Abstract
Objective
Alternative activation (M2) of adipose tissue resident macrophage (ATM) inhibits obesity-induced metabolic inflammation. The underlying mechanisms remain unclear. Recent studies have shown that dysregulated lipid homeostasis caused by increased lipolysis in white adipose tissue (WAT) in the obese state is a trigger of inflammatory responses. We investigated the role of M2 macrophages in lipotoxicity-induced inflammation.
Methods
We used microarray experiments to profile macrophage gene expression regulated by two M2 inducers, interleukin-4 (Il-4), and peroxisome proliferator-activated receptor delta/gamma (Pparδ/Pparγ) agonists. Functional validation studies were performed in bone marrow-derived macrophages and mice deprived of the signal transducer and activator of transcription 6 gene (Stat6; downstream effector of Il-4) or Pparδ/Pparγ genes (downstream effectors of Stat6). Palmitic acid (PA) and β-adrenergic agonist were employed to induce macrophage lipid loading in vitro and in vivo, respectively.
Results
Profiling of genes regulated by Il-4 or Pparδ/Pparγ agonists reveals that alternative activation promotes the cell survival program, while inhibiting that of inflammation-related cell death. Deletion of Stat6 or Pparδ/Pparγ increases the susceptibility of macrophages to PA-induced cell death. NLR family pyrin domain containing 3 (Nlrp3) inflammasome activation by PA in the presence of lipopolysaccharide is also increased in Stat6−/− macrophages and to a lesser extent, in Pparδ/γ−/− macrophages. In concert, β-adrenergic agonist-induced lipolysis results in higher levels of cell death and inflammatory markers in ATMs derived from myeloid-specific Pparδ/γ−/− or Stat6−/− mice.
Conclusions
Our data suggest that ATM cell death is closely linked to metabolic inflammation. Within WAT where concentrations of free fatty acids fluctuate, M2 polarization regulated by the Stat6-Ppar axis enhances ATM's tolerance to lipid-mediated stress, thereby maintaining the homeostatic state.
Keywords: Obesity, Adipose tissue macrophage, Alternative activation, Lipotoxicity, Meta-inflammation
Abbreviations: M2, alternative activation of macrophage; M1, classic activation of macrophage; WAT, white adipose tissue; ATM, adipose tissue resident macrophage; Ppar, peroxisome proliferator-activated receptor; Stat6, signal transducer and activator of transcription 6; PA, palmitic acid; Nlrp3, NLR family pyrin domain containing 3; BMDM, bone marrow derived macrophage; ILC2, type 2 innate lymphoid cell; SVF, stromal vascular fraction; OCR, oxygen consumption rate; Angptl4, angiopoietin-like 4; Sgms1, sphingomyelin synthase 1; Plin2, perilipin 2; Acadvl, acyl-CoA dehydrogenase, very long chain; Slc25a20, solute carrier family 25 member 20; Mogat1, monoacylglycerol O-acyltransferase 1; Mgl1, macrophage galactose-type lectin-1; Arg1, arginase 1; Bcl2, B-cell lymphoma 2; H2-Eb1, histocompatibility 2, class II antigen E beta
Highlights
-
•
Cell survival is transcriptionally regulated by macrophage alternative activation.
-
•
Fatty acid-triggered cell death is increased in Pparδ/γ−/− or Stat6−/− macrophages.
-
•
Il-4-Stat6 signaling suppresses lipotoxicity-induced inflammasome activation.
-
•
The Stat6-Pparδ/γ axis protects ATMs against lipolysis-induced cell death in vivo.
1. Introduction
Obesity and related metabolic syndrome are major medical and economic burdens worldwide. Chronic low-grade inflammation is often observed in obesity and is a key contributor to associated pathologies such as insulin resistance, type 2 diabetes, and atherosclerosis [1], [2], [3]. White adipose tissue (WAT) is a primary energy storing tissue that has been recognized as an important endocrine organ modulating metabolic-related inflammation (or meta-inflammation) through crosstalk with resident immune cells, such as macrophages and lymphocytes. In lean individuals, adipose tissue macrophages (ATMs) have an alternatively activated (M2) phenotype that limits inflammation and sustains homeostasis [1], [3]. With the onset of obesity, classically activated M1 macrophages preferentially accumulate in WAT and exhibit a pro-inflammatory phenotype characterized by the production of cytokines such as interleukin-1β (Il-1β) and tumor necrosis factor-α (Tnf-α) [1], [2], [3]. These M1 derived pro-inflammatory mediators have been shown to cause metabolic dysregulation and insulin resistance [1], [2], [3]. While this M1/M2 paradigm is instrumental for describing immune phenotypes in physiological and pathological states, recent studies have implicated a dynamic and complex ATM polarization process [4]. In addition, it has been shown that metabolically activated macrophages driven primarily by free fatty acids (FA) do not express M1 surface markers, despite increased production of Il-1β and Tnf-α in these cells [5].
Several stimuli from the adipose microenvironment have been reported to regulate the ATM phenotype. It has been suggested that at the lean state, eosinophils and type 2 innate lymphoid cells (ILC2s) constitutively produce Th2 cytokines interleukin-4 (Il-4) and interleukin-13 (Il-13), respectively, which promote M2 polarization [6], [7]. In the pathological context of obesity, adipocyte hypertrophy and cell death increase the production of chemokines, such as C-X-C motif chemokine ligand 12 (Cxcl12) and monocyte chemotactic protein-1 (Mcp-1) to recruit Ly6ChiCcr2+ monocytes into WAT [8], [9], where CD8+ cytotoxic T cells have been found to secrete interferon-γ (Ifn-γ) that contributes to macrophage M1 polarization [3], [10], [11]. In addition to the classical M1 activation, dysregulated lipid homeostasis caused by increased lipolysis in WAT has also been shown to be a trigger of inflammatory responses in ATMs as described above. Recent studies have demonstrated that saturated FA and their metabolites, notably ceramides, can act on NLR family pyrin domain containing 3 (Nlrp3) inflammasome formation, resulting in autocatalytic activation of caspase-1 and production of mature Il-1β to promote inflammation [12], [13]. Interestingly, WAT lipolysis by fasting or β-adrenergic stimulation can also drive ATM accumulation in lean individuals [14]. This raises the question of how M2 ATMs regulate FA-induced immune responses.
Macrophage M2 polarization requires activation and cooperation of several transcriptional factors [3], [15]. Signal transducer and activator of transcription 6 (Stat6), the canonical effector of Th2 signaling, has been proposed to regulate mitochondrial oxidative metabolism to fuel M2 activation [3], [15], [16]. In mice, Stat6 controls the expression of nuclear receptors peroxisome proliferator-activated receptor delta (Pparδ or Pparβ) and gamma (Pparγ), both of which can further modulate the function of M2 macrophages [9], [17], [18]. Pparδ/Pparγ are well-known lipid sensing nuclear receptors. A variety of endogenous lipids, including unsaturated FAs, saturated FAs and hydroxyeicosatetraenoic acids can physically bind to Pparδ and Pparγ, which, in turn, activate the transcriptional programs for FA oxidation, mitochondrial biogenesis, and anti-inflammatory response in macrophages [15], [19]. Stat6−/−, myeloid-Pparδ−/−, and myeloid-Pparγ−/− mice are more prone to high fat diet induced WAT inflammation and insulin resistance [17], [20], [21], [22]. The metabolic function of the Stat6-Ppar signaling cascade implicates a role for M2 macrophages in mediating FA homeostasis within WAT.
The current study aims to identify potential physiological functions of M2 polarization in ATMs. Through expression profiling and genetic approaches, we find that the Stat6-Ppar axis plays an important role in protecting macrophages against lipotoxicity-induced cellular dysfunction. This is mediated by transcriptional regulation of cell death/pro-survival genes, in addition to their known function in FA metabolism and mitochondrial respiration. Dysregulation of M2 signaling, such as in Stat6−/− and Pparδ/Pparγ−/− macrophages, increases susceptibility to palmitic acid (PA)-induced cell death, which contributes to the initiation of metabolic inflammation in WAT.
