Abstract
Objective
Several members of the angiopoietin-like (ANGPTL) family of proteins, including ANGPTL3 and ANGPTL8, regulate lipoprotein lipase (LPL) activity. Deficiency in either ANGPTL3 or ANGPTL8 reduces plasma triglyceride levels and increases LPL activity, whereas overexpression of either protein does the opposite. Recent studies suggest that ANGPTL8 may functionally interact with ANGPTL3 to alter clearance of plasma triglycerides; however, the nature of this interaction has remained elusive. We tested the hypothesis that ANGPTL8 forms a complex with ANGPTL3 and that this complex is necessary for the inhibition of vascular LPL by ANGPTL3.
Methods
We analyzed the interactions of ANGPTL3 and ANGPTL8 with each other and with LPL using co-immunoprecipitation, western blotting, lipase activity assays, and the NanoBiT split-luciferase system. We also used adenovirus injection to overexpress ANGPTL3 in mice that lacked ANGPTL8.
Results
We found that ANGPTL3 or ANGPTL8 alone could only inhibit LPL at concentrations that far exceeded physiological levels, especially when LPL was bound to its endothelial cell receptor/transporter GPIHBP1 (glycosylphosphatidylinositol-anchored high-density lipoprotein binding protein 1). Physical interaction was observed between ANGPTL3 and ANGPTL8 when the proteins were co-expressed, and co-expression with ANGPTL3 greatly enhanced the secretion of ANGPTL8. Importantly, ANGPTL3–ANGPTL8 complexes had a dramatically increased ability to inhibit LPL compared to either protein alone. Adenovirus experiments showed that 2-fold overexpression of ANGPTL3 significantly increased plasma triglycerides only in the presence of ANGPTL8. Protein interaction assays showed that ANGPTL8 greatly increased the ability of ANGPTL3 to bind LPL.
Conclusions
Together, these data indicate that ANGPTL8 binds to ANGPTL3 and that this complex is necessary for ANGPTL3 to efficiently bind and inhibit LPL.
Keywords: Plasma triglycerides, Lipoprotein metabolism, Lipolysis, Lipase inhibition
Abbreviations: ANGPTL, angiopoietin-like; GFP, green fluorescent protein; GPIHBP1, glycosylphosphatidylinositol-anchored high-density lipoprotein binding protein 1; LPL, lipoprotein lipase; NanoBiT, NanoLuc Binary Technology; RHMVECs, rat heart microvessel endothelial cells
Highlights
-
•
ANGPTL3 by itself is not a potent inhibitor of LPL.
-
•
When co-expressed, ANGPTL3 and ANGPTL8 form a complex.
-
•
ANGPTL3 facilitates the secretion of ANGPTL8.
-
•
ANGPTL3–ANGPTL8 complexes bind to LPL much better than either protein alone.
-
•
ANGPTL3 requires ANGPTL8 to efficiently inhibit LPL both in vitro and in vivo.
1. Introduction
Elevated plasma triglycerides are associated with several metabolic diseases including metabolic syndrome, type 2 diabetes mellitus, and atherosclerosis [1], [2], [3], [4], [5]. Clearance of plasma triglycerides is primarily mediated by the activity of lipoprotein lipase (LPL) [6]. LPL is expressed by the parenchymal cells of lipolytic tissues (e.g., adipocytes, myocytes) and is transported to capillary lumen by its endothelial cell transporter GPIHBP1 (glycosylphosphatidylinositol-anchored high-density lipoprotein binding protein 1) [7], [8]. After transport, LPL, which remains anchored to vascular endothelial cells by GPIHBP1 [9], hydrolyzes plasma triglycerides for local uptake into tissues.
The activity of LPL is critically regulated by several interacting proteins, including angiopoietin-like 3 (ANGPTL3). ANGPTL3 is thought to regulate triglyceride clearance by inhibiting LPL [10], [11], [12], [13]. ANGPTL3-deficient mice have significantly lower plasma triglycerides than wild-type mice and elevated levels of post-heparin plasma LPL activity, whereas mice overexpressing ANGPTL3 have elevated plasma triglycerides [10], [11], [14]. Humans with ANGPTL3 alleles predicted to disrupt ANGPTL3 function also have lower plasma triglyceride levels and increased post-heparin plasma LPL activity [15], [16]. ANGPTL3 is expressed exclusively in the liver [14], [17] but primarily regulates triglyceride uptake in peripheral tissues in the fed state [12], indicating that it acts in an endocrine manner. ANGPTL8, a more recently uncovered member of the ANGPTL family, also regulates triglyceride metabolism [18], [19], [20]. ANGPTL8 (also called lipasin, RIPL, and betatrophin) was also proposed as a regulator of beta cell proliferation, but this notion has been invalidated [21], [22], [23]. ANGPTL8 is most homologous to ANGPTL3, but lacks the fibrinogen-like domain of ANGPTL3 and other ANGPTL family members [14], [19], [20]. Similar to what has been found for ANGPTL3, ANGPTL8 overexpression increases plasma triglycerides [18], [19], and ANGPTL8-deficient mice have low plasma triglycerides [19], [20] and altered LPL activity in heart and skeletal muscle in the fed state [24]. Like ANGPTL3, ANGPTL8 is expressed highly in liver, but it is also expressed in adipose tissue [18], [19], [20]. Unlike ANGPTL3, which has similar gene expression levels and circulating protein levels in both the fed and fasted states [25], ANGPTL8 is highly induced by feeding [19], [20]. Beyond the similar phenotypes of ANGPTL3 and ANGPTL8-deficient mice, there is additional evidence that ANGPTL3 and ANGPTL8 function in concert. Overexpression of ANGPTL8 only increases plasma triglyceride levels when ANGPTL3 is present, and when ANGPTL8 is overexpressed, ANGPTL3 and ANGPTL8 can be pulled down together from the plasma [19]. These observations have led to the idea that ANGPTL8 may activate ANGPTL3 in some way [19]. However, the mechanism by which ANGPTL8 activates ANGPTL3, and whether that activation is actually necessary for ANGPTL3 function, remains unknown.
In this study, we investigated the ability of ANGPTL3, alone and in combination with ANGPTL8, to bind and inhibit LPL. We also investigated the mechanisms by which ANGPTL8 alters ANGPTL3 activity.
2. Material and methods
2.1. Production of LPL conditioned media
FLAG-tagged human LPL was concentrated from the medium of a Chinese hamster ovary cell line (CHO-K1) stably expressing FLAG-tagged human LPL as previously described [26], [27]. LargeBiT-human LPL (pEB12) was generated by cloning the linker and largeBiT sequence from pBiT1.1 (Promega) to the C-terminus of our FLAG-tagged LPL (pAH1) [27] using InFusion cloning (Clontech, catalog #638909). Lentiviruses containing this construct were produced by transfecting 293T cells with pEB12 and the lentiviral packaging vectors pMD2.G (Addgene #12259), pRSV-Rev (Addgene #12253), and pMDLg/pRRE (Addgene #12251). Conditioned media containing largeBiT-LPL expressing lentiviruses were collected and concentrated using Lenti-X Concentrator (Clontech, catalog #631231). 293T cells were then transduced with these lentiviruses and subjected to selection with puromycin (3 μg/ml) for 5 days. Conditioned media containing largeBiT-LPL was produced by culturing selected cells in serum-free DMEM for 72 h and then collecting media. LargeBiT-LPL conditioned media was concentrated before use with Amicon Ultra-15 Centrifugal Filter Units (EMD Millipore, catalog #UFC901024). The presence of LPL in the conditioned media was assessed by western blotting using a mouse antibody against the FLAG-tag (1:5000 Sigma–Aldrich; catalog #F1804). LPL activity was assessed by a lipase activity assay (see below).
2.2. Production of ANGPTL3 conditioned media
Constructs expressing full-length mouse ANGPTL3 (pXC3) or human ANGPTL3 (pXC4) were generated by amplifying the coding sequence of full-length mouse ANGPTL3 cDNA (Open Biosystems, MMM1013-202765361) or full-length human ANGPTL3 cDNA (Open Biosystems, MHS6278-202841262) and inserting it into a pCDNA6 vector using InFusion cloning (Clontech). A Strep-tag (WSHPQFEK; also known as Strep-tag II) was appended to the C-terminus of the open reading frame using Phusion site-directed mutagenesis (New England Biolabs) to create mouse ANGPTL3-Strep (pHS18) or human ANGPTL3-Strep (pHS15). Mouse smallBiT-ANGPTL3 (pEB14) was generated by adding a linker and the smallBiT sequence (GGGGSGGGGSSGVTGYRLFEEIL) to the C-terminus of pHS18 using Phusion site-directed mutagenesis. Strep-tagged truncated ANGPTL3 (pEB2) was generated by deleting the coding sequence for amino acids 222-455 using Phusion site-directed mutagenesis. A construct expressing the C-terminal domain of ANGPTL3 tagged with the Strep tag (pEB29) was generated by deleting the coding sequence for amino acids 17-224 (leaving the signal peptide intact) using Phusion site-directed mutagenesis. To generate conditioned media containing Strep-tagged ANGPTL3, smallBiT-Strep-ANGPTL3, Strep-tagged truncated ANGPTL3, or the C-terminal domain of Strep-ANGPTL3, 293T cells were transfected with pHS18, pHS15, pEB14, pEB2, or pEB29. 24 h post-transfection, media was switched to serum-free DMEM media containing protease inhibitors and grown for 2 days. Media were then collected, and the concentration of ANGPTL3 in conditioned media was determined using ANGPTL3 ELISA kits (mouse ANGPTL3: RayBiotech, catalog #ELM-ANGPTL3-1; human ANGPTL3: RayBiotech, catalog #ELH-ANGPTL3-1) following the manufacturer's instructions and using purified recombinant ANGPTL3 to generate a standard curve. The ELISA values for conditioned media prepared from untransfected cells were similar to blanks and this conditioned media was used as a control for all ANGPTL3 experiments.
