Abstract
The ITS (nuclear ribosomal internal transcribed spacer) RefSeq database at the National Center for Biotechnology Information (NCBI) is dedicated to the clear association between name, specimen and sequence data. This database is focused on sequences obtained from type material stored in public collections. While the initial ITS sequence curation effort together with numerous fungal taxonomy experts attempted to cover as many orders as possible, we extended our latest focus to the family and genus ranks. We focused on Trichoderma for several reasons, mainly because the asexual and sexual synonyms were well documented, and a list of proposed names and type material were recently proposed and published. In this case study the recent taxonomic information was applied to do a complete taxonomic audit for the genus Trichoderma in the NCBI Taxonomy database. A name status report is available here: https://www.ncbi.nlm.nih.gov/Taxonomy/TaxIdentifier/tax_identifier.cgi. As a result, the ITS RefSeq Targeted Loci database at NCBI has been augmented with more sequences from type and verified material from Trichoderma species. Additionally, to aid in the cross referencing of data from single loci and genomes we have collected a list of quality records of the RPB2 gene obtained from type material in GenBank that could help validate future submissions. During the process of curation misidentified genomes were discovered, and sequence records from type material were found hidden under previous classifications. Source metadata curation, although more cumbersome, proved to be useful as confirmation of the type material designation.
Database URL: http://www.ncbi.nlm.nih.gov/bioproject/PRJNA177353
Introduction
NCBI resources
Accurate fungal identification is often a challenge to non-experts. Morphological diagnostic characters can be scarce or invariable among genetically distinct lineages, resulting in a high abundance of cryptic species. The convergent evolutionary processes shaping characters like spore morphology and features of the reproductive structures are common in all Fungi but especially pronounced in the phylum Ascomycota. This often results in species complexes of cryptic species where DNA sequence comparisons are an essential distinguishing component. Over the last few decades the availability of associated sequences have increased and greatly facilitated comparative analyses. However, a sizable percentage of archived sequences remain incorrectly assigned or simply poorly identified (1). The additional need to assign taxonomic names to environmentally sampled sequences has made reference sequences especially valuable (2). This has resulted in the release of a number of specialized curated datasets. Examples include FUSARIUM-ID and Fusarium MLST, (3) addressing Fusarium taxonomy, the ISHAM ITS database of medical fungi (4) or TrichoBLAST (5) focused on Trichoderma reference sequences. Other databases such as UNITE (6) and the RDP Classifier (7) have a much broader focus on references covering all Fungi.
The National Center for Biotechnology Information (NCBI) maintains a number of interconnected databases, including GenBank, which functions as an archive of submissions to the International Nucleotide Sequence Database Collaboration (INSDC). A separate set of curated reference records (RefSeq) contains selected sequences copied from the public archives. RefSeq data can range from single-sequence records to complete genomes (8). The RefSeq targeted loci projects (https://www.ncbi.nlm.nih.gov/bioproject/224725) are focused on separate marker datasets for bacterial and fungal ribosomal RNA loci. This includes a dataset from the fungal nuclear ribosomal internal transcribed spacer (ITS) region. This marker set was selected after this region was formally proposed as the universal DNA barcode marker for Fungi (9). Although DNA barcode initiatives advocated standards for new sequence deposition (10), a large set of legacy submissions in the INSDC needs to be re-examined for use as references. These include many sequences obtained from type material, which were imprecisely annotated (11). Although qualifiers for biorepository details such as ‘specimen_voucher’ and ‘culture_collection’ have been available and promoted by GenBank since 1998, they remain inconsistently applied by submitters (12). More recently, NCBI databases began to explicitly annotate and track type material information (13).
At NCBI, an effort to re-annotate a focused, but phylogenetically diverse, set of ITS sequence accessions from type and reference material was recently completed. This was done in collaboration with numerous fungal taxonomic experts as well as curators at other widely used databases such as Index Fungorum, MycoBank and UNITE (14). This ITS dataset is housed under the RefSeq Targeted Loci ITS BioProject: http://www.ncbi.nlm.nih.gov/bioproject/PRJNA177353. While the initial RefSeq ITS project attempted to incorporate as many fungal orders as possible, we extended our latest focus to the family and genus ranks. This effort was prompted, in part, by changes in how fungal species are named (15). These nomenclatural changes increased the importance of reliable sequence data in dealing with the numerous names that require reassessment on the genus level (15).
Changes to fungal taxonomy and classification
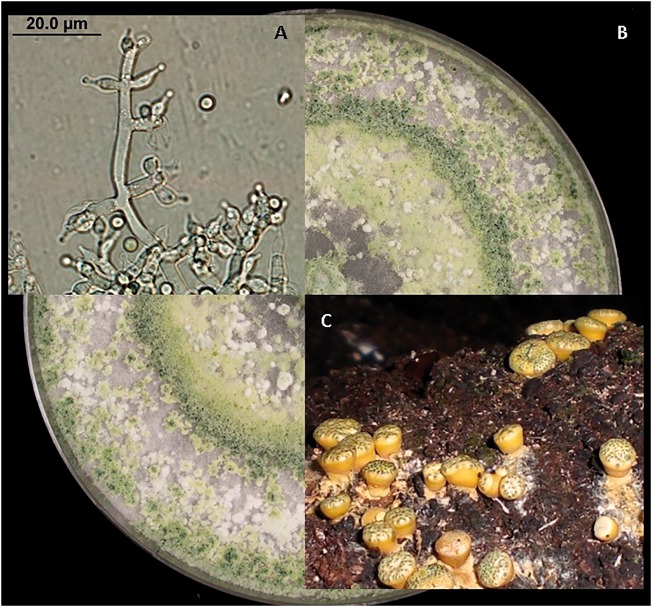
The prevalence of DNA sequence comparisons prompted the abolition of a nearly 150-year-old practice to use different names for some sexual and asexual morphological forms in Fungi. The latest version of the International Code of Nomenclature for algae, fungi and plants (ICN) was ratified in Melbourne 2011 and effectively ended this dual naming system (16). This means that often long standing names have to be merged so that related sexual and asexual morphs can be placed in a single genus. Although the phylogenetic boundaries for asexual and sexual morph genera do not always correlate perfectly in other groups, species in the genera Trichoderma and Hypocrea represent a well resolved clade. All species share a single common ancestor and the synonymy of asexual and sexual morph names are well documented. A choice was made to place all remaining Hypocrea species names, including those with and without recorded Trichoderma asexual morphs in Trichoderma (17–19). Figure 1 depicts morphology observed in the Trichoderma/Hypocrea clade.
Figure 1.
Morphology of specimens in the Trichoderma/Hypocrea clade: (A) asexual structures (conidiophore and conidia) of Trichoderma harzianum (FJ967806), (B) growth in culture of a specimen in the Trichoderma harzianum complex and (C) sexual reproduction structures of Hypocrea species.
Taxonomy of Trichoderma
The genus Trichoderma has broadly sampled sequence data tied to generally accessible cultures. It contains a number of species with remarkable ecological adaptability that are also important in industrial and biological control applications as well as opportunistic pathogens of humans (20). It has also garnered enough research interest (>15 000 references in Web of Science) to already have rudimentary comparative genomic data available (21).
We extensively relied on a number of recent papers to annotate type information and select representative INSDC sequence accessions to update the NCBI Taxonomy database. A recent publication listed >200 Trichoderma species recognized based on a molecular phylogenetic approach (22). Subsequently, a list of accepted species names as well as their associated type material have been proposed by Trichoderma taxonomists (19). We also utilized another study (23) that used DNA sequences from three protein coding genes (translation elongation factor 1 alpha, TEF1; RNA polymerase II second largest subunit, RPB2; and ATP citrate lyase, ACL1) and focused on a deep sampling of Trichoderma in southern Europe and Macaronesia. A third reference study (24) focused on samples from the tropics with deep sampling of the ‘Trichoderma harzianum’ complex. Another recent survey and discussion of Trichoderma species in New Zealand (25) provided additional data for comparison, although this was published after the main parts of our project was completed.