2. Materials and methods
2.1. Animal experiments
2.1.1. Mouse models
Stat6−/− mice and the C57BL/6J controls as well as lysozyme-cre mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). Pparδf/f and Pparγf/f in the C57BL/6J background were generated as described previously [23], [24]. Pparδ/γf/f mice were obtained by crossing the Pparδf/f and Pparγf/f alleles. These mice were used to create myeloid-specific Pparδ−/−, Pparγ−/− and Pparδ/γ−/− mice, respectively (Mac-Pparδ−/−, Mac-Pparγ−/− and Mac-Pparδ/γ−/−), by crossing to lysozyme-cre. All mice were housed in a 12 h light/12 h dark cycle with temperatures of 18–23 °C and 40–60% humidity on standard chow diet and allowed food and water ad libitum unless otherwise stated. Mice were sacrificed by CO2 asphyxiation. All animal studies were approved by the Harvard Medical Area Standing Committee on Animals.
2.1.2. Metabolic studies
In vivo studies were conducted in 3-month-old male mice on normal chow diet (n = 4–7). After 14 h fasting, animals were injected intraperitoneally with 1 mg/kg of CL316,243 (Santa Cruz Biotechnology, Dallas, TX, USA, Cat# sc-203895) or saline and sacrificed 2.5 h after injection.
2.1.3. Stromal vascular fraction (SVF) and ATMs isolation
For SVF isolation, perigonadal adipose tissue was excised and digested with digestion buffer (DMEM, 1 g/L glucose, 2 mg/ml collagenase type II and 2% albumin) at 37 °C for 30 min [17]. After filtering through 250 μm nylon mesh, digested adipose tissue was spun down at 400 g for 5 min. The supernatant was discarded and cells were resuspended in washing buffer (DMEM, 1 g/L glucose and 2% albumin). After filtering through 70 μm filter, cells were spun down at 400 g for 5 min and cell pellet was collected. Lysis buffer (150 mM NH4Cl, 1 mM KHCO3, 0.1 mM EDTA, pH 7.2–7.4) was added for 5 min to remove red blood cells. Cells were subsequently filtered through 40 μm filter, spun down, and collected for analysis.
For ATMs isolation, SVF was further incubated with an anti-F4/80 antibody (Invitrogen, Grand Island, NY, USA) for 30 min at 4 °C. After spinning down at 400 g for 5 min, SVF was resuspended in isolation buffer (PBS, 0.1% BSA, 2 mM EDTA, pH 7.4) containing Dynabeads (Invitrogen). After gently rotating for 30 min at 4 °C, F4/80 positive cells were selected by magnetic Dynabeads according to manufacturer's protocol.
2.1.4. Blood sampling and analysis
Blood glucose measurements were performed using the One Touch Ultra Blood Glucose Diabetic Test Strips (LifeScan, Milpitas, CA, USA). For blood chemistry, blood samples were taken from mouse hearts immediately after sacrifice and placed on ice for 30 min. Samples were subsequently centrifuged at 2,000 g for 10 min to remove the clot and serum was collected and store at −80 °C for analysis. Serum free FAs were measured using commercially kits from Wako Chemicals (Richmond, VA, USA) and triglycerides (TG) were tested by Infinity Triglycerides Reagents (Thermo Scientific, Waltham, MA, USA).
2.2. Cell culture
2.2.1. Differentiation and maintenance of mouse macrophages
RAW 264.7 macrophages were maintained in DMEM (1 g/L glucose) medium containing 10% FBS. For bone marrow-derived macrophages (BMDMs), bone marrow was flushed out in wash buffer (DMEM,1 g/L glucose, 10% FBS). After spinning down, cell pellet was resuspended in lysis buffer (150 mM NH4Cl, 1 mM KHCO3, 0.1 mM EDTA, pH 7.2–7.4) for 10 min to remove red blood cells. Remaining cells were then spun down and differentiated in differentiation medium (DMEM, 1 g/L glucose, 10% FBS, 20% L929 condition medium) [25]. Differentiation medium was replaced at day 3, 5, and 7. All experiments were finished between day 7–9. Similar results were obtained from marrow cells of male or female mice.
2.2.2. Treatment
Recombinant mouse Il-4 (rIl-4, 10 ng/ml, Peprotech, Rocky Hill, NJ, USA), GW501516, GW0742 (0.1 μM for both, Cayman Chemical, Ann Arbor, MI, USA), GW1929 (1 μM, Sigma, St. Louis, MO, USA), or vehicle (DMSO for Ppar agonists) were incubated with macrophages overnight in 10% FBS, DMEM. PA (Sigma) was prepared as previously described [26] and added to cells at final concentrations of 300 μM in 0.45% BSA, 2% double stripped FBS, DMEM for 16 h. For PA-induced inflammasome activation, lipopolysaccharide (LPS, 10 ng/ml, Sigma) was pre-incubated with macrophages for 3 h in 10% FBS, DMEM before the addition of PA.
2.3. Microarray and statistical analyses
For microarray analyses, samples were sent to the Yale Center for Genome Analysis and hybridized to a NimbleGen Eukaryotic Gene Expression 12 × 135 K microarray (Roche NimbleGen, Madison, WI, USA). The microarray data are available at the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under submission number GSE100237. Log transformed, normalized intensity for each sample was used for downstream analysis by MeV software suite (http://mev.tm4.org/). Statistical significance between treatment groups was determined using the linear models for microarray (limma) function within the MeV suite. Expression changes were considered significant with a nominal p value < 0.05 (see Supplementary Table 1–3 for significantly altered genes). Gene ontology over-representation analysis and clustering of significantly altered genes were performed in DAVID (https://david.ncifcrf.gov/) [27], [28] (see Supplementary Tables 4 and 5 for gene lists). Heat maps were generated by MultiExperiment Viewer.
Statistical analyses were performed using Student's t test (two tailed). Experiments were conducted with 3–4 replicates and repeated at least three times. In vivo studies were conducted in two cohorts (n = 4–7 per treatment per genotype). Values were presented as means ± SEM. p < 0.05 was considered significant.
2.4. Expression analyses and enzymatic assays
2.4.1. Real-time quantitative PCR (qPCR)
Total RNA was isolated from macrophages using PrepEase RNA Spin Kit (Affymetrix, Santa Clara, CA, USA, Cat# 78767) according to manufacturer's protocol. Isolated RNA (1 μg) was reversed transcribed into cDNA using Verso cDNA Synthesis Kit (Thermo Scientific, Cat# AB-1453/B) according to manufacturer's protocol. Quantitative PCR was performed with SYBR green-based assays. 36b4 was used a loading control. Expression of each gene transcript was calculated using a relative standard and normalize to 36b4 [29]. The primers used for qPCR were listed in Supplementary Table 6.
2.4.2. Caspase-3 activity/lactate dehydrogenase (LDH) release
Caspase-3 activity was measured with Caspase-Glo® 3/7 Assay kits (Promega, Madison, WI, USA). Lactate dehydrogenase (LDH) release was determined with CytoTox 96® Non-Radioactive Cytotoxicity Assay kits (Promega).
2.4.3. Flow cytometry
For flow cytometry, BMDMs were collected and stained with Alexa Fluor 647-conjugated anti-annexin-V antibody (Invitrogen, Cat. #A35110) for 15 min on ice in dark. Cells were then transferred into polystyrene test tubes and analyzed using FACS Caliber flow cytometer (BD Biosciences, San Jose, CA, USA). To determine ATM populations in WAT, SVF was isolated and stained with APC-conjugated anti-F4/80 antibody (Invitrogen, Cat. #MF48005) and Alexa Fluor 488-conjugated anti-annexin-V antibody (Invitrogen, Cat. #A13201) for 15 min on ice.
2.4.4. Immunoblotting
Protein samples were isolated from BMDMs using RIPA buffer containing protease and phosphatase inhibitors. The extracted proteins were frozen and stored at −80 °C. The prepared proteins were subsequently separated by SDS-PAGE and transferred on to membranes. We used PVDF Transfer Membranes (Thermo Scientific, Cat# 88518) for caspase-1 p45 and tubulin, and FluoroTrans® PVDF Transfer Membranes (Pall, Westborough, MA, USA, Cat# BSP0161) for caspase-1 p10. Membranes were incubated in the primary antibodies overnight at 4 °C and HRP-conjugated secondary antibodies for 1 h at room temperature. Tubulin was used as loading control. Antibody against caspase-1 was purchased from Santa Cruz Biotechnology (Cat. #SC-514) and tubulin was purchased from Cell Signaling Technology (Danvers, MA, USA, Cat# 2128).
