Abstract
Recent studies have revealed that the oxidative pentose phosphate pathway (PPP), malic enzyme (ME), and folate metabolism are the three major routes for generating cellular NADPH, a key cofactor involved in redox control and reductive biosynthesis. Many tumor cells exhibit altered NADPH metabolism to fuel their rapid proliferation. However, little is known about how NADPH metabolism is coordinated in tumor cells. Here we report that ME1 increases the PPP flux by forming physiological complexes with 6-phosphogluconate dehydrogenase (6PGD). We found that ME1 and 6PGD form a hetero-oligomer that increases the capability of 6PGD to bind its substrate 6-phosphogluconate. Through activating 6PGD, ME1 enhances NADPH generation, PPP flux, and tumor cell growth. Interestingly, although ME1 could bind either the dimer-defect mutant 6PGD (K294R) or the NADP+-binding defect 6PGD mutants, only 6PGD (K294R) activity was induced by ME1. Thus, ME1/6PGD hetero-complexes may mimic the active oligomer form of 6PGD. Together, these findings uncover a direct cross-talk mechanism between ME1 and PPP, may reveal an alternative model for signaling transduction via protein conformational simulation, and pave the way for better understanding how metabolic pathways are coordinated in cancer.
Keywords: enzyme, metabolism, pentose phosphate pathway (PPP), protein cross-linking, protein-protein interaction, 6PGD, NADPH, conformational simulation, malic enzyme
Introduction
PPP3 is a major glucose catabolic pathway that directs glucose metabolic flux into macromolecular biosynthesis by providing precursors such as ribose 5-phosphate and NADPH (1). Ribose 5-phosphate is a key precursor for nucleotides biosynthesis, whereas NADPH acts as an electron donor for reductive biosynthesis including nucleotides and lipid biosynthesis (1, 2). Besides, NADPH is an essential component of antioxidant system. In PPP, NADPH is generated by two reactions catalyzed by G6PD and 6PGD separately. Specifically, PPP (G6PD and 6PGD reactions) is the only source of reductant to reduce glutathione and prevent damage from reactive oxygen species in red blood cells (3, 4). Despite the key role in biosynthesis and redox control, PPP is significantly reprogrammed in many types of proliferating cells (4). Depletion of either G6PD or 6PGD leads to cell growth arrest, senescence, or even apoptosis (5–7). In many tumor cells G6PD and 6PGD are frequently dysregulated at both transcriptional and post-translational levels to promote tumor cell growth and/or survival (6–9).
NADPH can be produced from multiple metabolic pathways. However, growing evidence suggests that, in general, there are three major contributors for cellular NADPH pool: G6PD in the oxidative pentose phosphate pathway, malic enzymes, and the methylenetetrahydrofolate dehydrogenase in folate metabolism (10, 11). Interestingly, in adipocytes, most NADPH is produced from the oxidative PPP and particularly malic enzymes (12). Malic enzymes are the tricarboxylic acid (TCA) cycle-associated metabolic enzymes that convert malate to pyruvate and CO2, with the reduction of either NAD+ or NADP+. Three main isoforms of malic enzyme (ME) have been reported: cytoplasmic NADP+-dependent isoform (ME1), mitochondrial NADP+/NAD+-dependent isoform (ME2), and mitochondrial NADP+-dependent isoform (ME3). However, ME1 and ME2 appear to be main isoforms as ME3 is nearly undetectable in many mammalian cells assessed (10). Through sustaining cellular NADPH levels and adjusting TCA cycle flux, malic enzyme promotes tumor growth and prevents senescence (10). Although it has become increasing clear that alteration of NADPH metabolism is critical for cell proliferation and survival, the mechanisms underlying how NADPH metabolic pathways are coordinated remain poorly understood. Here we report that ME1 enhances PPP through direct binding and activating 6PGD, leading to tumor cell proliferation.