2.3. Production of ANGPTL8 conditioned media
A construct expressing full-length mouse ANGPTL8 (pBD219) was generated by amplifying the coding sequence of ANGPTL8 cDNA (Open Biosystems, MMM1013-202708024). As this cDNA clone was missing the first 49 bases of the coding sequence, we used a synthetic double stranded oligonucleotide containing this missing sequence and inserted it simultaneously with the coding sequence from the cDNA into a pCDNA6 vector containing a C-terminal V5-tag using InFusion cloning. A His-tag was appended to the C-terminus of the open reading frame using Phusion site-directed mutagenesis (New England Biolabs) to create a V5/His-tagged ANGPTL8 (pWL1). A construct expressing full-length V5/His-tagged human ANGPTL8 (pEB30) was generated by amplifying the coding sequence of human ANGPTL8 cDNA (R&D systems, RDC1636) and replacing the mouse ANGPTL8 coding sequence in pWL1 using InFusion cloning (Clontech). V5/His-tagged mouse ANGPTL8 with a C-terminal smallBiT (pEB22) was generated by inserting a linker and the smallBiT sequence (GGGGSGGGGSSGVTGYRLFEEIL) at the C-terminus of V5/His-tagged ANGPTL8 (pWL1) right before the stop codon using Phusion site-directed mutagenesis. V5/His-tagged mouse, smallBiT-tagged mouse, and V5/His-tagged human ANGPTL8 were also cloned into lentiviral vectors using InFusion cloning to generate pEB5, pEB27, and pEB31 respectively. Lentiviruses containing these constructs were produced by transecting 293T cells with pEB5, pEB31, or pEB27 and the lentiviral packaging vectors pMD2.G, pRSV-Rev, and pMDLg/pRRE (Addgene). Conditioned media containing V5/His-tagged ANGPTL8 lentiviruses were collected and concentrated using Lenti-X Concentrator (Clontech, catalog #631231). 293T cells were then transduced with concentrated lentiviruses containing the ANGPTL8 expression cassette. After 5 days of puromycin selection (3 μg/ml), surviving cells were then cultured in serum-free DMEM and conditioned media were collected 72 h later. Conditioned media prepared from untransduced cells was used as a control for all ANGPTL8 experiments. The concentrations of mouse and human ANGPTL8 protein in the conditioned media was determined by quantitative western blotting using known concentrations of purified recombinant ANGPTL8 protein produced in bacteria as a standard.
2.4. Production of ANGPTL4 conditioned media
ANGPTL4 conditioned media was produced as described previously [27]. To generate conditioned media containing both ANGPTL3 and ANGPTL4, 293T cells were transfected with pHS18 and pHS2. 24 h post-transfection, media was switched to serum-free DMEM media containing protease inhibitors and grown for 2 days.
2.5. Transfections and cell harvesting
293T cells in T75 flasks were transfected with 10 μg of DNA (ANGPTL3 + empty vector, ANGPTL3 + ANGPTL8, ANGPTL8 + empty vector, or ANGPTL3 + ANGPTL4) and 20 μl of 1 mg/ml PEI (polyethylenimine). HepG2 cells were transfected with the 4D-Nucleofector (Lonza, catalog #AAF-1002B) using the SF Cell Line 4D-Nucleofector® X Kit L (Lonza, catalog #V4XC-2012) following manufacturer's instructions. 24 h post transfection, cells were switched to serum-free DMEM for 293T cells and DMEM/F-12 media (ThermoFisher Scientific, catalog #11320-033) for HepG2 cells. 48–72 h later, conditioned media were collected and concentrated using Amicon Ultra-15 Centrifugal Filter Units (EMD Millipore, catalog #UFC901024). Cells were then washed twice with PBS and lysed with radioimmunoprecipitation assay buffer (1% Nonidet P-40 substitute, 0.5% sodium deoxycholate, and 0.1% SDS in PBS) with protease inhibitor. Cell lysates were cleared by centrifugation at 12,000×g at 4 °C for 5 min. Samples were then analyzed by western blotting.
2.6. LPL activity assay
Lipase activity was assayed using EnzChek lipase fluorescent substrate (Molecular Probes) as described previously [28]. Briefly, 50 μl of sample was mixed with 25 μl 4× assay buffer (0.6 M NaCl, 80 mM Tris–HCl pH 8.0, and 6% fatty acid–free BSA). 25 μl of substrate solution containing 2.48 μM EnzChek lipase fluorescent substrate and 0.05% 3-(N,N-Dimethylmyristylammonio) propanesulfonate zwittergent detergent (Acros Organics) in 1% methanol was then added to each sample. Samples were then incubated at 37 °C with fluorescence (485 nm excitation/528 nm emission) read every minute for 30 min with a Synergy Neo multimode plate reader (BioTek). Relative lipase activity was calculated by first subtracting background (calculated by reading fluorescence of a sample with no LPL) and then calculating the slope of the curve between 10 and 15 min using Microsoft Excel.
2.7. Detection of LPL activity on the cell surface
Rat heart microvessel endothelial cells (RHMVECs; VEC technologies) were grown in MCDB-131 base medium (Genedepot) supplemented with 10 mM l-glutamine, 1% PenStrep antibiotic solution (10,000 U/ml penicillin and 10,000 μg/ml streptomycin, Gibco), 5% Fetal Bovine Serum (Atlanta Biologicals), 1 μg/ml hydrocortisone (Sigma–Aldrich), 10 μg/ml human epidermal growth factor (Gibco, Life technologies), and 12 μg/ml bovine brain extract (Lonza). Lentiviruses encoding S-protein-tagged GPIHBP1 were transduced into endothelial cells and selected for stable transduction with puromycin as described previously [7]. To detect LPL activity on the cell surface, RHMVECs expressing GPIHBP1 were incubated with LPL at 4 °C for 3–4 h. After washing off unbound LPL, cells were treated with ANGPTL3 and/or ANGPTL8 or control conditioned media. After again washing with PBS, cells were treated with heparin (100 U/ml in H2O) for 10 min at 4 °C to release surface-bound LPL. Released LPL was collected and immediately assayed for lipase activity. Samples were also analyzed for LPL protein mass by western blotting.
2.8. Western blot
Protein samples were size fractionated on 12% SDS-PAGE gels and then transferred to a nitrocellulose membrane. Membranes were blocked with casein buffer (1% casein, Fisher Science Education). Primary antibodies were diluted in casein buffer. Primary antibody dilutions were 1:5000 for a mouse monoclonal antibody against FLAG-tag (Sigma–Aldrich), 1:5000 for a mouse monoclonal antibody against V5-tag (Invitrogen), 1:10,000 for a goat antibody against S-protein tag (Abcam), 1:1250 for a rabbit antibody against mouse ANGPTL3 (Thermo Scientific), 1:2000 for a rabbit antibody against Strep-tag (Abcam), and 1:2500 for a goat antibody against actin (Santa Cruz). After washing with PBS + 0.1% Tween (PBS-T), membranes were incubated with Dylight680- or Dylight800-labeled secondary antibodies (Thermo Scientific) diluted 1:5000 in casein buffer. After washing with PBS-T, antibody binding was detected using an Odyssey Infrared Scanner (Li-Cor).
2.9. Mice
To assess plasma ANGPTL3 concentrations, male C57BL/6 mice were either fasted for 12 h or fasted for 12 h and then re-fed for 4 h. Plasma samples were collected through cardiac puncture. ANGPTL3 concentrations in fasted/fed plasma were determined using a mouse ANGPTL3 ELISA kit (RayBiotech, catalog #ELM-ANGPTL3-1), the same mouse ANGPTL3 ELISA used for ANGPTL3 conditioned media. Gpihbp1−/− mice (B6;129S5-Gpihbp1tm1Lex/Mmucd) [29], [30] were used for the isolation of radiolabeled chylomicrons and were generated by breeding from strains obtained from the Mutant Mouse Resource and Research Center (MMRRC).
Angptl8−/− mice (B6;129S5-Gm6484tm1Lex/Mmucd) [29] were also obtained from MMRRC. These mice were housed at 22–24 °C with a 14-h light, 10-h dark cycle and provided with ad libitum water and a chow diet (6% calories from fat, 8664; Harlan Teklad, Indianapolis, IN).
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Iowa or Wayne State University and were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
2.10. HisTalon purification
To examine the interactions between ANGPTL3 and ANGPTL8, His-tagged ANGPTL8 was purified from conditioned media using a 1 ml gravity HisTalon purification column (Clontech). HisTalon columns were equilibrated with 20 ml of equilibration buffer (300 mM NaCl, 50 mM sodium phosphate, pH 7.4). Conditioned media were loaded onto the columns and flow-through was collected. Columns were then equilibrated with 7 ml of equilibration buffer, washed with 7 ml of wash buffer (10 mM imidazole in equilibration buffer), and bound proteins were then eluted from the column with elution buffer (150 mM imidazole in equilibration buffer). Eluted fractions were collected in 1.5 ml aliquots and analyzed by western blot (see above).
2.11. Co-immunoprecipitation
2 μg of anti–Strep tag antibody (Abcam; #ab76949) were incubated with 200 μl of conditioned media from cells expressing ANGPTL3, ANGPTL8, or both at 4 °C for 1 h with gentle rotation. For co-immunoprecipitation experiments in which ANGPTL3 and ANGPTL8 conditioned media were mixed (rather than co-transfection of both proteins), 2 μg of anti-Strep tag antibody were incubated with 100 μl of ANGPTL3 conditioned media and 300 μl of ANGPTL8 conditioned media at 4 °C for 1 h with gentle rotation. For co-immunoprecipitation experiments with cell lysates, cells were lysed with non-denaturing lysis buffer (20 mM Tris HCl, 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA, and protease inhibitor). 300 μl of cell lysates were incubated with 2 μg of anti-Strep tag antibody at 4 °C for 1 h with gentle rotation. For co-immunoprecipitation experiments with ANGPTL3 and ANGPTL4, 1.5 μg of anti-V5 tag antibody (Invitrogen; #46-0705) were incubated with 300 μl of conditioned media from cells expressing ANGPTL4 or both ANGPTL3 and ANGPTL4 at 4 °C for 1 h with gentle rotation. Antibody-antigen complex was then added to 25 μl of Protein A or Protein G Dynabeads (Thermo Scientific) and incubated at room temperature for 20 min with gentle rotation. Prior to incubation the Dynabeads were resuspended and washed with 1X PBS with 0.02% Tween. The Dynabeads-antibody-antigen complex was then placed on a magnet, and the supernatant was removed for analysis. The bead complex was washed three times with 1× PBS with 0.02% Tween, and the antibody–antigen complex was eluted by incubating the beads with 30 μl 0.1 M citrate (pH 2.3) for 2 min at room temperature with gentle rotation. Eluted samples were immediately neutralized by adding 8 μl 1 M Tris pH 8.0. Samples were then subjected to western blotting analysis as described above.
2.12. LPL activity assay with radiolabeled chylomicrons
To prepare radiolabeled chylomicrons, Gpihbp1−/− mice were fasted 4 h and then gavaged with 100 μCi of [9, 10-3H (N)]-Triolein (Perkin Elmer) suspended in olive oil. After 4 h, mice were anesthetized and blood was collected by cardiac puncture. Blood was diluted 1:10 with 0.5 M EDTA (pH 8.0) and centrifuged 1500×g for 15 min at 4 °C to pellet blood cells. The plasma was then transferred to ultracentrifuge tubes and mixed 1:1 with PBS. After centrifugation twice at 424,000×g for 2 h at 10 °C, the resulting upper layer containing chylomicrons was resuspended in PBS to the original plasma volume. Protein content was assayed using the BioRAD DC Protein Assay. Radiolabeled chylomicrons were then diluted 100-fold with 1× PBS.