2.4.5. ELISA
Supernatants were collected from BMDMs after treatment, and Il-1β concentrations were measured by using Il-1β ELISA kit (BD Biosciences, Cat. #559603) according to manufacturer's protocol.
2.5. Mitochondrial respiration
BMDMs were seeded in a 24-well holding plate (Seahorse Bioscience, Billerica, MA, USA). After PA treatment, macrophages were washed 3 times with running medium (DMEM without phenol red supplemented with 5 mM glucose and 1 mM sodium pyruvate pH 7.2–7.4) and subsequently kept in 560 μL running medium in CO2 37 °C incubator for an hour. The basal oxygen consumption rate (OCR) was determined on XF24 Seahorse extracellular flux analyzer (Agilent, Santa Clara, CA, USA).
2.6. FA oxidation
BMDMs were seeded in 6-well plates (Corning, NY, USA). After PA treatment, cells were washed with PBS once and replaced with 1 ml loading medium [DMEM, 1 g/L glucose and 2% BSA (Sigma, Cat. #A-6003)] for 30 min. After washing with PBS once, cells were replaced with 600 μL medium containing DMEM (1 g/L glucose), 2% BSA (Sigma Cat. #A6003), 0.25 mM carnitine (Sigma, Cat. #C-0158) and 2 uCi 3H-palmitic acid (American radiolabeled chemicals, St. Louis, MO, USA, Cat. #ART129) for 4 h. Supernatants were collected and the 3H radioactivity in the aqueous phase was quantified as described [30].
2.7. TG extraction
BMDMs were collected and lysed in lysis buffer [50 mM Tris (pH 8), 100 mM NaCl and 0.1% NP40]. After freezing and thawing 3 times, cellular lipid was extracted with chloroform: methanol (2:1 v/v) as described previously [31]. TG content was determined by Infinity Triglycerides Reagents (Thermo Scientific).
3. Results
3.1. Cell survival is transcriptionally regulated by macrophage alternative activation
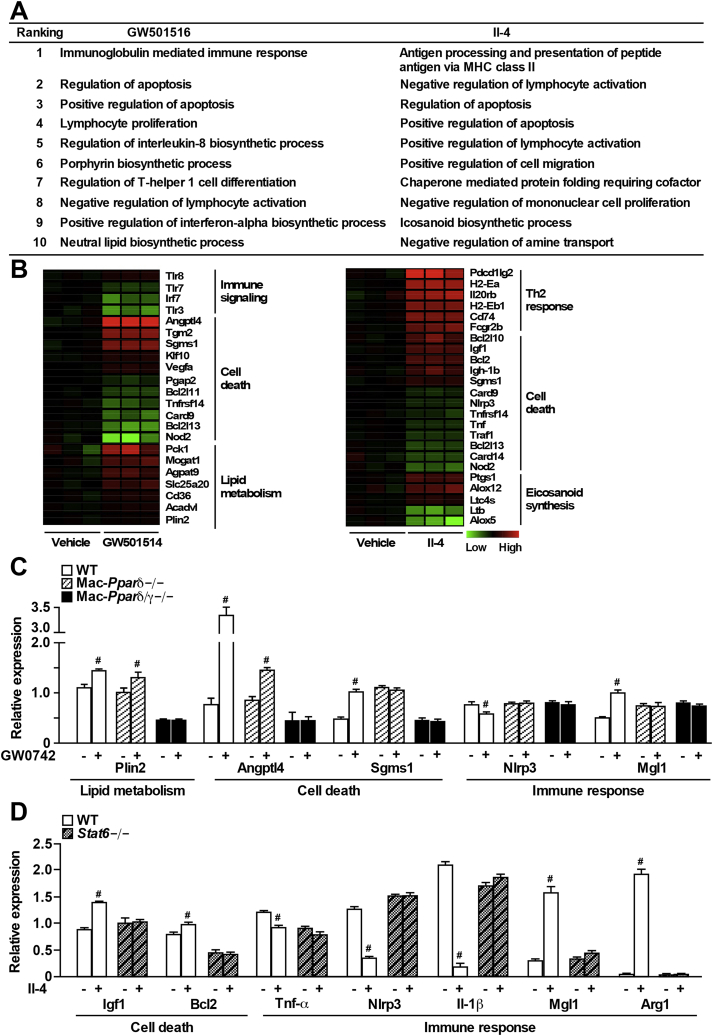
ATMs exhibit a unique phenotypic transition from M2 to M1 activation in the lean versus obese state. The Th2 cytokine-Stat6-Pparδ/γ axis has previous been shown to maintain the M2 phenotype of ATMs [3], [15]. To better understand the physiological function of M2 ATMs, we performed gene expression profiling using samples from RAW 264.7 cells stably overexpressing Pparδ (RAW-Pparδ) treated with vehicle, GW501516 (a synthetic Pparδ ligand), or Th2 cytokine Il-4 [see Supplementary Table 1–3 for significantly altered genes; original data in Gene Expression Omnibus database #GSE100237 (https://www.ncbi.nlm.nih.gov/geo/)]. DAVID functional annotation clustering analyses were employed to identify pathways significantly altered by either GW501516 or Il-4 treatment compared to vehicle control [27], [28]. As expected, GW501516 treatment led to enrichment of genes involved in lipid metabolism as well as certain immune signaling (e.g., T cell function), while Il-4 induced enrichment of genes related to Th2 response (e.g., major histocompatibility complex class II antigen presentation and lymphocyte activation) and eicosanoid synthesis (Figure 1A and B and Supplementary Tables 4 and 5). Interestingly, cell death was among the top 10 functional clusters enriched in either GW501516 or Il-4 treatment group.
Figure 1.
Pro-survival genes are up-regulated in alternatively activated macrophages. (A) List of top 10 DAVID functional annotation clusters for biological processes significantly altered by GW501516 or Il-4 treatment, compared to the vehicle control (see Supplementary Tables 4 and 5 for gene lists). Microarray analysis was conducted in Pparδ-overexpressing RAW 264.7 cells treated with GW501516 (0.1 μM) or rIl-4 (10 ng/ml) overnight. (B) Heat maps were shown for selected genes from the top 10 regulated pathways (Figure 1A) by GW501516 (Pparδ agonist, left) or Il-4 treatment (right) presented as fold change over vehicle control. (C) Validation of Ppar target genes identified through microarray using qPCR in WT, Mac-Pparδ−/− and Mac-Pparδ/γ−/− BMDMs treated with GW0742 (0.1 μM) overnight. (D) Validation of Il-4 regulated genes in WT and Stat6−/− BMDMs incubated with rIl-4 (10 ng/ml) overnight. Values were expressed as means ± SEM. #p < 0.05 between vehicle and GW0742 or rIl-4 treatment.
Selected genes from microarray results were first validated in RAW-Pparδ cells treated with another Pparδ agonist GW0742 or vehicle control. GW0742 significantly induced the expression of genes involved in cell death [angiopoietin-like 4 (Angptl4) and sphingomyelin synthase 1 (Sgms1)], lipid metabolism [perilipin 2 (Plin2, also called adipocyte differentiation-related protein, Adrp), acyl-CoA dehydrogenase, very long chain (Acadvl), CD36 molecule (Cd36), solute carrier family 25 member 20 (Slc25a20), and monoacylglycerol O-acyltransferase 1 (Mogat1)] and M2 markers [e.g., macrophage galactose-type lectin-1 (Mgl1)], while inhibiting genes involved in inflammation (e.g., Tnf-a) (Supplementary Figure 1A). In BMDMs from wild-type (WT) mice, GW0742 significantly increased the expression of genes identified by the microarray (e.g., Plin2, Angptl4, and Sgms1; Figure 1C). These effects were completely abolished in BMDMs from Mac-Pparδ/γ−/− but not from Mac-Pparδ−/− mice, suggesting that GW0742 at the dose used also activated Pparγ and that Pparγ shared common target genes with Pparδ. In concert, most of the Pparδ-controlled genes can also be regulated by Pparγ (Supplementary Figure 1B). Il-4 treated RAW-Pparδ cells showed significant induction of cell survival genes [insulin like growth factor 1 (Igf1), B-cell lymphoma 2 (Bcl2) and Sgms1] and M2 markers [histocompatibility 2, class II antigen E beta (H2-Eb1), Mgl1, Ym1 (also called chitinase-like 3, Chil3), and Arginase 1 (Arg1)], and reduced expression of pro-inflammatory genes (Tnf-a and Nlrp3) (Supplementary Figure 1C). Similar regulation was observed in BMDMs in a Stat6 dependent manner (Figure 1D).