Results
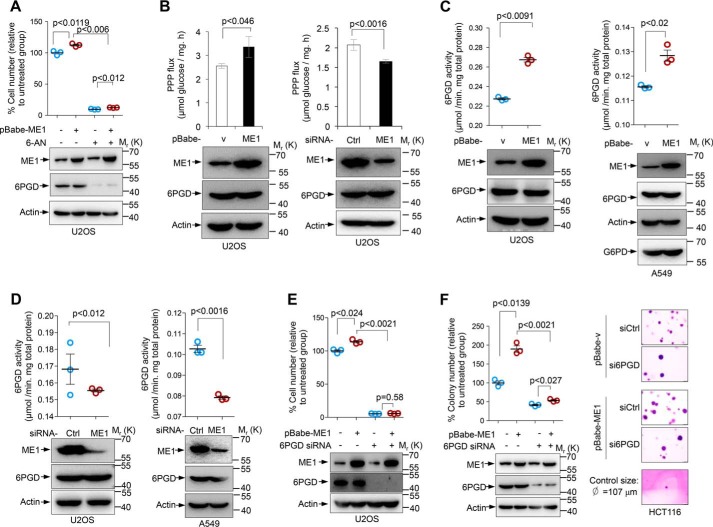
ME1 promotes PPP flux, cell growth, and cellular 6PGD activity
To explore the intrinsic relationship among NADPH metabolic pathways, we examined whether ME1 coordinates PPP to promote cell proliferation. The human osteosarcoma U2OS cells stably infected with retroviral vector expressing ME1 grew faster than control cells under in vitro culture condition. Interestingly, supplementation of 6-aminonicotinamide (6-AN), a competitive G6PD and 6PGD inhibitor (13, 14), blocked cell proliferation regardless of ME1 status (Fig. 1A). These findings raise a possibility that ME1-mediated cell proliferation may be dependent on the PPP activity. To directly investigate whether ME1 affects the PPP flux, we performed 13C isotope tracing experiments. U2OS cells stably expressing ME1 or control vector were cultured in medium containing [1,2-13C2] glucose, and the oxidative PPP flux was measured on the basis of the rate of glucose consumption and the generation of 13C-labeling lactate (M1) determined by liquid chromatography-mass spectrometry (LC-MS). Interestingly, we found that ME1 expression led to an increase in the oxidative PPP flux, whereas silencing of ME1 caused a significant drop in the oxidative PPP flux (Fig. 1B). We next investigated the mechanism by which ME1 promotes the PPP flux. By analyzing PPP enzymes activities, we observed that ME1 affected 6PGD activity. Stable overexpression of ME1 noticeably enhanced the enzymatic activity of 6PGD in U2OS cells (Fig. 1C, left panel). Similar results were observed in human lung cancer A549 cells (Fig. 1C, right panel). Conversely, knockdown of ME1 expression resulted in reduced 6PGD activity in these cell lines (Fig. 1D). To test whether the effect of ME1 on the cell proliferation is mediated by 6PGD, we knocked down 6PGD in control cells and ME1-overexpressed cells. Consistently, 6PGD silencing impaired cell proliferation and phenocopied 6-aminonicotinamide treatment (Fig. 1E). Moreover, in soft agar, an in vitro measure of tumorigenicity, tumor cells transduced with ME1 showed enhanced anchorage-independent growth, whereas silencing of 6PGD reduced the anchorage-independent growth of both control cells and ME1-overexpressed cells, minimizing the difference between these two cell types (Fig. 1F). Together, these data suggest that ME1 modulates PPP activity and promotes cell proliferation at least partially via 6PGD.
Figure 1.
