Abstract
A recent phase 1 trial of the fatty acid amide hydrolase (FAAH) inhibitor BIA 10-2474 led to the death of one volunteer and produced mild-to-severe neurological symptoms in four others. Although the cause of the clinical neurotoxicity is unknown, it has been postulated, given the clinical safety profile of other tested FAAH inhibitors, that off-target activities of BIA 10-2474 may have played a role. Here, we use activity-based proteomic methods to determine the protein interaction landscape of BIA 10-2474 in human cells and tissues. This analysis revealed that the drug inhibits several lipases that are not targeted by PF04457845, a highly selective and clinically tested FAAH inhibitor. BIA 10-2474, but not PF04457845, produced substantial alterations in lipid networks in human cortical neurons, suggesting that promiscuous lipase inhibitors have the potential to cause metabolic dysregulation in the nervous system.
Main text
In January 2016, a first-in-human study of the fatty acid amide hydrolase (FAAH) inhibitor BIA 10-2474 led to the death of one volunteer and the hospitalization of four others (1–4). All patients manifested mild-to-severe neurological symptoms (3). FAAH is a membrane-bound serine hydrolase that degrades the endocannabinoid anandamide and related amidated lipids (5–8). Three explanations for the clinical neurotoxicity of BIA 10-2474 have been proposed: (i) errors may have occurred in the clinical trial itself, either in the manufacturing or handling of the compound or in the conduct of the trial; (ii) through its inhibitory effects on FAAH, BIA 10-2474 may have produced high levels of long-chain fatty acid amides (e.g., anandamide) and their oxygenated metabolites, which could potentially overstimulate cannabinoid CB1 (8), TRPV1 (9), and/or NMDA receptors (10); or (iii) BIA 10-2474 and/or its metabolites might have off-target activities. The first hypothesis was dismissed by the French authorities (4). The second hypothesis is considered unlikely because other FAAH inhibitors, such as PF04457845, have exhibited favorable safety profiles in Phase 1 and 2 clinical trials (11, 12). The third hypothesis has not been directly evaluated, because little or no information is available regarding the protein interaction profile of BIA 10-2474 (1).
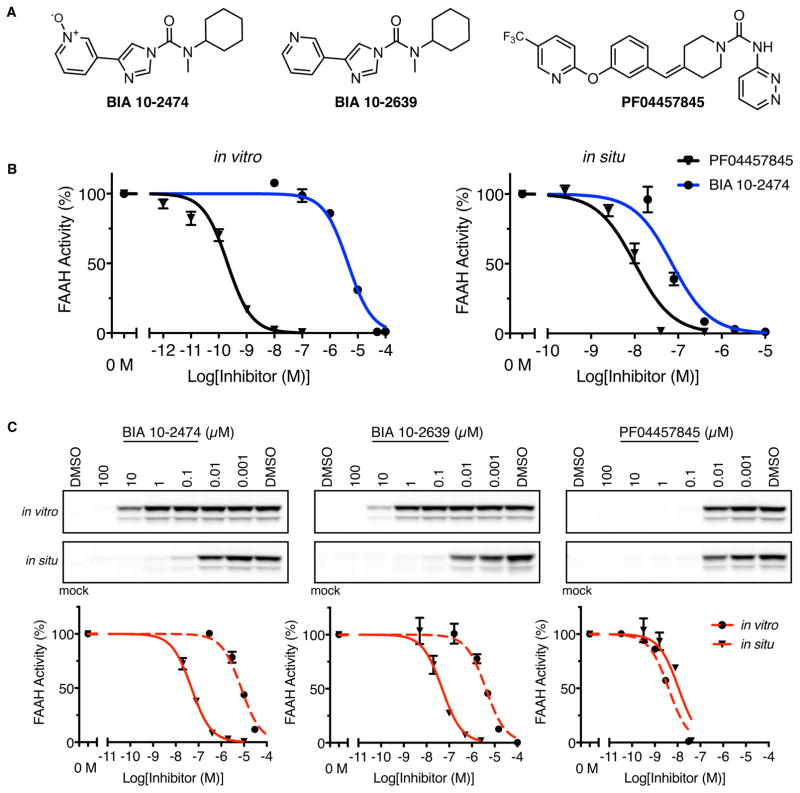
BIA 10-2474 (Fig. 1A) contains an electrophilic imidazole urea that may react with the nucleophilic serine of FAAH and other serine hydrolases to form covalent and irreversible adducts. We predicted that the serine hydrolase targets of BIA 10-2474 could be identified using chemical proteomic methods (13–15); this would allow us to compare its selectivity profile to that of PF04457845 (Fig. 1A), a FAAH inhibitor that progressed to Phase 2 trials without serious adverse events (16). We first synthesized BIA 10-2474, along with BIA 10-2639, a confirmed metabolite in which the N-oxide of BIA 10-2474 has been reduced to a pyridine (4) (Fig. 1A), in two independent labs and confirmed their structures by 1H- and 13C-NMR and high-resolution mass spectrometry (17). Both independently generated sets of compounds displayed equivalent activities in the biological assays described from here forward.