A database with tools to aid Trichoderma identification has been a long standing resource provided by the International Sub-commission on Trichoderma and Hypocrea Taxonomy (ISTH; http://www.isth.info/) (5, 26). This identification system uses the following markers: ITS, partial protein coding gene sequences from TEF1 and RPB2. It currently contains only 87 genetically characterized species and has not been updated recently. The project presented us with a chance to extend the ISTH resource by a complete nomenclatural audit of the Trichoderma/Hypocrea associated names in NCBI Taxonomy within a collaborative framework of database curators and taxonomists. The intention is to append and verify information on type material and sequence reliability. Additionally, to aid in the cross referencing of data from single loci and genomes we collected a list of quality records from RPB2 obtained from type material in GenBank that can act as a potential secondary barcode marker. We hope that this improved dataset can be used in addition to external tools to aid accurate fungal identification.
All the ITS marker records produced as part of this curation effort is available at the Bioproject (177353) for ITS RefSeq: http://www.ncbi.nlm.nih.gov/bioproject/PRJNA177353. The intention is to provide these as a reliable set of sequence markers that could benefit other efforts to track and verify Trichoderma species using DNA sequence data. We will address some of the implications in this paper.
Materials and methods
Processing and updating ITS records for RefSeq
Records in GenBank have been evaluated and those records that passed various quality control requirements (14) were propagated to the RefSeq database and given a RefSeq accession number. This accession is easily recognized by the inclusion of the underscore in the identifier. Since the 2014 publication a number of amendments to the workflow have been made. When the RefSeq ITS project was first reported (14), previous versions of the ITSx script were used. The script and its constituent HMM libraries have since been updated. Version: 1.0.11 was used to reassess all current RefSeq ITS sequences. Additionally all Refseq ITS records were rechecked for chimeras using UCHIME with UNITE’s latest library (version 7 of the date 01.01.2016) and also using our own RefSeq library: ((‘2012/10/01’[Publication Date]: ‘14 July 2016’[Publication Date])) AND 177353 [BioProject]). The length of potential ITS records were re-evaluated using new parameters where the ITS1 and ITS2 lengths were evaluated separately. Any sequences which met the following parameters: <0.75× or >2× of reference Order ITS1/2 min or max length were investigated. Those records with no order name in their lineage and with ITS1 or ITS2 lengths shorter than 100 bases or longer than a 1000 bases were investigated. If either the 18S and/or 28S region was longer than 150 bases it was trimmed to 100 bases, respectively.
The identity of ITS sequences were again evaluated as before (14) with a few adjustments in some alignment length and sequence identity cutoffs. Possible misidentifications or mislabeling of accessions were investigated for the following: >99.99% identical over >90% of the ITS region to type sequences of a different Taxonomy Database identifier (TaxID) or <99.5% identity to type sequence of the same TaxID and <97% identity to RefS/RepS from UNITE of the same TaxID. Potential additions to the RefSeq ITS database were also aligned to existing RefSeq records to ensure that near identical (>99.5% to RefSeq ITS) ITS records added were closely related to taxa based on identity of additional genes and not based solely on ITS sequence identity.
Periodically, the complete ITS RefSeq database was submitted to an all-vs-all BLAST search. BLAST alignments (longest hit >260 bases) of ITS sequences belonging to different genus/family/class were evaluated. This practice identified outliers which needed taxonomic and classification edits in the Taxonomy database and taxa that needed further phylogenetic study. Continuous taxonomic updates in the Taxonomy database resulted in several name changes and updates in their classification. Depending on the nature of the changes, records were verified for redundancy (synonymies) and the associated records were either suppressed because of type material becoming disassociated with the currently accepted name (by representing heterotypic synonyms) or the definition lines were updated. Sequence updates in the INSDC submitted records from which ITS RefSeq records originated were also monitored for sequence updates. Such updates were re-evaluated resulting in suppressed or updated RefSeq records. Since RefSeq records represent a copy of the original submitted sequence record, RefSeq curators can update, edit or suppress them as necessary. Third-party feedback is welcome via this page: https://www.ncbi.nlm.nih.gov/projects/RefSeq/update.cgi as mentioned in a previous paper (14). For the Trichoderma RefSeq ITS records source metadata information was collected from publications which are tabulated in Supplementary Material (Supplementary Table S1).
Identification of DNA-directed RNA polymerase II core subunit RPB2 records
To find protein records of the second largest subunit of polymerase II (RPB2) this Entrez query was used in the Protein database: RNA_pol_B_RPB2 + AND + Trichoderma + [orgn]. RNA_pol_B_RPB2 is the region name of the conserved domain CDD:305168 found in RPB2 protein sequences. Note, this search will also pull protein records of the second largest subunit of polymerase I (referred to in Saccharomyces cerevisiae S288C as DNA-directed RNA polymerase I core subunit RPA135) and III (referred to in S. cerevisiae S288C as DNA-directed RNA polymerase III core subunit RET1) but are filtered out in the next step.
A BLASTP search with the protein sequence of RPB2 (NP_014794.3) from the reference strain S. cerevisiae S288C as query against the protein sequences obtained in first search was performed and only those hits that met the following criteria were retained:
Protein sequence length 200 and longer;
blastp alignment start between positions 390 and 590 and end between positions 640 and 840 of the Sce RPB2 protein sequence;
more than 40% sequence identity.
As a result all accessions contained protein sequence that overlapped by at least 100 amino acids. To find the associated nucleotide accessions, the following ESearch utility was executed in a loop: epost -db protein -format acc -id $i | elink -target nuccore | efetch -format docsum | xtract -pattern DocumentSummary -element Caption TaxID Organism ≫ RPB2nuc.txt
Additional RPB2 nucleotide records that did not contain annotation but used in (23) were included and all records were restricted to sequences from type or synonym type material in the final list in Supplementary Material (Supplementary Table S2).
