Abstract
Background and purpose
To explore the integration of imaging and molecular data for response prediction to chemoradiotherapy (CRT) for rectal cancer.
Material and Methods
Eighty-five rectal cancer patients underwent preoperative CRT. 18F-FDG PET/CT and diffusion-weighted imaging (DWI) were acquired before (TP1) and during CRT (TP2) and prior to surgery (TP3). Inflammatory cytokines and gene expression were analyzed. Tumour response was defined as ypT0-1N0. Multivariate models were built combining the obtained parameters. Final models were calculated on the data combination with the highest AUC.
Results
Twenty-two patients (26%) achieved ypT0-1N0 response. 18F-FDG PET/CT had worse predictive performance than DWI and T2-volumetry (AUC 0.61 ± 0.04, 0.72 ± 0.03, and 0.72 ± 0.02, respectively). Combining all imaging parameters increased the AUC to 0.81 ± 0.03. Adding cytokines or gene expression did not improve the AUC (AUC of 0.72 ± 0.06 and 0.79 ± 0.04 respectively). Final models combining 18F-FDG PET/CT, DWI, and T2-weighted volumetry at all TPs and using only TP1 and TP3, allowed ypT0-1N0 prediction with a 75% sensitivity, 94% specificity and PPV of 80%.
Conclusions
Combining 18F-FDG PET/CT, DWI, and T2-weighted MRI volumetry obtained before CRT and prior to surgery may help physicians in selecting rectal cancer patients for organ-preservation.
Keywords: rectal cancer, chemoradiotherapy, response prediction, imaging, molecular markers
Introduction
With the implementation of total mesorectal excision (TME), local recurrence rates of rectal cancer decreased from above 20% to about 5% [1]. Local control of locally advanced rectal cancer further improved by the use of preoperative chemoradiotherapy (CRT) [2]. About 15–20% of the patients receiving preoperative CRT achieve a pathological complete response (pCR) [3]. These patients have an excellent outcome, regardless of their initial T- and N-stages [3,4]..
In the era of personalised medicine, the need for extensive surgery in well-responding patients has been questioned, and less invasive alternatives such as local excision and even a “watch-and-wait” policy have been suggested [5–7]. Adopting an organ-preserving strategy for good responders spares patients the morbidity (i.e. postoperative complications, long-term bowel, bladder and sexual dysfunction, or permanent stoma care) and the mortality associated with invasive surgery [8,9].
However, before such less invasive approach can be safely implemented, accurate assessment of response to chemoradiotherapy is of utmost importance. This is considered to be challenging since the concordance between mucosal appearance and pathological response has shown to be poor, and endoscopic biopsies are of limited value in ruling out persisting tumour after neoadjuvant CRT [10,11]. Because conventional morphologic imaging also lacks accuracy for restaging after CRT, alternative ways to assess response to CRT are needed [12,13]. There is growing interest in the use of functional imaging and molecular markers to improve clinical response assessment. Functional imaging techniques depict changes in tumour metabolism and microstructure before morphological changes become apparent. 18F-fluorodeoxyglucose positron emission tomography (18F-FDG PET) semi-quantitatively assesses tumour glucose metabolic activity through changes in FDG-uptake. A decrease in standardised uptake value (SUV) during treatment has been associated with pathological response in several tumour types, including rectal cancer [14]. Diffusion-weighted imaging (DWI) provides information on the microstructure of tissues through the assessment of differences in water diffusion [15]. By quantifying diffusion as the apparent diffusion coefficient (ADC), DWI can be used to monitor and to predict tumour response to CRT. An increase in ADC during and after CRT reflects a decreased cellularity and has been associated with tumour response to therapy [16]. Molecular markers have also been put forward as strategies to predict the response to CRT for rectal cancer. Some promising markers include inflammatory biomarkers and gene expression profiles [17–20].
In general, the predictive performance of 18F-FDG PET, DWI and molecular analysis as a single modality is insufficient to safely guide a patient-tailored treatment. Efforts have been made to investigate whether combining these markers contribute to a more accurate response prediction [21,22].
The purpose of this study was to explore the performance of integrating 18F-FDG PET, DWI, T2-weighted volumetry, inflammatory blood markers and gene expression profiles for prediction of ypT0-1N0 response to CRT.