Activity assays were performed by incubating 40 μl of LPL conditioned media at 37 °C for 30 min with 10 μl of ANGPTL3 alone, ANGPTL8 alone, ANGPTL3 mixed with ANGPTL8, or ANGPTL8 that had been co-transfected with ANGPTL3. At the end of the 30 min incubation, the samples were incubated with 50 μl of diluted radiolabeled chylomicrons at 37 °C for another hour. The reaction was stopped by adding 1.6 ml of heptane: chloroform: methanol (1:1.25:1.41) followed by 500 μl of 0.1 M potassium carbonate (pH 10.5). After centrifugation at 1800×g for 15 min, the radioactivity of the upper phase was determined in BioSafe II scintillation fluid on a Beckman-Coulter Liquid Scintillation Counter (BCLSC6500).
2.13. Adenovirus infections
An adenovirus expressing mouse ANGPTL3 tagged with a Strep-tag (Strep-tag II) at the C-terminus was generated by Cyagen Biosciences (Santa Clara, CA). Wild-type and ANGPTL8−/− mice were injected via the tail vein with 5 × 108 pfu (diluted in 200 μl of saline) of adenovirus expressing GFP or ANGPTL3. 4 days after injection, mice were fasted for 20 h and then re-fed for 4 h. Blood was collected by tail nick and serum triglyceride levels were determined using a triglyceride quantification kit (Biovision, Milpitas, CA).
The expression of the recombinant Strep-tagged ANGPTL3 in the livers of injected mice was confirmed by western blotting using an antibody against the Strep-tag (1:1000, Abcam, #ab76949). Total levels of circulating mouse ANGPTL3 were determined using a mouse ANGPTL3 ELISA kit (RayBiotech, catalog #ELM-ANGPTL3-1) following the manufacturer's instructions.
2.14. NanoBiT luciferase assays for protein-protein interactions
Protein-protein interactions were examined using the NanoLuc® Binary Technology (NanoBiT) system (Promega) [31]. In this system, Nano Luciferase [32] has been divided into a 17.6 kDa largeBiT and an 11 amino acid smallBiT. Only when the two BiTs come together is luciferase activity restored. The largeBiT was appended to the C-terminus of FLAG-tagged LPL and the smallBiT (11 aa) was appended to the C-terminus of the Strep-tagged ANGPTL3 or the V5/His-tagged ANGPTL8. Conditioned media containing largeBiT-LPL, smallBiT-ANGPTL3, or smallBiT-ANGPTL8 were produced in 293T cells as described above. LPL-GPIHBP1 complexes were formed by incubating RHMVECs stably expressing GPIHBP1 and grown on white 96-well plates with LPL conditioned media for 3 h at 4 °C. After washing cells three times with 1× PBS to remove all unbound LPL, cell bound LPL-GPIHBP1 complexes were incubated with 50 μl smallBiT-ANGPTL3, smallBiT-ANGPTL3 that had been co-transfected with ANGPTL8, or smallBiT-ANGPTL3 that had been mixed with ANGPTL8 for 15 min at 37 °C. In some experiments, cell bound LPL-GPIHBP1 complexes were incubated with 50 μl smallBiT-ANGPTL8, smallBiT-ANGPTL8 that had been co-transfected with ANGPTL3, or smallBiT-ANGPTL8 that had been mixed with ANGPTL3 for 15 min at 37 °C. A matched concentration of an unrelated secreted protein (ASIP, Agouti-signaling protein) with a C-terminal smallBiT was used as the negative control. After incubation, cells were washed twice with 1× PBS and 100 μl of serum-free DMEM was added to each well. 25 μl of 5× NanoLuc Live Cell substrate (Promega) was then added, resulting in a final volume of 125 μl. Luminescence from each sample was read with a SpectraMax i3 Multi-Mode Detection Platform.
2.15. Cell binding assays
To detect the binding of ANGPTL3 and ANGPTL8 to LPL on the surface of endothelial cells, RHMVECs expressing GPIHBP1 were incubated with LPL at 4 °C for 3–4 h. After washing off unbound LPL, cells were incubated with ANGPTL3 and/or ANGPTL8 conditioned media or control conditioned media at 37 °C for 10 min. After again washing the cells with PBS, cells were lysed using radioimmunoprecipitation assay buffer (1% Nonidet P-40 substitute, 0.5% sodium deoxycholate, and 0.1% SDS in PBS) with protease inhibitor. Cell lysates were cleared by centrifugation at 12,000×g at 4 °C for 5 min. Samples were then analyzed by western blotting.
2.16. Size-exclusion chromatography
Conditioned media from cells co-transfected with ANGPTL3 and ANGPTL8 was concentrated 10× using an Amicon Ultra-4 Centrifugal Filter Unit (Millipore, #UFC801024). 100 μl of concentrated conditioned media was loaded on a Superdex 200 5/150 size exclusion column (GE Health Life Sciences, #17517501). 150 μl fractions were collected and analyzed by western blotting.
3. Results
3.1. Ability of ANGPTL3 to inactivate LPL
Strep-tagged mouse ANGPTL3 conditioned media was generated from 293T cells as we have described previously for ANGPTL4 [27]. As expected, when incubated with LPL at 37 °C, ANGPTL3 inhibited LPL in a time- and dose-dependent manner (Figure 1A). While high concentrations of ANGPTL3 abolished LPL activity (Figure 1A and Figure S1A), LPL protein levels were left unchanged (Figure S1B). We next tested the ability of ANGPTL3-inactivated LPL to bind the endothelial cell LPL receptor/transporter GPIHBP1. LPL inactivated by heat or high concentrations of ANGPTL3 was not able to bind GPIHBP1 on endothelial cells (Figure S1C), whereas control treated LPL or LPL treated with a concentration of ANGPTL3 insufficient to robustly inhibit lipase activity (Figure S1A) bound GPIHBP1 avidly (Figure S1C). Thus, just as we have shown previously for ANGPTL4-inactivated LPL [27], ANGPTL3-inactivated LPL had little capacity to bind GPIHBP1 on the surface of endothelial cells.
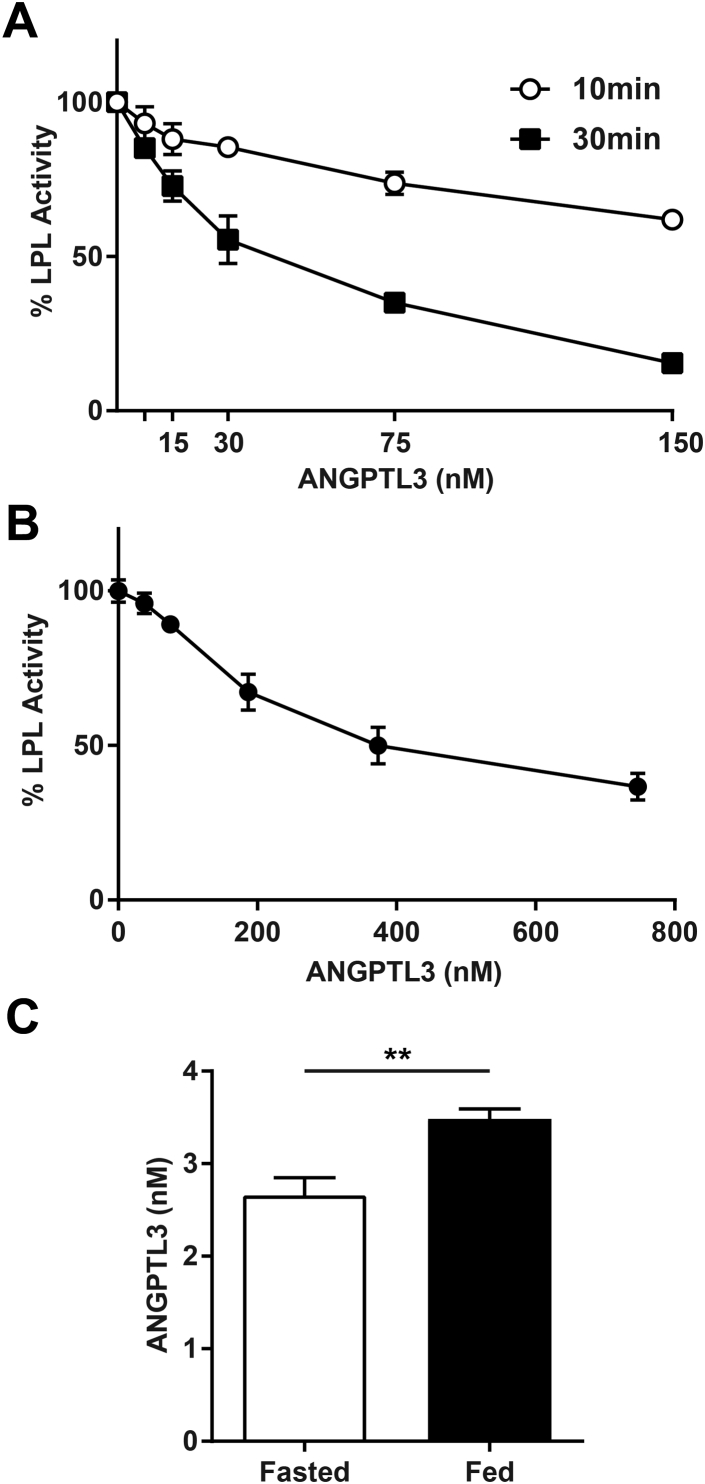
Figure 1.
ANGPTL3 inhibition of LPL. (A) Lipase activity of LPL conditioned media incubated with control media or the indicated concentrations of ANGPTL3 for 10 or 30 min at 37 °C. Activity was normalized to control treated LPL. Points represent mean (±SEM) of 2 independent experiments each done in duplicate. (B) GPIHBP1-expressing RHMVECs were incubated with LPL for 3.5 h at 4 °C. After washing off unbound LPL, cells were incubated with the indicated concentrations of ANGPTL3 or control media for 30 min at 37 °C and then washed again. Surface-bound LPL was then released from cells with 100 U/ml heparin and immediately assayed for lipase activity. Activity was normalized to control treated LPL. Points represent mean (±SEM) of 3 independent experiments each done in triplicate. (C) Plasma ANGPTL3 concentrations (mean ± SEM) in fasted and fed male C57BL/6 mice (n = 6/group; **p < 0.01).