Results and discussion
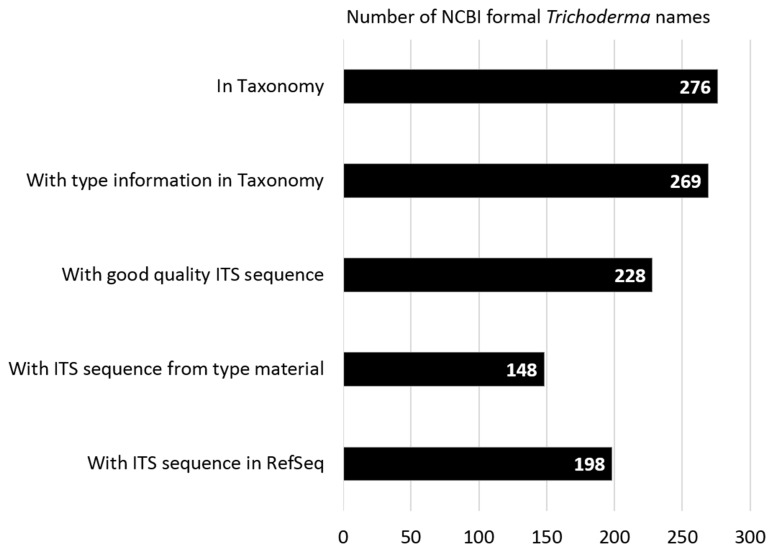
At the time of writing (March 2017) there were 276 validly published Trichoderma names (Trichoderma [orgn] AND specified [prop]) in the NCBI Taxonomy database of which 269 names were annotated with type information (Trichoderma [orgn] AND specified [prop] AND has type material [prop]) (Figure 2). The term ‘specified’ is used in the Taxonomy database to indicate names that exist as binomials and has a status under the ICN. In contrast the term ‘unspecified’ (‘unspecified’[prop]) refer to ‘informal names’ and exist in various formats but are not binomials and either reflect incomplete taxonomic knowledge or other environmental labels. The type information for a name included type material (e.g. format: type material: BPI 8543653) or type material from a taxonomic or heterotypic synonym (e.g. format: type material: BPI 843645 [[Hypocrea catoptron]]). Of the 276 names, 198 names were associated with acceptable quality ITS sequences, the identity verified, records curated and placed in the RefSeq ITS database (177353[BioProject] AND Trichoderma [orgn]). The Trichoderma records in the RefSeq ITS consisted of 136 from type material (177353[BioProject] AND Trichoderma [orgn] AND sequence from type [filter]), 37 from synonym type material (177353[BioProject] AND Trichoderma [orgn] AND sequence from synonym type [filter]) and 26 from material verified by expert Trichoderma taxonomists (177353[BioProject] AND Trichoderma [orgn] NOT sequence from synonym type [filter] NOT sequence from type [filter]).
Figure 2.
Bar graph showing the number of formal Trichoderma names associated with different attributes in databases at NCBI.
Trichoderma records in the ITS RefSeq database
Sequence quality
Most (96%) of the Trichoderma RefSeq ITS sequences contained no ambiguous characters and the small number (9) that did, had a maximum of 2 ambiguous characters. No chimeras were found compared against the UNITE library or the RefSeq library. Using ITSx annotation predictions (27), the complete ITS1 intergenic spacer ranged from 180 to 264 bases, the 5.8S region 158–159 bases and the complete ITS2 intergenic spacer 163 to 199 bases (Figure 3). Generally, ITS1 was longer than the ITS2, with more length and sequence variation as has been reported previously for other members of the Ascomycota (28). Currently, only 10 records have an ITS1 shorter than the minimum (180) as calculated from complete regions and only 5 records have an ITS2 region shorter than the minimum (163) as calculated from complete regions (Figure 3). Most of the Trichoderma sequences (163) had an identical 5.8S sequence, the same as found in NR_134342 (T. ovalisporum CBS 113299 from TYPE material, GenBank version: AY380897) (29). In the rest of the sequences, variations in the 5.8S region included mostly pyrimidine transition type differences (in 25 sequences) when compared with the most commonly seen 5.8S sequence.
Figure 3.
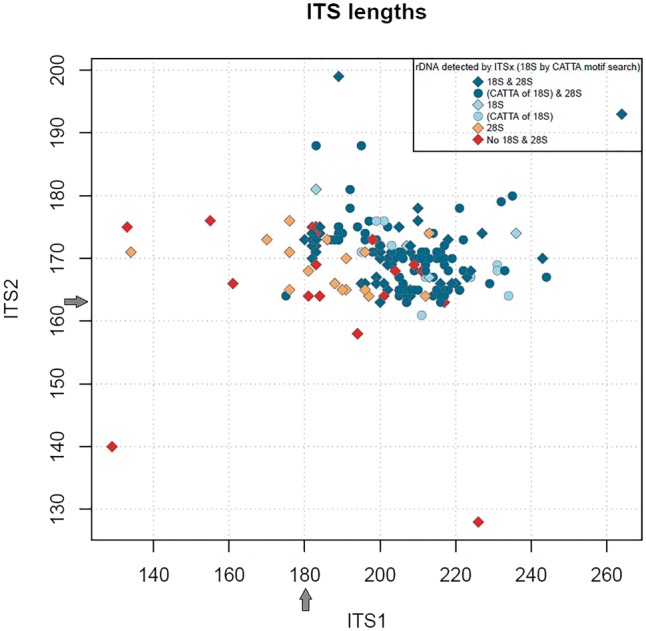
Graphical display of ITS1 length compared with ITS2 length from Trichoderma ITS RefSeq records. Grey arrows indicate the minimum lengths of ITS1 and ITS2 observed using ITSx annotation.
Sequence identity
In general, when the type material identifier appears in association with the current correct name in a record then the sequence–name association in GenBank has the potential to be more accurate than other sequence records, although, errors are always a concern. Since GenBank is an archival database it is always possible to see type material identifiers in records that are not in association with the currently accepted taxonomic name (Supplemental Table S3). These can be found by searching with the type identifier and genus name but excluding the currently accepted binomial name in the Entrez Nucleotide database (e.g.: G.J.S. 01-265 [Strain] NOT Trichoderma albofulvum[Organism] AND Trichoderma [orgn]). The ‘sequence from type [filter]’ query in the Entrez Nucleotide database will only list sequence records that have the latest taxonomic classification clearly annotated. This need for validation and corrections by the Trichoderma community of public data has already been pointed out (22). One approach is to diligently cite the original work which produced the original data and therefore incentivizing the production of good quality taxonomic data (30). In this case study, we have endeavored to apply this suggestion and cited and listed publications used to verify sequence and source metadata in Supplementary Material (Supplementary Table S1).
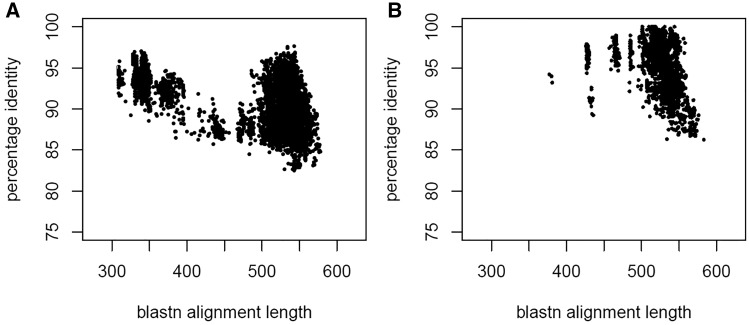
The presence of multiple sequence records obtained from the same biological material by independent research groups indicated data reliability. In the absence of this, a species phylogenetic analysis in concert with consultations from taxonomic experts helped resolve discrepancies. Clustering the ITS region at 100% identity generated a few clusters that contains two or more identical ITS sequences, but from different type source material (Table 1). For example, the ITS region of the following sequence records, NR_138444 [Trichoderma trixiae CBS 134702, GenBank version: DQ677647 (31)], NR_138447 [Trichoderma virilente DAOM 234234, GenBank version: EU280119 (31)] and NR_138429 [Trichoderma viridescens CBS 433.34, GenBank version: AF456922 (32)] are identical and these taxa are known to be part of a species complex. The ITS region, however, is not variable enough to discern these closely related lineages that can be resolved by other more variable markers (31). A graphical summary of RefSeq ITS sequence BLASTN search results (% identity and alignment length) between (Figure 4A) and within clades (Figure 4B), as clades defined by (23), shows different profiles. Between clades, the percentage identity of the ITS region were below 98%, whereas it is clear that within clades, the ITS region was identical or very close to being identical for some taxa, for example, members of the harzianum species complex in the RefSeq ITS database were 99–100% identical.
Table 1.
