Dear Editor,
The blaNDM-5 gene, which encodes New Delhi metallo-β-lactamase-5, was first reported in 2011 in Escherichia coli from a patient in the United Kingdom.1 NDM-5 has been reported in many other countries. A variety of plasmid types have been reported to contribute to the widespread dissemination of β-lactam resistance genes. These NDM carriers are mainly identified in Enterobacteriaceae, frequently in Klebsiella pneumoniae and E. coli. Although NDM-1 has been found in Salmonella enterica, to our knowledge, no other variants of NDM have been identified in S. enterica. Here we characterize the first identification of Salmonella enterica serovar Typhimurium (S. Typhimurium) strain SSH006 carrying blaNDM-5, through routine surveillance in China. This is the first report of the blaNDM-5-harboring S. Typhimurium.
Strain SSH006 was recovered from a fecal sample of a 71-year-old male patient in September 2015 in the enteric clinic of the Minhang district, Shanghai, China. The strain SSH006 was identified as S. Typhimurium by amplification and sequencing of the 16S rRNA gene. The serotype was also determined by using slide agglutination with commercial antiserum (S&A Reagents Laboratory, Bangkok, Thailand) according to the Kauffmann–White scheme. The presence of blaNDM-5 was determined by PCR screening and was confirmed by sequencing. Southern blotting showed that SSH006 contained two different plasmids (~45 and ~110 kb). The blaNDM-5 gene was located on the small plasmid, designated pNDM5-SSH006.
Antimicrobial susceptibility testing was performed with a Vitek 2 compact system, and the results were interpreted on the basis of the CLSI guidelines.2 The results showed that strain SSH006 was resistant to most tested antibiotics, including imipenem, but was susceptible to aztreonam, amikacin, ciprofloxacin, levofloxacin and nitrofurantoin (Supplementary Table S1). Transferability of the blaNDM-5 gene was assessed by conjugation experiments using the sodium azide-resistant E. coli strain J53 as the recipient. The transconjugants were selected on MacConkey agar plates with antimicrobial drugs. Antimicrobial susceptibility testing revealed that the transconjugants acquired resistance to imipenem, ampicillin and amoxicillin/clavulanic acid (Supplementary Table S1). Unexpectedly, resistance to cephalosporin antibiotics was also observed. Subsequent sequencing revealed that the cephalosporin resistance gene blaCTX-M-55 was located on the large plasmid, which was simultaneously transferred to the transconjugants. The presence of the blaNDM-5 and blaCTX-M-55 genes in the transconjugants was demonstrated by PCR amplification.
The whole genome of S. Typhimurium SSH006 was sequenced by the Novogene Company (Beijing, China) on the HiSeq 2500 platform. Paired-end reads of 350 bp were assembled using SOAPdenovo (v2.04) with 109-fold coverage. Multi-locus sequence typing analysis showed that SSH006 belongs to sequence-type 34 (ST34). In addition to blaNDM-5, multiple resistance genes were identified, including blaTEM-1, blaCTX-M-55, aadA1, sul1, sul2, sul3, aac(6’)-Iaa, tet(A), floR and dfrA12. Interestingly, there were two blaTEM-1 genes: one located on the chromosome and the other truncated and located adjacent to the blaCTX-M-55 gene. A BLAST search indicated that the blaCTX-M-55-carrying contig covered 5 kb and consisted of a novel combination of K. pneumoniae plasmid pKP090853 and Shewanella sp. ANA-3 plasmid 1 (CP000470, unpublished).