Fig. 1. Comparison of human FAAH inhibition by BIA 10-2474, BIA 10-2639, and PF04457845.
(A) The structures of BIA 10-2474, the metabolite BIA 10-2639, and PF04457845. (B) Inhibition of human FAAH in HEK293T cell lysates (in vitro) or intact cells (in situ) as measured using an anandamide substrate hydrolysis assay. (C) In vitro and in situ inhibition of human FAAH as measured by competitive gel-based ABPP. Top panels, HEK293T cell lysates (in vitro) or intact cells (in situ) recombinantly expressing human FAAH were pretreated with compound or DMSO (in vitro: 30 min, 37 °C in situ: 4 h, 37 °C). FAAH activity was measured by reactivity with the serine hydrolase-directed probe fluorophosponate-rhodamine and visualization of signals by gel-based ABPP. Bottom panels, Corresponding IC50 curves for gel-based ABPP data shown in Top Panels. N = 3 independent experiments per group.
Our initial experiments using substrate hydrolysis assays revealed that BIA 10-2474 showed weak in vitro inhibitory activity against human and rat FAAH, displaying IC50 values ≥ 1 μM (Fig. 1B, Fig. S1, and Table S1). Consistent with previous reports (6, 16), PF04457845 potently inhibited FAAH with IC50 values of ~1–10 nM (Fig. 1B and Table S1). In contrast, BIA 10-2474 exhibited greatly improved potency in cellular assays (in situ), blocking human FAAH activity in transfected HEK293T (human embryonic kidney) cells with IC50 values of 0.05–0.07 μM (Fig. 1B). BIA 10-2474 and PF04457845 did not interact with other proteins of the endocannabinoid system or with the endocannabinoid-binding TRP ion channels (Tables S2 and S3).
We also created alkynylated analogues of BIA 10-2474 – AJ167, AJ179 and AJ198 – and found that two of these compounds (AJ179 and AJ198) labeled mouse and human FAAH in brain lysates, as detected by coupling to azide fluorescent reporter groups via copper(I)-catalyzed azide-alkyne cycloaddition (“click”) chemistry (Fig. S2) (18). This finding, coupled with the time-dependent inhibition of FAAH displayed by BIA 10-2474 (Table S1), provides strong evidence that BIA 10-2474 and related imidazole ureas exhibit an irreversible mode of action.