3.2. Lipotoxicity-induced cell death is exacerbated in Pparδ/γ−/− or Stat6−/− macrophages
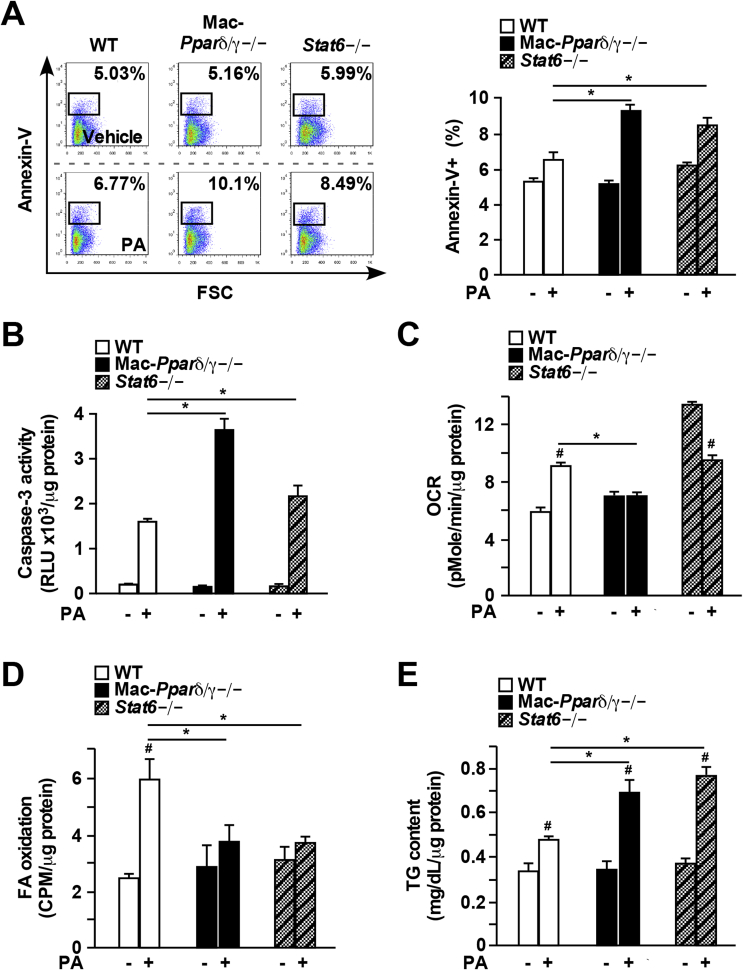
In obesity, increased free FAs released from insulin-resistant adipocytes result in lipotoxicity [8]. Ceramides, derived from FAs can cause cellular dysfunction at elevated concentrations, including cell death and inflammasome activation [12], [13]. Most of genes in the cell death category identified by the array were suppressed by GW501516 or Il-4 treatment (Supplementary Figure 1D and Supplementary Tables 7 and 8). 19 of those commonly down-regulated genes in both treatments included many cell death promoters, such as BCL2-like 13 (Bcl2l13) and caspase recruitment domain family, member 9 (Card9). Sgms1 was one of the 6 up-regulated (presumably pro-survival) genes shared by Pparδ/γ and Stat6 that converts ceramides to sphingomyelin and has been shown to suppress Bcl2-associated X Protein (BAX)-mediated apoptosis and prevent cell death induced by oxidative damage [32], [33]. To determine whether macrophage Ppars and Stat6 play a role in handling lipid-induced toxicity, we challenged BMDMs with PA, one of the most abundant FAs in the body. Flow cytometry assays showed increased annexin-V positive Pparδ/γ−/− and Stat6−/− cells after 16 h of PA treatment, compared to control cells (Figure 2A). There was a two-fold increase in cell death determined by caspase-3 activity in Pparδ/γ−/− BMDMs, compared to WT cells (Figure 2B). Interestingly, deletion of Pparδ or Pparγ alone in the macrophage was sufficient to promote PA-induced cell death (Supplementary Figure 2), suggesting that both Pparδ and Pparγ are required to maintain lipid homeostasis. Stat6−/− BMDMs also showed significantly increased caspase-3 activity, compared to WT cells (Figure 2B). These results demonstrate that macrophages lacking Pparδ/γ or Stat6 are more susceptible to PA-induced cell death.
Figure 2.
PA-induced cellular dysfunction is exacerbated in Mac-Pparδ/γ−/−and Stat6−/−macrophages. (A) and (B) Cell death quantified by annexin-V staining using flow cytometry or by caspase-3 activity in BMDMs treated with PA (300 μM) for 16 h. (C) Mitochondrial OCR measured by Seahorse bioenergetic analyzer in BMDMs treated with PA (300 μM) for 16 h (D) FA β-oxidation determined by using 3H-PA in control or PA-treated BMDMs. (E) Intracellular TG content in control or PA-treated BMDMs. Values were expressed as means ± SEM. *p < 0.05 between WT and Mac-Pparδ/γ−/− or Stat6−/−; #p < 0.05 between vehicle and PA treatment.
The Stat6/Ppar transcriptional cascade also regulates macrophage lipid metabolism and mitochondrial function [9], [15], which could limit lipotoxicity by reducing lipid burden. PA treatment increased mitochondrial respiration determined by OCR in WT BMDMs, and this effect was absent in Pparδ/γ−/− cells (Figure 2C). Although the basal OCR of Stat6−/− macrophages was higher than that of WT cells as reported previously [16], PA treatment suppressed, rather than enhanced mitochondrial respiration in Stat6−/− cells (Figure 2C). Similarly, FA-induced β-oxidation was abolished in Pparδ/γ−/− and Stat6−/− BMDMs (Figure 2D), while intracellular TG content was elevated in these cells (Figure 2E).
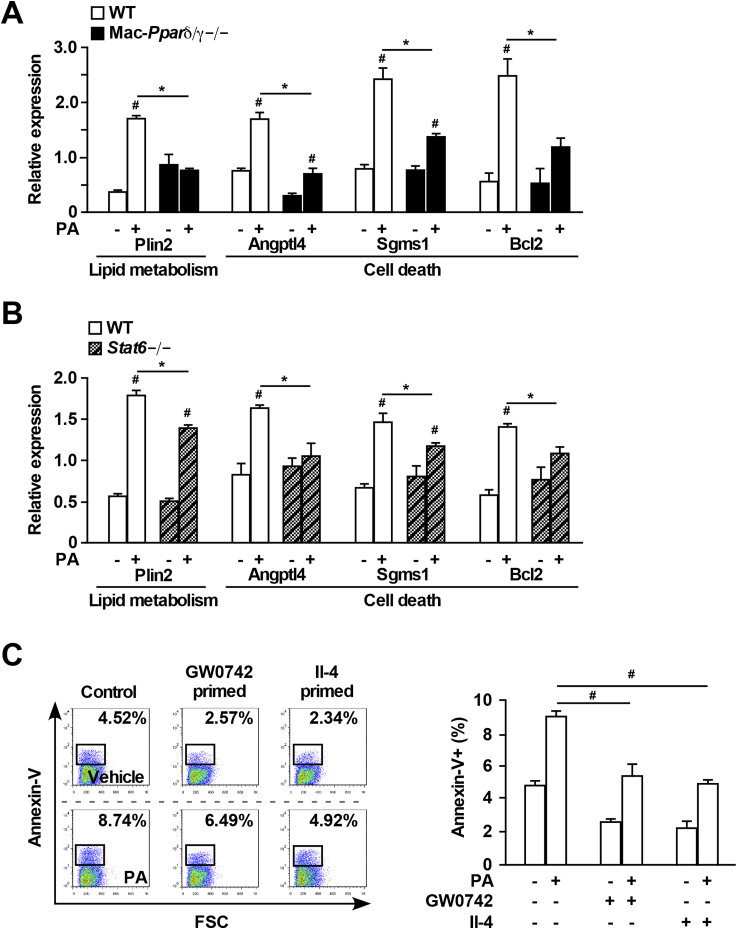
At the transcriptional level, PA induced expression of Plin2, a well-defined Ppar target gene in lipid droplet formation [34], was completely abolished in Pparδ/γ−/− macrophages (Figure 3A), consistent with the notion that FAs serve as Ppar agonists. The expression of cell survival genes (Angptl4, Sgms1, Bcl2) was also induced by PA treatment in WT macrophages. This response was blunted in Pparδ/γ−/− and Stat6−/− cells (Figure 3A and B).