Two or more Trichoderma RefSeq ITS records which had identical ITS1_5.8S_ITS2 sequences
| NR_138453(T. parareesei), NR_120297(T. reesei) |
| NR_134394(T. deliquescens), NR_134351(T. melanomagnum) |
| NR_137298(T. atrobrunneum), NR_137304(T. afroharzianum) – members of the harzianum species complex |
| NR_138434(T. brevicompactum), NR_138448(T. turrialbense),NR_134373(T. protrudens) |
| NR_137305(T. rifaii), NR_137301(T. afarasin)—members of the harzianum species complex |
| NR_131317(T. gamsii), NR_138446(T. neokoningii) |
| NR_144868(T. lentiforme), NR_137297(T. simmonsii)—members of the harzianum species complex |
| NR_138439(T. viridarium), NR_134367(T. paraviridescens) |
| NR_134371(T. hamatum), NR_138449(T. evansii) |
| NR_137308(T. texanum), NR_134363(T. martiale) |
| NR_138451(T. hispanicum), NR_138452(T. samuelsii), NR_138456(T. koningii), NR_144870(T. vinosum)—members of the koningii species complex |
| NR_137302(T. caribbaeum), NR_144874(T. taiwanense) |
| NR_131281(T. koningiopsis), NR_134342(T. ovalisporum), NR_137303 (T. dorotheae) |
| NR_138444(T. trixiae), NR_138447(T. virilente), NR_138429(T. viridescens) |
Figure 4.
A graphical summary of RefSeq ITS sequence BLASTn search results (% identity and alignment length) between (Figure 4A) and within clades (Figure 4B), where clades were defined by Jacklitsch and Voglmayr (23).
The ITS RefSeq sequences were compared with the clusters used by the UNITE database. UNITE is an ITS sequence database geared towards sequence-based identification and provides a number of reference and annotation tools (33). It is used for fungal classification by multiple widely used ecological applications, such as the QIIME pipeline (34). After quality control, UNITE divides the public ITS sequences into clusters based on different similarity thresholds (97–100%), each with a reference or representative sequence (either annotated by taxonomic experts or assigned by an algorithm), which represents a ‘species hypothesis’ (SH) (6). Depending on the resolving power of ITS sequences and an up-to-date taxonomy, one reference sequence can represent one or many differently described taxa. UNITE SH clusters were found for 189 of the Trichoderma RefSeq records and the labeled SH name matched the NCBI RefSeq organism name 78 times. Records that were not found in this release were either too new or formed a single sequence cluster. This represented 90 UNITE clusters (release public_20_11_2016), with 20 clusters containing more than one sequence (and species) from the RefSeq database (Table 2, Supplementary Table S1). Other discrepancies were because of name variations in the UNITE database. For example, the cluster SH181342.07FU, labeled Trichoderma viride contained the most (39) Trichoderma RefSeq sequences from type material (Table 2). Table 3 shows the % identity of the ITS region from T. viride [CBS 119325 from TYPE material, RefSeq NR_138441, GenBank version DQ323428 (32)] compared with other sequences from Trichoderma type material in the same cluster (SH181342.07FU). Table 3 also serves as a clear example of how the RefSeq ITS database differ from the UNITE ITS database. It underlines how several ITS records from type material from different taxa can be found in one UNITE SH cluster and why the labeled SH name differs from the RefSeq ITS organism name.
Table 2.
UNITE species hypothesis clusters with more than one RefSeq ITS record.
| UNITE_public_20_11_2016a | NCBI RefSeq accession count |
|---|---|
| SH181342.07FU | 39 |
| SH190868.07FU | 20 |
| SH207825.07FU | 10 |
| SH206530.07FU | 6 |
| SH177682.07FU | 6 |
| SH177683.07FU | 6 |
| SH177684.07FU | 6 |
| SH187755.07FU | 5 |
| SH177687.07FU | 4 |
| SH187759.07FU | 3 |
| SH177689.07FU | 3 |
| SH008832.07FU | 2 |
| SH177685.07FU | 2 |
| SH177686.07FU | 2 |
| SH002402.07FU | 2 |
| SH177694.07FU | 2 |
| SH217782.07FU | 2 |
| SH181351.07FU | 2 |
| SH187756.07FU | 2 |
| SH187757.07FU | 2 |
| SH177690.07FU | 2 |
https://unite.ut.ee/repository.php (Full ‘UNITE + INSD’ dataset, version no 7.1, release date 20 November 2016).
Table 3.
Percentage identity of the ITS region (bases 1–527) from NR_138441 (T. viride CBS 119325 from TYPE material) compared with other sequences from Trichoderma type material from the same UNITE cluster
| RefSeq records in cluster SH181342.07FUa | NCBI RefSeq name | % identityin BLASTn search |
|---|---|---|
| NR_138441.1 | T. viride CBS 119325; from TYPE material | 100 |
| NR_134363.1(a) | T. martiale BPI GJS 04-40; from TYPE material | 99.617 |
| NR_138440.1 | T. nothescens CBS 134882; from TYPE material | 99.616 |
| NR_138452.1(b) | T. samuelsii CBS 130537; from TYPE material | 99.432 |
| NR_138456.1(b) | T. koningii ATCC 64262; from verified material | 99.432 |
| NR_138451.1(b) | T. hispanicum CBS 130540; from TYPE material | 99.432 |
| NR_137308.1(a) | T. texanum CBS 139784; from TYPE material | 99.431 |
| NR_144870.1(b) | T. vinosum CBS 119087; from TYPE material | 99.419 |
| NR_134340.1 | T. appalachiense BPI GJS 97-243; from TYPE material | 99.242 |
| NR_138439.1(c) | T. viridarium CBS 120065; from verified material | 99.241 |
| NR_134367.1(c) | T. paraviridescens CBS 119321; from TYPE material | 99.237 |
| NR_134342.1(d) | T. ovalisporum CBS 113299; from TYPE material | 99.225 |
| NR_131281.1(d) | T. koningiopsis CBS 119075; from TYPE material | 99.225 |
| NR_137303.1(d) | T. dorotheae CBS 119089; from TYPE material | 99.225 |
| NR_138442.1 | T. petersenii CBS 119051; from TYPE material | 99.053 |
| NR_144874.1(e) | T. taiwanense CBS 119058; from TYPE material | 99.031 |
| NR_137302.1(e) | T. caribbaeum CBS 119093; from TYPE material | 99.031 |
| NR_138444.1 | T. trixiae CBS 134702; from TYPE material | 99.006 |
| NR_144876.1 | T. scalesiae CBS 120069; from TYPE material | 98.868 |
| NR_134362.1 | T. neosinense BPI GJS 94-11; from TYPE material | 98.851 |
| NR_131317.1 | T. gamsii; from verified material | 98.846 |
| NR_134343.1 | T. intricatum BPI GJS 97-88; from TYPE material | 98.837 |
| NR_111837.1 | T. erinaceus ATCC MYA-4844; from TYPE material | 98.491 |
| NR_138443.1 | T. dingleyae CBS 119056; from TYPE material | 98.45 |
| NR_134437.1 | T. strigosellum CBS 102817; from TYPE material | 97.938 |
| NR_134361.1 | T. stilbohypoxyli CBS 992.97; from TYPE material | 97.854 |
| NR_103571.1 | T. strigosum; from TYPE material | 97.736 |
| NR_134392.1 | T. junci CBS 120926; from TYPE material | 97.323 |
| NR_134419.1 | T. yunnanense CBS 121219; from TYPE material | 97.164 |
| NR_134360.1 | T. paucisporum BPI GJS 01-13; from TYPE material | 96.992 |
| NR_134359.1 | T. theobromicola CBS 119120; from TYPE material | 96.798 |
| NR_138438.1 | T. lieckfeldtiae CBS 123049; from TYPE material | 96.792 |
| NR_134446.1 | T. poronioideum BPI GJS 01-203; from TYPE material | 96.786 |
| NR_134447.1 | T. cerebriforme BPI GJS85-245; from verified material | 96.786 |
| NR_144875.1 | T. rogersonii CBS 119233; from TYPE material | 96.737 |
| NR_130668.1 | T. asperellum CBS 433.97; from TYPE material | 96.712 |
| NR_134371.1(f) | T. hamatum DAOM 167057; from TYPE material | 96.591 |
| NR_138449.1(f) | T. evansii CBS 123079; from TYPE material | 96.591 |
| NR_077179.1 | T. pubescens DAOM 166162; from TYPE material | 96.408 |
Identical letters in brackets indicate ITS regions with 100% identity to each other.
https://unite.ut.ee/repository.php (Full ‘UNITE + INSD’ dataset, version no 7.1, release date 20 November 2016).