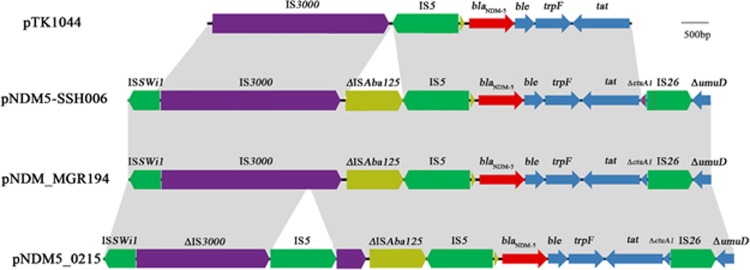
The complete sequence of the blaNDM-5-carrying plasmid pNDM5-SSH006 is 46 253 bp in length and shares >99% identity with the IncX3 plasmid pNDM_MGR194 that was isolated in India,4 with 17 nucleotide changes. Twelve of the 17 nucleotide changes are located within the truncated ctuA1 gene, and one is located in the insertion sequence IS26 downstream of the blaNDM-5 gene. In addition to these 13 nucleotide variations, the genetic context of blaNDM-5 in the two plasmids is identical (ISSwil-IS3000-ΔISAba125-IS5-blaNDM-5-ble-trpF-tat-ΔctuA1-IS26-ΔumuD). The shotgun whole-genome sequence and complete sequence of plasmid pNDM5-SSH006 have been deposited in GenBank under accession number MTKV00000000.
A variety of blaNDM-5-harboring plasmids have been identified and found to share similar sequences with plasmid pNDM_MGR194, such as pEc1929,5 pNDM5_0215,6 pECNDM101,7 and pNDM5-IncX3,8 pNDM-QD28 and pNDM-QD29.9 Most of these have been isolated in China (Supplementary Table S2). All of the above plasmids had the same genetic context of NDM except pNDM5_0215, which had an insertion of IS5 within the truncated insertion sequence IS3000 (Figure 1). Notably, the blaNDM-5-carrying IncN plasmid pTK1044 from Japan also shared a similar NDM genetic environment with a deletion of the truncated ISAba125 but was ~110 kb in length,10 thus suggesting a potential dissemination of NDM-5 via mobile genetic elements. Plasmids pJEG027 harboring blaNDM-411 and pKpN01-NDM7 harboring blaNDM-712 were also found to be almost identical to pNDM5-SSH006 except for the variation of the NDM gene. The blaNDM-7-harboring IncX3 plasmid pOM26-1 (KP776609, unpublished) is similar to pNDM5-SSH006, except that it has a deletion of ISAba125.
Figure 1.
Features of the genetic structure of pTK1044, pNDM5-SSH006, pNDM_MGR194 and pNDM5_0215. A 10.5-kb sequence of the genetic context of the NDM-5-harboring plasmid pNDM5-SSH006 is shown. The arrows indicate open reading frames. Light gray shading denotes homology regions. The blaNDM-5 gene is shown in red. IS3000, ISAba125 and the other insertion sequences are shown in purple, yellow and green, respectively. The left genes are shown in blue. Nucleotide changes between pNDM5-SSH006 and pNDM_MGR194 are indicated by red lines.
Given that blaNDM-5 and blaNDM-7 differ from blaNDM-4 by a single nucleotide change (G388A and G262T, respectively), the documentation of travel to India indicated that the blaNDM-5-harboring pNDM_MGR194 and the blaNDM-7 pKpN01-NDM7 plasmid may have evolved from the blaNDM-4-harboring pJEG027 plasmid,12 thus raising particular concern regarding their epidemic potential to mediate rapid spread of NDM. However, the pNDM5-SSH006-like plasmids were observed in diverse species at different geographical locations across China, in the absence of documented travel history or epidemiological linkage. The increased occurrence of this highly similar IncX3 plasmid indicated the existence of a natural reservoir and rapid transmission of the plasmid in China.