ANGPTL3 is expressed only in the liver and, thus, could only act on peripheral tissues in an endocrine manner [14], [17]. In this context, ANGPTL3 would primarily encounter LPL bound to GPIHBP1 on the luminal wall of capillaries [8], [9]. A previous study, which used soluble GPIHBP1 and a cell-free system, found that GPIHBP1-bound LPL was protected from ANGPTL3 inhibition [33]. We asked if this protection was still observed when using full-length GPIHBP1 anchored to live endothelial cells. LPL-GPIHBP1 complexes were formed on the surface of endothelial cells and then treated with 0–750 nM (0–40 μg/ml) ANGPTL3 conditioned media. After incubation, LPL was released from the endothelial cells surface by heparin (100 U/ml) and assayed immediately for lipase activity. ANGPTL3 inhibited GPIHBP1-bound LPL in a dose-dependent manner; however, the ANGPTL3 concentration required for inhibition was substantially higher for GPIHBP1-bound LPL than for unbound LPL (Figure 1B; compare to Figure 1A).
Because the concentrations of ANGPTL3 required to inhibit LPL were high, especially when LPL was bound to GPIHBP1, we assessed the physiological concentrations of ANGPTL3 in the plasma of fed and fasted mice. Employing the same ELISA used to determine the concentration of ANGPTL3 in conditioned media, we found that plasma ANGPTL3 concentrations in fasted male C57BL/6 mice ranged from 2.11 to 3.28 nM (mean 2.66 nM). Concentrations in fed mice were slightly higher (3.14–3.75 nM; mean 3.48 nM) (Figure 1C). Based on our inhibition data, these concentrations of ANGPTL3 would have minimal impact on LPL activity, yet ANGPTL3-deficient mice clearly have reduced plasma triglyceride levels and increased LPL activity [10], [11], [14]. Thus, we sought to determine what modifying factors might allow ANGPTL3 to inhibit LPL at physiological concentrations.
3.2. Ability of truncated ANGPTL3 to inactivate LPL
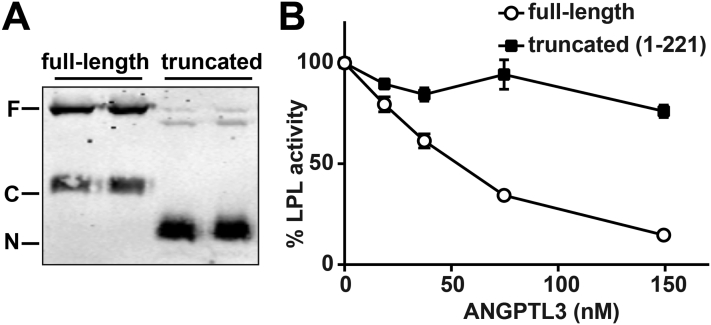
Like other members of the ANGPTL family, ANGPTL3 consists of a coiled-coil (CCD) N-terminal domain and a fibrinogen-like C-terminal domain [17]. In vivo, ANGPTL proteins can be cleaved by proprotein convertases, separating the coiled-coil domain from the fibrinogen-like domain [34], [35]. Like in ANGPTL family member ANGPTL4, the coiled-coil domain of ANGPTL3 has been reported to be both sufficient and necessary for LPL inhibition [34]. As it has been previously shown that a truncated form of ANGPTL4 containing only the coiled-coil domain is a much more potent LPL inhibitor than the full-length protein [27], [36], we asked if the same was true for ANGPTL3. Such a finding might explain why ANGPTL3 appeared to be more effective in vivo than in vitro. We generated Strep-tagged truncated ANGPTL3 (aa 1-221) conditioned media and tested its ability to inhibit LPL. Truncated ANGPTL3 inhibited LPL in a dose-dependent manner; however, unlike the case for ANGPTL4, truncated ANGPTL3 was less effective than full-length ANGPTL3 in inhibiting free LPL (Figure 2).
Figure 2.
Inhibitory activity of truncated ANGPTL3. (A) Western blot showing protein levels of full-length (F) and truncated (T) ANGPTL3 detected using the Strep-tag antibody. Cleavage of the C-terminally tagged full-length protein results in the detection of a C-terminal fragment (C). (B) Lipase activity of LPL conditioned media incubated with the indicated concentrations of full-length or truncated ANGPTL3 conditioned media for 30 min at 37 °C. Activity was normalized to control treated LPL. Points represent mean (±SEM) of 5 independent experiments each done in duplicate.
3.3. ANGPTL3 interacts with ANGPTL8
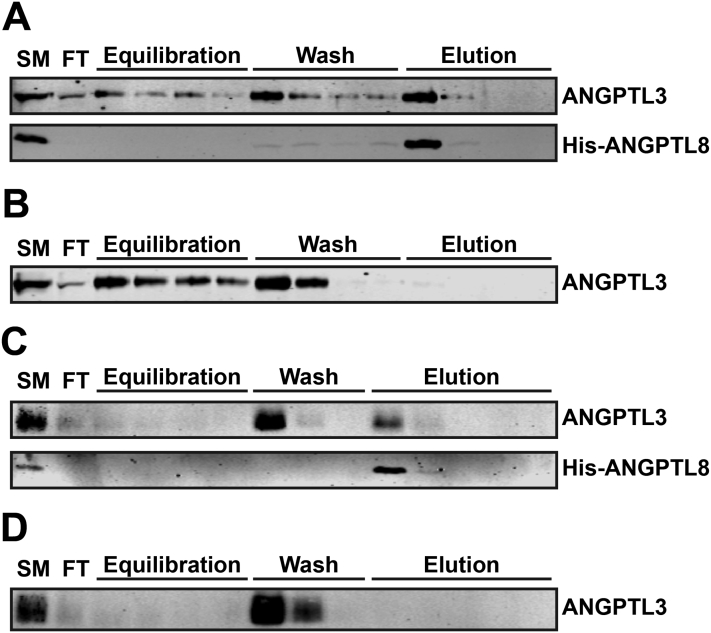
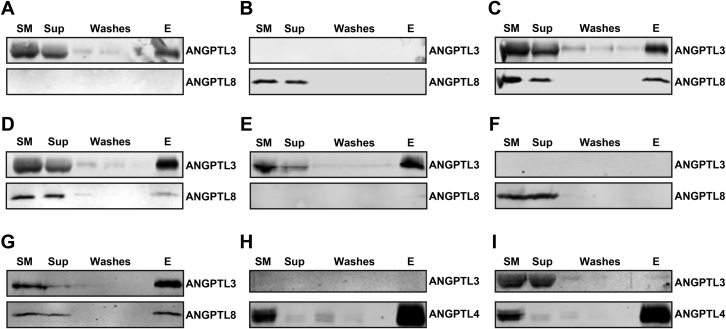
ANGPTL3 has been shown to reduce lipolysis in heart and skeletal muscle after feeding [12], but the levels of ANGPTL3 in mouse plasma increase only slightly during feeding (Figure 1). The expression of ANGPTL family member ANGPTL8, however, is highly induced by feeding, and it has been proposed that ANGPTL8 binds to and regulates the activity of ANGPTL3 [19], [24]. To test if ANGPTL8 physically interacts with ANGPTL3, Strep-tagged ANGPTL3 and His-tagged mouse ANGPTL8 were co-transfected into HEK 293T cells. After 72 h, conditioned media were collected and His-tagged ANGPTL8 was bound to and then eluted from a HisTalon column. Western blot analysis showed that when His-ANGPTL8 and Strep-ANGPTL3 were co-expressed, both proteins could be eluted from the column (Figure 3A); whereas, when ANGPTL3 was expressed by itself, no binding was observed (Figure 3B). The interaction between ANGPTL8 and ANGPTL3 appeared to depend on the coiled-coil region of ANGPTL3, as truncated ANGPTL3, when co-expressed with His-ANGPTL8, could also be pulled down (Figure 3C and D). In turn, when ANGPTL3 was precipitated from conditioned media using antibodies against the Strep-tag (Figure 4A–D), we also observed co-precipitation of ANGPTL8 (Figure 4C). Together, these data indicated that ANGPTL3 and ANGPTL8 form a complex when co-expressed. Interestingly, when ANGPTL3 and ANGPTL8 were transfected separately and then mixed, the binding between ANGPTL3 and ANGPTL8 was much less robust (Figure 4D), suggesting that these proteins may efficiently form a complex only when co-expressed. To determine whether ANGPTL3 and ANGPTL8 form a complex before secretion, we performed co-immunoprecipitation experiments on cell lysates rather than conditioned media. Again, we found that ANGPTL8 could be co-precipitated with ANGPTL3, indicating that the two proteins form a complex inside the cell (Figure 4E–G). To test if the formation of a complex between ANGPTL3 and ANGPTL8 was specific to these two proteins, we performed co-immunoprecipitation experiments with ANGPTL3 and the next most closely related family member, ANGPTL4. When ANGPTL3 and ANGPTL4 were co-expressed, ANGPTL3 was not co-precipitated with ANGPTL4, suggesting that these two proteins do not form a complex (Figure 4H and I).
Figure 3.
Pulldown of ANGPTL3 with ANGPTL8. The conditioned media from cells co-transfected with His-ANGPTL8 and full-length Strep-ANGPTL3 (A), full-length Strep-ANGPTL3 only (B), His-ANGPTL8 and truncated (aa 1-221) Strep-ANGPTL3 (C), or truncated Strep-ANGPTL3 only (D) were bound and eluted from a HisTalon column. Western blots show starting material (SM), column flow-through (FT), column equilibration washes (Equilibration), 10 mM imidazole washes (Wash), and 150 mM imidazole elutions (Elution).
Figure 4.
Pulldown of ANGPTL8 with ANGPTL3. (A–D) The conditioned media from cells transfected with Strep-ANGPTL3 (A), ANGPTL8 (B), or both Strep-ANGPTL3 and ANGPTL8 (C), as well as Strep-ANGPTL3 conditioned media mixed with ANGPTL8 conditioned media (D) were precipitated using Strep-tag antibody bound to protein A–coupled beads. (E–G) The lysates from cells transfected with Strep-ANGPTL3 (E), ANGPTL8 (F), or both Strep-ANGPTL3 and ANGPTL8 (G) were precipitated using Strep-tag antibody bound to protein A–coupled beads. (H–I) The conditioned media from cells transfected with V5-ANGPTL4 (H), or both V5-ANGPTL4 and Strep-ANGPTL3 (I) were precipitated using V5-tag antibody bound to protein G–coupled beads. All western blots show starting material (SM), unbound supernatant (Sup), washes, and elution (E).
Interestingly, when ANGPTL3 and ANGPTL8 were co-expressed, and ANGPTL8 was bound to the HisTalon column, significant amounts of ANGPTL3, but not ANGPTL8, came down in the flow-through, equilibration, and wash steps (Figure 3A and C). This appears to indicate that although ANGPTL8 and ANGPTL3 form a complex, a significant amount of ANGPTL3 is not in complex with ANGPTL8. To better understand the stoichiometry of the ANGPTL3–ANGPTL8 complexes, we ran conditioned media from cells transfected with both ANGPTL3 and ANGPTL8 through a size exclusion column. We found that both ANGPTL3 and ANGPTL8 were present in fractions representing a large range of molecular weights (Figure S2). These data suggest that there may be several species of complexes, with different stoichiometric ratios of ANGPTL3 and ANGPTL8.