Materials and Methods
Patients
Eighty-five rectal cancer patients were prospectively included between January 2012 and February 2015. Inclusion criteria were (1) primary histologically proven adenocarcinoma of the rectum, clinical stage T3-4N0 or T1-4N1-2, (2) WHO performance scale ≤2, and (3) adequate bone marrow, hepatic, and renal function assessed by biochemical examination. Exclusion criteria were (1) distant metastases (n=4), (2) prior chemotherapy or radiotherapy for rectal cancer (n=0), (3) previous or concurrent malignancies at other sites (n=0), and (4) known allergies to intravenous contrast agents or other contraindications for 18F-FDG PET/CT and MRI acquisition (n=0). Patients received CRT (45 Gy delivered in 1.8 Gy fractions, with a continuous infusion of 5-fluorouracil (225mg/m2/d)). Six patients received capecitabine (825 mg/m2 twice daily). TME was performed after an interval of eight weeks from completion of CRT (Figure 1). Patients underwent 18F-FDG PET/CT and DWI scans at regular time intervals. Blood and tissue samples were obtained. Endoscopy and digital rectal examination (DRE) were not consistently performed and were therefore not included as potential explanatory variables. This trial (NCT01171300) was approved by the institutional ethical committee and all patients gave written informed consent prior to study entry.
Figure 1. Study design.
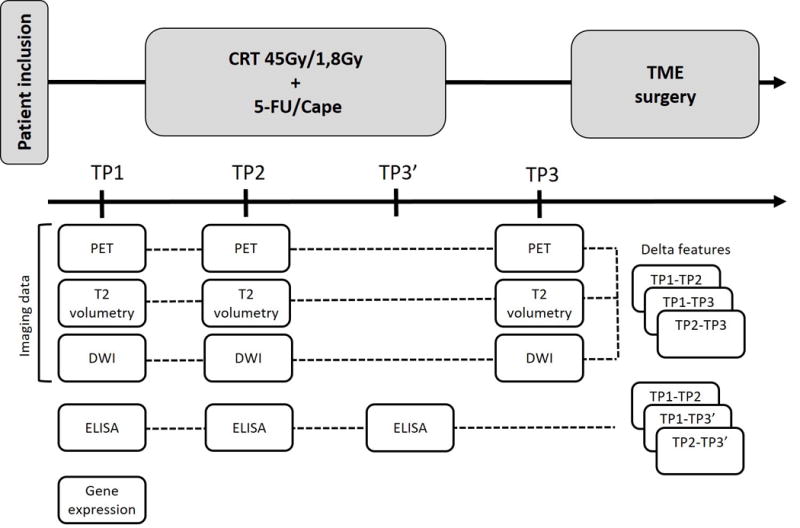
Abbreviations: 5-FU = 5-fluorouracil; Cape = capecitabine; CRT = chemoradiotherapy; TME = total mesorectal excision; TP = time point. Six patients received capecitabine (825 mg/m2 twice daily).
PET acquisition and evaluation
18F-FDG PET/CT scans were performed prior to CRT (TP1), after 10–15 fractions of CRT (TP2), and prior to surgery (TP3). Analyses were performed by a staff member of Nuclear Medicine (CD) who was unaware of the pathological and DWI results. Following PET parameters were extracted for each time point: SUVmax, SUVmean, SUVmin, SUVmedian, SUVpeak, metabolic tumour volume (MTV), metabolic diameter, and total lesion glycolysis (TLG = SUVmean*MTV). Absolute and relative changes in PET parameters between different time points were calculated, leading to a total of 72 PET variables. Details on PET acquisition can be found in Supplementary Material.
MRI acquisition and evaluation
MRI studies were acquired at the same time points as the 18F-FDG PET/CT scans. MRI analyses were performed by a staff member of Radiology (VV) who was blinded for pathological and 18F-FDG PET/CT results. Tumour volumetry was assessed by manually delineating tumour boundaries on the axial T2-weighted images. Besides the tumour volume in cm³, a diameter of the equivalent sphere was calculated. DWI images were acquired using six different b-values (b = 0, 50, 100, 300, 600, 1000 sec/mm2). Following DWI parameters were extracted for each time point: ADClow (b0-b300), ADCavg (b0-b1000), ADChigh (b600-b1000). Additionally, absolute and relative changes in T2-volumetry and relative changes in ADC between different time points were calculated, generating 18 T2-volumetry and 18 DWI variables. Details on MRI acquisition are described in Supplementary Material.