To investigate the serine hydrolase interaction landscape of BIA 10-2474, we used activity-based protein profiling (ABPP), a chemical proteomic method that employs active site-directed chemical probes (e.g., fluorophosphonates (FPs) or beta-lactones for serine hydrolases) to assess the functional state of entire enzyme classes directly in native biological systems (13, 15). When coupled to fluorescent reporter groups, ABPP probes enable visualization of enzyme activities in complex proteomes by SDS-PAGE and in-gel fluorescence scanning. When coupled to a biotin reporter group, ABPP probes enable affinity enrichment and identification of enzyme activities by mass spectrometry (MS)-based proteomics. In both formats, ABPP serves as a versatile method to assess target engagement and proteome-wide selectivity for small-molecule inhibitors. Gel-based ABPP using a fluorescent FP probe (FP-TAMRA) confirmed the relative in vitro and in situ potencies of BIA 10-2474 and PF04457845 for human FAAH in transfected HEK293T cell preparations (Fig. 1C). The reason for the increased cellular activity of BIA 10-2474 is unclear, but could reflect cellular accumulation of the compound, which has been observed for other types of enzyme inhibitors (19).
Initial ABPP studies were performed in the human colon carcinoma cell line SW620, which expresses a wide diversity of endogenous serine hydrolase activities, including FAAH and FAAH2. Isotopically heavy and light amino acid-labeled SW620 cells were treated with DMSO or drug (BIA 10-2474 or PF04457845; 0.2 or 10 μM each for 4 h, or 50 μM each for 24 h) and then lysed and treated with a biotinylated FP probe. The samples are then combined, and subjected to streptavidin enrichment and quantitative LC-MS analysis, in which proteins displaying heavy:light ratios of > 2.0 were designated as drug-inhibited targets. We focused on human cell studies and tested a broad range of inhibitor concentrations because the deleterious neurological effects of BIA 10-2474 were observed in humans, but not other mammals, and occurred at drug doses that were 10 to 50 times higher than that required for blockade of FAAH activity in the clinical trial participants (4).
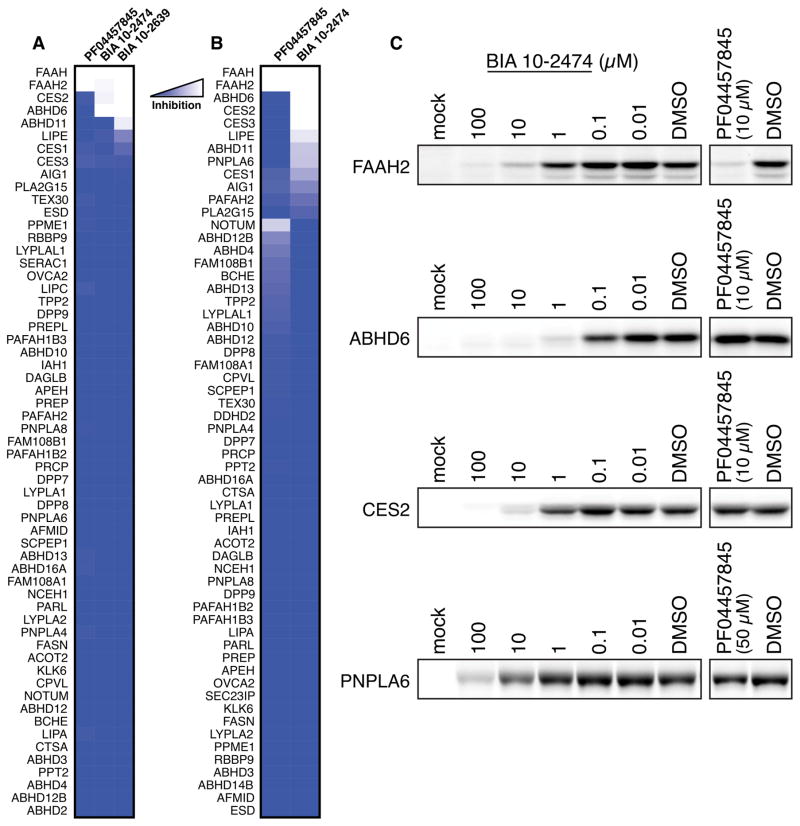
Our MS-based ABPP studies confirmed that both BIA 10-2474 and PF04457845 fully engaged human FAAH at all tested concentrations (0.2, 10, and 50 μM) (Fig. 2A, B and Fig. S3). Both drugs showed good selectivity for FAAH at the lowest concentration tested (0.2 μM; Fig. S3). PF04457845 maintained this selectivity profile at higher concentrations, displaying only a single major off-target – the homologous enzyme FAAH2 – among ~60 quantified serine hydrolases, consistent with previous studies (16). In contrast, BIA 10-2474 and its metabolite BIA 10-2639 exhibited numerous off-targets across the tested drug concentration range, including FAAH2 and several lipid hydrolases, such as ABHD6, ABHD11, LIPE, and PNPLA6 and xenobiotic drug-metabolizing enzymes CES1, CES2, and CES3 (Fig. 2A, B). Some of these off-targets, such as ABHD6 and CES2, were near-completely inhibited (>90%) at both 10 μM and 50 μM concentrations of BIA 10-2474.