ME1 promoted tumor cell growth, PPP flux, and 6PGD activation. A, U2OS cells stably overexpressing ME1 or control vector were cultured for 24 h and then treated with or without 100 μm 6-aminonicotinamide (6-AN) for another 16 h. Cell proliferation (% of control) was determined by counting. Data are the means ± S.D. (n = 3 independent experiments). The expression of ME1 and action was detected using Western blotting. B, U2OS cells stably overexpressing ME1 (left panel) or treated with ME1 siRNA or control siRNA (right panel) as indicated were cultured in medium containing 5 mm [1,2-13C2]glucose for 12 h. Oxidative PPP flux was measured on the basis of the rate of glucose consumption, and the generation of 13C-labeling lactate (M1 indicates PPP, and M2 indicates glycolysis) was determined by LC-MS. Data are the means ± S.D. (n = 3 independent experiments). Protein expression is shown below. C, 6PGD activity in U2OS cells (left panel) and A549 cells (right panel) stably overexpressing ME1 or control vector. Data are the means ± S.D. (n = 3 independent experiments). Protein expression was determined by Western blotting. D, 6PGD activity in U2OS cells (left panel) and A549 cells (right panel) treated with control or ME1 siRNA. Data are the means ± S.D. (n = 3 independent experiments). Protein expression was determined. E, proliferation (% of control) of U2OS cells stably overexpressing ME1 or control vector were transfected with 6PGD siRNA or control siRNA as indicated. Data are the means ± S.D. (n = 3 independent experiments). Protein expression is shown below. F, HCT116 cells stably overexpressing ME1 or control vector were treated with 6PGD or control siRNA as indicated. 24 h later cells were plated in soft agar and cultured for another 2 weeks. Numbers of colonies with a diameter >20 μm were quantified (means ± S.D., n = 3) (left panel). Results were normalized and represented as % of control. Representative images of colonies were stained with crystal violet (right panel).
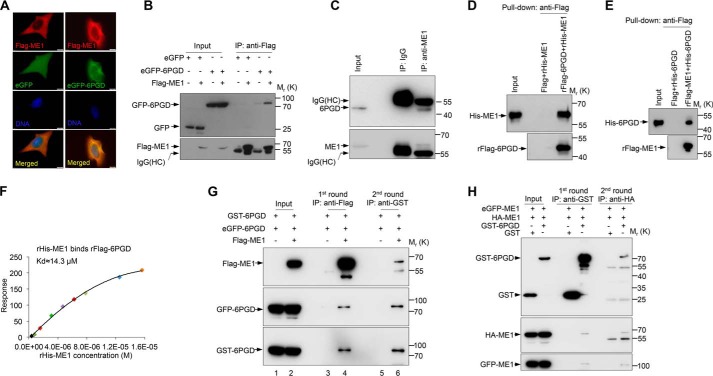
ME1 physically associates with 6PGD
As shown in Fig. 1, the levels of the 6PGD protein remained unchanged by either overexpression or depletion of ME1, suggesting that ME1-mediated stimulation of 6PGD activity is not due to the enhancement of protein synthesis or stabilization. To investigate the mechanism by which ME1 increases 6PGD activity, we performed immunofluorescence assay using cells co-expressing FLAG-tagged ME1 (FLAG-ME1) and enhanced green fluorescence protein-fused 6PGD (GFP-6PGD) or eGFP vector control. Results showed that ME1 and 6PGD co-localized in the cytoplasm (Fig. 2A). These findings led us to examine whether ME1 binds 6PGD. Immunoprecipitation assays revealed that FLAG-ME1 specifically associated with GFP-6PGD in 293T cells (Fig. 2B). Moreover, the interaction between ME1 and 6PGD occurred at endogenous levels (Fig. 2C). To investigate whether ME1 and 6PGD association is direct or influenced by other factors, we purified these proteins and performed the binding assay in vitro. As we expected, a notable association between ME1 and 6PGD was observed (Fig. 2, D and E). Moreover, analysis of the ME1–6PGD binding in real time using surface plasmon resonance (BIAcore) showed that ME1 and 6PGD bound each other with dissociation constant (Kd) of ∼14.3 μm (Fig. 2F). Together, these findings suggest that ME1 binds to 6PGD directly.
Figure 2.