ELISA assays
Blood samples were collected prior to CRT (TP1), two weeks into radiotherapy (TP2), and at the end of CRT (TP3′). Blood markers (interferon-gamma (IFN-γ), interleukin (IL) 10, IL12p70, IL13, IL1β, IL2, IL4, IL6, IL8, and tumour necrosis factor-alpha (TNF-α) were investigated by using a multiplex ELISA platform (Proinflammatory Human MSD assay, Mesoscale Diagnostics). The absolute and relative changes in cytokines between the time points were also calculated, leading to a total of 90 variables.
Microarray and gene expression profiling
Microarray analysis was performed on tumour biopsies obtained by endoscopy prior to CRT (details described in Supplementary Material). We used the following existing gene expression signatures for predicting rectal cancer outcomes: the Ghadimi et al. 54-gene signature, the Watanabe et al. 33 gene signature and the Kim et al. 95 gene signature [18–20]. Next, we added an Epithelial-to-Mesenchymal (EMT) signature and a hypoxia signature for colorectal cancer [23,24]. Finally, a five-gene signature for predicting metastasis of colorectal cancer was applied [25].
Pathology
Pathological TNM staging served as the gold standard. Histological evaluation of the resection specimen was independently performed by an expert pathologist (XS) according to the method described by Quirke et al [26]. The primary outcome measure for our study was tumour response defined as ypT0-1N0. Two patients (2%) did not undergo surgery due to strong clinical evidence of a complete response (repeated digital rectal examination, endoscopic evaluation, and DWI). These patients were strictly followed and were disease-free 42 and 38 months after the end of CRT, which we considered a surrogate endpoint for pCR.
Statistical analysis
The primary outcome measure was ypT0-1N0 response. Patients who had more than 30% missing variables in a particular data set (i.e. PET/CT, DWI, T2-weighted MR, ELISA and gene expression-specific features) were excluded from the analysis. Remaining missing data were estimated using a 15-Nearest Neighbour algorithm [27]. All variables were standardised to a zero mean and unit standard deviation. In a first step, models were built on each data set separately to identify the baseline performance of each modality. Secondly, we built models using different combinations of PET, T2-volumetry, DWI, ELISA, and microarray data applying logistic regression with lasso regularization. We used a ten-fold stratified cross validation strategy to assess the models’ performance on unseen data. We repeated this process ten times to randomise the process to split the data in folds. Performances were expressed as the area under the curve (AUC) of receiver operating characteristic (ROC) analysis, sensitivity, specificity, and positive predictive value (PPV). Final models were built on the complete data set after selecting the data combination with the highest AUC. All statistical analyses were done in the statistical language R version 3.1.1.
Results
Patients
Patient and tumour characteristics are depicted in Table 1. All patients completed their planned CRT schedule. All data were available for 58 patients (Supplementary Table 2). At histopathology, 11 patients had a ypT0 response, from which ten patients had ypT0N0 and one patient had ypT0N1. Ten patients had ypT1, 27 had ypT2 and 35 patients had ypT3. Fifty-six patients had no lymph node involvement at pathology, 22 had yN1 disease and five had yN2 disease. Additionally, two patients who did not undergo surgery and remained disease-free after a follow up of at least three years were also included. A total of 22 patients (26%) were considered to have ypT0-1N0 response.
Table 1.
Patient and tumour characteristics
| Total population | |
|---|---|
| Clinical stage | |
| T2N1 | 5 (6%) |
| T2N2 | 8 (9%) |
| T3N0 | 1 (1%) |
| T3N1 | 22 (26%) |
| T3N2 | 45 (53%) |
| T4N1 | 1 (1%) |
| T4N2 | 3 (4%) |
| Age (years) | 64.0 (38.3–85.7) |
| Male | 63.8 (38.3–85.7) |
| Female | 64.5 (46.2–84.3) |
| Gender | |
| Male | 60 (71%) |
| Female | 25 (29%) |
| Distance from anal verge (cm) | |
| <5 cm | 21 (25%) |
| 5–10 cm | 46 (54%) |
| >10 cm | 18 (21%) |
| Interval to TME (weeks) | 7.7 (3.6 – 11.9) |
| Type of surgery | |
| Sphincter-saving operation | 78 (92%) |
| Abdominoperineal excision | 5 (6%) |
| No surgery | 2 (2%) |
Data are n (%) or median (range).
Model performance of the individual datasets
The performance of the individual features extracted from the three imaging modalities can be found in Supplementary Table 3. The multivariate linear prediction models on each of the imaging data sets separately demonstrated that 18F-FDG PET/CT had a worse performance compared to DWI and T2-volumetry in predicting ypT0-1N0 (18F-FDG PET/CT AUC of 0.61 ± 0.04, DWI 0.72 ± 0.03, and T2-volumetry 0.72 ± 0.02) (Table 2). TP2 was redundant with all performances lower than using data at all time points or only TP1 and TP3 (Figure 2). Response prediction by the ten inflammatory cytokines tested at all TPs did not outperform the best imaging results (AUC 0.72 ± 0.06). None of the investigated gene expression signatures yielded an AUC of >0.60.