Fig. 2. Quantitative proteomic analysis of serine hydrolase targets of FAAH inhibitors in human cells.
(A, B) MS-based ABPP of serine hydrolase activities in SW620 cells treated with DMSO or FAAH inhibitor. Shown in (A) are BIA 10-2474, BIA 10-2639, and PF04457845 (10 μM, 4 h, 37 °C). Shown in (B) are BIA 10-2474 and PF04457845 (50 μM, 24 h, 37 °C). Data are expressed as median SILAC ratio values for all isotopic peptide pairs quantified per protein from two biological replicates. (C) Confirmation of representative off-targets of BIA 10-2474 by gel-based ABPP of recombinantly expressed enzymes in HEK293T cells.
Representative off-targets of BIA 10-2474 were recombinantly expressed in HEK293T cells and verified to engage BIA 10-2474 by gel-based ABPP (Fig. 2C). These experiments also confirmed the relative potency of off-targets mapped by MS-based ABPP, with BIA 10-2474 exhibiting greater inhibitory activity against ABHD6 and CES2 compared to PNPLA6. In contrast, none of the recombinantly expressed enzymes were inhibited by PF04457845 except FAAH2 (Fig. 2C).
Our chemical proteomic data, taken together, demonstrated that both BIA 10-2474 and its major metabolite BIA 10-2639 cross-react with several human serine hydrolases that do not interact with PF04457845 (Tables 1 and S4). One possible contributing factor to this broader interaction profile is the greater intrinsic reactivity of BIA 10-2474 compared to PF04457845, which is reflected in their respective rates of methanolysis (Fig. S4). We also note that human CES2 and ABHD6 were both more potently inhibited by BIA 10-2474 and BIA 10-2639 compared to the mouse orthologs of these enzymes (Table S2), indicating the potential for species differences in the off-target-mediated activities of these compounds.
Table 1. In vitro and in situ inhibitory potencies of BIA 10-2474, BIA 10-2639, and PF04457845 against FAAH and representative off-targets.
Measurements were made by gel-based ABPP of HEK293T cells recombinantly expressing the indicated human serine hydrolases. Data represent inhibitor treatment of cells for 4 h with the exception of PNPLA6, where data represent inhibitor treatment of cells for 24 h. Data represent average values from three independent experiments per group. See Table S4 for error measurements related to IC50 values.
| Enzyme | Treatment | IC50 (μM)
|
||
|---|---|---|---|---|
| BIA 10-2474 | BIA 10-2639 | PF04457845 | ||
| FAAH | in vitro | 7.5 | 4.1 | 0.0040 |
| FAAH | in situ (4 h) | 0.049 | 0.049 | 0.011 |
| FAAH2 | in situ (4 h) | 0.40 | 0.10 | 0.59 |
| ABHD6 | in situ (4 h) | 0.081 | 0.079 | >10 |
| CES2 | in situ (4 h) | 2.0 | 0.63 | >10 |
| ABHD11 | in situ (4 h) | >10 | 2.3 | >10 |
| PNPLA6 | in situ (24 h) | 11 | N.D. | >50 |
Many of the off-targets of BIA 10-2474 are involved in cellular lipid metabolism (20, 21) and most (with the exception of FAAH2) show substantial expression in human brain tissue (Fig. S5). While the poor in vitro activity displayed by BIA 10-2474 limited our ability to identify off-targets in brain tissue lysates, we were able to confirm cross-reactivity of this drug with both FAAH and ABHD6 in human frontal cortex proteome (post-mortem samples acquired from three male donors who were 49, 50, and 80 years of age and who were not associated with the BIA 10-2474 trial) (Fig. S6). We also observed several of the off-targets of BIA 10-2474 by ABPP of human cortical neurons derived from induced pluripotent stem cells (Fig. S7).