3.4. ANGPTL3 promotes the secretion of ANGPTL8
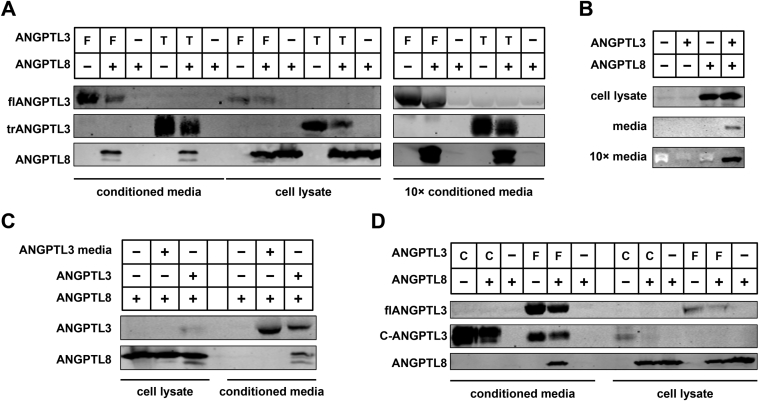
Although we were able to easily produce ANGPTL3 conditioned media simply by transfecting 293T cells, our initial attempts to do the same with ANGPTL8 met with limited success. Although ANGPTL8 protein was easily detectable in the cell lysate, little protein was secreted into the media (Figure 5A). Surprisingly, when ANGPTL8 was co-transfected with ANGPTL3, secreted ANGPTL8 could easily be detected in media (Figure 5A). ANGPTL8 was also secreted much more efficiently from HepG2 cells when co-expressed with ANGPTL3 (Figure 5B). Increased secretion of ANGPTL8 in the presence of ANGPTL3 depended on co-expression as adding ANGPTL3 exogenously (by growing ANGPTL8-transfected cells in ANGPTL3 conditioned media) did not facilitate ANGPTL8 secretion (Figure 5C). We found that truncated ANGPTL3 also promoted ANGPTL8 secretion (Figure 5A), again suggesting that the N-terminal region of ANGPTL3 mediates its interactions with ANGPTL8. Consistent with this notion, co-expression of the C-terminal domain of ANGPTL3 with ANGPTL8 did not promote the secretion of ANGPTL8 into the media (Figure 5D). Combined with our data showing that ANGPTL3 and ANGPTL8 interact only when co-expressed, these data support a hypothesis that ANGPTL3 and ANGPTL8 form a complex within the cell and are then secreted together.
Figure 5.
Secretion of ANGPTL8. (A) Western blots showing conditioned media (lanes 1–6) and cell lysates (lanes 7–12) from 293T cells transfected with full-length (F) or truncated (T) ANGPTL3 and/or with ANGPTL8. Blots on the right show media after 10-fold concentration using Millipore centrifugal filter units. (B) Western blots against ANGPTL8 showing cell lysates, conditioned media, and 10× concentrated conditioned media from HepG2 cells transfected with full-length ANGPTL3 and/or ANGPTL8. (C) Western blots showing cell lysate (lanes 1–3) and conditioned media (lanes 5–7) from 293T cells transfected with ANGPTL8 alone (lanes 1 and 5), ANGPTL8 alone and incubated in ANGPTL3 conditioned media for 48 h (lanes 2 and 6), or co-transfected with ANGPTL8 and ANGPTL3 (lanes 3 and 7). (D) Western blots showing conditioned media (lanes 1–6) and cell lysates (lanes 7–12) from 293T cells transfected with full-length ANGPTL3 (F), the C-terminal domain of ANGPTL3 (C) and/or with ANGPTL8. All western blots were performed with antibodies against the V5 tag for ANGPTL8 and against the Strep tag for ANGPTL3.
3.5. ANGPTL8 promotes the inhibitory efficiency of ANGPTL3
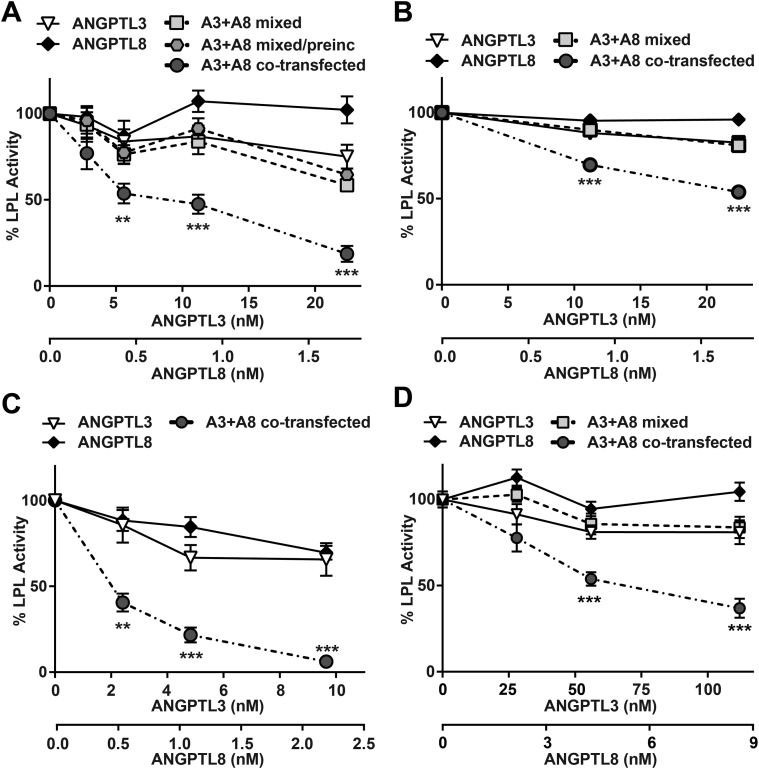
Previous studies have reported that overexpression of ANGPTL8 by adenoviral injection increased plasma TG levels in mice (presumably by decreasing LPL activity), but only when ANGPTL3 was present [19]. These data suggest that ANGPTL8 may function as a modulator of ANGPTL3 activity. We asked if ANGPTL8 could promote the inhibitory efficiency of ANGPTL3 ex vivo. To generate sufficient secreted ANGPTL8 for controls, ANGPTL8 conditioned media were collected from stable, lentivirally-transduced 293T cells. We incubated LPL with ANGPTL3 alone, ANGPTL8 alone, ANGPTL3 and ANGPTL8 that had been mixed post-secretion, and co-expressed ANGPTL3 and ANGPTL8. We found that ANGPTL8 had little ability to inhibit LPL on its own (Figure 6A). ANGPTL3 displayed some inhibitory activity, but this inhibitory activity was not enhanced when ANGPTL3 and ANGPTL8 were mixed post-secretion, even if ANGPTL8 and ANGPTL3 were incubated together before being adding to LPL (Figure 6A). However, when ANGPTL8 and ANGPTL3 were co-expressed, the inhibitory activity of ANGPTL3 was greatly enhanced (Figure 6A). Similar results were obtained when LPL activity was determined by measuring lipolysis of radiolabeled chylomicrons (Figure 6B) or when using human ANGPTL3 and ANGPTL8 (Figure 6C).
Figure 6.
Enhancement of LPL inhibition by ANGPTL8. Lipase activity of LPL conditioned media incubated with the conditioned media of cells expressing ANGPTL3 alone (ANGPTL3), ANGPTL8 alone (ANGPTL8), both ANGPTL3 and ANGPTL8 (A3 + A8 co-transfected), a mixture of ANGPTL3 and ANGPTL8 conditioned media (A3 + A8 mixed) or a mixture of ANGPTL3 and ANGPTL8 conditioned media that was incubated for 30 min at 37 °C before being added to LPL (A3 + A8 mixed and pre-incubated). (A) Lipase activity of LPL after incubation with the indicated concentrations of mouse ANGPTL3 and/or mouse ANGPTL8 conditioned media for 30 min at 37 °C as measured by fluorescence assay. Points represent mean (±SEM) of 3 independent experiments each done in duplicate. (B) Lipase activity of LPL after incubation with the indicated concentrations of mouse ANGPTL3 and/or mouse ANGPTL8 conditioned media for 30 min at 37 °C as measured by the lipolysis of radiolabeled chylomicrons. Points represent mean (±SEM) of 3 independent experiments each done in quadruplicate. (C) Lipase activity of LPL after incubation with the indicated concentrations of human ANGPTL3 and/or human ANGPTL8 conditioned media for 30 min at 37 °C as measured by fluorescence assay. Points represent mean (±SEM) of 3 independent experiments each done in duplicate. (D) Lipase activity of GPIHBP1-bound LPL (see Figure 1) incubated with the indicated concentrations of ANGPTL3 and/or ANGPTL8 conditioned media for 30 min at 37 °C and then washed. Points represent mean (±SEM) of 3 independent experiments each done in triplicate. For each graph, activity was normalized to control treated LPL. **p < 0.01; ***p < 0.001 compared to ANGPTL3 alone.
Sonnenburg et al. previously reported that GPIHBP1 protects LPL from ANGPTL3 inhibition [33], and our data with ANGPTL3 alone were consistent with that report (see Figure 1). Thus, we asked if ANGPTL8 could promote the inhibitory efficiency of ANGPTL3 when LPL was anchored to endothelial cells by GPIHBP1. As before, LPL-GPIHBP1 complexes were formed on the surface of endothelial cells. Cells were then incubated with ANGPTL3 alone, ANGPTL8 alone, ANGPTL3 and ANGPTL8 that had been mixed post-secretion, and co-expressed ANGPTL3 and ANGPTL8. After incubation, LPL was released into solution by heparin (100 U/ml) and immediately assayed for lipase activity. Consistent with our previous observations, ANGPTL3 alone, ANGPTL8 alone, and ANGPTL3 mixed with ANGPTL8 had little ability to inhibit GPIHBP1-bound LPL, whereas co-expressed ANGPTL3 and ANGPTL8 demonstrated significant inhibition (Figure 6D). It is important to note that although ANGPTL3–ANGPTL8 complexes could inhibit LPL, GPIHBP1 still provided substantial protection from inhibition (Figure 6D).
Although on its own, truncated ANGPTL3 was not more effective than full-length ANGPTL3 at inhibiting LPL, we considered the possibility that ANGPTL8 might be an even more potent activator of truncated ANGPTL3 than of full-length ANGPTL3. When using equivalent protein levels (Figure S3A), co-expression with ANGPTL8 did increase the ability of truncated ANGPTL3 to inhibit LPL; however, as before, this inhibition did not exceed that observed with full-length ANGPTL3 (Figure S3B). Similar results were observed when we tested the ability of truncated ANGPTL3 to inhibit LPL bound to endothelial cells by GPIHBP1 (Figure S3C).