A comparison of the Trichoderma ITS RefSeq dataset to reference sequences curated in the ISTH TrichoBLAST database (http://www.isth.info/tools/blast/index.php) was attempted but a number of discrepancies were found, including different names for the same GenBank accession identifier and sequence differences. Since the ISTH resource has not been updated recently (I. Druzhinina, personal communication), any further comparisons were not pursued. In addition to providing quality control to guide NCBI users, this case study can provide additional information to inform and extend external database resources like UNITE and ISTH.
Source metadata
The delineation of species serves a very practical purpose. It allows researchers to define populations having shared characteristics, not only on a genetic level, but also regarding to its environmental or ecological functions. Ultimately, associations beyond a single specimen and barcode sequence (30, 36) can guide hypotheses on population structure, ecological relevance and presence of pathogenicity factors, despite strain variability (30, 36). Important work has already been done at the UNITE database to facilitate third party metadata annotations for ITS sequence data using the PlutoF workbench (37). This resulted in annotation efforts focused on specific standards, such the MIxS-Built Environmental standard (38) and a call for additional taxonomic information on the most commonly found unidentified ITS sequence clusters from the environment (39). To compliment these efforts in the NCBI public databases we decided to append basic information such as host, isolation source and location for Trichoderma RefSeq records. Before metadata was added, the majority of this information was lacking with only a minority of records containing these qualifiers: collection-date (4%), country (47%), isolation-source (33%) and nat-host (22%). After curation, this was increased to the following levels: collection-date (75%), country (93%) and isolation-source (76%).
Collection date, country and location information are not only highly valuable for Trichoderma research but also for any other microbes. For instance, recording and studying new and emerging diseases include basic questions that rely on geographic data to record first reports and inform the extent of a pathogen’s distribution. In our case study, the nat-host field, however, proved challenging to append with additional data because whenever there was doubt that the isolated source was also the natural host, no information could be added. In specific groups such as viruses, the isolation source is very tightly linked to the host information and as such less ambiguous. In Trichoderma, however, species are frequently reported as inhabitants of decaying wood, but, upon further examination, these same species have been found obtaining nutrients by parasitizing other fungal species that are co-inhabiting the same substrate. Even though there are several species reported as saprobes (for example T. reseei), phylogenetic and genomic approaches have suggested that the ancestor of Trichoderma was a mycoparasite (35, 40). To account for this, authors of taxonomy descriptions have often opted to give detailed information on the other fungi found co-inhabiting the Trichoderma species being described. For example, for Trichoderma bavaricum (NR_134385.1, GenBank version FJ860737) (41), the following isolation source is given: ‘on corticated branches and twigs cut from a tree of Betula pendula 0.3–2 cm thick, emergent through and on bark and on/soc. Diatrypella favacea, also overgrowing long-necked effete pyrenomycete in the bark, soc. Tubeufia cerea’ (41). In this case, an unambiguous isolation source could not be inferred and so the information was not added to the sequence record of the type material. Including as much information as possible in the original description is a good practice, because it is often necessary to consider the possibility that a Trichoderma species may be parasitising a co-inhabiting fungal species.
Apart from making source metadata available, the curation process also uncovered discrepancies between source information in the species description and source information in the sequence record. For example, Trichoderma arundinaceum with RefSeq sequence record NR_134372.1 (GenBank version: EU330927.1) (42) has been suppressed and the record AY154921 will only be considered as the representative of the type material of Trichoderma arundinaceum once the unspecific species label (‘Trichoderma sp. Ir.500’) could be verified and updated. At the time of writing, AY154921 was associated with an unspecified name and it was only known via a single publication (42) that the sequence record belongs to the type material. Another example where source information was informative is NR_134424 (GenBank version: HM769754.1) (43) from Trichoderma pseudogelatinosum which has already been replaced with NR_144878 (GenBank version: JQ797389.1).
Access and search tools
Curation of ITS RefSeq records also include checking of lineage information and the effort benefits any Entrez database and BLAST search, so that results will be appropriately filtered for any sequence or genome associated with these taxa. ITS RefSeq records can be accessed directly from the NCBI Nucleotide database, or indirectly via a number of other resources summarized in Table 4. From the ITS RefSeq BioProject, the Nucleotide (total) link in the Project Data box leads to the Nucleotide database. By appending this query: ‘AND Trichoderma [organism]’ to the BioProject ID already in the search window retrieves Trichoderma ITS RefSeq records only. From the Trichoderma page in the Taxonomy Browser the Nucleotide link also leads to the Nucleotide database and adding the following: AND ‘ITS region AND RefSeq [filter]’ to the TaxID information already in the search box retrieves Trichoderma ITS RefSeq records only.
Table 4.
Accessing ITS RefSeq records at NCBI
| NCBI Resource | URL | Additional Entrez Query text |
|---|---|---|
| Nucleotide | https://www.ncbi.nlm.nih.gov/nuccore/ | ‘ITS region’ AND Trichoderma [organism] AND RefSeq [filter] |
| BioProject | https://www.ncbi.nlm.nih.gov/bioproject/177353 | |
| (Nucleotide via BioProject) | (https://www.ncbi.nlm.nih.gov/nuccore/?term=177353[BioProject]+AND+Trichoderma + [orgn]) | AND Trichoderma [orgn] |
| Taxonomy Browser | https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=5543 | |
| (Nucleotide via Taxonomy browser) | (https://www.ncbi.nlm.nih.gov/nuccore/?term=txid5543[Organism%3Aexp]+AND+%22ITS+region%22+AND+RefSeq+[filter]) | AND ‘ITS region’ AND RefSeq [filter] |
| NCBI BLAST | https://blast.ncbi.nlm.nih.gov/Blast.cgi | ‘ITS region’ AND Trichoderma [organism] AND RefSeq [filter] |
| Targeted Loci Blast | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=TargLociBlast | |
| NCBI FTP | ftp://ftp.ncbi.nlm.nih.gov/refseq/TargetedLoci/Fungi/ |
As the definition line (title) of the RefSeq ITS records are very different from archival records, it is also important to comment on the use of the search term ‘5.8 S’ to find ITS records. The simplest search, which also finds most of the submitted ITS records to the INSDC of the genus, and not many unrelated records, is: 5.8S [ti] AND Trichoderma [orgn] However, this search will also find 5.8S sequence only records annotated in genomes from the RefSeq database. To exclude those 5.8S sequence only records the following can be added to the query: NOT (5.8S [ti] AND RefSeq [filter]) The simplest way to find the curated and reviewed RefSeq sequence records (associated with the Targeted Loci BioProject) that includes the ITS1,5.8S and ITS2 sequence, is: ITS region [ti] AND Trichoderma [orgn] To be explicit about which database to search, the following can be added to the query: AND RefSeq [filter] However, it is not necessary since the RefSeq ITS records are the only set that use this phrase in the definition line. To find sequences from type material in both archival records and those in the RefSeq database, the following search is needed: (ITS region [ti] OR 5.8S [ti]) AND Trichoderma [orgn] AND sequence from type [filter]. Again, the unwanted 5.8S only sequence records can be excluded from the search results by adding to the query: NOT (5.8S [ti] AND RefSeq [filter]) Users should be aware that occasionally only the RefSeq version of a sequence from type material for a particular organism will be in the search results since submitters do not consistently provide the type identifier information (as provided in the protolog) in their sequence records submitted to the INSDC.