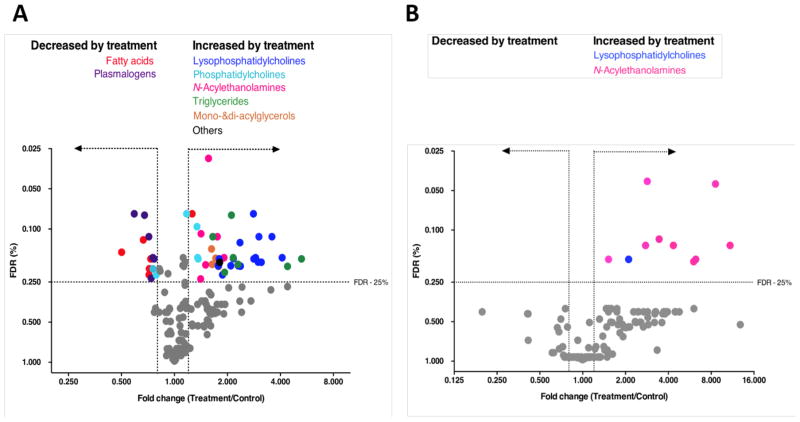
We next tested whether prolonged exposure to BIA 10-2474 altered lipid metabolism in human cortical neurons. We performed targeted lipidomic analysis of human cortical neurons cultures treated with vehicle (DMSO) or BIA 10-2474 (50 μM) at a concentration that was ~20x above the Cmax observed in the human clinical trial (22). In total the levels of 161 lipid species were quantified, of which 54 showed a fold change of ≥ 1.20 or ≤ 0.80 when using a Benjamini–Hochberg false discovery rate ≤ 25% (Fig. 3A and Table S5). The lipids affected by BIA 10-2474 included FAAH substrates (N-acylethanolamines), as well as several other lipid classes, including triglycerides, monoacylglycerols, (lyso)phosphatidylcholines, free fatty acids and plasmalogens. In contrast, treatment of human cortical neuron cultures with PF04457845 (1 μM), also tested at a concentration that was 20x above the clinical Cmax for the drug (11), produced a more restricted profile of lipid changes, predominantly corresponding to the expected elevations of N-acylethanolamines (Fig. 3B).
Fig. 3. BIA 10-2474, but not PF04457845, causes substantial alterations in lipid metabolism in human cortical neurons.
(A, B) Cortical neurons were treated with DMSO (A, B), BIA 10-2474 (50 μM) (A) or with PF04457845 (1 μM) (B) and analyzed by MS-based lipidomics after 48 h. X-axis denotes fold change of lipid species in the inhibitor-treated cells vs DMSO-treated cells. Lipidomic data are presented as a volcano plot and lipids with a fold change (FC) threshold of ≥1.20 or ≤0.80 and Benjamini–Hochberg false discovery rate (FDR) ≤ 25% are represented by colored circles distinguished by lipid class. Data represent average values from at least two independent experiments.
In summary, we have found that BIA 10-2474 acts as an irreversible inhibitor of FAAH that displays greater cross-reactivity with human serine hydrolases than the clinically tested FAAH inhibitor PF04457845. Many of the off-targets of BIA 10-2474 are lipolytic enzymes, raising the possibility that disruption of cellular lipid networks may have contributed to the compound’s neurotoxicity. Notably, disruption of neuronal lipid metabolism by inhibition of PNPLA6, one of the off-target proteins of BIA 10-2474 identified herein, has previously been linked to organophosphate-based neurotoxicity in humans (21, 23–25), and recessive loss-of-function mutations in the pnpla6 gene are responsible for a broad spectrum of neurodegenerative disorders (26, 27).
