Abstract
Soil salinization and the associated land degradation are major and growing ecological problems. Excess salt in soil impedes plant photosynthetic processes and root uptake of water and nutrients such as K+. Arbuscular mycorrhizal (AM) fungi can mitigate salt stress in host plants. Although, numerous studies demonstrate that photosynthesis and water status are improved by mycorrhizae, the molecular mechanisms involved have received little research attention. In the present study, we analyzed the effects of AM symbiosis and salt stress on photosynthesis, water status, concentrations of Na+ and K+, and the expression of several genes associated with photosynthesis (RppsbA, RppsbD, RprbcL, and RprbcS) and genes coding for aquaporins or membrane transport proteins involved in K+ and/or Na+ uptake, translocation, or compartmentalization homeostasis (RpSOS1, RpHKT1, RpNHX1, and RpSKOR) in black locust. The results showed that salinity reduced the net photosynthetic rate, stomatal conductance, and relative water content in both non-mycorrhizal (NM) and AM plants; the reductions of these three parameters were less in AM plants compared with NM plants. Under saline conditions, AM fungi significantly improved the net photosynthetic rate, quantum efficiency of photosystem II photochemistry, and K+ content in plants, but evidently reduced the Na+ content. AM plants also displayed a significant increase in the relative water content and an evident decrease in the shoot/root ratio of Na+ in the presence of 200 mM NaCl compared with NM plants. Additionally, mycorrhizal colonization upregulated the expression of three chloroplast genes (RppsbA, RppsbD, and RprbcL) in leaves, and three genes (RpSOS1, RpHKT1, and RpSKOR) encoding membrane transport proteins involved in K+/Na+ homeostasis in roots. Expression of several aquaporin genes was regulated by AM symbiosis in both leaves and roots depending on soil salinity. This study suggests that the beneficial effects of AM symbiosis on the photosynthetic capacity, water status, and K+/Na+ homeostasis lead to the improved growth performance and salt tolerance of black locust exposed to salt stress.
Keywords: arbuscular mycorrhizal fungi, black locust, photosynthesis, water status, ion homeostasis, salt stress
Introduction
Soil salinization and the associated land degradation are major and growing ecological problems, particularly in arid and semiarid areas (Porcel et al., 2012, 2015). Excess salt in the soil impedes plant growth and development (Evelin et al., 2009) by causing a series of physiological stresses in plants (Aroca et al., 2013). Photosynthesis, the key process of primary metabolism (Pinheiro and Chaves, 2011), is affected by salinity in several ways (Chaves et al., 2009). First, CO2 availability declines because of the limitations of diffusion through the stomata and mesophyll (Chaves et al., 2009), and second, salt stress hinders the photosynthetic electron flow through photosystems II and I (PSII and PSI) (Lu and Vonshak, 2002; Allakhverdiev and Murata, 2008). Moreover, the D1 and D2 proteins, which are in the PSII reaction center and are pivotal in the phosphorylation-coupled linear electron flow (Jansen et al., 1996), degrade under salt stress (Sudhir et al., 2005; Neelam and Subramanyam, 2013).
Another challenge faced by plants exposed to salt stress is acquiring sufficient amounts of water from the soil (Ouziad et al., 2006). Root uptake of water is limited by a diminished water potential due to the accumulation of salt in the soil (Ruiz-Lozano et al., 2012). Aquaporins are proposed to play a pivotal role in plant water equilibrium and water use efficiency (WUE) (Sade et al., 2010). The passive transport of water molecules across membranes is facilitated by these intrinsic membrane proteins (Maurel et al., 2008). Plant aquaporins are categorized into five subfamilies (Ruiz-Lozano et al., 2012), of which plasma membrane intrinsic proteins (PIPs) and tonoplast intrinsic proteins (TIPs) are the most abundant in plant plasma and vacuolar membranes, respectively (Luu and Maurel, 2005), and serve as the primary path for transcellular and intracellular water movement, respectively (Maurel et al., 2008).
Plants exposed to salt stress also suffer from Na+ toxicity and K+ deficiency, since the acquisition of K+ is disrupted by excess Na+ in the soil (Porcel et al., 2016). Over-accumulation of Na+ in plants damages cellular organelles, impairs photosynthesis, and competes with K+ for protein biosynthesis and enzyme activation (Shabala and Cuin, 2008; Porcel et al., 2016). The regulation of several genes encoding membrane transport proteins responsible for K+ and/or Na+ uptake, translocation or compartmentalization is included among the strategies of plants to deal with excessive accumulation of Na+ and a deficit in K+. Plasma membrane Na+/H+ antiporter SOS1 and tonoplast (Na+, K+)/H+ antiporter NHX1 are responsible for efflux of Na+ from cells and sequestration of this cation in the vacuole, respectively (Qiu et al., 2002; Deinlein et al., 2014). They are two major mechanisms that maintain low Na+ accumulation in the cytosol (Deinlein et al., 2014). Moreover, class I HKT transporters are involved in removing Na+ from the xylem in order to prevent Na+ from transport to the leaves (Sunarpi et al., 2005; Davenport et al., 2007), while the outward-rectifying K+ channel SKOR controls K+ loading into the xylem and K+ transport to shoots (Zhang et al., 2017).
Arbuscular mycorrhizal (AM) fungi symbiotically associate with most terrestrial plants (Ruiz-Lozano et al., 2012). AM symbiosis not only improves the water and nutrient uptake (Smith and Read, 2008) but also mitigates environmental stresses (e.g., salinity) in the host plants (Porcel et al., 2012). Previous studies have demonstrated that mycorrhizal symbiosis can increase photosynthesis in plants exposed to salt stress (Sheng et al., 2008; Hajiboland et al., 2010; Porcel et al., 2015). However, the molecular mechanisms involved remain to be determined. First, the regulation of psbA (encoding D1 protein) and psbD (encoding D2 protein) by AM fungi can be expected because of a frequently observed increase in the quantum yield of PSII photochemistry from AM symbiosis (Sheng et al., 2008; Hajiboland et al., 2010; Porcel et al., 2015). Additionally, Porcel et al. (2015) found that the expression of rbcL (encoding the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, rubisco) and rbcS (encoding the small subunit of rubisco) was not upregulated by mycorrhizal colonization in the leaves of rice. Therefore, we conducted an additional investigation to explore the influence of AM fungi on gene expression when plants are subjected to salt stress.
Arbuscular mycorrhizal symbiosis also improves the water uptake and water status in plants exposed to salt stress (Aroca et al., 2007; Sheng et al., 2008; Wu et al., 2015), and such improvement is likely related to changes in the expression of aquaporin genes (Ruiz-Lozano et al., 2012). Under salt stress, the regulation of plant aquaporin genes by AM symbiosis has been observed (Ouziad et al., 2006; Aroca et al., 2007; Jahromi et al., 2008). However, most of these studies focused on the regulation of aquaporin genes in roots, and the regulation in leaves has rarely been studied. The regulation of aquaporin genes in leaves may mediate membrane permeability to water and CO2 and further affect cellular water conservation and the mesophyll conductance of CO2 diffusion (Jang et al., 2004; Saibo et al., 2009; Yang and Cui, 2009). Thus, the regulation of aquaporin genes in leaves must be considered in the further exploration of the effects of AM symbiosis on plant water status and photosynthesis. Additionally, the response of aquaporin genes in roots to AM symbiosis and salt stress is inconsistent. For example, mycorrhizal colonization downregulates LePIP1 under salt stress (Ouziad et al., 2006), whereas mycorrhizae upregulates LsPIP1 under saline conditions (Jahromi et al., 2008). Such contradictions may be a consequence of the different plant and fungi species used and the different methods of salt application (Ouziad et al., 2006). However, most studies investigated AM symbiosis and the regulation of aquaporins in response to salinity using agronomic plant species. The regulation of these genes by AM fungi in response to salinity is unknown in a tree species.
Moreover, mycorrhizal symbiosis can facilitate K+ uptake while preventing Na+ absorption and translocation to the shoots (Evelin et al., 2012; Garcia et al., 2017). Exploring the regulation of genes encoding membrane transport proteins involved in K+/Na+ homeostasis in AM plants subjected to salinity is an essential step in understanding the mechanisms underlying salt tolerance conferred by AM fungi, but such research is lacking. Porcel et al. (2016) found that the expression of OsSOS1, OsNHX3, OsHKT2;1, and OsHKT1;5 was upregulated by mycorrhizal colonization in plants subjected to salt stress, concomitantly with the reduced roots to shoots distribution of Na+ in AM plants. Estrada et al. (2013) found that AM fungi native to saline areas were better able to regulate root ZmAKT2, ZmSKOR, and ZmSOS1 expression levels than non-native AM fungi, as demonstrated by stronger correlation with higher K+/Na+ ratios in plants inoculated with native AM fungi than in plants inoculated with non-native AM fungi and non-mycorrhizal (NM) plants. Ouziad et al. (2006) found that the expression of LeNHX1 and LeNHX2 was not altered by AM symbiosis.