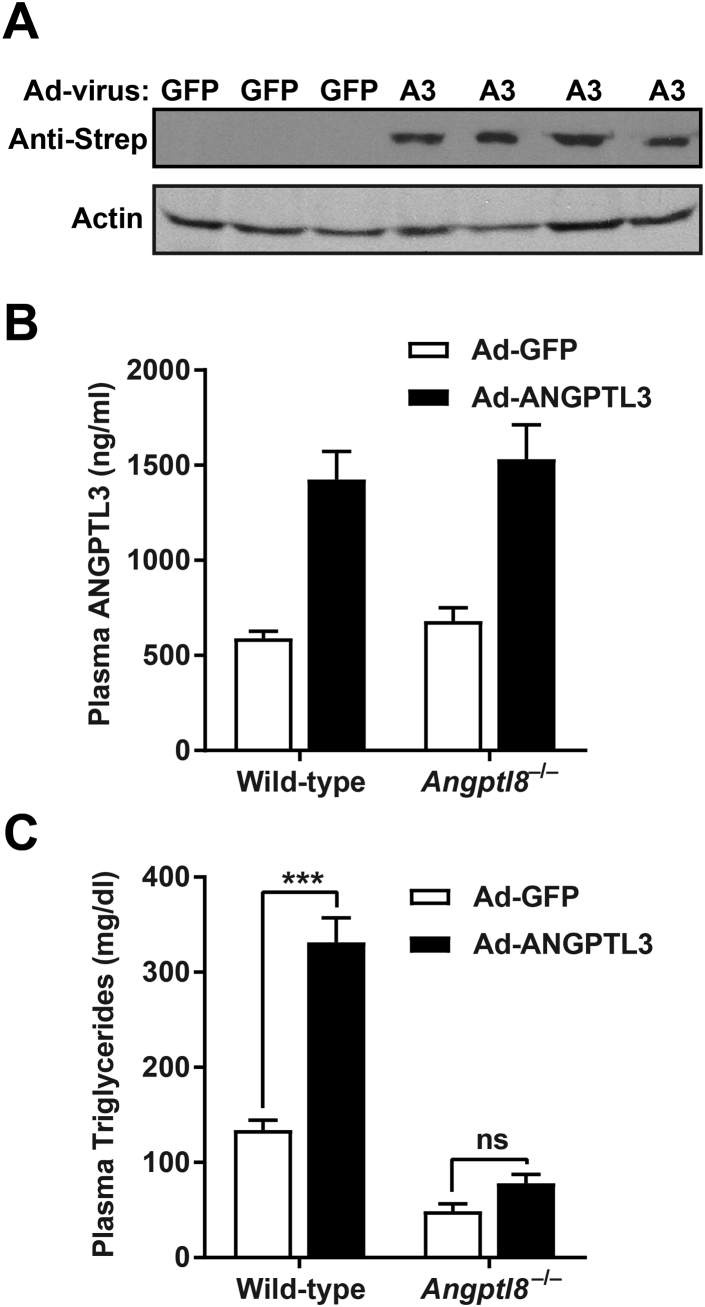
Previous studies have shown that ANGPTL8 requires ANGPTL3 to modulate plasma triglyceride levels. Our data suggest that the converse is also true, that at physiological concentrations, ANGPTL3 also requires ANGPTL8 to inhibit LPL. To test this notion in vivo we injected wild-type and Angptl8−/− mice with adenovirus expressing either mouse ANGPTL3 or GFP. After 5 days we measured liver and plasma protein levels and plasma triglycerides in the fed state. Western blot analysis of liver samples showed successful expression of exogenous ANGPTL3 (Figure 7A). Total plasma concentrations of ANGPTL3 were increased 2–2.5-fold in both wild-type and Angptl8−/− mice injected with ANGPTL3 adenovirus (Figure 7B). Plasma triglycerides in wild-type mice injected with ANGPTL3-adenovirus were significantly increased compared to GFP-adenovirus injected controls, and this increase in triglyceride levels was proportional to the increase in plasma ANGPTL3 (Figure 7C). Injection of ANGPTL3-adenovirus into Angptl8−/− mice, however, led only to a marginal, and not statistically significant, increase in plasma triglyceride levels (Figure 7C), despite the fact that the increase in plasma ANGPTL3 was similar to that observed in wild-type mice. These data suggest that ANGPTL8 is required for efficient inhibition of LPL by ANGPTL3 in vivo.
Figure 7.
Overexpression of ANGPTL3 in ANGPTL8-deficient mice. 14–15 week old male wild-type and Angptl8−/− mice (n = 3–5/group) were injected via the tail vein with adenovirus expressing either Strep-tagged mouse ANGPTL3 (A3) or GFP. After 4 days, mice were fasted for 20 h and then re-fed for 4 h. (A) Western blot of liver lysates from Angptl8−/− mice using an antibody against the Strep-tag. (B) Total plasma ANGPTL3 levels as measured by ELISA. Bar graphs show mean ± SEM. C. Plasma triglyceride levels (mean ± SEM; ***p < 0.001; ns, not significant).
3.6. ANGPTL8 promotes the binding of ANGPTL3 to LPL
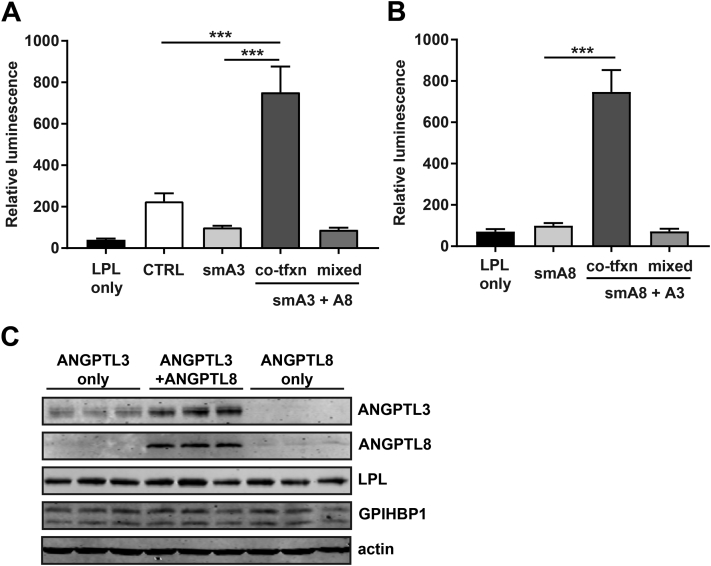
We next examined the mechanism by which ANGPTL8 increased the inhibitory activity of ANGPTL3. We hypothesized that ANGPTL8 might increase LPL inhibition by increasing the ability of ANGPTL3 to bind LPL. To test this hypothesis, we used Promega's split-luciferase NanoBiT system, wherein two parts of NanoLuciferase (the largeBiT and the smallBiT) are appended to proteins, and luciferase activity is only reconstituted when the target proteins interact [31]. The largeBiT was appended to the C-terminus of LPL while ANGPTL3 was tagged with the smallBiT. Thus, functional luciferase activity would only be reconstituted when LPL and ANGPTL3 interact. As a negative control we attached the smallBiT to ASIP (Agouti-signaling protein), an unrelated secreted protein that has no known role in LPL metabolism [37]. We asked if ANGPTL8 could enhance the ability of ANGPTL3 to bind LPL on the surface of endothelial cells by incubating smallBiT-ANGPTL3 alone or in combination with ANGPTL8 with largeBiT-LPL bound to endothelial cells by GPIHBP1. When LPL-GPIHBP1 complexes were incubated with ANGPTL3 alone, we did not observe luciferase activity above that of the negative control, indicating little binding to LPL (Figure 8A). However, when ANGPTL3 was co-expressed with ANGPTL8, significant luciferase activity was observed, indicating that ANGPTL8 could greatly enhance the ability of ANGPTL3 to bind LPL (Figure 8A). Consistent with our previous observations, mixing ANGPTL3 and ANGPTL8 that had been expressed separately did not enhance ANGPTL3 binding (Figure 8A).
Figure 8.
Binding of ANGPTL3 and ANGPTL8 to LPL. GPIHBP1-expressing RHMVECs were incubated with largeBiT-LPL (A and B) or flag-LPL (C) for 3.5 h at 4 °C and unbound LPL was washed away. (A) Relative luminescence of cells incubated with control (smallBiT-ASIP), smallBiT-ANGPTL3 (smA3), smallBiT-ANGPTL3 and ANGPTL8 co-transfected (co-tfxn), or smallBiT-ANGPTL3 and ANGPTL8 separately transfected and then mixed (mixed) for 15 min at 37 °C, and then washed. Bar graphs show mean (±SEM) of 4 independent experiments each done in triplicate. (B) Relative luminescence of cells incubated with smallBiT-ANGPTL8 (smA8), smallBiT-ANGPTL8 and ANGPTL3 co-transfected (co-tfxn), or smallBiT-ANGPTL8 and ANGPTL3 separately transfected and then mixed (mixed) for 15 min at 37 °C, and then washed. Bar graphs show mean (±SEM) of 3 independent experiments each done in triplicate. (C) Western blot of lysates from cells incubated with ANGPTL3 only, ANGPTL8 only, or ANGPTL3 and ANGPTL8 co-transfected conditioned media for 10 min at 37 °C and then washed. Western blot is representative of 3 independent experiments done in triplicate.
To determine if the binding of ANGPTL3–ANGPTL8 complexes to LPL was primarily mediated by ANGPTL8, we performed experiments with smallBiT-tagged ANGPTL8. We found that smallBiT-ANGPTL8 alone bound poorly to LPL, but that binding was greatly enhanced when smallBiT-ANGPTL8 was co-expressed with ANGPTL3 (Figure 8B), These data suggest that neither ANGPTL3 nor ANGPTL8 alone binds efficiently to LPL, but binding is greatly enhanced when the two proteins form a complex.
To confirm these findings, we used western blot analysis to analyze the binding of ANGPTL3 and ANGPTL8 to LPL-GPIHBP1 complexes on the surface of endothelial cells. After incubating GPIHBP1-expressing endothelial cells with LPL to generate cell-bound LPL-GPIHBP1 complexes, these cells were incubated with ANGPTL3, ANGPTL8, or both. After washing off unbound protein, cells were harvested and analyzed by western blot to determine ANGPTL3 and ANGPTL8 binding. ANGPTL8 alone did not bind to cells (Figure 8C; lanes 7–9), and only weak binding of ANGPTL3 was detected when cells were incubated with ANGPTL3 alone (Figure 8C; lanes 1–3). However, binding of both ANGPTL3 and ANGPTL8 was observed when cells were incubated with ANGPTL3 and ANGPTL8 that had been co-expressed (Figure 8C; lanes 4–6), again indicating that co-expression of both proteins is necessary for efficient binding to GPIHBP1-bound LPL.