While our data provide information about the selectivity of BIA 10-2474, they do not allow us to conclude that inhibition of one or more of the identified off-target proteins is responsible for the clinical neurotoxicity caused by this drug. Nor can we exclude the possibility that non-covalent interactions of BIA 10-2474 or its metabolites with other proteins might have contributed to the reported clinical effects (28). Regardless, our study highlights the general utility of ABPP as a versatile chemical proteomic method to assess on-target engagement and off-target activity of covalent drugs to guide therapeutic development.
Supplementary Material
Acknowledgments
The human brain samples were obtained from The Netherlands Brain Bank (NBB), Netherlands Institute for Neuroscience, Amsterdam (open access: www.brainbank.nl). All material was collected from donors for whom, or from whom, the NBB had obtained written informed consent for a brain autopsy and the use of the material and clinical information for research purposes. We thank R. M. Suciu for assistance with the computational analysis of ABPP data. This work was supported by a Dutch Research Council– Chemical Sciences ECHO grant (to A.P.A.J. and M.v.d.S.); an ECHO-STIP Grant (to M.S. and M.v.d.S.) and Leiden University, Faculty of Science (“Profiling Programme: Endocannabinoids”; M.v.d.S., E.M., T.H., V.K.) grants from the Chinese Scholarship Council (to H.D.); the NIH (DA033760 to B.F.C.).; Netherlands Organisation for Scientific Research (NWO 024.003.001 to S.A.K.). The work of F.F. and M.M. was partly supported by the Italian Ministry of Education, University and Research (competitive PRIN 2015 grant to M.M.). B.F.C. is a founder and advisor to Abide Therapeutics, a biotechnology company interested in developing serine hydrolase inhibitors as therapeutics. G.H. is founder of Toxys B.V., a company that performs cytotoxicity testing of compounds. The ToxTracker platform used for toxicological profiling is available for research purposes from Toxys B.V under a material transfer agreement.
Footnotes
References and Notes
- 1.Eddleston M, Cohen AF, Webb DJ. Implications of the BIA-102474-101 study for review of first-into-human clinical trials. Br J Clin Pharmacol. 2016;81:582–586. doi: 10.1111/bcp.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler D, Callaway E. Scientists in the dark after French clinical trial proves fatal. Nature. 2016;529:263–264. doi: 10.1038/nature.2016.19189. [DOI] [PubMed] [Google Scholar]
- 3.Kerbrat A, et al. Acute Neurologic Disorder from an Inhibitor of Fatty Acid Amide Hydrolase. N Engl J Med. 2016;375:1717–1725. doi: 10.1056/NEJMoa1604221. [DOI] [PubMed] [Google Scholar]
- 4.Bégaud B, et al. Report by the Temporary Specialist Scientific Committee (TSSC), “FAAH (Fatty Acid Amide Hydrolase)”, on the causes of the accident during a Phase 1 clinical trial. 2016:1–28. [Google Scholar]
- 5.Cravatt BF, et al. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 6.Kathuria S, et al. Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med. 2002;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- 7.Devane W, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science (80-) 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 8.Long JZ, et al. Dual blockade of FAAH and MAGL identifies behavioral processes regulated by endocannabinoid crosstalk in vivo. Proc Natl Acad Sci U S A. 2009;106:20270–20275. doi: 10.1073/pnas.0909411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Stelt M, et al. Anandamide acts as an intracellular messenger amplifying Ca2+ influx via TRPV1 channels. EMBO J. 2005;24:3026–3037. doi: 10.1038/sj.emboj.7600784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampson AJ, et al. Dual effects of anandamide on NMDA receptor-mediated responses and neurotransmission. J Neurochem. 1998;70:671–676. doi: 10.1046/j.1471-4159.1998.70020671.x. [DOI] [PubMed] [Google Scholar]