Black locust (Robinia pseudoacacia L.), a woody legume, has been widely planted as a pioneer tree in arid and semiarid regions in China (Tian et al., 2003). The species is fast growing, is tolerant to drought and salt stress and can improve soil through N-fixation in root nodules (Taniguchi et al., 2007; Jin et al., 2011; Gu et al., 2012). Black locust also has great economic value because the leaves, flowers, and wood can be used to feed livestock, to produce honey and as lumber, respectively (Yang et al., 2014; He et al., 2016). AM fungi can establish a mutualistic symbiosis with black locust and can alleviate drought stress or metal toxicity (He et al., 2017; Huang et al., 2017); however, little is known about the influence of the mutualism under salt stress, particularly the molecular mechanisms involved.
Therefore, the purposes of the present study were the following: (1) to determine the effects of AM symbiosis on photosynthesis, gas-exchange parameters, PSII operating efficiency and the expression pattern of four genes (RppsbA, RppsbD, RprbcL, and RprbcS) associated with photosynthesis in the leaves of black locust subjected to salt stress, (2) to examine the responses of six aquaporin genes to AM symbiosis and salinity in the leaves and roots of black locust, and (3) to determine the regulation of K+ and Na+ content and the K+/Na+ ratio, as well as the expression of four genes (RpSOS1, RpHKT1, RpNHX1, and RpSKOR) encoding transport proteins involved in K+ and/or Na+ uptake, translocation or compartmentalization by AM symbiosis in the leaves and roots of black locust exposed to salt stress.
Materials and Methods
Experimental Design
A pot experiment was conducted at Northwest A&F University using a randomized complete block design with three salinity levels (0, 100, and 200 mM NaCl) and two inoculation status (NM and mycorrhizal plants) for six treatments total, with 30 replicates per treatment (one plant per pot for a total of 180 pots).
Growth Substrate
The growth substrate was soil (<2 mm) mixed with fine sand (<2 mm) in the proportion of 1:1 (v/v). The soil/sand mixture was autoclaved (0.11 MPa, 121°C) for 2 h. The original soil collected from Northwest A&F University had a pH of 7.6 (1:5, soil:water, w/v) and contained 34.9 mg kg-1 available nitrogen, 15.8 mg kg-1 available phosphorous, 165.7 mg kg-1 available potassium and 17.5 g kg-1 organic matter.
Plant Material
Seeds of R. pseudoacacia acquired from Northwest A&F University (Yangling, China) were surface sterilized in a 5% NaClO solution for 10 min, washed three times with sterile water, pre-germinated on sterilized moist filter paper in petri dishes and transplanted into pots (10 cm × 12 cm, three germinated seeds per pot) containing 630 g of the growth substrate. After 1 week, the plants were thinned to one seedling per pot.
Inoculation Treatment
The AM inoculum, Rhizophagus irregularis (BGC BJ109), was obtained from the Beijing Academy of Agriculture and Forestry Sciences (Beijing, China) and was multiplied in pot cultures of Zea mays L. (Yang et al., 2014). The inoculum contained sand, spores (approximately 50 spores per gram), mycelia, and colonized root fragments. Each AM plant received 8 g of the inoculum at the time of the transplant, whereas each NM plant received the same amount of the autoclaved inoculum and the filtrate (<20 μm) of the AM inoculum to obtain a native microbial population free of mycorrhizal propagules.
Growth Conditions
The experiment was conducted in a greenhouse with a relative humidity of 50-75% and temperatures ranging from 20 to 35°C. The plants were watered daily and fertilized every 3 weeks with 50 ml per pot of Hoagland solution containing 1/2 phosphate throughout the experimental period. Plants were grown for 10 weeks before salinization treatment. Then, three levels (0, 100, and 200 mM NaCl) of saline solution were added to the corresponding pots for 7 days (15 ml/day; a total of 105 ml was added). The electrical conductivity values were 1.12, 5.63, and 8.95 dS m-1 for the growth substrate treated with 0, 100, and 200 mM NaCl, respectively. After an additional 3 weeks of growth, the plants were harvested.
Parameters Measured
Mycorrhizal Colonization
Fresh roots, used to determine mycorrhizal colonization (five plants per treatment, n = 5), were gently washed with distilled water, cut into 1-cm pieces, cleared in 5% KOH, then acidified with 1% HCl and stained in 0.05% trypan blue in lactophenol following the method of Phillips and Hayman (1970) before observation under a microscope. The gridline intersect method (Giovannetti and Mosse, 1980) was used to determine the mycorrhizal colonization.
Plant Biomass and Leaf Water Status
At harvest, the leaves, stems and roots of plants (five plants per treatment, n = 5) were separated, and the fresh weights (FWs) were measured. Then, the leaves were soaked in distilled water for 24 h for the turgid weight (TW) determination. Finally, the three plant parts were oven-dried at 70°C to a constant weight to measure the dry weights (DWs). The shoot dry mass was calculated as the sum of the DWs of leaves and stems. The relative water content (RWC) and water saturation deficit (WSD) were calculated using the following formulas (Gao, 2006):
Photosynthesis and Gas Exchange Parameters
The net photosynthetic rate (Pn), stomatal conductance (Gs), intercellular CO2 concentration (Ci), and transpiration rate (Tr) were measured from 8:30 to 11:30 a.m. with a Li-6400 portable open flow gas-exchange system (Li-Cor Inc., Lincoln, NE, United States) before harvest. The fifth or sixth youngest leaf of each plant (five plants per treatment, n = 5) was used for the measurements. The measurement conditions were as follows: photosynthetically active irradiation, 1000 μmol m-2 s-1; temperature, 25°C; relative humidity, 60%; and CO2 concentration, 400 μmol mol-1. The WUE was calculated as the ratio of Pn to Tr (Sheng et al., 2008).
Chlorophyll Fluorescence Parameters
Chlorophyll fluorescence parameters were measured with a modulated chlorophyll fluorimeter (Mini-Imaging-PAM, Walz, Germany). The fifth or sixth youngest leaf of each plant (four plants per treatment, n = 4) was used for the measurements. Plants were adapted to the dark for 30 min before the measurement of F0 and Fm. The maximum quantum efficiency of PSII photochemistry (Fv/Fm), PSII operating efficiency (ΦPSII), PSII maximum efficiency (Fv′/Fm′), photochemical quenching coefficient (qP), and non-photochemical quenching (NPQ) were calculated with F0, Fm and the light-adapted parameters using the following formulas (Baker, 2008):
Concentration of Na+ and K+
Oven-dried leaves and roots (five plants per treatment, n = 5) were ground to powder and samples (0.1 g) were acid-digested with 8 ml HNO3 + 2 ml HClO4, and heated to 220°C for extraction of Na+ and K+ ions. After cooling to room temperature, extraction solution was diluted before determination of the Na+ and K+ content with a Z-2000 atomic absorption spectrophotometer (Shimadzu, Japan).
RNA Extraction and cDNA Synthesis
At harvest, the leaves and roots of plants (three plants per treatment, n = 3) were frozen in liquid nitrogen and stored at -80°C until use. After the frozen leaves and roots were ground to a fine powder in liquid nitrogen, RNA was extracted with an E.Z.N.ATM plant RNA kit (Omega Bio-Tek, Norcross, GA, United States). RNA integrity and purity were checked by 1% agarose gel electrophoresis and spectrophotometric analysis using a NanoDrop 2000 (Thermo Scientific, Pittsburgh, PA, United States). Reverse transcription to complementary DNA (cDNA) was conducted using a PrimeScriptTM RT reagent kit with gDNA eraser (TaKaRa Bio, Dalian, China).
Cloning of Partial Coding Sequences (CDSs) of RprbcS, RpSOS1, RpHKT1, and RpSKOR
After ClustalW alignment of published CDSs for rbcS (Ammopiptanthus mongolicus, Arachis hypogaea, Phaseolus vulgaris, Vigna radiata, Pisum sativum, Cicer arietinum, Medicago truncatula, Glycine max, and Medicago sativa), SOS1 (Artemisia japonica, Glycine max, Vigna angularis, and Vigna radiate), HKT1 (Arachis ipaensis, Arachis duranensis, Lupinus angustifolius, and Medicago truncatula), and SKOR (Arachis ipaensis, Arachis duranensis, Cicer arietinum, Lupinus angustifolius, Vigna angularis, and Vigna radiata) (all from NCBI1) using MEGA 5.0 (Tamura et al., 2011), four pairs of degenerate primers (Supplementary Table S1) were designed based on nucleotide sequence conservation. Then, the degenerate primers were used to amplify the cDNA in a gradient PCR reaction (20 μl) containing 10 μl of Premix Taq®Version 2.0, 0.4 μM of each primer, 1.6 μl of a 1:5 dilution of cDNA and 6.8 μl of ddH2O. The PCR reaction was conducted using a C1000 thermocycler (Bio-Rad Laboratories, Hercules, CA, United States) with the following procedure: a 2 min denaturation at 95°C, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 40 to 60°C for 30 s and extension at 72°C for 30 s, followed by a final extension at 72°C for 10 min. PCR products were inserted into the pEASY-T1 cloning vector (TransGen Biotech, Beijing, China) and transformed into Trans5α chemically competent cells (TransGen Biotech, Beijing, China). Then, LB agar plates containing 15 mg ml-1 agarose, 10 mg ml-1 NaCl, 10 mg ml-1 peptone, 5 mg ml-1 yeast extract and 100 μg ml-1 ampicillin were used to select the transformants. Beijing AuGCT DNA-SYN Biotechnology Co., Ltd. (Beijing, China) sequenced the clones with inserts, and the obtained sequences were blasted using blastx on NCBI2. Putative partial CDSs of RprbcS (MF948148), RpSOS1 (MF948149), RpHKT1 (MF948150), and RpSKOR (MF948151) have been deposited in GenBank.