To the best of our knowledge, this is the first report of S. Typhimurium carrying the blaNDM-5 gene. The blaNDM-5-carrying plasmids have been reported in a narrow host range in E. coli, K. pneumoniae and recently P. mirabilis.13 Our work further expands the host range and provides additional evidence of the rapid dissemination of blaNDM-5 among different species of Enterobacteriaceae in China. Given that NDM-5 differs from NDM-1 by two amino-acid substitutions and confers increased resistance to expanded-spectrum cephalosporins and carbapenems,1 the ability of this blaNDM-5-harboring plasmid to transfer across species boundaries may pose a great threat to humans and is quite worrisome. S. Typhimurium ST34 clones have raised international concern regarding its rapid spread and multi-drug resistance. A previous study has revealed that S. Typhimurium ST34 clones experienced a rapid expansion in China and exhibit a low percentage susceptibility to cephalosporin antibiotics.14 However, S. Typhimurium ST34 SSH006 exhibited higher resistance to all tested cephalosporin antibiotics.
Here we report the first case of S. Typhimurium ST34 SSH006 harboring the blaNDM-5 gene. The co-existence of two transferable plasmids carrying blaCTX-M-55 and blaNDM-5 in S. Typhimurium highlights the urgent need for more extensive surveillance and effective action to control its further dissemination.
Acknowledgments
This work was supported by grants from the Mega-projects of Science and Technology Research (NO 2012ZX10004215 and AWS15J006) and the National Nature Science Foundation of China (NO 81373053, 81371854 and 81673237).
Footnotes
Supplementary Information for this article can be found on the Emerging Microbes & Infections website (http://www.nature.com/emi)
Supplementary Material
References
- Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 2011; 55: 5952–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Third Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute, 2013. [Google Scholar]
- Shin J, Soo Ko K. Single origin of three plasmids bearing blaCTX-M-15 from different Klebsiella pneumoniae clones. J Antimicrob Chemother 2014; 69: 969–972. [DOI] [PubMed] [Google Scholar]
- Krishnaraju M, Kamatchi C, Jha AK et al. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J Med Microbiol 2015; 33: 30–38. [DOI] [PubMed] [Google Scholar]
- Chen D, Gong L, Walsh TR et al. Infection by and dissemination of NDM-5-producing Escherichia coli in China. J Antimicrob Chemother 2016; 71: 563–565. [DOI] [PubMed] [Google Scholar]
- Yang P, Xie Y, Feng P et al. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother 2014; 58: 7548–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong LH, Lei CW, Ma SZ et al. Various sequence types of Escherichia coli co-harboring blaNDM-5 and mcr-1 genes from a commercial swine farm in China. Antimicrob Agents Chemother 2017;61. pii: e02167–16. [DOI] [PMC free article] [PubMed]
- Li A, Yang Y, Miao M et al. Complete sequences of mcr-1-harboring plasmids from extended-spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 2016; 60: 4351–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YQ, Zhao JY, Xu C et al. Identification of an NDM-5-producing Escherichia coli sequence type 167 in a neonatal patient in China. Sci Rep 2016; 6: 29934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano R, Nakano A, Hikosaka K et al. First report of metallo-β-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob Agents Chemother 2014; 58: 7611–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espedido BA, Dimitrijovski B, van Hal SJ et al. The use of whole-genome sequencing for molecular epidemiology and antimicrobial surveillance: identifying the role of IncX3 plasmids and the spread of blaNDM-4-like genes in the Enterobacteriaceae. J Clin Pathol 2015; 68: 835–838. [DOI] [PubMed] [Google Scholar]
- Chen L, Peirano G, Lynch T et al. Molecular characterization by using next-generation sequencing of plasmids containing blaNDM-7 in Enterobacteriaceae from Calgary, Canada. Antimicrob Agents Chemother 2015; 60: 1258–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Xie L, Wang X et al. Further spread of bla NDM-5 in Enterobacteriaceae via IncX3 plasmids in Shanghai, China. Front Microbiol 2016; 7: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MHY, Yan M, Chan EWC et al. Expansion of Salmonella Typhimurium ST34 clone carrying multiple resistance determinants in China. Antimicrob Agents Chemother 2013; 57: 4599–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.