Table 2.
Comparison of predictive performance of all multivariate linear regression models in terms of area under the ROC curve (AUC) and standard deviation (SD) for predicting response to CRT.
| AUC (mean) | SD | |
|---|---|---|
| PET | 0.61 | 0.04 |
| DWI | 0.72 | 0.03 |
| T2-MRI | 0.72 | 0.02 |
| Combo | 0.81 | 0.03 |
| PET12 | 0.57 | 0.05 |
| DWI12 | 0.61 | 0.02 |
| T2-MRI12 | 0.55 | 0.03 |
| Combo12 | 0.63 | 0.06 |
| PET13 | 0.67 | 0.03 |
| DWI13 | 0.74 | 0.04 |
| T2-MRI13 | 0.69 | 0.02 |
| Combo13 | 0.83 | 0.03 |
| MA | 0.55 | 0.04 |
| Combo + MA | 0.79 | 0.04 |
| ELISA | 0.72 | 0.06 |
| ELISA + Combo | 0.72 | 0.04 |
Abbreviations: ROC = Receiver Operating Characteristic, AUC = area under the ROC curve, SD = standard deviation
Figure 2. Models’ performance in prediction of ypT0-1N0 response.
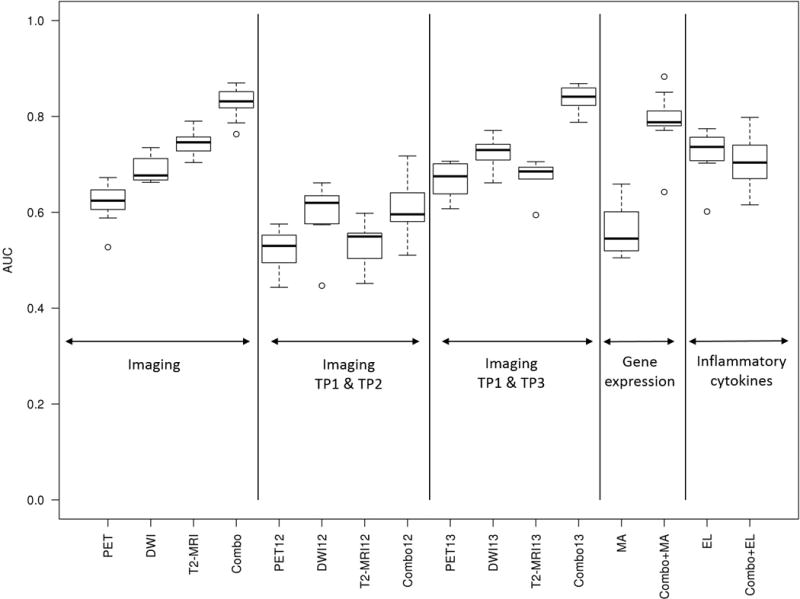
Box plots showing the performance in terms of the AUC of the multivariate linear prediction models for several combinations of imaging, clinical and molecular data at different time points. Abbreviations: AUC = area under the ROC curve; EL = ELISA; MA = microarray gene expression signatures; ROC = Receiver Operating Characteristic; TP = time point; TP12 = combination of data obtained at TP1 and TP2; TP13 = combination of data obtained at TP1 and TP3.
Model performance with combined datasets
A model combining the imaging modalities outperformed any of the single imaging modality models (AUC 0.81 ± 0.03) (Table 2, Figure 2). Adding protein biomarkers or gene expression signatures did not improve this result (AUC of 0.72 ± 0.04 and 0.79 ± 0.04 respectively). Although the performance of the combined imaging and gene expression model is similar to combined imaging only, further exploration of these models shows that in virtually all iterations, none of the gene expression signatures was selected in the models. Similar to the models of the individual datasets, TP2 was redundant whereas TP3 appeared essential (AUC of Combo12 0.63 ± 0.06 and Combo13 0.83 ± 0.03 respectively).