Gene Expression
The primers used in the qRT-PCR were designed with Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA, United States) and are listed in Supplementary Table S2. The qRT-PCR reaction was conducted using the CFX96 real-time PCR detection system (Bio-Rad Laboratories) and contained 10 μl of SYBR®Premix Ex TaqTM II (TakaRa Bio, Dalian, China), 0.4 μM of each primer, 1.6 μl of a 1:5 dilution of cDNA and 6.8 μl of ddH2O. The PCR procedure consisted of a 30 s denaturation at 95°C, followed by 40 cycles of denaturation at 95°C for 5 s and annealing/elongation at the annealing temperature (Supplementary Table S2) for 60 s. For RprbcS, the PCR procedure was switched to a 30 s denaturation at 95°C, followed by 40 cycles of 5 s at 95°C, 30 s at 57°C and 60 s at 72°C. A heat melt curve (from 65 to 95°C) was used to check the specificity of the PCR amplification. All samples were technically replicated three times. Negative controls without cDNA were run within each analysis. Normalization was performed based on the expression of the R. pseudoacacia actin gene (Meng et al., 2016). The relative quantity of transcripts was determined using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Statistical Analyses
The SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, United States) was used for statistical analyses. Data were subjected to a one-way ANOVA and post hoc comparisons (Duncan’s test, P < 0.05), in addition to a two-way ANOVA with three sources of variation (salt, AM fungi inoculation and their interaction). Figures were drawn with SigmaPlot 10.0 (Systat Software Inc., San Jose, CA, United States) and the package “pheatmap” in R.
Results
Mycorrhizal Colonization and Plant Biomass
Arbuscular mycorrhizal plants showed 89.29% mycorrhizal colonization under non-saline conditions, which decreased to 79.16% under the 200 mM NaCl treatment (Figure 1A). No significant differences in colonization rate were detected between the 0 and 100 mM NaCl treatments. No mycorrhizal colonization was found in NM plants.
FIGURE 1.
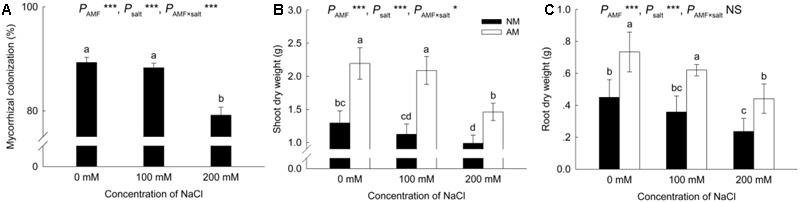
Effects of salinity and Rhizophagus irregularis inoculation on mycorrhizal colonization (A) and shoot (B) and root (C) dry mass in Robinia pseudoacacia. Different letters indicate significant differences (P < 0.05). NS not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. AM, arbuscular mycorrhizal; NM, non-mycorrhizal.
Increased salinity decreased the shoot and root dry mass in both NM and AM plants (Figures 1B,C). Nevertheless, the shoot and root DWs of AM plants were greater than those of NM plants at all salinity levels.
Photosynthesis and Gas-Exchange Parameters
Salinity negatively affected the Pn in NM and AM plants, causing reductions in the Pn of NM plants of 56% and 60% at 100 and 200 mM NaCl, respectively (Figure 2A). However, in AM plants, the reductions due to salinity were only significant (33%) at 200 mM NaCl. AM symbiosis improved the Pn by 106% at 100 mM NaCl and by 81% at 200 mM NaCl.
FIGURE 2.
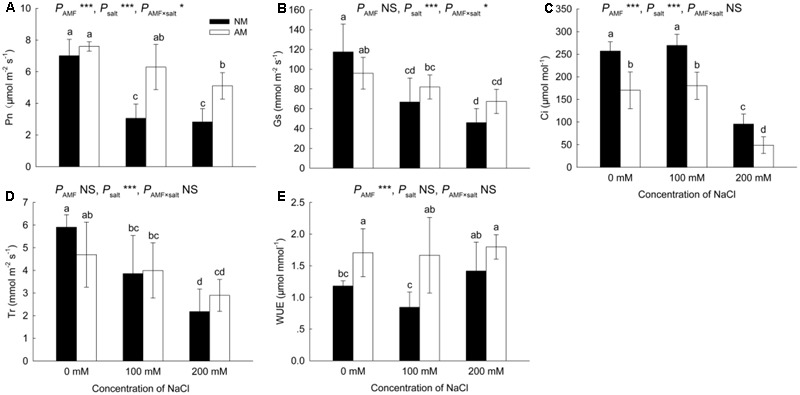
Effects of salinity and R. irregularis inoculation on net photosynthetic rate (Pn) (A), stomatal conductance (Gs) (B), intercellular CO2 concentration (Ci) (C), transpiration rate (Tr) (D), and water use efficiency (WUE) (E) in leaves of R. pseudoacacia. Different letters indicate significant differences (P < 0.05). NS not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. AM, arbuscular mycorrhizal; NM, non-mycorrhizal.
Salinity reduced the Gs and Tr in NM and AM plants (Figures 2B,D). Similar to the results for the Pn, salinity caused a greater reduction in the Gs and Tr in NM plants than in AM plants. However, the differences in the Tr and Gs were not significant between NM and AM plants.
The application of 200 mM NaCl reduced the Ci in NM and AM plants (Figure 2C), although the Ci was higher in NM plants than in AM plants at all salinity levels.
The increases in salinity had little effect on the WUE (Figure 2E). However, the WUE of AM plants was higher than that of NM plants at 0 and 100 mM NaCl.
Chlorophyll Fluorescence Parameters
Increased salinity reduced the Fv/Fm, ΦPSII, Fv′/Fm′ and qP in NM plants; however, in AM plants, increased salinity did not cause a significant decrease in these four parameters (Figures 3A–D). Values of Fv/Fm, ΦPSII and Fv′/Fm′ were higher in AM plants than in NM plants under saline conditions, and qP was increased by AM symbiosis at 200 mM NaCl.
FIGURE 3.
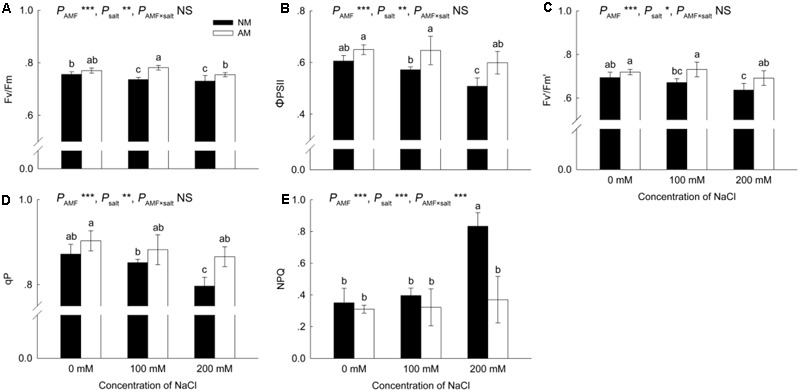
Effects of salinity and R. irregularis inoculation on maximum quantum efficiency of PSII photochemistry (Fv/Fm) (A), PSII operating efficiency (ΦPSII) (B), PSII maximum efficiency (Fv′/Fm′) (C), photochemical quenching coefficient (qP) (D) and non-photochemical quenching (NPQ) (E) in leaves of R. pseudoacacia. Different letters indicate significant differences (P < 0.05). NS not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. AM, arbuscular mycorrhizal; NM, non-mycorrhizal.
The NPQ was similar in NM and AM plants under 0 and 100 mM NaCl, but at 200 mM NaCl, the NPQ increased in NM plants and was higher (125%) than that in AM plants (Figure 3E).
Leaf Water Status
The RWC declined with exposure to salinity (Figure 4A). In NM plants, the reduction in the RWC ranged from 12% at 100 mM NaCl to 17% at 200 mM NaCl, whereas in AM plants, the reduction was 11 and 12% under 100 and 200 mM NaCl, respectively. AM fungi led to a 9% increase in the RWC at 200 mM NaCl.