Figure 3.
Alternative activation protects macrophages from lipotoxicity-induced cell death. (A) Gene expression determine by qPCR in WT and Mac-Pparδ/γ−/− BMDMs treated with vehicle or PA (300 μM) for 16 h. (B) Gene expression in WT and Stat6−/− BMDMs ± PA (300 μM) for 16 h (C) WT BMDMs were pre-incubated with GW0742 (0.1 μM) or rIl-4 (10 ng/ml) overnight before the addition of PA (300 μM) for 16 h. Percentage of annexin-V+ cells was measured by flow cytometry. Values were presented as means ± SEM. *p < 0.05 between WT and Mac-Pparδ/γ−/− or Stat6−/−; #p < 0.05 between vehicle and PA, GW0742 or rIl-4 treatment.
We next sought to determine if activation of Ppars or Stat6 could alleviate lipotoxicity-induced cell death. WT BMDMs were primed with GW0742 or Il-4 overnight followed by 16 h of PA treatment. Both GW0742 and Il-4 pretreatment significantly reduced the percentages of annexin-V+ cells (Figure 3C). Collectively, these data suggest that the Stat6-Pparδ/γ axis in the macrophage protects against lipotoxicity-triggered cell death through regulation of genes involved in cell survival, in addition to mitochondrial function and FA metabolism.
3.3. Il-4-Stat6 signaling suppresses lipotoxicity-induced inflammasome activation
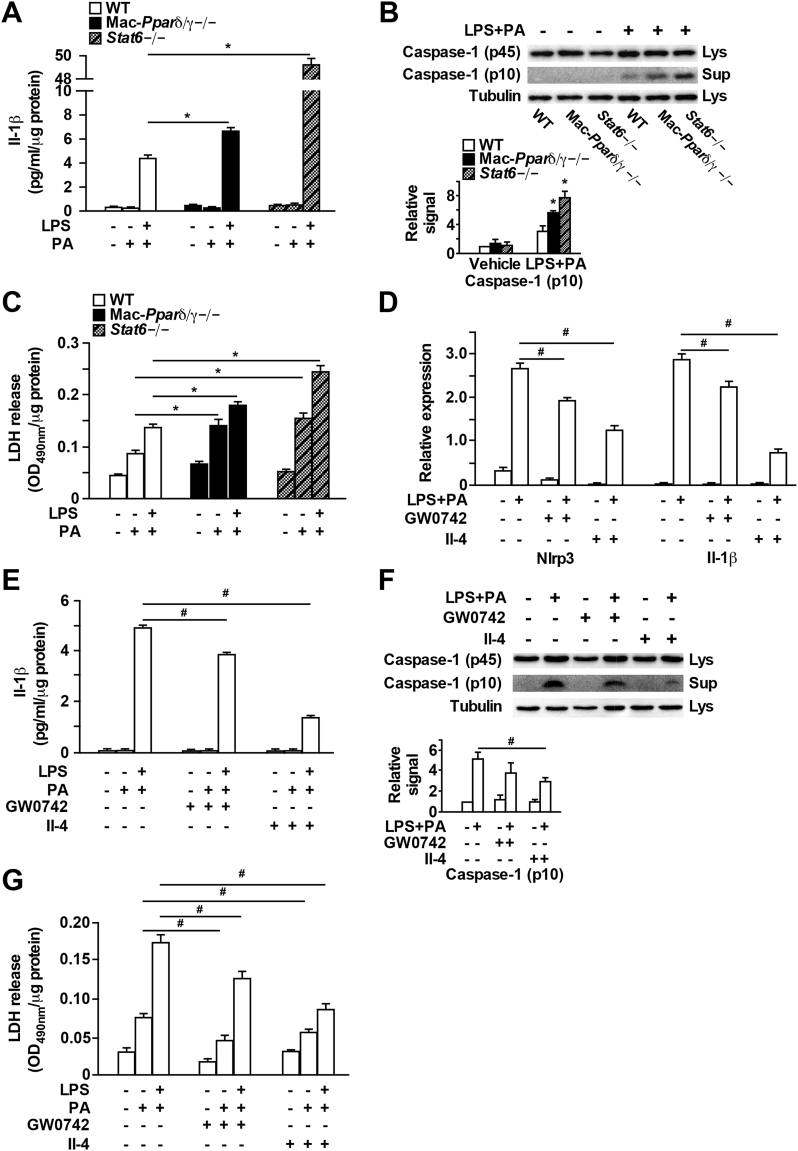
Lipotoxicity-mediated inflammasome activation has emerged as a key pathogenic contributor to WAT inflammation and its metabolic sequelae [2]. Previous studies suggest PA and its metabolite ceramide can cause NLRP3 inflammasome activation in LPS-primed macrophages [12], [13]. Our microarray results indicated that expression of inflammasome-related genes such as Nlrp3 were inhibited by Il-4 treatment (Figure 1B). Furthermore, qPCR in Raw 264.7 cells and BMDMs has demonstrated that Il-4 treatment substantially down-regulated Nlrp3 and its downstream effector Il-1β (Figure 1D and Supplementary Figure 1C). Based on these observations, we tested if Stat6-Ppar signaling modulates lipotoxicity-induced inflammasome activation. WT, Pparδ/γ−/− and Stat6−/− BMDMs were primed with LPS for 3 h followed by 16 h of LPS + PA treatment. Cell lysates and conditioned media were collected for analysis. Il-1β release in the media determined by ELISA was significantly increased after LPS + PA stimulation (Figure 4A). Pparδ/γ deficiency resulted in a moderate increase in Il-1β release, while Stat6 disruption led to ∼10-fold higher Il-1β production (Figure 4A). In line with this observation, the level of cleaved caspase-1 (P10) was increased in Stat6−/− macrophages, and to a lesser extent in Pparδ/γ−/− macrophages, compared to WT control (Figure 4B). Similar results were observed with the level of LDH protein in the media, which serves as a marker for inflammasome-induced cell death, or pyroptosis (Figure 4C). Reciprocally, activating Stat6 in WT BMDMs through Il-4 pretreatment suppressed LPS + PA-induced Nlrp3 and Il-1β expression, Il-1β production, caspase-1 processing and LDH release (Figure 4D–G). Pparδ/γ activation by GW0742 could also suppress inflammasome activation. However, the effect was not as strong as Il-4 treatment (Figure 4D–G). These findings provide a mechanistic explanation for how M2 activation inhibits metabolic inflammation.
Figure 4.
Il-4-Stat6 signaling inhibits lipotoxicity-induced inflammasome activation. (A–C) WT, Mac-Pparδ/γ−/− and Stat6−/− BMDMs were treated with or without LPS (10 ng/ml) for 3 h before the addition of PA (300 μM) for 16 h. (A) Supernatant Il-1β concentrations measured by ELISA. (B) Immunoblotting for activated p10 caspase-1 in supernatant (Sup) and for unprocessed p45 caspase-1 and tubulin in cell lysates (Lys). Bottom panel: relative western signal of caspase-1 (p10) normalized to tubulin and shown as fold change to vehicle treated WT sample (set as 1). Results were the average of three independent experiments. (C) LDH release in supernatant. (D–G) WT BMDMs were pretreated with GW0742 (0.1 μM) or rIl-4 (10 ng/ml) overnight followed by PA or LPS + PA treatment. (D) Expression of genes involved in inflammasome activation. (E) Supernatant Il-1β concentrations. (F) Caspase-1 processing was measured by western blotting. Bottom panel: relative western signal of caspase-1 (p10) normalized to tubulin and shown as fold change to vehicle treated control sample (set as 1). (G) Quantification of LDH release in supernatants. Values were expressed as means ± SEM. *p < 0.05 between WT and Mac-Pparδ/γ−/− or Stat6−/−; #p < 0.05 between vehicle and GW0742 or rIl-4 treatment.