Final prediction models
Two final models were built. The first one integrated the imaging parameters of all time points (model Combo) and the second one only consisted of the imaging parameters that were obtained at TP1 and TP3 (model Combo13) (Table 3). Whether a patient has achieved ypT0-1N0 response is predicted by multiplying the weight of the retained parameters by their values obtained in an individual patient. Using a minimum PPV of 80%, thresholds for binary classification were set at −0.54 and −0.58 for model Combo and model Combo13 respectively. This resulted in a prediction of ypT0-1N0 response with a 75% sensitivity, a94% specificity and a negative predictive value of 92% for both models. The positive and negative likelihood ratios were 12.5 and 0.27 respectively.
Table 3.
Final predictive models with retained parameters and their weights.
| Combo | Combo13 | ||
|---|---|---|---|
|
| |||
| Parameter | Weight | Parameter | Weight |
| SUVpeak_TP3 | −0.74 | SUVpeak_TP3 | −0.46 |
| RISUVpeak_TP1TP2 | 0.14 | ADC_TP3_avg | 0.21 |
| ADC_TP3_avg | 0.08 | ADCratioavg_TP1TP3 | 0.73 |
| ADCratiohigh_TP1TP2 | −0.18 | Sphere_TP3 | −0.07 |
| ADCratioavg_TP1TP3 | 0.96 | DeltaSphere_TP1TP3perc | 0.46 |
| ADCratioavg_TP2TP3 | 0.13 | ||
| DeltaSphere_TP2TP3 | 0.09 | ||
| DeltaSphere_TP2TP3perc | 0.71 | ||
Final prediction models with integration of all imaging parameters at all time points (Model Combo) and with imaging parameters that were obtained at TP1 and TP3 (Model Combo13). Whether a patient has achieved ypT0-1N0 response is predicted by multiplying the weight of the retained parameters by their values obtained in an individual patient.
Discussion
Response prediction to CRT is challenging since clinical assessment, functional imaging and molecular analysis as stand-alone modalities are not accurate enough to safely defer well responding patients from a less invasive approach. Our study demonstrates that a combination of 18F-FDG PET/CT, DWI, and T2-weighted volumetry allows prediction of ypT0-1N0 response with 75% sensitivity, 94% specificity, and a PPV of 80%.
The strategy of combining clinical factors with imaging parameters has shown to promising before. Using a bi-institutional database, van Stiphout and co-workers developed a nomogram based on cT-stage, cN-stage, response index of SUVmean and maximal tumour diameter during CRT [21]. The nomogram allowed pCR prediction with an AUC of 0.78. The added value of DWI was demonstrated by Maas et al. who found that a combination of DWI, T2-weighted MRI and clinical response assessment increased predictive performance compared to clinical assessment alone and allowed for a post-test probability for a complete response to CRT of 98% [22].
Some groups reported that 18F-FDG PET/CT and DWI during CRT might be helpful in predicting tumour response [28]. In the present study however, imaging and molecular marker acquisition at TP1 and TP2 did not allow for an accurate response prediction with AUCs ranging from 0.55 to 0.63. Moreover, a model based on imaging acquired before CRT and prior to surgery had a slightly better performance than a model taking all time points into account. These findings demonstrate that imaging during CRT can be omitted as it does not contribute substantially to the prediction of the final tumour response.
The analyzed inflammatory blood markers did not show an added value for response prediction and none of the previously developed gene expression signatures could be validated. Although many studies reported gene expression signatures that appear highly accurate in predicting response to CRT, the composition of these signatures typically differ widely with little gene overlap [29]. Even with a more accurate prediction and extensive validation of predictive classifiers in prospective clinical trials, non-uniformity regarding blood and tissue storage and processing, costs and workload impede incorporation of gene expression profiling into future clinical practice.
In our study, only 14% of the patients achieved a complete response to CRT, partly due to the fact that the majority of patients had locally advanced disease and partly because of the meticulous pathological analysis. Thorough examination of the pathological specimen has shown to make a difference in pCR rate [30]. Moreover, restaging imaging was already performed at six weeks after the end of CRT, while a recent meta-analysis of 13 studies demonstrated that an interval of more than the “usual” 6–8 weeks resulted in more patients obtaining a pCR (relative risk 1.42, absolute increase of 14 to 20%) [31]. Sloothaak et al. showed that patients who underwent surgery 15–16 weeks after start of CRT had the highest chance on pCR. Waiting longer than 11 weeks after end of CRT had no added benefit [32]. The low number of patients with a complete response did not allow building a robust pCR prediction model. Since tumour regression after CRT is an ongoing process, the viability of residual tumour cells can be questioned. Sporadic tumour cells found in the resection specimen at eight weeks post-CRT may undergo apoptosis and mitotic catastrophe and might disappear when the interval to surgery is prolonged to 12 weeks.