FIGURE 4.
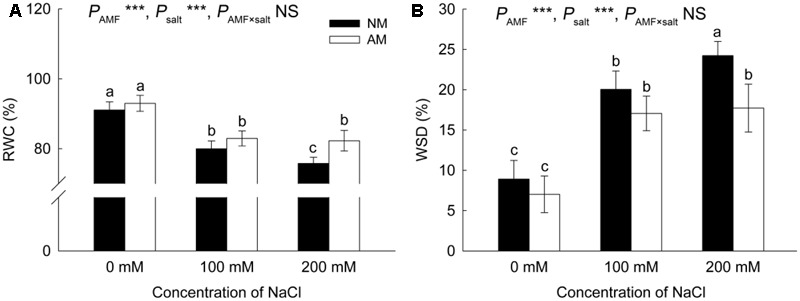
Effects of salinity and R. irregularis inoculation on relative water content (RWC) (A) and water saturation deficit (WSD) (B) in leaves of R. pseudoacacia. Different letters indicate significant differences (P < 0.05). NS not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. AM, arbuscular mycorrhizal; NM, non-mycorrhizal.
The WSD increased when plants were exposed to salt stress (Figure 4B). Under the 200 mM NaCl treatment, the WSD was lower (27%) in AM plants than in NM plants.
Concentration of Na+ and K+, K+/Na+ Ratio, and Shoot/Root Na+ Ratio
Salinity increased the Na+ concentration in leaves and roots of both NM and AM plants (Figures 5A,B). Under non-saline conditions, AM plants accumulated a similar amount of Na+ in leaves to NM plants, however, under saline conditions, AM plants accumulated less Na+ in leaves than NM plants. Meanwhile, in roots, Na+ accumulation was similar in NM and AM plants at all salinity levels.
FIGURE 5.
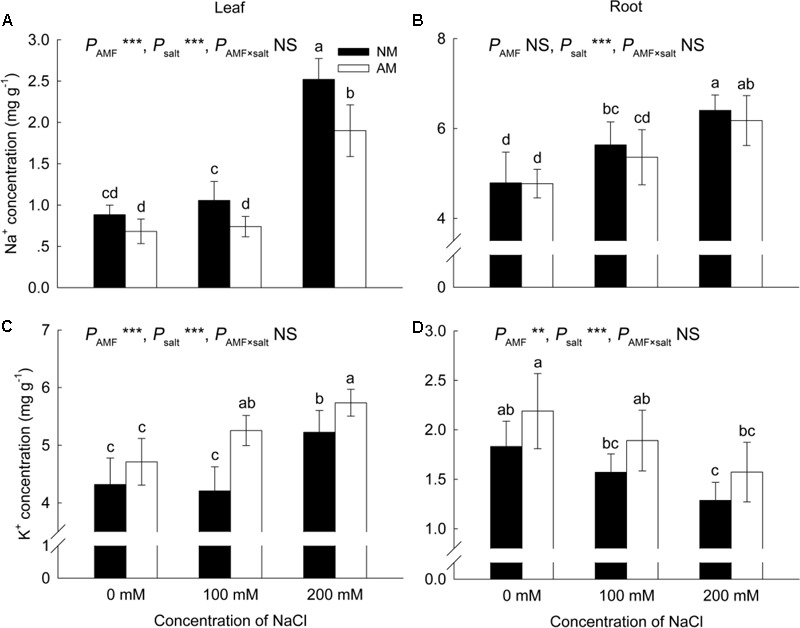
Effects of salinity and R. irregularis inoculation on concentration of Na+ (A,B) and K+ (C,D) in leaves and roots of R. pseudoacacia. Different letters indicate significant differences (P < 0.05). NS not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. AM, arbuscular mycorrhizal; NM, non-mycorrhizal.
Upon increasing salinity, the concentration of K+ decreased in the roots of both NM and AM plants, but increased in the leaves of both plants (Figures 5C,D). In leaves, K+ accumulation was similar in NM and AM plants under non-saline conditions, but under saline conditions, the K+ concentration in AM plants was higher than that in NM plants. Meanwhile in roots, AM plants accumulated a similar amount of K+ as NM plants at all salinity levels.
Salinity negatively affected the K+/Na+ ratio in the leaves and roots of NM and AM plants (Figures 6A,B). In roots, the K+/Na+ ratio was similar in NM and AM plants at all salinity levels, while in leaves, the ratio was improved by AM symbiosis at 0 and 100 mM NaCl.
FIGURE 6.
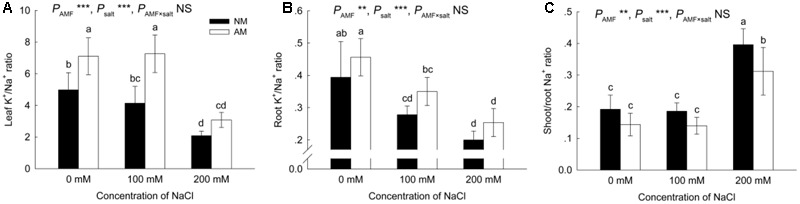
Effects of salinity and R. irregularis inoculation on leaf (A) and root (B) K+/Na+ ratio and shoot/root Na+ ratio (C) in R. pseudoacacia. Different letters indicate significant differences (P < 0.05). NS not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. AM, arbuscular mycorrhizal; NM, non-mycorrhizal.
The shoot/root Na+ ratio increased upon exposure to salinity (Figure 6C), and AM symbiosis reduced the shoot/root Na+ ratio at 200 mM NaCl.
Expression of RppsbA, RppsbD, RprbcL, and RprbcS
The expression of RppsbA and RppsbD was downregulated in NM and AM plants in the salinity treatments (Figure 7). AM fungi upregulated the expression of RppsbA under non-saline and saline conditions, and the expression of RppsbD was upregulated by mycorrhizae at 100 mM NaCl.
FIGURE 7.

Effects of salinity and R. irregularis inoculation on expression of RppsbA, RppsbD, RprbcL, and RprbcS in leaves of R. pseudoacacia. Expression of the R. pseudoacacia actin gene was used as an internal control for normalization. Different letters within each gene indicate significant differences (P < 0.05). AM, arbuscular mycorrhizal; NM, non-mycorrhizal.
Salinity also downregulated the expression of RprbcL and RprbcS in NM plants, but only reduced the expression of RprbcL in AM plants (Figure 7). The expression level of RprbcL was higher in AM plants than in NM plants at 0 and 100 mM NaCl, whereas the expression of RprbcS was similar in NM and AM plants at all salinity levels.
Expression of Aquaporin Genes
In the roots of NM plants, almost all aquaporin genes tested (except for RpPIP1;1, which was unaffected by salinity) were downregulated in response to salinity (Figure 8A), whereas in the roots of AM plants, the expression was reduced less (RpTIP1;3), was unaffected (RpPIP1;1 and RpTIP2;1) or was even increased (RpPIP1;3, RpPIP2;1, and RpTIP1;1) with increasing salinity. Under non-saline conditions, mycorrhizal fungi caused a downregulation of almost all aquaporin genes (except for RpPIP1;1) in roots. However, at 200 mM NaCl, the expression of four aquaporins (RpPIP1;1, RpPIP1;3, RpPIP2;1, and RpTIP1;1) was higher in AM plants than in NM plants.
FIGURE 8.
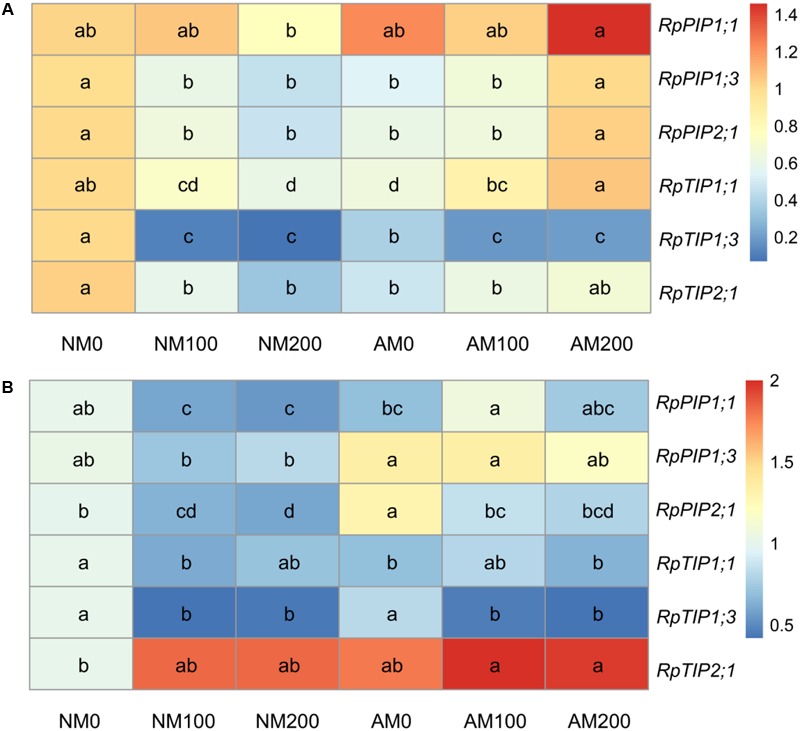
Effects of salinity and R. irregularis inoculation on expression of aquaporin genes in roots (A) and leaves (B) of R. pseudoacacia. Expression of the R. pseudoacacia actin gene was used as an internal control for normalization. Different letters within each gene indicate significant differences (P < 0.05). AM, arbuscular mycorrhizal; NM, non-mycorrhizal.