- 11.Huggins JP, Smart TS, Langman S, Taylor L, Young T. An efficient randomised, placebo-controlled clinical trial with the irreversible fatty acid amide hydrolase-1 inhibitor PF-04457845, which modulates endocannabinoids but fails to induce effective analgesia in patients with pain due to osteoarthritis of th. Pain. 2012;153:1837–1846. doi: 10.1016/j.pain.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 12.Li GL, et al. Assessment of the pharmacology and tolerability of PF-04457845, an irreversible inhibitor of fatty acid amide hydrolase-1, in healthy subjects. Br J Clin Pharmacol. 2012;73:706–716. doi: 10.1111/j.1365-2125.2011.04137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niphakis MJ, Cravatt BF. Enzyme Inhibitor Discovery by Activity-Based Protein Profiling. Annu Rev Biochem. 2014;83:341–377. doi: 10.1146/annurev-biochem-060713-035708. [DOI] [PubMed] [Google Scholar]
- 14.Baggelaar MP, et al. Highly Selective, Reversible Inhibitor Identified by Comparative Chemoproteomics Modulates Diacylglycerol Lipase Activity in Neurons. J Am Chem Soc. 2015;137:8851–8857. doi: 10.1021/jacs.5b04883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: The serine hydrolases. Proc Natl Acad Sci. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn K, et al. Mechanistic and Pharmacological Characterization of PF-04457845: A Highly Potent and Selective Fatty Acid Amide Hydrolase Inhibitor That Reduces Inflammatory and Noninflammatory Pain. J Pharmacol Exp Ther. 2011;338:114–124. doi: 10.1124/jpet.111.180257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiss LE, et al. Pharmaceutical Compounds. 2010074588 A2 WO. 2010
- 18.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew Chemie Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Lanning BR, et al. A road map to evaluate the proteome-wide selectivity of covalent kinase inhibitors. Nat Chem Biol. 2014;10:760–767. doi: 10.1038/nchembio.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas G, et al. The serine hydrolase ABHD6 Is a critical regulator of the metabolic syndrome. Cell Rep. 2013;5:508–520. doi: 10.1016/j.celrep.2013.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang PA, Wu YJ. Neuropathy target esterase: An essential enzyme for neural development and axonal maintenance. Int J Biochem Cell Biol. 2010;42:573–575. doi: 10.1016/j.biocel.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Patat A. Safety & Regulation in Early Clinical Drug Development. Ghent, Belgium: 2016. http://www.bapu.be/autres/86_the_bial_10-2474.pdf. [Google Scholar]
- 23.Richardson RJ, Hein ND, Wijeyesakere SJ, Fink JK, Makhaeva GF. Neuropathy target esterase (NTE): overview and future. Chem Biol Interact. 2013;203:238–44. doi: 10.1016/j.cbi.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 24.Read DJ, Li Y, Chao MV, Cavanagh JB, Glynn P. Neuropathy target esterase is required for adult vertebrate axon maintenance. J Neurosci. 2009;29:11594–600. doi: 10.1523/JNEUROSCI.3007-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moser M, et al. Cloning and expression of the murine sws/NTE gene. Mech Dev. 2000;90:279–82. doi: 10.1016/s0925-4773(99)00239-7. [DOI] [PubMed] [Google Scholar]