By selecting the appropriate databases and filters as listed in Table 4, ITS RefSeq records can be searched via the general Nucleotide Web BLAST portal or via a specialized search using the Targeted Loci Blast database. When using the general Nucleotide Web BLAST portal, the summary page, by default, should show a non-redundant list that are most similar to the query sequence (ordered by Total score) and representatives (one for each identical sequence) picked according to a hierarchical order (with RefSeq prioritized). Identical records can be found when scrolling down to the details section of each alignment hit or using the Taxonomy Report link at the top of the page. Also, hits from the archival database may contain a name different from that in the organism field of the primary INSDC record, since definition lines (also known as sequence titles) of archival records are not always up to date and thus a reason for some discrepancies in Web BLAST results. The Taxonomy Report displays the organism field names ordered by name, not total score. Identical accessions (if present) are listed in the Organism Report section directly below each representative record. The Targeted Loci Blast database, however, contains only curated RefSeq records with sequence titles frequently updated. Finally, BLAST should not be the only sequence comparison analysis used when trying to determine the identity of a new sequence. It can be used as a first filter to find similar sequences. The ranked nature of the BLAST results in the web interface has the potential to mislead a naïve user in thinking that the top hit will always carry the correct identification for their query sequence of interest. Guidelines on how to verify newly generated fungal ITS sequences have been published (44) as well as more general guidelines (45) with suggested procedures for identification of Fungi (46). All the ITS RefSeq data can be downloaded from the NCBI FTP site, and an ftp link is also available in the BioProject page (Table 4).
Impact on genome data
The ITS region is the proposed official universal DNA barcode of Fungi (9). However, it remains limited in being able to distinguish all specific species in highly speciose genera, including Trichoderma (47). With this in mind, TEF1 has been proposed as secondary barcode for Trichoderma (24). TEF1 also remains the most popular marker for Trichoderma species identification. However, the PCR fragments that different researchers employ do not always overlap and it can be a challenge to obtain a reliable alignment in order to complete a genus phylogeny. Despite this, TEF1 was recently employed in a genus-wide phylogenetic study of Trichoderma (25). Based on the broad usage of RPB2 (although not as good as TEF1 to distinguish species) in other fungal species (48, 49) as well as its demonstrable ability to provide full genus phylogenetics (23) we decided to utilize and expand the list of accessions nominated by Bissett et al. (19).
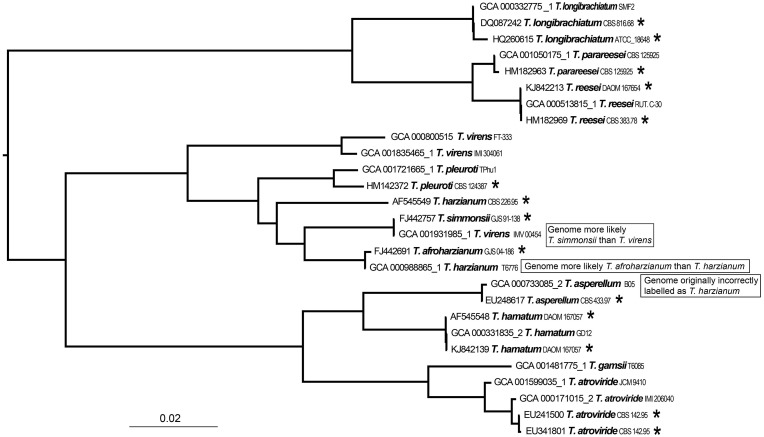
One important argument in favor of having more expansive non-ribosomal markers is their utility in validating genome sequences. Genome assemblies frequently do not include sequence data from the ribosomal cistron. In addition, if the region is included, it is not always correctly assembled due to its highly repetitive nature. A marginally contaminated culture that is targeted for genome sequencing will often include a contig of the contaminants’ ITS region in its genome assembly. Genome verification via the ITS sequence may therefore be inaccurate. These problems are expected to become less important as more genome assemblies rely on longer read technologies, but the ability to utilize type material verification still remains important to genome data quality going forward. All 16 Trichoderma assemblies evaluated at NCBI contain the RPB2 gene sequence in the genome but only 10 assemblies contain the ITS region. In addition, one assembly contained two different ITS regions, one that matched to the intended targeted organism (JNNP01000802.1, range: 6152-6666) but one that did not, it belonged to a Penicillium species instead (JNNP01000511.1, range: 1-1206). RPB2 sequences within genomes matched to RPB2 sequences of type material as expected, except in cases were the genome assemblies have likely been identified incorrectly (Figure 5 with matrix and tree provided as Supplementary Material) (17, 24). The current rate at which fungal genomes from type material are sequenced, assembled and submitted to NCBI is more or less the same rate at which incorrectly labeled genome assemblies are being submitted to NCBI. Therefore, it will be useful to transition to alternative markers such as single copy protein coding genes for genome verification and inclusion into RefSeq in the future.
Figure 5.
A phylogenetic tree generated by a FastTree analysis using a MAFFT alignment of RPB2 nucleotide sequences from type material (with asterisk) of Trichoderma and genomes labeled as Trichoderma.
We made an effort to identify a number of RPB2 records from type material of 708–1223 bp in length with all having 517 bases overlapping when aligned (Supplementary Table S2). Together with a whole genome k-mer analysis this list will be useful in confirming the identity of future Trichoderma genome submissions. Sequences from type material and a k-mer analysis have already helped to identify an incorrectly labeled genome (and confirmed by a Trichoderma expert) which was subsequently corrected (Figure 5). NCBI has started to undertake corrections for genome data in bacteria where reliable type material comparisons can be made (50). The next phase could be to expand this to other practical groups when enough data is available.
Additional taxonomic questions
At the time of writing (March 2017), there were 2007 taxids under the Trichoderma genus name in the NCBI Taxonomy database of which only 276 belong to validly published names. The rest of the taxids are mostly associated with unspecified names and a few names which included a strain identifier in combination with a valid name. At the time of writing (March 2017), Index Fungorum contained 749 Trichoderma/Hypocrea names and compared with NCBI Taxonomy, 245 were preferred names, 164 were annotated as synonyms and 340 names were not known to NCBI. The unknown names to NCBI from Index Fungorum were mostly under Hypocrea (283 names) and a few under Trichoderma (57 names) but also included synonyms of other current names as indicated by Species Fungorum. MycoBank contained 94 Trichoderma/Hypocrea binomial names and compared with NCBI Taxonomy, 38 were preferred names, 10 were annotated as synonyms and 45 names were not known to NCBI. Similarly, the unknown names to NCBI from MycoBank were mostly under Hypocrea (40 names) and a few under Trichoderma (5 names). From Index Fungorum and MycoBank combined, 342 names (58 Trichoderma and 284 Hypocrea) were unknown to NCBI Taxonomy.