In the leaves of NM plants, the expression of RpPIP1;1, RpPIP2;1, RpTIP1;1, and RpTIP1;3 was downregulated by the application of 100 or 200 mM NaCl, while in the leaves of AM plants, salinity only reduced the expression of RpPIP2;1 and RpTIP1;3 (Figure 8B). The opposite trend was observed in the leaf expression of RpPIP1;1, which was upregulated by the application of 100 mM NaCl in AM plants. Under non-saline conditions, mycorrhizal fungi increased the leaf expression of RpPIP2;1, whereas the expression of RpTIP1;1 decreased in response to mycorrhizae. Under saline conditions, the upregulation in leaf expression of RpPIP1;1 and RpPIP1;3 by mycorrhizae was observed at 100 mM NaCl. No significant differences in the leaf expression of RpTIP1;3 or RpPIP2;1 were found between NM and AM plants.
Expression of RpSOS1, RpHKT1, RpNHX1, and RpSKOR
In roots, salinity upregulated the expression of RpSOS1 and RpHKT1 in AM plants but only increased the expression of RpSOS1 in NM plants (Figure 9A). Salinity decreased the expression of RpSKOR in the roots of NM plants, but had no effect on the expression this gene in the roots of AM plants. Mycorrhizal colonization upregulated the expression of RpSOS1 and RpSKOR under saline conditions, and increased the expression of RpHKT1 at 200 mM NaCl. Neither salinity nor AM symbiosis had a significant effect on the expression of RpNHX1 in roots.
FIGURE 9.
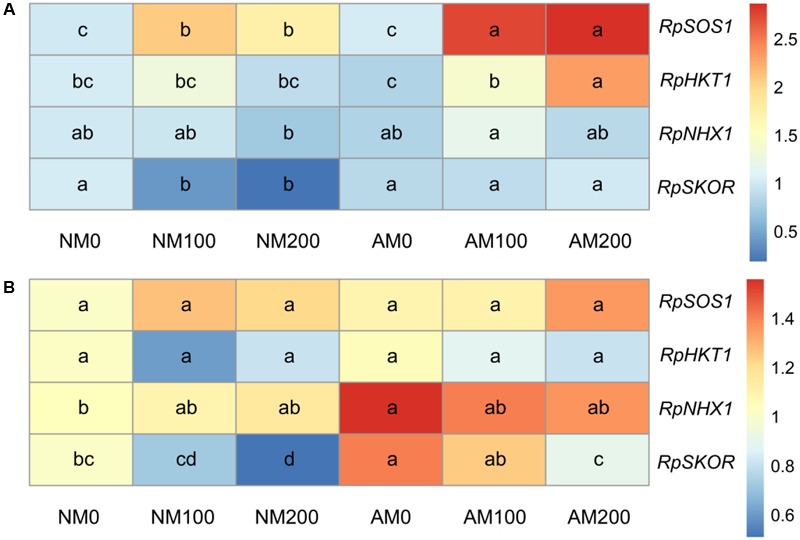
Effects of salinity and R. irregularis inoculation on expression of RpSOS1, RpHKT1, RpNHX1, and RpSKOR in roots (A) and leaves (B) of R. pseudoacacia. Expression of the R. pseudoacacia actin gene was used as an internal control for normalization. Different letters within each gene indicate significant differences (P < 0.05). AM, arbuscular mycorrhizal; NM, non-mycorrhizal.
In leaves, salinity downregulated the expression RpSKOR in NM and AM plants, but did not affect the expression of RpSOS1, RpHKT1, or RpNHX1 in both plants (Figure 9B). AM symbiosis upregulated the expression of RpSKOR at all salinity levels, and enhanced the expression of RpNHX1 under non-saline conditions. No significant differences in the leaf expression of RpSOS1 or RpHKT1 were found between NM and AM plants.
Discussion
Salinity decreases the colonization capacity of AM fungi by suppressing hyphal growth, sporulation, and spore germination (Juniper and Abbott, 2006; Jahromi et al., 2008). In the present research, salinity caused a reduction in mycorrhizal colonization, consistent with previous studies on lettuce and Trigonella foenum-graecum (Aroca et al., 2013; Evelin and Kapoor, 2014).
Plant biomass production is the most obvious trait reflecting plant performance under abiotic stress and the symbiosis efficiency of AM fungi (Evelin et al., 2009; Ruiz-Lozano et al., 2012; Porcel et al., 2016; Zhang et al., 2017). In the present study, although the dry masses of the shoots and roots of both NM and AM plants were reduced by salt stress, AM plants grew better than NM plants at all salinity levels, which suggests that the AM symbiosis mitigated salt stress in black locust and indicates a high symbiosis efficiency of R. irregularis. Beneficial influence of AM symbiosis on biomass production in plants exposed to salt stress are also found in other plant species, including tomato, rice, lettuce, and maize (Hajiboland et al., 2010; Sheng et al., 2011; Aroca et al., 2013; Porcel et al., 2016).
The reduction in plant growth under saline conditions is attributed to a reduction in photosynthetic capacity caused by salinity (Porcel et al., 2015). In the present study, although salinity reduced the Pn in both NM and AM plants, the Pn of AM plants was higher than that of NM plants under saline conditions, which correlated with the improved biomass production in the shoots and roots of AM plants exposed to saline conditions. Salt stress affects CO2 diffusion in leaves through a decline in stomatal and mesophyll conductance (Chaves et al., 2009), and the Pn enhancement by mycorrhizal fungi is likely related to a higher Gs in AM plants than in NM plants (Hajiboland et al., 2010; Porcel et al., 2015). However, Wu et al. (2015) showed that under saline conditions, mycorrhizal fungi improved the Pn in male Populus cathayana but did not significantly improve the Gs in host plants, although the Gs decreased less in AM plants compared with that in NM plants when salinity increased (Wu et al., 2015). Consistent with the results of Wu et al. (2015), we also observed less reduction in the Gs in AM plants than in NM plants with increasing levels of salinity and similar Gs values in AM and NM plants under saline conditions. The lower reduction in Gs in AM plants than in NM plants with increasing salinity suggests CO2 diffusion through the stomata was diminished less, hence the water status may be better in AM plants. Moreover, in this study, AM plants had a lower Ci than NM plants under all salinity levels, which is consistent with a previous study of AM-positive effects on plant photosynthesis by Sheng et al. (2008). Under saline conditions, a high Ci may indicate the destruction of the photosynthetic apparatus and passivation of the enzymes, which could both decrease CO2 assimilation and induce CO2 accumulation in intercellular areas (Sheng et al., 2008). In the present study, RprbcL expression was upregulated in AM plants at 0 and 100 mM NaCl. The RprbcL gene encodes the large subunit of rubisco, which catalyzes the first step of photosynthetic carbon assimilation in C3 plants (Sudhir and Murthy, 2004), and its upregulation may contribute to improved CO2 assimilation. Thus, the lower Ci in AM plants might be a consequence of a lower metabolic limitation of CO2 assimilation in these plants compared with the NM plants.
Photochemical reactions reflect the photosynthetic capacity and energy use efficiency in plants and are sensitive to abiotic stresses (Hu et al., 2017). A correlation between maintenance of PSII activity and adaptation to abiotic stresses has been reported in previous studies (Sheng et al., 2008; Hajiboland et al., 2010; Porcel et al., 2016; Hu et al., 2017). For the chlorophyll fluorescence parameters measured in this study, AM symbiosis had a positive effect on the Fv/Fm and ΦPSII under saline conditions, and on the qP in the presence of 200 mM NaCl, which implies that AM plants had a less stressed PSII, a higher quantum efficiency of PSII photochemistry and a greater ability to drive electron transport in an excited PSII reaction center under the stress conditions (Baker, 2008). Moreover, the NPQ increased in NM plants with increasing salinity but was unaffected in AM plants. Normally, an increase in the NPQ is a photoprotective mechanism used to dissipate the excess excitation energy as heat, but this increase can lead to a decrease in the quantum efficiency of PSII photochemistry (Baker, 2008). Therefore, AM symbiosis increased the use of absorbed light in photochemical processes with minimal dissipation of light energy (Hu et al., 2017). In addition, mycorrhizal colonization upregulated the expression of RppsbA at all salinity levels, and increased the expression of RppsbD at 100 mM NaCl. This upregulation by AM symbiosis may make AM plants more capable of repairing PSII following degradation of D1 and D2 proteins by salinity. Thus, our results suggest that plants inoculated with AM fungi were more tolerant to salt stress due to a higher PSII efficiency under saline conditions, which increased photosynthetic capacity in these plants.