3.4. Increased cell death in ATMs from macrophage-specific Pparδ/γ−/− or Stat6−/− mice
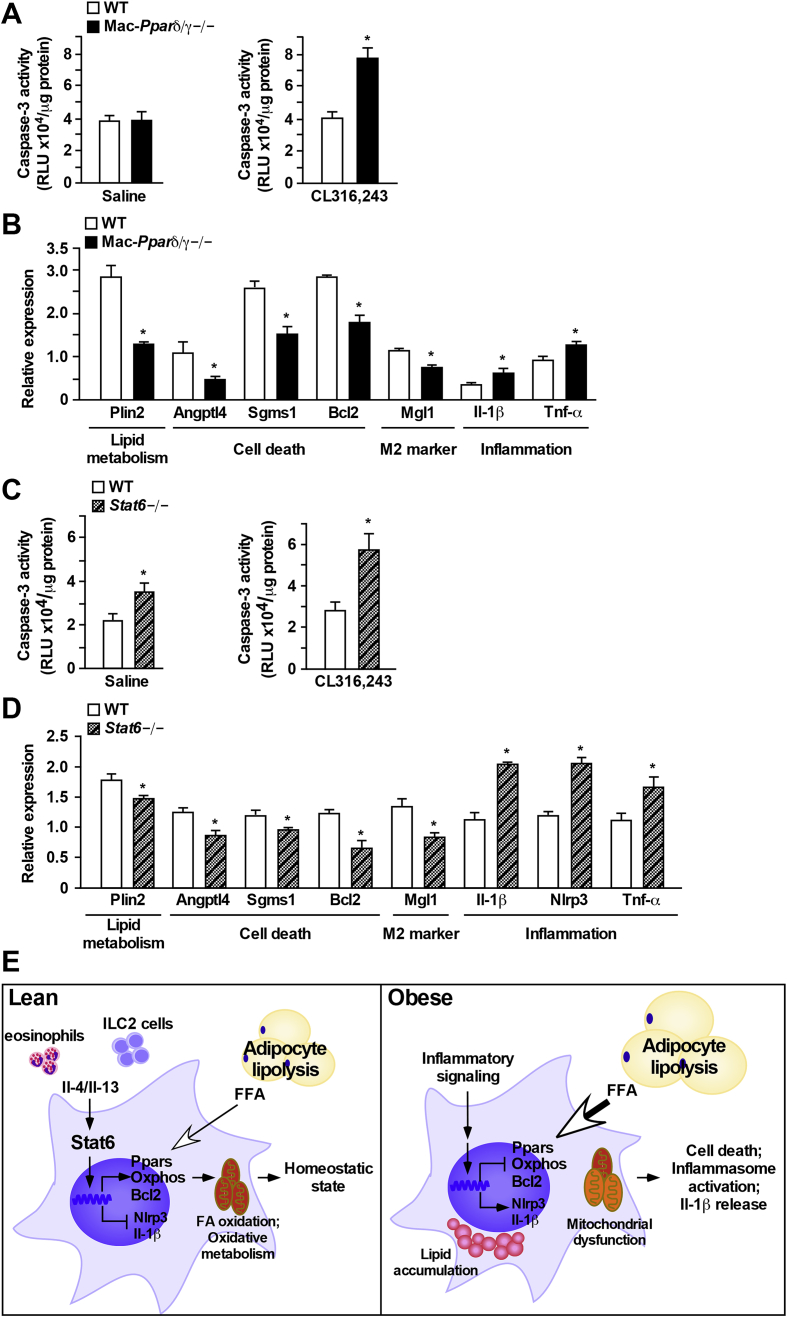
Results described thus far implicate a critical role of Pparδ/Pparγ and Stat6 in modulating macrophage survival ex vivo. To validate in vivo functional relevance, mice were fasted for 14 h, followed by injection with a β3-adrenergic agonist CL316,243 to induce lipolysis. The SVF was isolated from perigonadal adipose tissue to assess the ATM phenotype. CL316,243 treatment in fasted mice induced a ∼2-fold increase in serum free FA levels (Table 1). Pparδ/γ depletion led to an increase in macrophage infiltration in WAT demonstrated by higher percentage of F4/80+ cells in SVF of CL316,243-injected Mac-Pparδ/γ−/− mice, and a trend towards an increase in macrophage infiltration in vehicle (saline)-injected Mac-Pparδ/γ−/− mice (Supplementary Figure 3A). There was no difference in ATM caspase-3 activity between genotypes after 14 h fasting (Figure 5A, left panel). CL316,243 treatment led to increased caspase-3 activity in Mac-Pparδ/γ−/− ATMs, compared to WT cells (Figure 5A, right panel). ATMs from Mac-Pparδ/γ−/− mice in both saline and CL316,243 injected groups had reduced expression of FA metabolism, cell survival and M2 genes, while pro-inflammatory genes (e.g., Il-1β and Tnf-α) were up-regulated (Figure 5B and Supplementary Figure 3B). Consistent with the increased caspase 3 activity, CL316,243 treatment further suppressed survival genes, such as Sgms1 and Bcl2.
Table 1.
Metabolic parameters of mouse studies.
| Saline injection |
CL316,243 injection |
|||
|---|---|---|---|---|
| WT | Mac-Pparδ/γ−/− | WT | Mac-Pparδ/γ−/− | |
| Body weight (g) | 31.6 ± 1.6 | 29.6 ± 1.9 | 30.6 ± 1.5 | 30.4 ± 1.3 |
| WAT/body weight | 0.040 ± 0.006 | 0.036 ± 0.008 | 0.046 ± 0.004 | 0.040 ± 0.004 |
| Glucose (mg/dl) | 141.5 ± 13.5 | 136.5 ± 12.1 | 72.4 ± 2.3# | 73.3 ± 3.1# |
| Free FA (mmol/l) | 1.03 ± 0.11 | 1.02 ± 0.12 | 2.23 ± 0.24# | 2.17 ± 0.24# |
| TG (mg/dl) | 120.3 ± 15.8 | 104.8 ± 12.8 | 190.0 ± 28.2 | 232.8 ± 23.9# |
| Insulin (ng/ml) | 0.83 ± 0.10 | 1.02 ± 0.25 | 1.03 ± 0.19 | 1.05 ± 0.11 |
| WT | Stat6−/− | WT | Stat6−/− | |
|---|---|---|---|---|
| Body weight (g) | 30.5 ± 0.6 | 31.1 ± 0.8 | 28.6 ± 1.2 | 27.9 ± 2.0 |
| WAT/body weight | 0.044 ± 0.003 | 0.041 ± 0.001 | 0.034 ± 0.002# | 0.032 ± 0.004# |
| Glucose (mg/dl) | 197.8 ± 27.8 | 130.0 ± 9.9 | 73.0 ± 2.2# | 87.8 ± 4.6*# |
| Free FA (mmol/l) | 1.03 ± 0.08 | 1.16 ± 0.03 | 1.79 ± 0.15# | 2.01 ± 0.05# |
| TG (mg/dl) | 103.7 ± 4.8 | 110.5 ± 7.8 | 156.2 ± 20.1# | 157.8 ± 18.9 |
| Insulin (ng/ml) | 0.89 ± 0.14 | 0.79 ± 0.15 | 0.87 ± 0.09 | 1.72 ± 0.29*# |
Values are expressed as means ± SEM. *p < 0.05 between genotypes; #p < 0.05 between saline and CL316,243 injection.
Figure 5.
Increased cell death and inflammation triggered by lipolysis in adipose tissue macrophages from Mac-Pparδ/γ−/−or Stat6−/−mice. (A–D) Chow fed WT and Mac-Pparδ/γ−/− or Stat6−/− mice (3-month-old males) were fasted for 14 h, followed by intraperitoneal injection of 1 mg/kg CL316,243 or saline (vehicle). Animals were sacrificed 2.5 h after injection and SVF and ATMs were isolated from WAT. (A) Quantification of caspase-3 activity in isolated ATMs. Left and right panels were from saline and CL316,243 injection, respectively. (B) Gene expression determined by qPCR in ATMs collected from WT and Mac-Pparδ/γ−/− mice with CL316,243 injection. (C) Quantification of caspase-3 activity of ATMs from WT and Stat6−/− mice. (D) Gene expression in ATMs collected from WT and Stat6−/− mice with CL316,243 injection. Studies were conducted in two cohorts (n = 4–7 per treatment per genotype). Values were presented as means ± SEM. *p < 0.05 between WT and Mac-Pparδ/γ−/− or Stat6−/− mice. (E) Model for the role of M2 polarization in protecting ATMs against lipotoxicity-induced cell death and metabolic inflammation.
