Abstract
Repair enzymes must communicate across hundreds of nucleotides to undo errors made during DNA replication. Imaging reveals that the enzymes do this by forming a series of ring-like clamps that diffuse along the DNA.
As we write up results for publication, any spelling errors are quickly identified by our computer’s built-in spellchecker. However, this is not an invention of the modern world; nature has been using DNA spellcheckers for millions of years to avoid genetic errors that arise during DNA replication, which it corrects through a process called mismatch repair. In the bacterium Escherichia coli, repair requires communication between enzymes across long stretches of DNA, and how this occurs has been hotly debated for decades1–3. On page 583, Liu et al.4 help to solve this mystery by using state-of-the-art techniques to analyse mismatch repair at the single-molecule level.
DNA exists as a double-stranded duplex, connected across the strands by complementary base-pairing — guanine (G) with cytosine (C) and adenine (A) with thymine (T). During DNA replication, the duplex unwinds, and each strand is copied using new nucleotides to create a freshly synthesized daughter strand. Errors occur when incorrect bases are incorporated into the daughter strand, creating a mismatch that requires subsequent repair to prevent mutations from arising. In E. coli, DNA with the base sequence GATC is normally tagged with a methyl group, but the newly synthesized DNA is temporarily unmethylated. These ‘hemi-methylated’ regions provide a means for mismatch-repair enzymes to distinguish between parent and daughter DNA.
Hemi-methylated sequences are also the sites at which the endonuclease enzyme MutH generates a single-strand break in the erroneous DNA strand during mismatch repair. Because of the rarity of GATC sequences, MutH-generated breaks can occur hundreds of bases from the site of the mismatch. Following breakage, the damaged strand is excised by an exonuclease enzyme that works from the break to the error, and the DNA is resynthesized to incorporate the correct base. A key aspect of this process is the coordination of mismatch detection with loading of the DNA-excision machinery, to ensure that excision occurs towards, rather than away from, the mismatch.
For the past decade, it has been thought5,6 that the protein MutS, which recognizes DNA mismatches, acts as a sliding clamp — a ring around DNA that can efficiently move up or down the two strands, enabling communication between the mismatch and the hemi-methylated sites. A third protein, MutL, activates the cutting activity of MutH. But how MutS links mismatch detection to the activation of MutH has been unclear.
To study initiation of mismatch repair, Liu et al. added fluorescent tags to MutH, MutS and MutL and analysed the movements of the individual proteins along single DNA molecules. They found that MutL is loaded onto DNA through interactions with a MutS molecule that has already located a mismatch. This observation builds on a structural study7 showing that the DNA duplex is positioned in the open channel of the MutS ring following mismatch verification, and that MutL binding sites on MutS are revealed during this process, permitting formation of the MutS–MutL complex.
Next, the authors confirmed another previous observation6 — that the MutS–MutL complex forms a single sliding clamp. Extending these observations, they showed that MutL can detach from MutS to act as a clamp on its own. In this role, MutL undertakes short excursions away from MutS. Surprisingly, some of these excursions pass behind MutS, an observation that Liu et al. took to imply that MutL might pass through the MutS channel. Therefore, MutL can be thought of as acting like a yo-yo, separately scouting the DNA. But scouting for what?
The answer comes from the group’s demonstration that MutL can recruit MutH to the DNA, thereby setting the stage for the excision phase of mismatch repair. The MutL–MutH complex behaves similarly to MutL alone, detaching from MutS and diffusing along DNA more quickly than when the three proteins are bound together — presumably in search of hemi-methylated sites. The authors propose that the MutS–MutL–MutH complex is the dominant species, because the sliding-clamp nature of MutS means that this complex is tethered to the DNA. However, efficient searching is also possible with MutL– MutH, which Liu and colleagues posit hops along the DNA.
Together, these observations might have solved the long-standing mystery of how the mismatch and excision sites interact, superseding previous models in which the DNA is looped through the MutS clamp or in which the clamp moves in a targeted manner along the duplex1,8. The model that emerges from these data suggests that MutS acts as a guiding clamp, sending out scouting clamps to search for hemi-methylated GATC sequences in a highly energy-efficient manner (Fig. 1).
