Abstract
Despite a wealth of biochemical data, the mechanism by which targets of the nonsense‐mediated mRNA decay (NMD) pathway are recognized during translation termination remains elusive. A new study by Neu‐Yilik et al (2017) using a fully reconstituted in vitro translation termination system reveals new roles for the NMD factor UPF3B in translation termination and a direct interaction between UPF3B and ribosome release factors.
Subject Categories: Protein Biosynthesis & Quality Control, RNA Biology
Nonsense‐mediated mRNA decay (NMD) is a cellular surveillance pathway recognizing and degrading mRNAs that fail to terminate translation properly. However, at which termination codons (TCs) and under which circumstances NMD is triggered, and how normal translation termination differs mechanistically from proper termination is still poorly understood. To study the influence of the evolutionarily conserved NMD factors UPF1, UPF2, and UPF3B on translation termination, Neu‐Yilik et al (2017) adopted a fully reconstituted in vitro translation termination system previously developed in the Pestova laboratory (Alkalaeva et al, 2006). In this system, so‐called pre‐termination complexes (pre‐TCs) are assembled from purified mammalian ribosomal subunits, aminoacylated tRNAs, and initiation and elongation factors. These pre‐TCs contain the peptidyl‐tRNA in the P‐site, and the TC is aligned to the A‐site of the ribosome. Ribosomal occupancy at the termination codon can be visualized at single nucleotide resolution by the toeprinting technique, and these complexes yield toeprints 13 nucleotides downstream of the U nucleotide of the TC. Addition of the eukaryotic release factors eRF1 and eRF3a leads to conformational changes in the pre‐TC that shifts the toeprint 1–2 nucleotides forward and subsequent GTP hydrolysis by eRF3 triggers the release of the nascent peptide, converting the pre‐TC into the post‐termination complex (post‐TC). Based on the evidence that termination events resulting in NMD are slower than normal termination events (Amrani et al, 2004; Peixeiro et al, 2011; Ivanov et al, 2016), Neu‐Yilik et al (2017) attempted to mimic this situation by adding only limiting amounts of eRFs to their translation termination assays. Under these conditions, neither UPF1 nor UPF2 had an effect on pre‐TC‐to‐post‐TC transition. By contrast, addition of UPF3B reduced the efficiency of post‐TC formation. Interestingly, addition of UPF2 together with UPF3B abolished the capacity of UPF3B to delay translation termination, indicating that binding to UPF2 prevents UPF3B from interfering with translation termination. Monitoring peptide release showed that UPF3B reduced the peptidyl‐tRNA hydrolysis, a process that is catalyzed by the conserved GGQ motif of eRF1 and strongly enhanced by GTP hydrolysis of eRF3a.
Previous co‐immunoprecipitation (co‐IP) experiments had indicated a physical link between the NMD apparatus – specifically UPF1 – and the release factors (Kashima et al, 2006; Ivanov et al, 2008; Singh et al, 2008), but these co‐IPs give no information on whether the detected interactions are direct or indirect. With purified recombinant proteins in hand, Neu‐Yilik et al (2017) re‐examined the interactions between UPF1, UPF2, UPF3B, and eRF1/eRF3a. Unexpectedly and inconsistent with the previous interpretations of the co‐IP data, recombinant UPF1 neither bound eRF1 nor eRF3a individually, nor the eRF1/eRF3a complex. By contrast, UPF3B was found to interact with eRF3a and to a lesser extent with eRF1 and a stable trimeric complex consisting of UPF3B, eRF3a, and eRF1 was detected by size exclusion chromatography, suggesting that the observed effect of UPF3B on translation termination was mediated by a direct interaction of this protein with the release factors. The authors further showed that the N‐terminal part of eRF3a, which is dispensable for its function in translation termination, interacts with the so far uncharacterized middle domain of UPF3B (amino acids 147–419). Interestingly, most of the reported UPF3B mutations associated with neurodevelopmental effects are located in this region (Alrahbeni et al, 2015). A further surprise from these interaction studies was that UPF1 and UPF3B also appear to interact directly with each other, an interaction that hitherto was thought to be bridged by UPF2. These findings support a role of UPF3B as a hub that directly binds to the ribosome, UPF1, and RNA and also takes part in the termination complex by interacting with release factors, providing opportunities for an immediate link to NMD activation.
Using the in vitro translation termination assays, Neu‐Yilik et al (2017) also examined effects of UPF1‐3 in the presence of saturating release factor concentrations. Under these conditions, essentially all pre‐TCs were converted to post‐TCs as indicated by the toeprint shifts of 1–2 nucleotides. Addition of UPF1, irrespectively of its phosphorylation status or ATPase activity, or UPF2 had no effect on the toeprint pattern, suggesting that these two factors have no impact on translation termination. Addition of UPF3B still led to a small retention of pre‐TCs, implying that even when release factors were not limiting, UPF3B caused some delay in termination. However, a more prominent effect of UPF3B observed under these assay conditions was a decrease in the post‐TC toeprint signal and a concomitant increase in the full‐length mRNA toeprint signal, indicating that UPF3B promotes the release of post‐TCs from the mRNA. This apparent UPF3B‐mediated destabilization of post‐TCs is independent of UPF1 but prevented by UPF2. Neither the N‐terminal RNA recognition motif (RRM) nor the middle domain of UPF3B were sufficient on their own to exert post‐TC dissociation, while the exon junction complex binding motif (EBM) was dispensable. The authors further showed that the presence of nonhydrolyzable GTP or a peptide release‐deficient eRF1 mutant (eRF1 AGQ) in the reaction abolished the UPF3B‐promoted post‐TC dissociation, showing that UPF3B exerts its function after release of the nascent peptide. Curiously, UPF3B also dissociated post‐TCs in the absence of eRF3a, indicating that the above‐characterized UPF3B–eRF3a interaction is not required for UPF3B‐mediated ribosome dissociation. Similar to the previously reported ribosome recycling activities of eIF3, eIF1, and eIF1A (Pisarev et al, 2007), the UPF3B activity was only observed at low (physiological) Mg2+ concentrations, suggesting that UPF3B may promote ribosome release by accessing the ribosome subunit interface, which is stabilized at elevated Mg2+ concentrations.