On the other hand, fasting-induced lipolysis was sufficient to cause an ∼60% increase in caspase-3 activity in Stat6−/− ATMs compared to WT control (Figure 5C left panel). This difference was exacerbated by CL316,243 injection (Figure 5C right panel), which was accompanied by a reduction in ATMs (Supplementary Figure 3C). As expected, Stat6 deficiency impaired macrophage M2 polarization, as evidenced by a lower percentage of F4/80+Mgl1+ population (data not shown) and decreased Mgl1 expression in Stat6−/− ATMs under control and CL316,243 treatment, compared to WT ATMs (Figure 5D and Supplementary Figure 3D). In contrast, inflammatory markers, including Il-1β, Nlrp3, and Tnf-α, were up-regulated. CL316,243 treatment similarly exerted a greater effect on decreasing the expression of Sgms1 and Bcl2 in Stat6−/− ATMs (Figure 5D). Furthermore, concentrations of fasting glucose and insulin were significantly higher in Stat6−/− than in WT mice after CL316,243 injection (Table 1). Altogether, these data suggest that an intact Stat6-Pparδ/γ signaling is required for maintaining ATM homeostasis by protecting macrophages against lipotoxicity-mediated cellular dysfunction.
4. Discussion
In this study, we describe a mechanism through which M2 polarization protects macrophages from lipotoxicity-mediated cellular dysfunction and consequently maintains adipose tissue homeostasis. Gene expression profiling studies reveal that both Stat6 and Ppar signaling up-regulates pro-survival genes while inhibiting inflammation-related cell death pathways. Along this line, cell death triggered by elevated free FA levels is exacerbated in Pparδ/γ−/− and Stat6−/− macrophages both in vitro and in vivo. In addition, Il-4-Stat6 suppresses Nlrp3 inflammasome activation and Il-1β production by PA in LPS-primed macrophages. Whereas M2 macrophages are thought to promote metabolic homeostasis through an anti-inflammatory mechanism, our data indicate that the Th2 cytokine-Stat6-Ppar signaling cascade also increases tolerance to lipid stress within WAT where ATMs encounter fluctuating FA flux. Interestingly, obesity has been shown to increase the viability of pro-inflammatory ATMs mediated by NF-κB [35], suggesting that selective activation of macrophage survival program through the Stat6-Ppar axis over that of NF-κB is a critical step to limit metabolic inflammation.
It has been reported that increased lipolysis either by fasting/β-adrenergic agonists or obesity promotes macrophage infiltration to WAT [1], [9], [14]. These ATMs accumulate TG-enriched lipid droplets leading to a foamy morphology analogous to cholesterol-laden foam cells in atherosclerosis [9]. In the context of obesity, ATMs are the main source of inflammatory mediators, such as Mcp-1, Tnf-α and Il-1β [1], [9], implicating a role for FA-mediated metabolic stress in macrophage dysfunction and pro-inflammatory activation. Our results suggest that a physiological function of M2 ATMs is to help alleviate such stress (Figure 5E). Immediately downstream of Th2 cytokines is the master M2 regulator Stat6 that directly controls mitochondrial fitness, including oxidative metabolism and pro-survival gene Bcl2, and up-regulates Pparγ and Pparδ, both of which promote FA β-oxidation and mitochondrial biogenesis. In addition, Stat6 suppresses the expression of Nlrp3 gene. Enhanced mitochondrial function is expected to also reduce the production of reactive oxygen species, a stimulant of Nlrp3 inflammasome activation [36]. Obesity or over-nutrition leads to deprivation of Th2 cytokine producing cells [1], [2], [3] and enrichment of pro-inflammatory macrophages, which have increased glycolytic metabolism and dampened mitochondrial function [9], [16], [20]. The inability of these pro-inflammatory macrophages to help relieve excess FAs within WAT likely contributes to systemic lipid dysregulation and the chronic, unresolvable meta-inflammation. In concert, studies have demonstrated that Ppar-mediated lipid metabolism plays a key role in limiting inflammation in metabolically activated macrophages [5].
Previous studies have demonstrated that Stat6−/−, myeloid-Pparδ−/− and myeloid-Pparγ−/− mice are more insulin resistance due to M1 skewing of ATMs [17], [20], [21], [22]. Findings from the current work indicate that increased lipotoxicity-induced macrophage death may also contribute to this phenotype. In support of this notion, β-adrenergic agonist CL316,243 triggered lipolysis induces more caspase-3 activity, accompanied by up-regulation of pro-inflammatory gene expression in ATMs isolated from Mac-Pparδ/γ−/− or Stat6−/− mice (Figure 5). The pro-survival effect is mediated by transcriptional regulation of genes related to cell death/survival, in addition to the well-established function of the Stat6-Ppar pathway in mitochondrial integrity and activity. The array data reveal that pro-survival genes regulated by Ppars and Sta6 are largely non-overlapping (Supplementary Figure 1D and Supplementary Tables 7 and 8), although the expression of some of those genes is similarly affected in Mac-Pparδ/γ−/− or Stat6−/− macrophages, supporting a Stat6-Ppar signaling cascade. Angptl4, a Ppar target, inhibits TG hydrolysis thereby preventing macrophages from lipid overload [37]. This is consistent with a previous report suggesting that ATMs take up excess lipids and secrete anti-lipolytic factors to reduce local free FA concentrations [14]. On the other hand, Stat6 up-regulates Bcl2, a mitochondrial anti-apoptotic gene [38]. The pro-survival effect of Ppars and Stat6 could also be mediated by suppression of cell death promoting genes.
Among the cell death/survival genes, Sgms1 is a commonly regulated gene by Pparδ/γ and Stat6 that reduces levels of ceramide by converting it to sphingomyelin [32], [33]. Ceramide synthesis is elevated by increasing cellular FA uptake [39] and serves as a trigger for cell death and inflammasome activation [1], [13], [15]. This may explain why deletion and activation of Stat6/Ppars enhances and suppresses Nlrp3 inflammasome activation/Il-1β release, respectively. Il-4/Stat6 exhibits a substantially stronger effect likely due to the ability of Stat6 to down-regulate Nlrp3 expression. Stat6−/− macrophages have increased basal mitochondrial oxygen consumption (Figure 2C) and, at same time, show more cell death in the fasted state (Figure 5C). It is possible that the increased OCR is a compensatory response to defective substrate metabolism (e.g., fat oxidation). In addition, the uncoupled respiration (proton leak) is higher in Stat6−/− (3.2 ± 0.16 pMole/min/μg), compared to WT (1.68 ± 0.13) and Pparδ/γ−/− (1.73 ± 0.13) macrophages, implicating mitochondrial dysfunction. Coupled with the elevated Nlrp3 (Supplementary Figure 3D) that can mediate inflammasome-induced cell death (pyroptosis, Figure 4C), Stat6−/− ATMs would be expected to be more susceptible to cell death by fasting-induced lipolysis.
Collectively, these observations indicate that FA-induced cell death and inflammation is intimately linked. In the microenvironment of adipose tissues where local free FA concentrations fluctuate, M2 polarization regulated by the Stat6-Ppar axis affords ATMs the metabolic capacity to reduce lipid burden and tolerate lipotoxicity, thereby maintaining the homeostatic state within WAT. It is worth noting that macrophages perform diverse functions dynamically regulated by external stimuli through multiple signaling pathways. The current study focuses on the Stat6-Ppar axis in the context of excess lipid-induced metabolic stress and as such, does not exclude contributions from other regulatory mechanisms. In addition, gene deletion does not truly reflect a pathological condition. Nevertheless, gain-of-function studies using Il-4 and Ppar agonists do support a protective role for Stat6-Ppars in free FA-mediated metabolic inflammation and cell death.
Author contributions
K.J.S and S.L. conducted the initial array experiment, validation and data analysis; P.B. generated the mouse models and stable cell lines and assisted in macrophage experiments; Y.-H.L. was involved in FACS assays; D.J. was involved in plasma insulin measurement; R.K.A, and N.H.K. assisted in mitochondrial respiration assays and writing the manuscript; A.H and M.R.G prepared reagents; L.D. performed majority of the experiments. L.D. and C.H.L designed the study and wrote the manuscript. C.H.L. has full access to all data and takes responsibility for the accuracy of data analysis and interpretation
Acknowledgments
We thank Dr. Tiffany Horng (Harvard T.H. Chan School of Public Health) for help with the inflammasome studies. This work was supported by the National Natural Science Foundation of China (No. 81603205), Hunan Provincial Natural Science Foundation of China (No. 2016JJ3179) and Research Funds from Xiangya Hospital of Central South University (No. 2014Q03) (L.D.); Research Funds from Ministry of Science and Technology of Taiwan (Y-H.L.); NIH F31GM117854 (R.K.A.) and F31DK107256 (N.H.K.); and NIH R01DK113791 and American Heart Association 16GRNT31460005 (C.H.L.).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.08.001.