Figure 1. Scouting for errors.
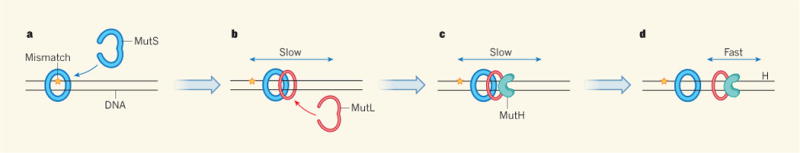
a, Following DNA replication, the protein MutS clamps around DNA at mismatches — sites of incorrect pairing between the two complementary DNA strands. b, Liu et al.4 report that, once a mismatch is detected, MutS recruits the protein MutL, and the two proteins slowly slide together up and down DNA. c, MutL in turn recruits the enzyme MutH, and the three move as one complex. d, MutL and MutH can detach from MutS to move rapidly along DNA in search of hemi-methylated sites (H) — sequences at which the parent strand is tagged with a methyl group but the mismatched daughter strand is not. The daughter strand is subsequently cleaved by MutH, initiating DNA excision and repair.
In 2015, Paul Modrich received the Nobel Prize in Chemistry for his seminal contributions to our understanding of mismatch repair1. However, many aspects of the process remain unresolved. One is that mismatch repair in eukaryotic cells (those that have a nucleus) involves more-complex protein– protein and protein–DNA interactions that have yet to be fully described. For instance, the eukaryotic equivalent of MutL has endonuclease activity, and eukaryotes use different mechanisms for excision depending on whether the break point is 3′ or 5′ of the mismatch1,3. Given this complexity, a better understanding of eukaryotic mismatch repair will surely present new mechanistic surprises. The use of single-molecule approaches to enable analysis of the eukaryotic process, already under way in several laboratories, will be central to elucidating these systems.
The principles outlined by Liu et al. might apply to enzymes involved in other types of DNA repair. A recent report9 revealed that the yeast protein Rad4, which is involved in nucleotide-excision repair, does not directly bind damaged sites, but instead diffuses for up to 1 kilobase around the damage, providing a dynamic platform for the recruitment of other repair proteins. Furthermore, the protein Mfd, which dislodges RNA polymerase enzymes that become stalled while transcribing DNA, then scouts ahead for DNA damage on the transcribed strand10.
The next chapter of the mismatch-repair story will surely see experiments that follow the entire process of repair, enabling observation of every protein in real time in a single assay. With single-molecule fluorescence techniques developing rapidly, such experiments are in sight, and should provide a molecular understanding of biology in vitro that can be adapted to systems in vivo.
Contributor Information
NEIL M. KAD, School of Biosciences, University of Kent, Canterbury CT2 7NJ, UK
BENNETT VAN HOUTEN, Department of Pharmacology and Chemical Biology, University of Pittsburgh, University of Pittsburgh Cancer Institute, Hillman Cancer Center, Pittsburgh, Pennsylvania 15213, USA.
References
- 1.Modrich P. Angew Chem Int Edn. 2016;55:8490–8501. doi: 10.1002/anie.201601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fishel R. J Biol Chem. 2015;290:26395–26403. doi: 10.1074/jbc.R115.660142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kunkel TA, Erie DA. Annu Rev Genet. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, et al. Nature. 2016;539:583–587. doi: 10.1038/nature20562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeong C, et al. Nature Struct Mol Biol. 2011;18:379–385. doi: 10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorman J, et al. Proc Natl Acad Sci USA. 2012;109:E3074–E3083. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groothuizen FS, et al. eLife. 2015;4:e06744. doi: 10.7554/eLife.06744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer RR, Pluciennik A, Burdett V, Modrich PL. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 9.Kong M, et al. Mol Cell. 2016;64:376–387. doi: 10.1016/j.molcel.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haines NM, Kim YIT, Smith AJ, Savery NJ. Proc Natl Acad Sci USA. 2014;111:4037–4042. doi: 10.1073/pnas.1322350111. [DOI] [PMC free article] [PubMed] [Google Scholar]