Collectively, the data presented by Neu‐Yilik et al (2017) beg for some adjustments in the currently prevailing mechanistic models for NMD. Foremost, their data suggest that UPF3B rather than UPF1 may be the key player in functionally and physically interacting with the translation termination machinery. Neither UPF1 per se nor its helicase activity or phosphorylation status affected the termination reaction in the reconstituted in vitro system and the previously reported interaction between UPF1 and the release factors appears to be indirect, mediated primarily by UPF3B. While it cannot be ruled out that the employed in vitro assay fails to faithfully recapitulate the in vivo situation, the new results seem to indicate that UPF1 exerts its essential role in NMD at a step after translation termination, possibly in recruiting the RNA decay machinery and in dissociating the NMD complex from the 3′ decay fragment after SMG6‐mediated endonucleolytic cleavage of the mRNA (Franks et al, 2010). Instead, UPF3B appears to influence translation termination at two steps: first by further delaying pre‐TC‐to‐post‐TC transition and peptide release when termination is inefficient and second by promoting dissociation of post‐TCs after peptide release. Based on their results, Neu‐Yilik et al (2017) propose a modified NMD model (Fig 1), according to which UPF3B binds ribosomes that stall at TCs in the absence of termination‐promoting PABPC1 and facilitates the recruitment of the release factors. After peptide release has occurred and now independent of its interaction with the release factors, UPF3B promotes ribosome release. Subsequently, it nucleates assembly of an NMD‐triggering complex, by associating with UPF1 and UPF2, which leads to the phosphorylation of UPF1 and in turn the recruitment of the decay factors (e.g., SMG6 and SMG5/SMG7). This modified NMD model makes a number of predictions that need to be tested in the future, for example, that UPF3B would be associated specifically with terminating ribosomes on mRNAs targeted by NMD and that UPF3B requires the presence of UPF1 to link aberrant translation termination to NMD activation. Regardless of some inconsistencies and contradictions between the results of this study and previously published data that eventually need to be clarified, the new data by Neu‐Yilik et al (2017) constitute an important and thought‐stimulating contribution to the intricate challenge of deciphering the molecular mechanism of NMD. Clearly, the in vitro translation termination assay represents a powerful and promising tool for succeeding in this endeavor.
Figure 1. The role of the NMD factor UPF3B in aberrant translation termination.
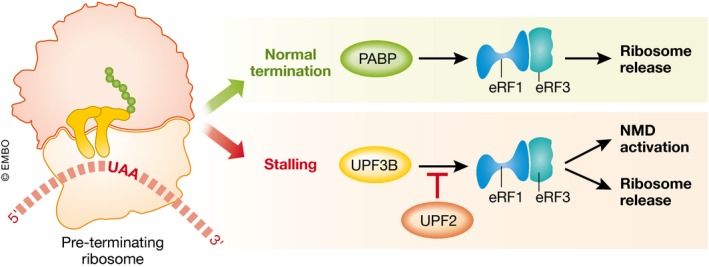
In the context of normal translation termination, PABP (PolyA‐binding protein) facilitates recruitment of the release factors (eRF1/eRF3) to the pre‐TC. When translation termination is aberrant, ribosome stalling is further enhanced by the NMD factor UPF3B, which aids the recruitment of release factors and subsequently promotes ribosome release and the assembly of NMD factors. UPF2 antagonizes the UPF3B‐enhanced recruitment of the eRF1/eRF3 complex to the pre‐TC.
See also: G Neu‐Yilik et al (October 2017)
References
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV (2006) In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Alrahbeni T, Sartor F, Anderson J, Miedzybrodzka Z, McCaig C, Muller B (2015) Full UPF3B function is critical for neuronal differentiation of neural stem cells. Mol Brain 8: 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′‐UTR promotes aberrant termination and triggers nonsense‐mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke‐Andersen J (2010) Upf1 ATPase‐dependent mRNP disassembly is required for completion of nonsense‐ mediated mRNA decay. Cell 143: 938–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE (2008) Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Mikhailova T, Eliseev B, Yeramala L, Sokolova E, Susorov D, Shuvalov A, Schaffitzel C, Alkalaeva E (2016) PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res 44: 7766–7776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG‐1‐Upf1‐eRF1‐eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense‐mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu‐Yilik G, Raimondeau E, Eliseev B, Yeramala L, Amthor B, Deniaud A, Huard K, Kerschgens K, Hentze MW, Schaffitzel C, Kulozik AE (2017) Dual function of UPF3B in early and late translation termination. EMBO J 36: 2968–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixeiro I, Inacio A, Barbosa C, Silva AL, Liebhaber SA, Romao L (2011) Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG‐proximal nonsense mutations. Nucleic Acids Res 40: 1160–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV (2007) Recycling of eukaryotic posttermination ribosomal complexes. Cell 131: 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke‐Andersen J (2008) A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense‐mediated mRNA decay. PLoS Biol 6: e111 [DOI] [PMC free article] [PubMed] [Google Scholar]