Although DRE and endoscopy are practical strategies to assess the tumour response to CRT, they were not taken into account for modeling. Findings obtained by DRE and endoscopy cannot be quantified which makes these tools difficult to incorporate into a mathematical model. Since the aim of this study was predictive modeling and not organ-preservation, endoscopy was not routinely performed and highly-seated tumours were included which are not accessible by DRE. Moreover, evaluation of the submucosal layers and the mesorectum is not possible with DRE and endoscopy and requires MR imaging. The predictive models demand DWI and 18F-FDG PET/CT acquisition before CRT and prior to surgery. Since MR staging and restaging are standard practice and many patients undergo PET-imaging prior to CRT for distant staging, the burden of one additional PET-scan prior to surgery is minimal. In the future, hybrid PET/MR techniques likely will contribute to a more efficient assessment of the tumour response to CRT.
The predictive models described in this study are built on parameters obtained from prospectively included patients. Patients who missed more than 30% of the features in a specific dataset were omitted from further analysis. Since missing data appeared to be a random event (e.g. metal artifact impeding correct MR analysis, missing data because patients did not show up…), it is unlikely that omitting these patients would introduce a selection bias. Histopathology, 18F-FDG PET/CT and DWI analyses were performed by a pathologist, nuclear doctor and radiologist respectively, blinded for each other’s results. All had specific expertise in colorectal cancer. Although we did not check for inter- and intraobserver variability, we believe the standardised and widely-used method described by Quirke et al. allows for a robust evaluation of tumoural response to CRT [26].
The obtained models in this study are based on multiple covariates. Overfitting was avoided by using a multivariate criterium for feature selection and by applying a ten-fold cross-validation strategy. Hence, the model was never tested on the same data that it was trained on. A limitation of this study is the lack of external validation. Although the models depicted in Figure 2 were obtained after ten times applying a ten-fold cross-validation strategy, which makes the estimation of the performance of each parameters combination very robust, this cannot replace model validation in an independent patient cohort. Therefore, we are currently conducting a prospective study in which surgical treatment is tailored to the patient’s individual response to CRT based on clinical examination (with DRE and endoscopy) and on the predictive models. Patients with a clinical complete response are offered the possibility to enter a ‘”watch and wait” protocol with a strict follow-up. This follow-up study will also allow us to evaluate whether the prediction models can be optimised and whether they add to current tumoural restaging in clinical practice.
Nowadays, response evaluation is typically based on digital rectal examination, endoscopy (with or without biopsy) and pelvic MRI. The models described in this study are based on a unique dataset of markers which were prospectively obtained at different time points in a homogeneous population of rectal cancer patients, treated in the same centre by the same group of experts involved in rectal cancer treatment. The added value of this study is that it investigates the changes in tumour metabolism and microstructure throughout the treatment course, and that it explores the use of inflammatory blood markers and gene expression signatures. In this hypothesis-generating study, we showed that integrating 18F-FDG PET/CT, DWI, and T2-weighted volumetry before CRT and prior to surgery allowed prediction of ypT0-1N0 response with a higher sensitivity, specificity, and PPV compared to inflammatory bloodmarkers and gene expression profiles. The obtained models will be validated in an ongoing prospective study in which surgical treatment is tailored to the patient’s individual response to CRT.
Supplementary Material
Acknowledgments
We thank all participating patients and data managers who were involved in this project.
Role of the Funding Source
This work was funded by the Belgian Government Agency for Innovation by Science and Technology (IWT). The funding source had no role in the study design, in the collection, analysis and interpretation of data, in the writing of the manuscript and in the decision to submit the manuscript for publication.
Footnotes
Conflict of Interest Statement
There are no conflicts of interest to declare.