The beneficial effects of AM symbiosis on plant growth and photosynthesis can also be ascribed to an improvement in water status with mycorrhizal colonization. A positive effect on the RWC due to AM symbiosis in the 200 mM NaCl treatment was observed in this study. Moreover, the lower reduction in RWC in AM plants exposed to salinity correlated with the lower reduction in Gs in these plants. Given that aquaporins mediate root hydraulic conductance and plant water homeostasis (Luu and Maurel, 2005; Sade et al., 2010), mycorrhizal regulation of aquaporin genes could be expected.
Plasma membrane intrinsic proteins are expressed in abundance in roots and regulate most of the water absorption by roots, whereas TIPs regulate water exchange between the vacuole and cytosol and are important in cellular osmoregulation (Luu and Maurel, 2005). Downregulation of PIPs and TIPs under salt stress may be a mechanism to reduce membrane permeability for water and to conserve cellular water (Jang et al., 2004; He et al., 2016). In the present study, we also observed decreased expression of aquaporin genes in both the leaves and roots of NM plants, as well as RpPIP2;1 (in leaves) and RpTIP1;3 (in leaves and roots) in AM plants under salt stress. However, salinity also resulted in the upregulation of one TIP (RpTIP1;1 at 100 and 200 mM NaCl) and two PIPs (RpPIP1;3 and RpPIP2;1 at 200 mM NaCl) in AM roots, and one PIP (RpPIP1;1 at 100 mM NaCl) in AM leaves. The upregulation of PIPs under saline conditions likely assists in water uptake to sustain water homeostasis in cells with high salinity levels (Jang et al., 2004), and the upregulation of TIPs under such conditions may facilitate water transport from the cytosol to the vacuole to increase osmotic pressure (Hu et al., 2017). Variable responses by different members of the aquaporins to salt or drought stress have been observed in previous studies (Jang et al., 2004; Ouziad et al., 2006; Bárzana et al., 2014; He et al., 2016), which may reflect the complexity of the transcriptional regulation of different members of the large aquaporin family (Ouziad et al., 2006).
In the present study, AM fungi regulated several aquaporin genes in both leaves and roots. The root expression of RpPIP1;1, RpPIP1;3, RpPIP2;1, and RpTIP1;1 was upregulated by AM symbiosis at 200 mM NaCl. In previous studies, under short-term drought (Bárzana et al., 2014) and salt stress (Aroca et al., 2007), an increase in the root expression of PIPs and TIPs by mycorrhizae was correlated with higher free-exuded sap flow rate and osmotic root hydraulic conductance in AM plants than in NM plants. Therefore, the upregulation of aquaporin genes by mycorrhizal fungi at 200 mM NaCl may reflect the stress conditions under which AM roots take up more water than NM roots, which would contribute to the improved water status of AM plants compared with NM plants. In the present study, mycorrhizal fungi increased the leaf expression of RpPIP1;1 and RpPIP1;3 at 100 mM NaCl. Aquaporins can transport CO2 and are involved in CO2 diffusion across the plasma membrane of mesophyll cells (Saibo et al., 2009; Yang and Cui, 2009). In plants overexpressing NtAQP1, the membrane permeability for CO2 diffusion was increased (Uehlein et al., 2003), while the presence of HgCl2, which inhibits most aquaporins, reduced the permeability (Terashima and Ono, 2002). Therefore, the upregulation of aquaporin genes by mycorrhizal fungi in leaves suggests that AM plants have a better supply of water from the roots (Ouziad et al., 2006) and a higher mesophyll permeability for CO2 diffusion, which would reduce CO2 accumulation in intercellular areas and contribute to a higher Pn in these plants compared with NM plants.
Salt tolerance in plants depends to a great extent on the absorption and distribution of Na+ within the plant (Porcel et al., 2016). Previous studies reported a reduction in Na+ accumulation (Talaat and Shawky, 2011; Garcia et al., 2017) and reduced root-to-shoot allocation of Na+ (Evelin et al., 2012; Porcel et al., 2016) in AM plants. We also found that AM plants accumulated less Na+ in leaves compared with NM plants under salt conditions, concomitantly with a reduced shoot/root Na+ ratio in AM plants at 200 mM NaCl. Preventing Na+ transport from roots to shoots is a strategy for protecting photosynthetic organs against the toxic effects of Na+ (Zhu et al., 2016). Ion selection can be controlled by AM fungi during nutrient absorption from the soil (Hammer et al., 2010). In the present study, leaf K+ levels were significantly higher in AM plants than in NM plants under saline conditions, and the leaf K+/Na+ ratio was improved by AM symbiosis at 0 and 100 mM NaCl. Increased accumulation of K+ and reduced Na+ concentration in AM plants have also been reported in other studies (Evelin et al., 2012; Garcia et al., 2017). K+ is a macronutrient that is essential for many plant processes including osmoregulation, protein biosynthesis, enzyme activation, and photosynthesis (Shabala and Cuin, 2008; Wang and Wu, 2013). Na+ competes with K+ for metabolic processes requiring K+ (Shabala and Cuin, 2008), but Na+ cannot replace the function of K+ (Estrada et al., 2013). Thus, maintenance of a higher concentration of K+ and a higher K+/Na+ ratio in AM plants exposed to salinity was presumably crucial for salt tolerance.
Based on the better performance of AM plants in reducing the shoot/root Na+ ratio and in improving the K+/Na+ ratio in leaves, we decided to investigate the response of four genes involved in K+ and/or Na+ uptake, translocation, or compartmentalization. The results showed that the root expression of RpSOS1 was upregulated under increased salinity in both NM and AM plants, and the expression was also enhanced by AM symbiosis under saline conditions. Upregulation of RpSOS1 in roots promotes Na+ export back to the soil or to the apoplastic spaces (Shi et al., 2002). As a consequence, AM plants may be more capable of reducing Na+ influx into roots and/or restricting the toxicity of Na+, since Na+ is less toxic in the apoplastic spaces. In the present study, we also observed that mycorrhizal colonization increased the root expression of RpHKT1 at 200 mM NaCl, which correlated with the lower shoot/root Na+ ratio in AM plants at 200 mM NaCl and suggests that unloading of Na+ from the xylem was enhanced in AM plants in order to prevent allocation of the cation to the shoots (Deinlein et al., 2014). The upregulation of RpSOS1 and RpHKT1 in roots by AM symbiosis contributed to reduced Na+ accumulation in the leaves of AM plants. The results of the present study also showed that the root expression of RpSKOR was increased by AM symbiosis under saline conditions. This upregulation increased K+ loading into the xylem (Zhang et al., 2017) and correlated with improved K+ accumulation in the leaves of AM plants observed in the present study. In addition, the enhanced expression of RpSKOR in combination with RpSOS1 and RpHKT1 in AM plants can account for the higher K+/Na+ ratio in the leaves of these plants. Curiously, AM symbiosis had on effect on the expression of RpNHX1 in leaves or roots under saline conditions, consistent with research on tomato by Ouziad et al. (2006). However, Porcel et al. (2016) found that AM symbiosis considerably upregulated OsNHX3 expression in leaves under saline conditions. Indeed, in rice, tonoplast-localized OsNHX1 - 4 function as vacuolar (Na+, K+)/H+ antiporters (Porcel et al., 2016). Therefore, the regulation of additional genes encoding for NHX proteins in R. pseudoacacia by mycorrhizal fungi requires further exploration in order to illustrate their contribution to Na+ compartmentalization in AM plants. Interestingly, it seems that mycorrhizal colonization had a more evident influence on the expression of RpSOS1 and RpHKT1 in roots than in leaves, since no significant induction by AM symbiosis was detected in leaves. This is inconsistent with the study on rice plants by Porcel et al. (2016) in which the expression of genes involved in Na+ uptake or translocation in leaves was influenced by AM symbiosis to a greater extent than that in roots, and this apparent contradiction may be a consequence of the different plant and fungi species used in the experiments.
Conclusion
Arbuscular mycorrhizal fungi improved salt tolerance of black locust by increasing photosynthetic capacity, water status and K+/Na+ homeostasis, as well as reducing the root-to-shoot allocation of Na+. The improvement in photosynthesis by AM fungi was related to a lower reduction in Gs, a higher PSII efficiency and higher expression of three chloroplastic genes (RprbcL, RppsbA, and RppsbD) compared with NM plants. Mycorrhizal fungi regulated the expression of several aquaporin genes, depending on the salinity level applied and the plant tissue. Under saline conditions, the upregulation of aquaporin genes by AM symbiosis in leaves and roots might facilitate water uptake by roots, water transport to special tissues and CO2 diffusion across the plasma membrane. AM symbiosis also facilitated Na+ secretion from root cells, Na+ unloading from the xylem, and K+ translocation to the shoots. This is associated with the upregulation of RpSOS1, RpHKT1, and RpSKOR expression in roots by AM fungi under stress conditions. The beneficial effects of AM symbiosis on the photosynthetic capacity, water status and ionic homeostasis resulted in the improved growth performance of black locust exposed to salt stress.