- 26.Hufnagel RB, et al. Neuropathy target esterase impairments cause Oliver–McFarlane and Laurence–Moon syndromes. J Med Genet. 2015;52:85–94. doi: 10.1136/jmedgenet-2014-102856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Topaloglu AK, et al. Loss-of-function mutations in PNPLA6 encoding neuropathy target esterase underlie pubertal failure and neurological deficits in Gordon Holmes syndrome. J Clin Endocrinol Metab. 2014;99:E2067–75. doi: 10.1210/jc.2014-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertrand D, Bertrand S, Neveu E, Fernandes P. Molecular characterization of off-target activities of telithromycin: a potential role for nicotinic acetylcholine receptors. Antimicrob Agents Chemother. 2010;54:5399–402. doi: 10.1128/AAC.00840-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baggelaar MP, et al. Development of an activity-based probe and in silico design reveal highly selective inhibitors for diacylglycerol lipase-α in brain. Angew Chem Int Ed Engl. 2013;52:12081–5. doi: 10.1002/anie.201306295. [DOI] [PubMed] [Google Scholar]
- 30.Bachovchin DA, et al. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci U S A. 2010;107:20941–6. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cognetta AB, et al. Selective N-Hydroxyhydantoin Carbamate Inhibitors of Mammalian Serine Hydrolases. Chem Biol. 2015;22:928–937. doi: 10.1016/j.chembiol.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inloes JM, et al. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc Natl Acad Sci U S A. 2014;111:14924–9. doi: 10.1073/pnas.1413706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu C, et al. RPLC-ion-trap-FTMS method for lipid profiling of plasma: method validation and application to p53 mutant mouse model. J Proteome Res. 2008;7:4982–91. doi: 10.1021/pr800373m. [DOI] [PubMed] [Google Scholar]
- 34.Gattinoni S, et al. Enol carbamates as inhibitors of fatty acid amide hydrolase (FAAH) endowed with high selectivity for FAAH over the other targets of the endocannabinoid system. ChemMedChem. 2010;5:357–360. doi: 10.1002/cmdc.200900472. [DOI] [PubMed] [Google Scholar]
- 35.Ligresti A, et al. Prostamide F2a receptor antagonism combined with inhibition of FAAH may block the pro-inflammatory mediators formed following selective FAAH inhibition. Br J Pharmacol. 2014;171:1408–1419. doi: 10.1111/bph.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssen FJ, et al. Discovery of Glycine Sulfonamides as Dual Inhibitors of sn-1- Diacylglycerol Lipase α and α/β-Hydrolase Domain 6. J Med Chem. 2014;57:6610–22. doi: 10.1021/jm500681z. [DOI] [PubMed] [Google Scholar]
- 37.van der Wel T, et al. A natural substrate-based fluorescence assay for inhibitor screening on diacylglycerol lipase α. J Lipid Res. 2015;56:927–935. doi: 10.1194/jlr.D056390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peppard JV, Mehdi S, Li Z, Duguid MS. Assay methods for identifying agents that modify the activity of nape-pld or abhd4. 2008 [Google Scholar]
- 39.Mukhopadhyay P, et al. The novel, orally available and peripherally restricted selective cannabinoid CB 2 receptor agonist LEI-101 prevents cisplatin-induced nephrotoxicity. Br J Pharmacol. 2016;173:446–458. doi: 10.1111/bph.13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogasawara D, et al. Rapid and profound rewiring of brain lipid signaling networks by acute diacylglycerol lipase inhibition. Proc Natl Acad Sci. 2016;113:26–33. doi: 10.1073/pnas.1522364112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandtorv AH, Bjørsvik H-R. Fast Halogenation of Some N - Heterocycles by Means of N, N′-Dihalo-5,5-dimethylhydantoin. Adv Synth Catal. 2013;355:499–507. [Google Scholar]
- 42.Roumen L, et al. Synthesis, Biological Evaluation, and Molecular Modeling of 1-Benzyl-1 H -imidazoles as Selective Inhibitors of Aldosterone Synthase (CYP11B2) J Med Chem. 2010;53:1712–1725. doi: 10.1021/jm901356d. [DOI] [PubMed] [Google Scholar]
- 43.Bial-Portela SA, Russo CaD, Wahnon JBR, Maton W, Eszenyi T. Process For The Synthesis Of Substituted Urea Compounds. 2014017938 A2 WO. 2014
- 44.Johnson DS, et al. Discovery of PF-04457845: A Highly Potent, Orally Bioavailable, and Selective Urea FAAH Inhibitor. ACS Med Chem Lett. 2011;2:91–96. doi: 10.1021/ml100190t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.