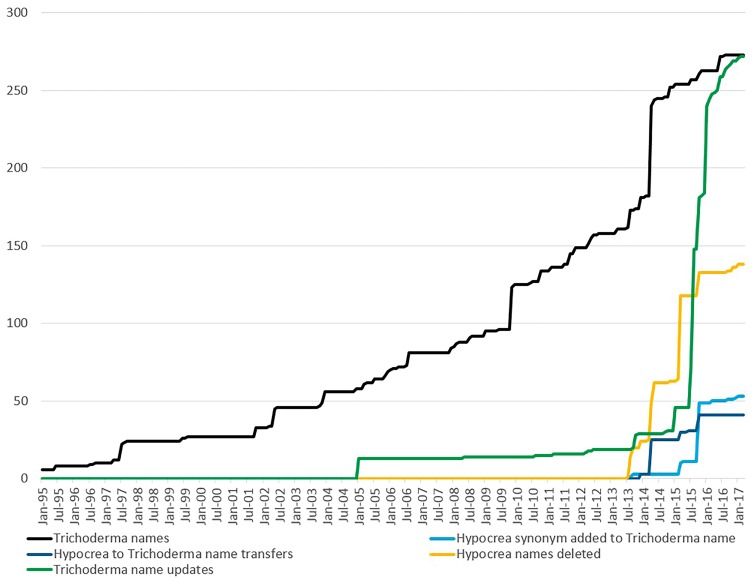
The sexual Hypocrea state has long been recognized, resulting in >1000 species names (18). As part of this project, the majority of Hypocrea names in the NCBI Taxonomy could be synonymized with a Trichoderma synonym with unspecified Hypocrea (‘Hypocrea sp.’) names, listed under their Trichoderma equivalents (Figure 6). Forty specified Hypocrea names were merged into Trichoderma names and 12 Hypocrea sp. names were moved to Trichoderma sp. names. However, a small number of names in the NCBI Taxonomy database remain problematic. Hypocrea fomitopsis was updated to Trichoderma fomitopsis in NCBI Taxonomy based on a new combination (51), but it was excluded by Bissett et al. (19) under ‘names not currently in use’. Another name, Hypocrea muroiana has not been treated in Bissett et al. (19) and remains without a Trichoderma synonym, although it is possibly a taxonomic synonym of another name excluded by Bissett et al. (19), Trichoderma corrugata (51). An additional name present in NCBI Taxonomy, and indicated by Bissett et al. (19) to not be in current use is Trichoderma subsulphureum (52). As valid names these will be included in GenBank, but sequences from these are excluded from RefSeq. Hypocrea coprosma, like Hypocrea muroiana is a valid name without a clearly assigned Trichoderma synonym. Currently, this will be listed in square brackets under Trichoderma in the NCBI Taxonomy: ‘[Hypocrea] muroiana’ and ‘[Hypocrea] coprosma’. We follow the square bracket format in NCBI Taxonomy to indicate cases of valid names that may be in need of re-classification.
Figure 6.
Additions and updates to Trichoderma and Hypocrea binomial names in the NCBI Taxonomy database over the past 22 years.
Conclusions
Although multiple electronic resources are available to update the NCBI taxonomic classification this activity is still, to a large extent, reliant on taxonomic curators’ ability to verify changes in the publication record and directly interact with specialists. Increased usage of automatic processes is inevitable and intensive manual curation is not scalable. In this study we selected a particular genus where impactful changes could be made. We extended the taxonomic classification for public sequence data and produced a set of reference markers that could enhance other sequence verification efforts.
Although some recent Trichoderma species descriptions only generated molecular data for RPB2 and TEF1 markers (53), the ITS region was still included in other descriptions (54). Despite the clear utility of RPB2 and TEF1 markers for Trichoderma taxonomy we still advocate that an ITS sequence be provided as an additional marker to complete the comparative sequence record. Importantly, the ITS region is still widely used in microbiome and environmental biodiversity studies and thus this sequence region connects the organism to environmental metadata that would otherwise not be accessible via other molecular markers.
The RefSeq ITS database highlights the relationship between classic and sequence based taxonomy. It is essential that type material information is paired with up to date taxonomic names and accurate sequence data in the public sequence databases and act as part of an extensive network of online databases informing modern mycology (55). To do this effectively curators are dependent on comprehensive taxonomic publications and effective communication with expert researchers. The request to researchers in the scientific community to maintain public sequence data (22) is strongly endorsed here, especially where it involves type material. As fungal taxonomy makes a transition to genome comparisons, it is also important to have a set of reliable markers such as RPB2 ready together with whole genome analyses to help validate these more complex datasets.
Supplementary data
Supplementary data are available at Database Online.
Funding
The Intramural Research Programs of the National Center for Biotechnology Information, National Library of Medicine
Conflict of interest. None declared.
Supplementary Material
Acknowledgements
We thank the input of experts in the Trichoderma community as well the work by members of the International Subcommission on Trichoderma and Hypocrea (ISTH) as part of the International Commission on the Taxonomy of Fungi (ICTF). In addition, we are grateful for helpful comments by the anonymous reviewers of this article.
References
- 1. Nilsson R.H., Ryberg M., Kristiansson E.. et al. (2006) Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. Plos One, 1, e59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagy L.G., Petkovits T., Kovács G.M.. et al. (2011) Where is the unseen fungal diversity hidden? A study of Mortierella reveals a large contribution of reference collections to the identification of fungal environmental sequences. New Phytol., 191, 789–794. [DOI] [PubMed] [Google Scholar]
- 3. O'Donnell K., Humber R.A., Geiser D.M.. et al. (2012) Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia, 104, 427–445. [DOI] [PubMed] [Google Scholar]
- 4. Irinyi L., Serena C., Garcia-Hermoso D.. et al. (2015) International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database–the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med. Mycol., 53, 313–337. [DOI] [PubMed] [Google Scholar]
- 5. Kopchinskiy A., Komon M., Kubicek C.P.. et al. (2005) TrichoBLAST: a multilocus database for Trichoderma and Hypocrea identifications. Mycol. Res., 109, 658–660. [DOI] [PubMed] [Google Scholar]
- 6. Kõljalg U., Nilsson R.H., Abarenkov K.. et al. (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol., 22, 5271–5277. [DOI] [PubMed] [Google Scholar]
- 7. Deshpande V., Wang Q., Greenfield P.. et al. (2015) Fungal identification using a Bayesian classifier and the Warcup training set of internal transcribed spacer sequences. Mycologia, 108, 1–5. [DOI] [PubMed] [Google Scholar]
- 8. O'Leary N.A., Wright M.W., Brister J.R.. et al. (2015) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res., 44, D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schoch C.L., Seifert K.A., Huhndorf S.. et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U. S. A., 109, 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ratnasingham S., Hebert P.D. (2007) Bold: The barcode of life data system. Mol. Ecol. Notes, 7, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tedersoo L., Abarenkov K., Nilsson R.H.. et al. (2011) Tidying up international nucleotide sequence databases: Ecological, geographical and sequence quality annotation of its sequences of mycorrhizal fungi. PLoS One, 6, e24940.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Federhen S., Hotton C., Mizrachi I. (2009) Comments on the paper by Pleijel et al. (2008): vouching for GenBank. Mol. Phylogenet. Evol., 53, 357–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Federhen S. (2015) Type material in the NCBI Taxonomy Database. Nucleic Acids Res., 43, D1086–D1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schoch C.L., Robbertse B., Robert V.. et al. (2014) Finding needles in haystacks: linking scientific names, reference specimens and molecular data for Fungi. Database, bau061, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hawksworth D.L. (2011) A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. Mycokeys, 1, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McNeill J., Barrie F.R., Buck W.R.. et al. (2012) International Code of Nomenclature for algae, fungi, and plants (Melbourne Code) In McNeill (ed). Regnum Vegetabile. Koeltz Scientific Books, Königstein, Vol. 154, pp. 240. [Google Scholar]