Author Contributions
MT conceived the study. JC conducted the experiments, analyzed the data and drafted manuscript. XZ participated in the experiment. HZ and MT reviewed and edited the manuscript. All authors read and approved the manuscript for final submission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (41671268, http://www.nsfc.gov.cn/), the Shaanxi Science and Technology Innovation Project Plan (2016KTCL02-07).
Abbreviations
- AM
arbuscular mycorrhizal
- Ci
intercellular CO2 concentration
- DW
dry mass
- Fv/Fm
maximum quantum efficiency of PSII photochemistry
- Fv′/Fm′
PSII maximum efficiency
- FW
fresh weight
- Gs
stomatal conductance
- NM
non-mycorrhizal
- NPQ
non-photochemical quenching
- Pn
net photosynthetic rate
- qP
photochemical quenching coefficient
- RWC
relative water content
- Tr
transpiration rate
- TW
turgid weight
- WSD
water saturation deficit
- WUE
water use efficiency
- ΦPSII
PSII operating efficiency
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01739/full#supplementary-material
References
- Allakhverdiev S. I., Murata N. (2008). Salt stress inhibits photosystems II and I in cyanobacteria. Photosynth. Res. 98 529–539. 10.1007/s11120-008-9334-x [DOI] [PubMed] [Google Scholar]
- Aroca R., Porcel R., Ruiz-Lozano J. M. (2007). How does arbuscular mycorrhizal symbiosis regulate root hydraulic properties and plasma membrane aquaporins in Phaseolus vulgaris under drought, cold or salinity stresses? New Phytol. 173 808–816. 10.1111/j.1469-8137.2006.01961.x [DOI] [PubMed] [Google Scholar]
- Aroca R., Ruiz-Lozano J. M., Zamarreño Á. M., Paz J. A., García-Mina J. M., Pozo M. J., et al. (2013). Arbuscular mycorrhizal symbiosis influences strigolactone production under salinity and alleviates salt stress in lettuce plants. J. Plant Physiol. 170 47–55. 10.1016/j.jplph.2012.08.020 [DOI] [PubMed] [Google Scholar]
- Baker N. R. (2008). Chlorophyll fluorescence, a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59 89–113. 10.1146/annurev.arplant.59.032607.092759 [DOI] [PubMed] [Google Scholar]
- Bárzana G., Aroca R., Bienert G. P., Chaumont F., Ruiz-Lozano J. M. (2014). New insights into the regulation of aquaporins by the arbuscular mycorrhizal symbiosis in maize plants under drought stress and possible implications for plant performance. Mol. Plant Microbe Interact. 27 349–363. 10.1094/MPMI-09-13-0268-R [DOI] [PubMed] [Google Scholar]
- Chaves M. M., Flexas J., Pinheiro C. (2009). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103 551–560. 10.1093/aob/mcn125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport R. J., Muñoz-Mayor A., Jha D., Essah P. A., Rus A., Tester M. (2007). The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 30 497–507. 10.1111/j.1365-3040.2007.01637.x [DOI] [PubMed] [Google Scholar]
- Deinlein U., Stephan A. B., Horie T., Luo W., Xu G., Schroeder J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19 371–379. 10.1016/j.tplants.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B., Aroca R., Maathuis F. J. M., Barea J. M., Ruiz-Lozano J. M. (2013). Arbuscular mycorrhizal fungi native from a Mediterranean saline area enhance maize tolerance to salinity through improved ion homeostasis. Plant Cell Environ. 36 1771–1782. 10.1111/pce.12082 [DOI] [PubMed] [Google Scholar]
- Evelin H., Giri B., Kapoor R. (2012). Contribution of Glomus intraradices inoculation to nutrient acquisition and mitigation of ionic imbalance in NaCl-stressed Trigonella foenum-graecum. Mycorrhiza 22 203–217. 10.1007/s00572-011-0392-0 [DOI] [PubMed] [Google Scholar]
- Evelin H., Kapoor R. (2014). Arbuscular mycorrhizal symbiosis modulates antioxidant response in salt-stressed Trigonella foenum-graecum plants. Mycorrhiza 24 197–208. 10.1007/s00572-013-0529-4 [DOI] [PubMed] [Google Scholar]
- Evelin H., Kapoor R., Giri B. (2009). Arbuscular mycorrhizal fungi in alleviation of salt stress: a review. Ann. Bot. 104 1263–1280. 10.1093/aob/mcp251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. F. (2006). Experimental Guidance for Plant Physiology 1st Edn. Beijing: Higher Education Press. [Google Scholar]
- Garcia K., Chasman D., Roy S., Ane J. M. (2017). Physiological responses and gene co-expression network of mycorrhizal roots under K+ deprivation. Plant Physiol. 173 1811–1823. 10.1104/pp.16.01959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti M., Mosse B. (1980). An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84 489–500. 10.1111/j.1469-8137.1980.tb04556.x [DOI] [Google Scholar]
- Gu J., Weina L., Akinnagbe A., Wang J., Jia L., Yang M. (2012). Effect of salt stress on genetic diversity of Robinia pseudoacacia seedlings. Afr. J. Biotechnol. 11 1838–1847. 10.5897/AJB11.1369 [DOI] [Google Scholar]
- Hajiboland R., Aliasgharzadeh N., Laiegh S. F., Poschenrieder C. (2010). Colonization with arbuscular mycorrhizal fungi improves salinity tolerance of tomato (Solanum lycopersicum L.) plants. Plant Soil 331 313–327. 10.1007/s11104-009-0255-z [DOI] [Google Scholar]
- Hammer E. C., Nasr H., Pallon J., Olsson P. A., Wallander H. (2010). Elemental composition of arbuscular mycorrhizal fungi at high salinity. Mycorrhiza 21 117–129. 10.1007/s00572-010-0316-4 [DOI] [PubMed] [Google Scholar]
- He F., Sheng M., Tang M. (2017). Effects of Rhizophagus irregularis on photosynthesis and antioxidative enzymatic system in Robinia pseudoacacia L. under drought stress. Front. Plant Sci. 8:183. 10.3389/fpls.2017.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Zhang H., Tang M. (2016). Aquaporin gene expression and physiological responses of Robinia pseudoacacia L. to the mycorrhizal fungus Rhizophagus irregularis and drought stress. Mycorrhiza 26 311–323. 10.1007/s00572-015-0670-3 [DOI] [PubMed] [Google Scholar]
- Hu W., Zhang H., Chen H., Tang M. (2017). Arbuscular mycorrhizas influence Lycium barbarum tolerance of water stress in a hot environment. Mycorrhiza 27 451–463. 10.1007/s00572-017-0765-0 [DOI] [PubMed] [Google Scholar]
- Huang L., Zhang H., Song Y., Yang Y., Chen H., Tang M. (2017). Subcellular compartmentalization and chemical forms of lead participate in lead tolerance of Robinia pseudoacacia L. with Funneliformis mosseae. Front. Plant Sci. 8:517. 10.3389/fpls.2017.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahromi F., Aroca R., Porcel R., Ruiz-Lozano J. M. (2008). Influence of salinity on the in vitro development of Glomus intraradices and on the in vivo physiological and molecular responses of mycorrhizal lettuce plants. Microb. Ecol. 55 45–53. 10.1007/s00248-007-9249-7 [DOI] [PubMed] [Google Scholar]
- Jang J. Y., Kim D. G., Kim Y. O., Kim J. S., Kang H. (2004). An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol. Biol. 54 713–725. 10.1023/B:PLAN.0000040900.61345.a6 [DOI] [PubMed] [Google Scholar]
- Jansen M. A. K., Gaba V., Greenberg B. M., Mattoo A. K., Edelman M. (1996). Low threshold levels of ultraviolet-B in a background of photosynthetically active radiation trigger rapid degradation of the D2 protein of photosystem-II. Plant J. 9 693–699. 10.1046/j.1365-313X.1996.9050693.x [DOI] [Google Scholar]
- Jin T., Liu G., Fu B., Ding X., Yang L. (2011). Assessing adaptability of planted trees using leaf traits: a case study with Robinia pseudoacacia L. in the Loess Plateau, China. Chin. Geogr. Sci. 21 290–303. 10.1007/s11769-011-0470-4 [DOI] [Google Scholar]
- Juniper S., Abbott L. K. (2006). Soil salinity delays germination and limits growth of hyphae from propagules of arbuscular mycorrhizal fungi. Mycorrhiza 16 371–379. 10.1007/s00572-006-0046-9 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu C., Vonshak A. (2002). Effects of salinity stress on photosystem II function in cyanobacterial Spirulina platensis cells. Physiol. Plant. 114 405–413. 10.1034/j.1399-3054.2002.1140310.x [DOI] [PubMed] [Google Scholar]