References
- 1.Heald RJ, Ryall RD. Recurrence and survival after total mesorectal excision for rectal cancer. Lancet. 1986;1(8496):1479–82. doi: 10.1016/s0140-6736(86)91510-2. [DOI] [PubMed] [Google Scholar]
- 2.Sauer R, Liersch T, Merkel S, et al. Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J Clin Oncol. 2012;30(16):1926–33. doi: 10.1200/JCO.2011.40.1836. [DOI] [PubMed] [Google Scholar]
- 3.Maas M, Nelemans PJ, Valentini V, et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol. 2010;11(9):835–44. doi: 10.1016/S1470-2045(10)70172-8. [DOI] [PubMed] [Google Scholar]
- 4.Rödel C, Martus P, Papadoupolos T, et al. Prognostic significance of tumor regression after preoperative chemoradiotherapy for rectal cancer. J Clin Oncol. 2005;23(34):8688–96. doi: 10.1200/JCO.2005.02.1329. [DOI] [PubMed] [Google Scholar]
- 5.Lezoche G, Baldarelli M, Guerrieri M, et al. A prospective randomized study with a 5-year minimum follow-up evaluation of transanal endoscopic microsurgery versus laparoscopic total mesorectal excision after neoadjuvant therapy. Surg Endosc. 2008;22(2):352–58. doi: 10.1007/s00464-007-9596-y. [DOI] [PubMed] [Google Scholar]
- 6.Habr-Gama A, Perez RO, Nadalin W, et al. Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: long-term results. Ann Surg. 2004;240(4):711–17. doi: 10.1097/01.sla.0000141194.27992.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maas M, Beets-Tan RG, Lambregts DM, et al. Wait-and-see policy for clinical complete responders after chemoradiation for rectal cancer. J Clin Oncol. 2011;29(35):4633–40. doi: 10.1200/JCO.2011.37.7176. [DOI] [PubMed] [Google Scholar]
- 8.Marijnen CA, Kapiteijn E, van de Velde CJ, et al. Acute side effects and complications after short-term preoperative radiotherapy combined with total mesorectal excision in primary rectal cancer: report of a multicenter randomized trial. J Clin Oncol. 2002;20(3):817–25. doi: 10.1200/JCO.2002.20.3.817. [DOI] [PubMed] [Google Scholar]
- 9.Sartori CA, Sartori A, Vigna S, Occhipinti R, Baiocchi GL. Urinary and sexual disorders after laparoscopic TME for rectal cancer in males. J Gastrointest Surg. 2011;15(4):637–43. doi: 10.1007/s11605-011-1459-0. [DOI] [PubMed] [Google Scholar]
- 10.Smith FM, Wiland H, Mace A, Pai RK, Kalady MF. Clinical criteria underestimate complete pathological response in rectal cancer treated with neoadjuvant chemoradiotherapy. Dis Colon Rectum. 2014;57(3):311–15. doi: 10.1097/DCR.0b013e3182a84eba. [DOI] [PubMed] [Google Scholar]
- 11.Perez RO, Habr-Gama A, Pereira GV, et al. Role of biopsies in patients with residual rectal cancer following neoadjuvant chemoradiation after downsizing: can they rule out persisting cancer? Colorectal Dis. 2012;14(6):714–20. doi: 10.1111/j.1463-1318.2011.02761.x. [DOI] [PubMed] [Google Scholar]
- 12.Hanly AM, Ryan EM, Rogers AC, et al. Multicenter Evaluation of Rectal cancer ReImaging pOst Neoadjuvant (MERRION) Therapy. Ann Surg. 2014;259(4):723–27. doi: 10.1097/SLA.0b013e31828f6c91. [DOI] [PubMed] [Google Scholar]
- 13.Huh JW, Park YA, Jung EJ, Lee KY, Sohn SK. Accuracy of endorectal ultrasonography and computed tomography for restaging rectal cancer after preoperative chemoradiation. J Am Coll Surg- 2008;207(1):7–12. doi: 10.1016/j.jamcollsurg.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Perez RO, Habr-Gama A, Gama-Rodrigues J, et al. Accuracy of positron emission tomography/computed tomography and clinical assessment in the detection of complete rectal tumor regression after neoadjuvant chemoradiation: long-term results of a prospective trial (National Clinical Trial 00254683) Cancer. 2012;118(14):3501–11. doi: 10.1002/cncr.26644. [DOI] [PubMed] [Google Scholar]
- 15.Patterson DM, Padhani AR, Collins DJ. Technology insight: water diffusion MRI–a potential new biomarker of response to cancer therapy. Nat Clin Pract Oncol. 2008;5(4):220–33. doi: 10.1038/ncponc1073. [DOI] [PubMed] [Google Scholar]
- 16.Lambrecht M, Vandecaveye V, De Keyzer F, et al. Value of diffusion-weighted magnetic resonance imaging for prediction and early assessment of response to neoadjuvant radiochemotherapy in rectal cancer: preliminary results. Int J Radiat Oncol Biol Phys. 2012;82(2):863–70. doi: 10.1016/j.ijrobp.2010.12.063. [DOI] [PubMed] [Google Scholar]