- 17. Jaklitsch W.M., Voglmayr H. (2013) New combinations in Trichoderma (Hypocreaceae, Hypocreales). Mycotaxon, 126, 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rossman A.Y., Seifert K.A., Samuels G.J.. et al. (2013) Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus, 4, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bissett J., Gams W., Jaklitsch W.M.. et al. (2015) Accepted Trichoderma names in the year 2015. IMA Fungus, 6, 263–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sandoval-Denis M., Sutton D.A., Cano-Lira J.F.. et al. (2014) Phylogeny of the clinically relevant species of the emerging fungus Trichoderma and their antifungal susceptibilities. J. Clin. Microbiol., 52, 2112–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Druzhinina I.S., Seidl-Seiboth V., Herrera-Estrella A.. et al. (2011) Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol., 9, 749–759. [DOI] [PubMed] [Google Scholar]
- 22. Atanasova L., Druzhinina I., Jaklitsch W.M., (2013) Two hundred Trichoderma species recognized on the basis of molecular phylogeny In: Mukherjee P.K., Horwitz B.A., Singh U.S.. et al. (eds). Trichoderma: Biology and Applications. CAB International, London, UK, pp. 10–42. [Google Scholar]
- 23. Jaklitsch W.M., Voglmayr H. (2015) Biodiversity of Trichoderma (Hypocreaceae) in Southern Europe and Macaronesia. Stud. Mycol., 80, 1–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaverri P., Branco-Rocha F., Jaklitsch W.. et al. (2015) Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia, 107, 558–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Braithwaite M., Johnston P.R., Ball S.L.. et al. (2016) Trichoderma down under: species diversity and occurrence of Trichoderma in New Zealand. Aust.. Plant Pathol., 46, 1–20. [Google Scholar]
- 26. Druzhinina I.S., Kopchinskiy A.G., Komon M.. et al. (2005) An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal. Genet. Biol., 42, 813–828. [DOI] [PubMed] [Google Scholar]
- 27. Bengtsson-Palme J., Ryberg M., Hartmann M.. et al. (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol., 4, 914–919. [Google Scholar]
- 28. Blaalid R., Kumar S., Nilsson R.H.. et al. (2013) ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol. Ecol. Resour., 13, 218–224. [DOI] [PubMed] [Google Scholar]
- 29. Holmes K.A., Schroers H.-J., Thomas S.E.. et al. (2004) Taxonomy and biocontrol potential of a new species of Trichoderma from the Amazon basin of South America. Mycol. Prog., 3, 199–210. [Google Scholar]
- 30. Hibbett D., Abarenkov K., Koljalg U.. et al. (2016) Sequence-based classification and identification of Fungi. Mycologia, 108, 1049–1068. [DOI] [PubMed] [Google Scholar]
- 31. Jaklitsch W.M., Samuels G.J., Ismaiel A.. et al. (2013) Disentangling the Trichoderma viridescens complex. Persoonia, 31, 112–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jaklitsch W.M., Samuels G.J., Dodd S.L.. et al. (2006) Hypocrea rufa/Trichoderma viride: a reassessment, and description of five closely related species with and without warted conidia. Stud. Mycol., 56, 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kõljalg U., Larsson K.H., Abarenkov K.. et al. (2005) UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol., 166, 1063–1068. [DOI] [PubMed] [Google Scholar]
- 34. Caporaso J.G., Kuczynski J., Stombaugh J.. et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chaverri P., Samuels G.J. (2013) Evolution of habitat preference and nutrition mode in a cosmopolitan fungal genus with evidence of interkingdom host jumps and major shifts in ecology. Evolution, 67, 2823–2837. [DOI] [PubMed] [Google Scholar]
- 36. Crous P.W., Groenewald J.Z., Slippers B.. et al. (2016) Global food and fibre security threatened by current inefficiencies in fungal identification. Philos. Trans. R. Soc. B, 371, 20160024.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abarenkov K., Tedersoo L., Nilsson R.H.. et al. (2010) PlutoF-a web based workbench for ecological and taxonomic research, with an online implementation for fungal ITS sequences. Evol. Bioinform., 6, 189–196. [Google Scholar]
- 38. Abarenkov K., Adams R.I., Irinyi L.. et al. (2016) Annotating public fungal ITS sequences from the built environment according to the MIxS-Built Environment standard - a report from a May 23-24, 2016 workshop (Gothenburg, Sweden). Mycokeys, 16, 1–15. [Google Scholar]
- 39. Nilsson R.H., Wurzbacher C., Bahram M.. et al. (2016) Top 50 most wanted fungi. Mycokeys, 12, 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kubicek C.P., Herrera-Estrella A., Seidl-Seiboth V.. et al. (2011) Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol., 12, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jaklitsch W.M. (2011) European species of Hypocrea part II: species with hyaline ascospores. Fungal Divers, 48, 1–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Degenkolb T., Dieckmann R., Nielsen K.F.. et al. (2008) The Trichoderma brevicompactum clade: a separate lineage with new species, new peptaibiotics, and mycotoxins. Mycol. Prog., 7, 177–219. [Google Scholar]
- 43. Kim C.S., Yu S.H., Nakagiri A.. et al. (2012) Re-evaluation of Hypocrea pseudogelatinosa and H. pseudostraminea isolated from shiitake mushroom (Lentinula edodes) cultivation in Korea and Japan. Plant Pathol. J., 28, 341–356. [Google Scholar]
- 44. Nilsson R.H., Tedersoo L., Abarenkov K.. et al. (2012) Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. MycoKeys, 4, 37–63. [Google Scholar]
- 45. Hyde K.D., Udayanga D., Manamgoda D.S.. et al. (2013) Incorporating molecular data in fungal systematics: a guide for aspiring researchers. Curr. Res. Environ. Appl. Mycol., 3, 1–32. [Google Scholar]
- 46. Raja H.A., Miller A.N., Pearce C.J.. et al. (2017) Fungal identification using molecular tools: a primer for the natural products research community. J. Nat. Prod., 80, 756–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stielow J.B., Lévesque C.A., Seifert K.A.. et al. (2015) One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia, 35, 242–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. James T.Y., Kauff F., Schoch C.L.. et al. (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature, 443, 818–822. [DOI] [PubMed] [Google Scholar]
- 49. Schoch C.L., Sung G.H., Lopez-Giraldez F.. et al. (2009) The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol., 58, 224–239. [DOI] [PubMed] [Google Scholar]
- 50. Federhen S., Rossello-Mora R., Klenk H.-P.. et al. (2016) Meeting report: GenBank microbial genomic taxonomy workshop (12-13 May, 2015). Stand. Genomic Sci., 11, 15. [Google Scholar]
- 51. Zhao-Xiang Z., Wen-Ying Z. (2014) Twelve species of Trichoderma (Hypocreaceae) new to China and three new combinations. Mycosystema, 33, 1175–1209. [Google Scholar]
- 52. Overton B.E., Stewart E.L., Geiser D.M.. et al. (2006) Systematics of Hypocrea citrina and related taxa. Stud. Mycol., 56, 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Qin W.T., Zhuang W.Y. (2016) Seven wood-inhabiting new species of the genus Trichoderma (Fungi, Ascomycota) in Viride clade. Sci. Rep., 6, 27074.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sun J.Z., Pei Y.F., Li E.W.. et al. (2016) A new species of Trichoderma hypoxylon harbours abundant secondary metabolites. Sci. Rep., 6, 37369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Triebel D., Hagedorn G., Rambold G. (2012) An appraisal of megascience platforms for biodiversity information. Mycokeys, 5, 45–63. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.