4. Discussion
In this study, we investigated the mechanism by which ANGPTL8 promotes the inhibitory efficiency of ANGPTL3. Our major findings were that (1) neither full-length nor truncated ANGPTL3 was a potent inhibitor of LPL. (2) ANGPTL3-inactivated LPL was not able to bind GPIHBP1. (3) ANGPTL8 can form a complex with both truncated and full-length ANGPTL3, but only when the proteins are co-expressed. (4) ANGPTL3 facilitates the secretion of ANGPTL8. (5) ANGPTL8 enhances the ability of ANGPTL3 to inhibit LPL and does so in part by improving the binding of ANGPTL3 to LPL.
The ability of ANGPTL3 to modulate LPL activity and regulate triglyceride metabolism is supported by a preponderance of physiological data. ANGPTL3-deficient mice have lower plasma triglycerides and increased post-heparin plasma LPL activity, whereas ANGPTL3 overexpression increases plasma triglycerides [10], [11], [12], [14], [16]. Moreover, biochemical studies have reported that ANGPTL3 can directly inhibit LPL in vitro [13], [33], [34]. However, each of these studies required supraphysiologic concentrations of ANGPTL3 to inhibit LPL. Likewise, our studies required high concentrations of ANGPTL3, and this issue was further compounded when LPL was bound to GPIHBP1 on the surface of endothelial cells, the context in which ANGPTL3 most likely encounters LPL. In aggregate, these results suggest that ANGPTL3 must be activated in some way before it can effectively regulate LPL. A previous study found that ANGPTL3 cleavage improved ANGPTL3 action in vivo, but the same study found that preventing ANGPTL3 cleavage had no effect on LPL inhibition in vitro [34]. Our data are consistent with the latter finding, and, furthermore, we show that truncated ANGPTL3 does not improve inhibition of GPIHBP1-bound LPL. The physiological role of cleaved ANGPTL3 is not yet clear, but it is possible that removal of the fibrinogen-like domain improves its endocrine qualities in some way, perhaps by improving its circulation.
Despite previous evidence that ANGPTL3 and ANGPTL8 may act together [19], the nature of this interaction has remained elusive. Our data suggest that ANGPTL3 and ANGPTL8 form a complex and do so much more efficiently when the two proteins are co-expressed. It is not clear why complex formation is more efficient when the proteins are co-expressed, but there are several possibilities. Glycosylation or other post-translational modifications may inhibit complex formation after secretion. It is also possible that, in the absence of ANGPTL8, ANGPTL3 forms homooligomers [25], and that formation of these oligomeric structures prevents oligomerization with ANGPTL8. Finally, it is possible that co-translation and co-folding of the proteins, perhaps in the presence of a chaperone, is necessary for the proteins to adopt a confirmation conducive to complex formation.
As a complex, ANGPTL3 and ANGPTL8 exhibited a greatly enhanced ability to bind LPL compared to either protein alone, thus providing a clear mechanism by which ANGPTL8 augments ANGPTL3's inhibitory action. While this manuscript was in preparation, Haller et al. published their findings, which, like ours, strongly support the hypothesis that ANGPTL8 requires ANGPTL3 to inhibit LPL activity [38]. Their data suggested a model wherein the binding of ANGPTL3 to ANGPTL8 unmasks an inhibitory motif in ANGPTL8, thus allowing ANGPTL8 to act on LPL. This model is compatible with our data, and it could be that the unmasking of a motif in ANGPTL8 is what allows the complex as a whole to bind and inhibit LPL.
Our study also indicates that ANGPTL3, unless highly overexpressed, cannot significantly inhibit LPL in the absence of ANGPTL8, and provides clear evidence that ANGPTL8 is responsible for the activation of ANGPTL3. Only when ANGPTL3 and ANGPTL8 were co-expressed could they robustly inhibit LPL. Moreover, in vivo overexpression of ANGPTL3 significantly increased plasma triglycerides only in wild-type mice but had little effect on triglycerides in Angptl8−/− mice. As ANGPTL8 is induced in the fed state [19], [20], our results explain why ANGPTL3, despite exhibiting consistent expression across feeding states [25], primarily modulates LPL activity in the fed state [12]. Extreme overexpression of ANGPTL3 may increase plasma triglycerides in the absence of ANGPTL8, as ANGPTL3 at high concentrations can inhibit LPL in the absence of ANGPTL8 (see Figure 1), but there is no evidence that, physiologically, such high levels of ANGPTL3 ever occur.
It had been previously proposed that ANGPTL8 may function to promote the cleavage of ANGPTL3 [19]. However, a later study showed that ANGPTL8 was not required for ANGPTL3 cleavage [21], and our own data suggest that ANGPTL3 cleavage alone would not be sufficient to render ANGPTL3 into an effective LPL inhibitor. Instead, we found that ANGPTL8 forms a complex with ANGPTL3 and it is this complex that is necessary to efficiently bind and inhibit LPL.
We found that ANGPTL8 required ANGPTL3 co-expression to be efficiently secreted from cells. This finding explains an initial study showing that ANGPTL8 could not be secreted from cells [20] and is consistent with a more recent study showing that ANGPTL8 was secreted from CHO cells co-transfected with ANGPTL3 [39]. This observation also has important physiological implications. Both ANGPTL3 and ANGPTL8 are expressed in liver [14], [17], [18], [19], [20] and thus both could be secreted from liver as a complex, enter into the circulation, and inhibit vascular LPL. In addition to the liver, ANGPTL8 is also expressed in adipose tissue. Despite this expression, ANGPTL8 appears to inhibit LPL activity primarily in heart and skeletal muscle, not in adipose tissue [24]. Our study implies that ANGPTL8 would not be secreted efficiently from adipose tissue, as adipose lacks ANGPTL3 expression. Because ANGPTL8 must interact with ANGPTL3 to bind or inhibit LPL, the role of ANGPTL8 in adipose tissue is likely LPL-independent. It is important to note that although ANGPTL8 required ANGPTL3 for efficient secretion, the reverse was not true. ANGPTL3 was efficiently secreted from cells even when it was not co-transfected with ANGPTL8, and circulating levels of ANGPTL3 in Angptl8−/− mice were not lower than those in wild-type mice.
Our data strongly suggest that ANGPTL3 requires ANGPTL8 to inhibit LPL. However, our data also suggest that far more ANGPTL3 is secreted than ANGPTL8, a result consistent with human data; studies have found that circulating ANGPTL3 levels are 10- to 500-fold higher than circulating ANGPTL8 levels [40], [41], [42]. Moreover, our pull-down experiments suggest that even when ANGPTL3 and ANGPTL8 are co-expressed, a significant amount of ANGPTL3 is not complexed with ANGPTL8. Together, these data suggest that ANGPTL3 may have an ANGPTL8-independent function. Although further studies will be needed to elucidate this function, one possibility is the inhibition of endothelial lipase. ANGPTL3 has been shown to inhibit endothelial lipase [43], but the role of ANGPTL8 in this inhibition, if any, is not known. It is possible that ANGPTL3 inhibits endothelial lipase in the absence of ANGPTL8, but when ANGPTL8 is expressed, a subset of ANGPTL3 forms a complex with ANGPTL8 allowing efficient inhibition of LPL.
In summary, the current study is consistent with a model in which ANGPTL8, expressed in the fed state, activates the ability of ANGPTL3 to inhibit LPL [12], [19], [44] and provides a clear mechanism by which that activation occurs. Suppressing ANGPTL3 activity has been shown to reduce triglyceride levels in mice and monkeys [45], and recently reported studies found that targeting ANGPTL3 in humans also lowered plasma triglycerides [46], [47]. Recent studies have also shown that antibodies that block ANGPTL8 likewise reduce plasma triglycerides in mice, humanized ANGPTL8 mice, and dyslipidemic cynomolgus monkeys [24], [48]. Our studies provide mechanistic insight into the interactions of ANGPTL3 and ANGPTL8 and thus provide critical information that could be used for development of therapeutics targeting ANGPTL8 or the interactions between ANGPTL8 and ANGPTL3. Going forward, it will be increasingly important to understand how modulating ANGPTL8 function may affect metabolism and human health.
Acknowledgements
We thank Catherine Musselman for expert technical assistance. This work was supported by grants from the National Heart, Lung, and Blood Institute of the National Institutes of Health (R01HL130146 to BSJD, R01HL134787 to RZ) and a Carver Medical Research Initiative grant from the Carver Trust (to BSJD).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.06.014.