Contributor Information
Lingling Dai, Email: ldai@hsph.harvard.edu.
Chih-Hao Lee, Email: clee@hsph.harvard.edu.
Conflict of interest
The authors declare no competing financial interests.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Lackey D.E., Olefsky J.M. Regulation of metabolism by the innate immune system. Nature Reviews Endocrinology. 2016;12(1):15–28. doi: 10.1038/nrendo.2015.189. [DOI] [PubMed] [Google Scholar]
- 2.Hotamisligil G.S. Inflammation, metaflammation and immunometabolic disorders. Nature. 2017;542(7640):177–185. doi: 10.1038/nature21363. [DOI] [PubMed] [Google Scholar]
- 3.Brestoff J.R., Artis D. Immune regulation of metabolic homeostasis in health and disease. Cell. 2015;161(1):146–160. doi: 10.1016/j.cell.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knudsen N.H., Lee C.H. Identity crisis: CD301b(+) mononuclear phagocytes blur the m1–m2 macrophage line. Immunity. 2016;45(3):461–463. doi: 10.1016/j.immuni.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Kratz M., Coats B.R., Hisert K.B., Hagman D., Mutskov V., Peris E. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metabolism. 2014;20(4):614–625. doi: 10.1016/j.cmet.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molofsky A.B., Nussbaum J.C., Liang H.E., Van Dyken S.J., Cheng L.E., Mohapatra A. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. The Journal of Experimental Medicine. 2013;210(3):535–549. doi: 10.1084/jem.20121964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu D., Molofsky A.B., Liang H.E., Ricardo-Gonzalez R.R., Jouihan H.A., Bando J.K. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332(6026):243–247. doi: 10.1126/science.1201475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kusminski C.M., Bickel P.E., Scherer P.E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nature Reviews Drug Discovery. 2016;15(9):639–660. doi: 10.1038/nrd.2016.75. [DOI] [PubMed] [Google Scholar]
- 9.Chawla A., Nguyen K.D., Goh Y.P. Macrophage-mediated inflammation in metabolic disease. Nature Reviews Immunology. 2011;11(11):738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Revelo X.S., Tsai S., Lei H., Luck H., Ghazarian M., Tsui H. Perforin is a novel immune regulator of obesity-related insulin resistance. Diabetes. 2015;64(1):90–103. doi: 10.2337/db13-1524. [DOI] [PubMed] [Google Scholar]
- 11.Nishimura S., Manabe I., Nagasaki M., Eto K., Yamashita H., Ohsugi M. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nature Medicine. 2009;15(8):914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 12.Wen H., Gris D., Lei Y., Jha S., Zhang L., Huang M.T. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nature Immunology. 2011;12(5):408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandanmagsar B., Youm Y.H., Ravussin A., Galgani J.E., Stadler K., Mynatt R.L. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nature Medicine. 2011;17(2):179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosteli A., Sugaru E., Haemmerle G., Martin J.F., Lei J., Zechner R. Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. The Journal of Clinical Investigation. 2010;120(10):3466–3479. doi: 10.1172/JCI42845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhargava P., Lee C.H. Role and function of macrophages in the metabolic syndrome. The Biochemical Journal. 2012;442(2):253–262. doi: 10.1042/BJ20111708. [DOI] [PubMed] [Google Scholar]
- 16.Vats D., Mukundan L., Odegaard J.I., Zhang L., Smith K.L., Morel C.R. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metabolism. 2006;4(1):13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang K., Reilly S.M., Karabacak V., Gangl M.R., Fitzgerald K., Hatano B. Adipocyte-derived Th2 cytokines and myeloid PPARdelta regulate macrophage polarization and insulin sensitivity. Cell Metabolism. 2008;7(6):485–495. doi: 10.1016/j.cmet.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szanto A., Balint B.L., Nagy Z.S., Barta E., Dezso B., Pap A. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity. 2010;33(5):699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chawla A. Control of macrophage activation and function by PPARs. Circulation Research. 2010;106(10):1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odegaard J.I., Ricardo-Gonzalez R.R., Goforth M.H., Morel C.R., Subramanian V., Mukundan L. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447(7148):1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ricardo-Gonzalez R.R., Red Eagle A., Odegaard J.I., Jouihan H., Morel C.R., Heredia J.E. IL-4/STAT6 immune axis regulates peripheral nutrient metabolism and insulin sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(52):22617–22622. doi: 10.1073/pnas.1009152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odegaard J.I., Ricardo-Gonzalez R.R., Red Eagle A., Vats D., Morel C.R., Goforth M.H. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metabolism. 2008;7(6):496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barak Y., Nelson M.C., Ong E.S., Jones Y.Z., Ruiz-Lozano P., Chien K.R. PPAR gamma is required for placental, cardiac, and adipose tissue development. Molecular Cell. 1999;4(4):585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 24.Barak Y., Liao D., He W., Ong E.S., Nelson M.C., Olefsky J.M. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(1):303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C.H., Chawla A., Urbiztondo N., Liao D., Boisvert W.A., Evans R.M. Transcriptional repression of atherogenic inflammation: modulation by PPARdelta. Science. 2003;302(5644):453–457. doi: 10.1126/science.1087344. [DOI] [PubMed] [Google Scholar]
- 26.Holzer R.G., Park E.J., Li N., Tran H., Chen M., Choi C. Saturated fatty acids induce c-Src clustering within membrane subdomains, leading to JNK activation. Cell. 2011;147(1):173–184. doi: 10.1016/j.cell.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee C.H., Olson P., Hevener A., Mehl I., Chong L.W., Olefsky J.M. PPARdelta regulates glucose metabolism and insulin sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(9):3444–3449. doi: 10.1073/pnas.0511253103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reilly S.M., Bhargava P., Liu S., Gangl M.R., Gorgun C., Nofsinger R.R. Nuclear receptor corepressor SMRT regulates mitochondrial oxidative metabolism and mediates aging-related metabolic deterioration. Cell Metabolism. 2010;12(6):643–653. doi: 10.1016/j.cmet.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S., Brown J.D., Stanya K.J., Homan E., Leidl M., Inouye K. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502(7472):550–554. doi: 10.1038/nature12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lafont E., Milhas D., Carpentier S., Garcia V., Jin Z.X., Umehara H. Caspase-mediated inhibition of sphingomyelin synthesis is involved in FasL-triggered cell death. Cell Death and Differentiation. 2010;17(4):642–654. doi: 10.1038/cdd.2009.130. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z., Khoury C., Jean-Baptiste G., Greenwood M.T. Identification of mouse sphingomyelin synthase 1 as a suppressor of Bax-mediated cell death in yeast. FEMS Yeast Research. 2006;6(5):751–762. doi: 10.1111/j.1567-1364.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- 34.Son S.H., Goo Y.H., Chang B.H., Paul A. Perilipin 2 (PLIN2)-deficiency does not increase cholesterol-induced toxicity in macrophages. PLoS One. 2012;7(3):e33063. doi: 10.1371/journal.pone.0033063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill A.A., Anderson-Baucum E.K., Kennedy A.J., Webb C.D., Yull F.E., Hasty A.H. Activation of NF-kappaB drives the enhanced survival of adipose tissue macrophages in an obesogenic environment. Molecular Metabolism. 2015;4(10):665–677. doi: 10.1016/j.molmet.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nathan C., Cunningham-Bussel A. Beyond oxidative stress: an immunologist's guide to reactive oxygen species. Nature Reviews Immunology. 2013;13(5):349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lichtenstein L., Mattijssen F., de Wit N.J., Georgiadi A., Hooiveld G.J., van der Meer R. Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell Metabolism. 2010;12(6):580–592. doi: 10.1016/j.cmet.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang J., Cao R., Wang X., Zhang Y., Wang P., Gao H. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2. Cell Research. 2017;27(3):329–351. doi: 10.1038/cr.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samuel V.T., Shulman G.I. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148(5):852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.