- Luu D. T., Maurel C. (2005). Aquaporins in a challenging environment: molecular gears for adjusting plant water status. Plant Cell Environ. 28 85–96. 10.1111/j.1365-3040.2004.01295.x [DOI] [Google Scholar]
- Maurel C., Verdoucq L., Luu D. T., Santoni V. (2008). Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59 595–624. 10.1146/annurev.arplant.59.032607.092734 [DOI] [PubMed] [Google Scholar]
- Meng F., Luo Q., Wang Q., Zhang X., Qi Z., Xu F., et al. (2016). Physiological and proteomic responses to salt stress in chloroplasts of diploid and tetraploid black locust (Robinia pseudoacacia L.). Sci. Rep. 6:23098. 10.1038/srep23098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelam S., Subramanyam R. (2013). Alteration of photochemistry and protein degradation of photosystem II from Chlamydomonas reinhardtii under high salt grown cells. J. Photochem. Photobiol. B Biol. 124 63–70. 10.1016/j.jphotobiol.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Ouziad F., Wilde P., Schmelzer E., Hildebrandt U., Bothe H. (2006). Analysis of expression of aquaporins and Na+/H+ transporters in tomato colonized by arbuscular mycorrhizal fungi and affected by salt stress. Environ. Exp. Bot. 57 177–186. 10.1016/j.envexpbot.2005.05.011 [DOI] [Google Scholar]
- Phillips J. M., Hayman D. S. (1970). Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 55 158–161. 10.1016/S0007-1536(70)80110-3 [DOI] [Google Scholar]
- Pinheiro C., Chaves M. M. (2011). Photosynthesis and drought: Can we make metabolic connections from available data? J. Exp. Bot. 62 869–882. 10.1093/jxb/erq340 [DOI] [PubMed] [Google Scholar]
- Porcel R., Aroca R., Azcon R., Ruiz-Lozano J. M. (2016). Regulation of cation transporter genes by the arbuscular mycorrhizal symbiosis in rice plants subjected to salinity suggests improved salt tolerance due to reduced Na+ root-to-shoot distribution. Mycorrhiza 26 673–684. 10.1007/s00572-016-0704-5 [DOI] [PubMed] [Google Scholar]
- Porcel R., Aroca R., Ruiz-Lozano J. M. (2012). Salinity stress alleviation using arbuscular mycorrhizal fungi. A review. Agron. Sustain. Dev. 32 181–200. 10.1007/s13593-011-0029-x [DOI] [Google Scholar]
- Porcel R., Redondo-Gómez S., Mateos-Naranjo E., Aroca R., Garcia R., Ruiz-Lozano J. M. (2015). Arbuscular mycorrhizal symbiosis ameliorates the optimum quantum yield of photosystem II and reduces non-photochemical quenching in rice plants subjected to salt stress. J. Plant Physiol. 185 75–83. 10.1016/j.jplph.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Qiu Q. S., Guo Y., Dietrich M. A., Schumaker K. S., Zhu J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99 8436–8441. 10.1073/pnas.122224699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lozano J. M., Porcel R., Azcón C., Aroca R. (2012). Regulation by arbuscular mycorrhizae of the integrated physiological response to salinity in plants, new challenges in physiological and molecular studies. J. Exp. Bot. 63 4033–4044. 10.1093/jxb/ers126 [DOI] [PubMed] [Google Scholar]
- Sade N., Gebretsadik M., Seligmann R., Schwartz A., Wallach R., Moshelion M. (2010). The role of tobacco Aquaporin1 in improving water use efficiency, hydraulic conductivity, and yield production under salt stress. Plant Physiol. 152 245–254. 10.1104/pp.109.145854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibo N. J. M., Lourenco T., Oliveira M. M. (2009). Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann. Bot. 103 609–623. 10.1093/aob/mcn227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S., Cuin T. A. (2008). Potassium transport and plant salt tolerance. Physiol. Plant. 133 651–669. 10.1111/j.1399-3054.2007.01008.x [DOI] [PubMed] [Google Scholar]
- Sheng M., Tang M., Chen H., Yang B., Zhang F., Huang Y. (2008). Influence of arbuscular mycorrhizae on photosynthesis and water status of maize plants under salt stress. Mycorrhiza 18 287–296. 10.1007/s00572-008-0180-7 [DOI] [PubMed] [Google Scholar]
- Sheng M., Tang M., Zhang F., Huang Y. (2011). Influence of arbuscular mycorrhiza on organic solutes in maize leaves under salt stress. Mycorrhiza 21 423–430. 10.1007/s00572-010-0353-z [DOI] [PubMed] [Google Scholar]
- Shi H., Quintero F. J., Pardo J. M., Zhu J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14 465–477. 10.1105/tpc.010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. E., Read D. J. (2008). Mycorrhizal Symbiosis 3rd Edn. London: Academic Press. [Google Scholar]
- Sudhir P., Murthy S. D. S. (2004). Effects of salt stress on basic processes of photosynthesis. Photosynthetica 42 481–486. 10.1007/S11099-005-0001-6 [DOI] [Google Scholar]
- Sudhir P. R., Pogoryelov D., Kovács L., Garab G., Murthy S. D. S. (2005). The effects of salt stress on photosynthetic electron transport and thylakoid membrane proteins in the cyanobacterium Spirulina platensis. J. Biochem. Mol. Biol. 38 481–485. 10.5483/BMBRep.2005.38.4.481 [DOI] [PubMed] [Google Scholar]
- Sunarpi, Horie T., Motoda J., Kubo M., Yang H., Yoda K., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 44 928–938. 10.1111/j.1365-313X.2005.02595.x [DOI] [PubMed] [Google Scholar]
- Talaat N. B., Shawky B. T. (2011). Influence of arbuscular mycorrhizae on yield, nutrients, organic solutes, and antioxidant enzymes of two wheat cultivars under salt stress. J. Plant Nutr. Soil Sci. 174 283–291. 10.1002/jpln.201000051 [DOI] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Kanzaki N., Tamai S., Yamanaka N., Futai K. (2007). Does ectomycorrhizal fungal community structure vary along a Japanese black pine (Pinus thunbergii) to black locust (Robinia pseudoacacia) gradient? New Phytol. 173 322–334. 10.1111/j.1469-8137.2006.01910.x [DOI] [PubMed] [Google Scholar]
- Terashima I., Ono K. (2002). Effects of HgCl2 on CO2 dependence of leaf photosynthesis: evidence indicating involvement of aquaporins in CO2 diffusion across the plasma membrane. Plant Cell Physiol. 43 70–78. 10.1093/pcp/pcf001 [DOI] [PubMed] [Google Scholar]
- Tian C., He X., Zhong Y., Chen J. (2003). Effect of inoculation with ecto- and arbuscular mycorrhizae and Rhizobium on the growth and nitrogen fixation by black locust, Robinia pseudoacacia. New For. 25 125–131. 10.1023/A:1022675915627 [DOI] [Google Scholar]
- Uehlein N., Lovisolo C., Siefritz F., Kaldenhoff R. (2003). The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425 734–737. 10.1038/nature02027 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wu W. H. (2013). Potassium transport and signaling in higher plants. Annu. Rev. Plant Biol. 64 451–476. 10.1146/annurev-arplant-050312-120153 [DOI] [PubMed] [Google Scholar]
- Wu N., Li Z., Liu H., Tang M. (2015). Influence of arbuscular mycorrhiza on photosynthesis and water status of Populus cathayana Rehder males and females under salt stress. Acta Physiol. Plant. 37 183–196. 10.1007/s11738-015-1932-6 [DOI] [PubMed] [Google Scholar]
- Yang S., Cui L. (2009). The action of aquaporins in cell elongation, salt stress and photosynthesis. Chin. J. Biotechnol. 25 321–327. [PubMed] [Google Scholar]
- Yang Y., Tang M., Sulpice R., Chen H., Tian S., Ban Y. (2014). Arbuscular mycorrhizal fungi alter fractal dimension characteristics of Robinia pseudoacacia L. seedlings through regulating plant growth, leaf water status, photosynthesis, and nutrient concentration under drought stress. J. Plant Growth Regul. 33 612–625. 10.1007/s00344-013-9410-0 [DOI] [Google Scholar]
- Zhang H., Wei S., Hu W., Xiao L., Tang M. (2017). Arbuscular mycorrhizal fungus Rhizophagus irregularis increased potassium content and expression of genes encoding potassium channels in Lycium barbarum. Front. Plant Sci. 8:440. 10.3389/fpls.2017.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Shabala L., Cuin T. A., Huang X., Zhou M., Munns R., et al. (2016). Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J. Exp. Bot. 67 835–844. 10.1093/jxb/erv493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