Conflicts of interest
None.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.De Man F.H., Cabezas M.C., Van Barlingen H.H., Erkelens D.W., de Bruin T.W. Triglyceride-rich lipoproteins in non-insulin-dependent diabetes mellitus: post-prandial metabolism and relation to premature atherosclerosis. European Journal of Clinical Investigation. 1996;26(2):89–108. doi: 10.1046/j.1365-2362.1996.114256.x. [DOI] [PubMed] [Google Scholar]
- 2.Subramanian S., Chait A. Hypertriglyceridemia secondary to obesity and diabetes. Biochimica Et Biophysica Acta. 2012;1821(5):819–825. doi: 10.1016/j.bbalip.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Do R., Willer C.J., Schmidt E.M., Sengupta S., Gao C., Peloso G.M. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nature Genetics. 2013;45(11):1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kannel W.B., Vasan R.S. Triglycerides as vascular risk factors: new epidemiologic insights. Current Opinion in Cardiology. 2009;24(4):345–350. doi: 10.1097/HCO.0b013e32832c1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller M., Stone N.J., Ballantyne C., Bittner V., Criqui M.H., Ginsberg H.N. Triglycerides and cardiovascular disease a scientific statement from the American heart association. Circulation. 2011;123(20):2292–2333. doi: 10.1161/CIR.0b013e3182160726. [DOI] [PubMed] [Google Scholar]
- 6.Korn E.D. Clearing factor, a heparin-activated lipoprotein lipase i.e. isolation and characterization of the enzyme from normal rat heart. Journal of Biological Chemistry. 1955;215(1):1–14. [PubMed] [Google Scholar]
- 7.Davies B.S.J., Beigneux A.P., Barnes R.H., II, Tu Y., Gin P., Weinstein M.M. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metabolism. 2010;12(1):42–52. doi: 10.1016/j.cmet.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beigneux A.P., Davies B.S.J., Gin P., Weinstein M.M., Farber E., Qiao X. Glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metabolism. 2007;5(4):279–291. doi: 10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulbourne C.N., Gin P., Tatar A., Nobumori C., Hoenger A., Jiang H. The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metabolism. 2014;19(5):849–860. doi: 10.1016/j.cmet.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujimoto K., Koishi R., Shimizugawa T., Ando Y. ANGPTL3-null mice show low plasma lipid concentrations by enhanced lipoprotein lipase activity. Experimental Animals. 2006;55(1):27–34. doi: 10.1538/expanim.55.27. [DOI] [PubMed] [Google Scholar]
- 11.Köster A., Chao Y.B., Mosior M., Ford A., Gonzalez-DeWhitt P.A., Hale J.E. Transgenic angiopoietin-like (ANGPTL)4 overexpression and targeted disruption of ANGPTL4 and ANGPTL3: regulation of triglyceride metabolism. Endocrinology. 2005;146(11):4943–4950. doi: 10.1210/en.2005-0476. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., McNutt M.C., Banfi S., Levin M.G., Holland W.L., Gusarova V. Hepatic ANGPTL3 regulates adipose tissue energy homeostasis. Proceedings of the National Academy of Sciences. 2015;112(37):11630–11635. doi: 10.1073/pnas.1515374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizugawa T., Ono M., Shimamura M., Yoshida K., Ando Y., Koishi R. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. The Journal of Biological Chemistry. 2002;277(37):33742–33748. doi: 10.1074/jbc.M203215200. [DOI] [PubMed] [Google Scholar]
- 14.Koishi R., Ando Y., Ono M., Shimamura M., Yasumo H., Fujiwara T. ANGPTL3 regulates lipid metabolism in mice. Nature Genetics. 2002;30(2):151–157. doi: 10.1038/ng814. [DOI] [PubMed] [Google Scholar]
- 15.Romeo S., Yin W., Kozlitina J., Pennacchio L.A., Boerwinkle E., Hobbs H.H. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. The Journal of Clinical Investigation. 2009;119(1):70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robciuc M.R., Maranghi M., Lahikainen A., Rader D., Bensadoun A., Öörni K. ANGPTL3 deficiency is associated with increased insulin sensitivity, lipoprotein lipase activity, and decreased serum free fatty acids. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33(7):1706–1713. doi: 10.1161/ATVBAHA.113.301397. [DOI] [PubMed] [Google Scholar]
- 17.Conklin D., Gilbertson D., Taft D.W., Maurer M.F., Whitmore T.E., Smith D.L. Identification of a mammalian angiopoietin-related protein expressed specifically in liver. Genomics. 1999;62(3):477–482. doi: 10.1006/geno.1999.6041. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R. Lipasin, a novel nutritionally-regulated liver-enriched factor that regulates serum triglyceride levels. Biochemical and Biophysical Research Communications. 2012;424(4):786–792. doi: 10.1016/j.bbrc.2012.07.038. [DOI] [PubMed] [Google Scholar]
- 19.Quagliarini F., Wang Y., Kozlitina J., Grishin N.V., Hyde R., Boerwinkle E. Atypical angiopoietin-like protein that regulates ANGPTL3. Proceedings of the National Academy of Sciences. 2012;109(48):19751–19756. doi: 10.1073/pnas.1217552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren G., Kim J.Y., Smas C.M. Identification of RIFL, a novel adipocyte-enriched insulin target gene with a role in lipid metabolism. American Journal of Physiology – Endocrinology and Metabolism. 2012;303(3):E334–E351. doi: 10.1152/ajpendo.00084.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Quagliarini F., Gusarova V., Gromada J., Valenzuela D.M., Cohen J.C. Mice lacking ANGPTL8 (Betatrophin) manifest disrupted triglyceride metabolism without impaired glucose homeostasis. Proceedings of the National Academy of Sciences. 2013;110(40):16109–16114. doi: 10.1073/pnas.1315292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi P., Park J.-S., Melton D.A. Betatrophin: a hormone that controls pancreatic β cell proliferation. Cell. 2013;153(4):747–758. doi: 10.1016/j.cell.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Gusarova V., Alexa C.A., Na E., Stevis P.E., Xin Y., Bonner-Weir S. ANGPTL8/betatrophin does not control pancreatic beta cell expansion. Cell. 2014;159(3):691–696. doi: 10.1016/j.cell.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Z., Abou-Samra A.B., Zhang R. A lipasin/ANGPTL8 monoclonal antibody lowers mouse serum triglycerides involving increased postprandial activity of the cardiac lipoprotein lipase. Scientific Reports. 2015;5 doi: 10.1038/srep18502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge H., Cha J.-Y., Gopal H., Harp C., Yu X., Repa J.J. Differential regulation and properties of angiopoietin-like proteins 3 and 4. Journal of Lipid Research. 2005;46(7):1484–1490. doi: 10.1194/jlr.M500005-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Beigneux A.P., Gin P., Davies B.S.J., Weinstein M.M., Bensadoun A., Fong L.G. Highly conserved cysteines within the ly6 domain of gpihbp1 are crucial for the binding of lipoprotein lipase. Journal of Biological Chemistry. 2009;284(44):30240–30247. doi: 10.1074/jbc.M109.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi X., Shetty S.K., Shows H.W., Hjelmaas A.J., Malcolm E.K., Davies B.S.J. Angiopoietin-like 4 modifies the interactions between lipoprotein lipase and its endothelial cell transporter gpihbp1. Journal of Biological Chemistry. 2015;290(19):11865–11877. doi: 10.1074/jbc.M114.623769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basu D., Manjur J., Jin W. Determination of lipoprotein lipase activity using a novel fluorescent lipase assay. Journal of Lipid Research. 2011;52(4):826–832. doi: 10.1194/jlr.D010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang T., Li L., Tang J., Li Y., Lin W.Y., Martin F. A mouse knockout library for secreted and transmembrane proteins. Nature Biotechnology. 2010;28(7):749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- 30.Young S.G., Davies B.S.J., Voss C.V., Gin P., Weinstein M.M., Tontonoz P. GPIHBP1, an endothelial cell transporter for lipoprotein lipase. Journal of Lipid Research. 2011;52(11):1869–1884. doi: 10.1194/jlr.R018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon A.S., Schwinn M.K., Hall M.P., Zimmerman K., Otto P., Lubben T.H. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chemical Biology. 2016;11(2):400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 32.Hall M.P., Unch J., Binkowski B.F., Valley M.P., Butler B.L., Wood M.G. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chemical Biology. 2012;7(11):1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnenburg W.K., Yu D., Lee E.-C., Xiong W., Gololobov G., Key B. GPIHBP1 stabilizes lipoprotein lipase and prevents its inhibition by angiopoietin-like 3 and angiopoietin-like 4. Journal of Lipid Research. 2009;50(12):2421–2429. doi: 10.1194/jlr.M900145-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono M., Shimizugawa T., Shimamura M., Yoshida K., Noji-Sakikawa C., Ando Y. Protein region important for regulation of lipid metabolism in angiopoietin-like 3 (ANGPTL3) ANGPTL3 is cleaved and activated in vivo. Journal of Biological Chemistry. 2003;278(43):41804–41809. doi: 10.1074/jbc.M302861200. [DOI] [PubMed] [Google Scholar]
- 35.Essalmani R., Susan-Resiga D., Chamberland A., Asselin M.-C., Canuel M., Constam D. Furin is the primary in vivo convertase of angiopoietin-like 3 and endothelial lipase in hepatocytes. Journal of Biological Chemistry. 2013;288(37):26410–26418. doi: 10.1074/jbc.M113.501304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei X., Shi F., Basu D., Huq A., Routhier S., Day R. Proteolytic processing of angiopoietin-like protein 4 by proprotein convertases modulates its inhibitory effects on lipoprotein lipase activity. Journal of Biological Chemistry. 2011;286(18):15747–15756. doi: 10.1074/jbc.M110.217638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu D., Willard D., Patel I.R., Kadwell S., Overton L., Kost T. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature. 1994;371(6500):799–802. doi: 10.1038/371799a0. [DOI] [PubMed] [Google Scholar]
- 38.Haller J.F., Mintah I.J., Shihanian L.M., Stevis P., Buckler D., Alexa-Braun C.A. ANGPTL8 requires ANGPTL3 to inhibit lipoprotein lipase and plasma triglyceride clearance. Journal of Lipid Research. 2017 doi: 10.1194/jlr.M075689. jlr.M075689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nidhina Haridas P.A., Soronen J., Sädevirta S., Mysore R., Quagliarini F., Pasternack A. Regulation of angiopoietin-like proteins (ANGPTLs) 3 and 8 by insulin. The Journal of Clinical Endocrinology & Metabolism. 2015;100(10):E1299–E1307. doi: 10.1210/jc.2015-1254. [DOI] [PubMed] [Google Scholar]
- 40.Abu-Farha M., Al-Khairi I., Cherian P., Chandy B., Sriraman D., Alhubail A. Increased ANGPTL3, 4 and ANGPTL8/betatrophin expression levels in obesity and T2D. Lipids in Health and Disease. 2016;15 doi: 10.1186/s12944-016-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung H.S., Lee M.J., Hwang S.Y., Lee H.J., Yoo H.J., Seo J.-A. Circulating angiopoietin-like protein 8 (ANGPTL8) and ANGPTL3 concentrations in relation to anthropometric and metabolic profiles in Korean children: a prospective cohort study. Cardiovascular Diabetology. 2016;15 doi: 10.1186/s12933-015-0324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haridas N.A.P., Soronen J., Sädevirta S., Mysore R. Regulation of angiopoietin-like proteins (ANGPTLs) 3 and 8 by insulin. The Journal of Clinical Endocrinology & Metabolism. 2015;100(10):E1299–E1307. doi: 10.1210/jc.2015-1254. [DOI] [PubMed] [Google Scholar]
- 43.Shimamura M., Matsuda M., Yasumo H., Okazaki M., Fujimoto K., Kono K. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arteriosclerosis, Thrombosis, and Vascular Biology. 2007;27(2):366–372. doi: 10.1161/01.ATV.0000252827.51626.89. [DOI] [PubMed] [Google Scholar]
- 44.Zhang R. The ANGPTL3-4-8 model, a molecular mechanism for triglyceride trafficking. Open Biology. 2016;6(4) doi: 10.1098/rsob.150272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gusarova V., Alexa C.A., Wang Y., Rafique A., Kim J.H., Buckler D. ANGPTL3 blockade with a human monoclonal antibody reduces plasma lipids in dyslipidemic mice and monkeys. Journal of Lipid Research. 2015;56(7):1308–1317. doi: 10.1194/jlr.M054890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graham M.J., Lee R.G., Brandt T.A., Tai L.-J., Fu W., Peralta R. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. New England Journal of Medicine. 2017;0(0) doi: 10.1056/NEJMoa1701329. null. [DOI] [PubMed] [Google Scholar]
- 47.Dewey F.E., Gusarova V., Dunbar R.L., O'Dushlaine C., Schurmann C., Gottesman O. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. New England Journal of Medicine. 2017;0(0) doi: 10.1056/NEJMoa1612790. null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gusarova V., Banfi S., Alexa-Braun C.A., Shihanian L.M., Mintah I.J., Lee J.S. ANGPTL8 blockade with a monoclonal antibody promotes triglyceride clearance, energy expenditure and weight loss in mice. Endocrinology. 2017 doi: 10.1210/en.2016-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.