- 17.Debucquoy A, Goethals L, Geboes K, Roels S, Mc Bride WH, Haustermans K. Molecular responses of rectal cancer to preoperative chemoradiation. Radiother Oncol. 2006;80(2):172–77. doi: 10.1016/j.radonc.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 18.Ghadimi BM, Grade M, Difilippantonion MJ, et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol. 2005;23(9):1826–38. doi: 10.1200/JCO.2005.00.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe T, Kumoro Y, Kiyomatgsu T, et al. Prediction of sensitivity of rectal cancer cells in response to preoperative radiotherapy by DNA microarray analysis of gene expression profiles. Cancer Res. 2006;66(7):3370–74. doi: 10.1158/0008-5472.CAN-05-3834. [DOI] [PubMed] [Google Scholar]
- 20.Kim IJ, Lim SB, Kang HC, et al. Microarray gene expression profiling for predicting complete response to preoperative chemoradiotherapy in patients with advanced rectal cancer. Dis Colon Rectum. 2007;50(9):1342–53. doi: 10.1007/s10350-007-277-7. [DOI] [PubMed] [Google Scholar]
- 21.van Stiphout RG, Valentini V, Buijsen J, et al. Nomogram predicting response after chemoradiotherapy in rectal cancer using sequential PETCT imaging: a multicentric prospective study with external validation. Radiother Oncol. 2014;113(2):215–22. doi: 10.1016/j.radonc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Maas M, Lambregts DM, Nelemans PJ, et al. Assessment of Clinical Complete Response After Chemoradiation for Rectal Cancer with Digital Rectal Examination, Endoscopy, and MRI: Selection for Organ-Saving Treatment. Ann Surg Oncol. 2015;22(12):3873–80. doi: 10.1245/s10434-015-4687-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schell MJ, Yang M, Missiaglia E, et al. A Composite Gene Expression Signature Optimizes Prediction of Colorectal Cancer Metastasis and Outcome. Clin Cancer Res. 2016;22(3):734–45. doi: 10.1158/1078-0432.CCR-15-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dekervel J, Hompes D, van Malenstein H, et al. Hypoxia-driven gene expression is an independent prognostic factor in stage II and III colon cancer patients. Clin Cancer Res. 2014;20(8):2159–68. doi: 10.1158/1078-0432.CCR-13-2958. [DOI] [PubMed] [Google Scholar]
- 25.Hao JM, Chen JZ, Sui HM, et al. A five-gene signature as a potential predictor of metastasis and survival in colorectal cancer. J Pathol. 2010;220(4):475–89. doi: 10.1002/path.2668. [DOI] [PubMed] [Google Scholar]
- 26.Quirke P, Durdey P, Dixon MF, Williams NS. Local recurrence of rectal adenocarcinoma due to inadequate surgical resection. Histopathological study of lateral tumour spread and surgical excision. Lancet. 1986;2(8514):996–9. doi: 10.1016/s0140-6736(86)92612-7. [DOI] [PubMed] [Google Scholar]
- 27.Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17(6):520–5. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 28.Lambrecht M, Deroose C, Roels S, et al. The use of FDG-PET/CT and diffusion-weighted magnetic resonance imaging for response prediction before, during and after preoperative chemoradiotherapy for rectal cancer. Acta Oncol. 2010;49(7):956–63. doi: 10.3109/0284186X.2010.498439. [DOI] [PubMed] [Google Scholar]
- 29.Akiyoshi T, Kobunai T, Watanabe T. Predicting the response to preoperative radiation or chemoradiation by a microarray analysis of the gene expression profiles in rectal cancer. Surg Today. 2012;42(8):713–9. doi: 10.1007/s00595-012-0223-8. [DOI] [PubMed] [Google Scholar]
- 30.Glynne-Jones R, Anyamene N. Just how useful and endpoint is complete pathological response after neoadjuvant chemoradiation in rectal cancer? Int J Radiat Oncol Biol Phys. 2006;66:319–20. doi: 10.1016/j.ijrobp.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 31.Petrelli F, Sgroi G, Sarti E, Barni S. Increasing the interval between neoadjuvant chemoradiotherapy and surgery in rectal cancer: A meta-analysis of published studies. Ann Surg. 2016;263(3):458–64. doi: 10.1097/SLA.0000000000000368. [DOI] [PubMed] [Google Scholar]
- 32.Sloothaak DA, Geijsen DE, van Leersum NJ, et al. Optimal time interval between neoadjuvant chemoradiotherapy and surgery for rectal cancer. Br J Surg. 2013;100:933–9. doi: 10.1002/bjs.9112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


