcripts. However, recent cancer genomic data indicate that CDKN3 mutations are rare and likely non-disruptive. Aberrant transcripts, when detected, were always present as minor transcripts co-existing with the more abundant full-length normal CDKN3 transcript. Truncated proteins are often rapidly degraded in the cells (Lykke-Andersen and Jensen, 2015). Whether any of the reported aberrant transcripts of CDKN3 yield a detectable endogenous protein remains to be seen.
While CDKN3 is often overexpressed in human cancer, few cases have CDKN3 gene amplification. Gene amplification is predicted to result in constitutively elevated expression. Thus, the fact that the CDKN3 gene is rarely amplified in human cancer suggests that a constitutive increase in CDKN3 level does not give cancer cells an advantage. We have found that CDKN3 transcript and protein levels fluctuate during the cell cycle and peak at mitosis. Since rapidly growing tumors have more mitotic cells, the high level of CDKN3 in mitotic phase provides the best plausible explanation for the frequent CDKN3 overexpression in human cancer. While inactivating CDK1 to ensure mitotic exit may explain the need of a high level CDKN3 in the mitotic phase, the mechanisms of dynamic control of CDKN3 mRNA and protein expression during cell cycle remain to be investigated.
Acknowledgments
This work was supported by NIH grant R01CA178456. We thank our colleagues who have contributed to CDKN3 research. We have used publicly available data from TCGA Research Network via cBioPortal (www.cbioportal.org).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM, Sung CO, Baek D, Haq F, Ansari AA, Lee SY, Chun SM, Choi S, Choi HJ, Kim J, Kim S, Hwang S, Lee YJ, Lee JE, Jung WR, Jang HY, Yang E, Sung WK, Lee NP, Mao M, Lee C, Zucman-Rossi J, Yu E, Lee HC, Kong G. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60(6):1972–1982. doi: 10.1002/hep.27198. [DOI] [PubMed] [Google Scholar]
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117(6):699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Barron EV, Roman-Bassaure E, Sanchez-Sandoval AL, Espinosa AM, Guardado-Estrada M, Medina I, Juarez E, Alfaro A, Bermudez M, Zamora R, Garcia-Ruiz C, Gomora JC, Kofman S, Perez-Armendariz EM, Berumen J. CDKN3 mRNA as a Biomarker for Survival and Therapeutic Target in Cervical Cancer. PloS one. 2015;10(9):e0137397. doi: 10.1371/journal.pone.0137397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Lawrie AM, Morris MC, Tunnah P, Divita G, Johnson LN, Endicott JA. Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity. The Journal of biological chemistry. 1999;274(13):8746–8756. doi: 10.1074/jbc.274.13.8746. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa AM, Alfaro A, Roman-Basaure E, Guardado-Estrada M, Palma I, Serralde C, Medina I, Juarez E, Bermudez M, Marquez E, Borges-Ibanez M, Munoz-Cortez S, Alcantara-Vazquez A, Alonso P, Curiel-Valdez J, Kofman S, Villegas N, Berumen J. Mitosis is a source of potential markers for screening and survival and therapeutic targets in cervical cancer. PloS one. 2013;8(2):e55975. doi: 10.1371/journal.pone.0055975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Chen L, Huang Q, Shen T, Welsh EA, Teer JK, Cai J, Cress WD, Wu J. Overexpression of major CDKN3 transcripts is associated with poor survival in lung adenocarcinoma. British journal of cancer. 2015;113(12):1735–1743. doi: 10.1038/bjc.2015.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11(11):3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Casso D, Beach D. KAP: a dual specificity phosphatase that interacts with cyclin-dependent kinases. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(5):1731–1735. doi: 10.1073/pnas.91.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Reimer CL, Fang L, Iruela-Arispe ML, Aaronson SA. Overexpression of kinase-associated phosphatase (KAP) in breast and prostate cancer and inhibition of the transformed phenotype by antisense KAP expression. Molecular and cellular biology. 2000;20(5):1723–1732. doi: 10.1128/mcb.20.5.1723-1732.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nature reviews. Molecular cell biology. 2015;16(11):665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- Nalepa G, Barnholtz-Sloan J, Enzor R, Dey D, He Y, Gehlhausen JR, Lehmann AS, Park SJ, Yang Y, Yang X, Chen S, Guan X, Chen Y, Renbarger J, Yang FC, Parada LF, Clapp W. The tumor suppressor CDKN3 controls mitosis. The Journal of cell biology. 2013;201(7):997–1012. doi: 10.1083/jcb.201205125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon RY, Hunter T. Dephosphorylation of Cdk2 Thr160 by the cyclin-dependent kinaseinteracting phosphatase KAP in the absence of cyclin. Science. 1995;270(5233):90–93. doi: 10.1126/science.270.5233.90. [DOI] [PubMed] [Google Scholar]
- Song H, Hanlon N, Brown NR, Noble ME, Johnson LN, Barford D. Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2. Molecular cell. 2001;7(3):615–626. doi: 10.1016/s1097-2765(01)00208-8. [DOI] [PubMed] [Google Scholar]
- Srinivas V, Kitagawa M, Wong J, Liao PJ, Lee SH. The Tumor Suppressor Cdkn3 Is Required for Maintaining the Proper Number of Centrosomes by Regulating the Centrosomal Stability of Mps1. Cell Rep. 2015;13(8):1569–1577. doi: 10.1016/j.celrep.2015.10.039. [DOI] [PubMed] [Google Scholar]
- Yang C, Sun JJ. Mechanistic studies of cyclin-dependent kinase inhibitor 3 (CDKN3) in colorectal cancer. Asian Pac J Cancer Prev. 2015;16(3):965–970. doi: 10.7314/apjcp.2015.16.3.965. [DOI] [PubMed] [Google Scholar]
- Yeh CT, Lu SC, Chen TC, Peng CY, Liaw YF. Aberrant transcripts of the cyclindependent kinase-associated protein phosphatase in hepatocellular carcinoma. Cancer research. 2000;60(17):4697–4700. [PubMed] [Google Scholar]
- Yu Y, Jiang X, Schoch BS, Carroll RS, Black PM, Johnson MD. Aberrant splicing of cyclin-dependent kinase-associated protein phosphatase KAP increases proliferation and migration in glioblastoma. Cancer research. 2007;67(1):130–138. doi: 10.1158/0008-5472.CAN-06-2478. [DOI] [PubMed] [Google Scholar]
- Zang X, Chen M, Zhou Y, Xiao G, Xie Y, Wang X. Identifying CDKN3 Gene Expression as a Prognostic Biomarker in Lung Adenocarcinoma via Meta-analysis. Cancer informatics. 2015;14(Suppl 2):183–191. doi: 10.4137/CIN.S17287. [DOI] [PMC free article] [PubMed] [Google Scholar]