ME1 physically and directly interacted with 6PGD. A, co-localization of transfected ME1 and 6PGD in the cytosol. U2OS cells were transfected with FLAG-ME1 together with GFP-6PGD or eGFP control vector. FLAG-ME1 was immune-stained with an anti-FLAG antibody followed by an Alexa Fluor®594-conjugated secondary antibody, and DNA was stained with DAPI. The individual and merged images are shown. Scale bars, 10 μm. B, 293T cells were transfected with eGFP-6PGD and FLAG-ME1 as indicated. Cell lysates were immunoprecipitated with an anti-FLAG antibody and followed by Western blotting analysis. C, lysates from U2OS cells were immunoprecipitated with anti-ME1 antibody or isotype-matching control antibody (IgG). Immunoprecipitates and input were analyzed by Western blotting. D, purified recombinant His-ME1 (His-ME1) was incubated with FLAG-peptide or recombinant FLAG-6PGD (rFLAG-6PGD) conjugated to anti-FLAG M2 beads separately. Beads-bound and input proteins were analyzed by Western blotting using anti-His antibody and anti-FLAG antibody as indicated. E, purified recombinant His-6PGD (His-6PGD) was incubated with FLAG-peptide or recombinant FLAG-ME1 (rFLAG-ME1) conjugated to anti-FLAG M2 beads separately. Beads-bound and input proteins were analyzed by Western blotting. F, purified FLAG-6PGD was coupled to CM5 chip, and His-ME1 functioned as a moving phase. The dissociation constant (Kd) for ME1 from immobilized 6PGD was determined by surface plasmon resonance (BIAcore). G, 293T cells were transfected with GFP-6PGD, GST-6PGD, and FLAG-ME1 or control vector as indicated. Cell lysates were immunoprecipitated with anti-FLAG M2 beads (lanes 3 and 4). FLAG-ME1 and the associated proteins were eluted with FLAG peptide. Twenty percent of the eluent was subject to Western blotting analysis using the indicated antibodies. The remaining eluent was used for secondary immunoprecipitation with anti-GST beads (lanes 5 and 6). H, 293T cells were transfected with GFP-ME1, HA-ME1, and GST-6PGD or GST vector control as indicated. Two rounds of immunoprecipitations were performed using anti-GST and anti-HA beads sequentially as described in Fig. 2G. Immunoprecipitates and input were analyzed by Western blotting using indicated antibodies. All results are representative of at least three independent experiments.
To test whether ME1 can form complexes with 6PGD, we employed cell lysates from 293T cells transfected with FLAG-ME1 and 6PGD fused with either GST or GFP to perform sequential immunoprecipitation experiments. An initial immunoprecipitation assay using an anti-FLAG antibody against FLAG-ME1 pulled down both GST-tagged and eGFP-tagged 6PGD (Fig. 2G, lane 4). The immunocomplexes were eluted, and GST-6PGD was subsequently precipitated by an anti-GST antibody. ME1 and GFP-6PGD were present in the anti-GST-6PGD precipitates (Fig. 2G, lane 6). Conversely, using anti-GST antibody to pull down GST-6PGD in the first round of immunoprecipitation and anti-HA antibody to precipitate HA-ME1, we could subsequently observe that 6PGD is able to form complexes with both HA-ME1 and GFP-ME1 (Fig. 2H). Thus, these data reveal that ME1 physically forms complexes with 6PGD.
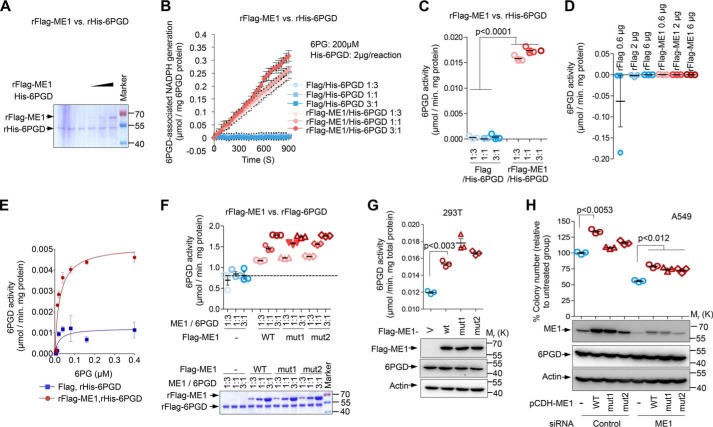
ME1 directly enhances 6PGD enzymatic activity independent of its own activity
To investigate whether the direct association underlines the mechanism for the regulation of 6PGD activity by ME1, we purified recombined FLAG-ME1 and His-tagged 6PGD (rHis-6PGD) proteins and carried out an enzymatic activity assay in vitro. However, protein may undergo structural or conformational changes during purification, leading to inhibition or even loss of activity. To exclude this possibility, we examined the enzymatic activity of each of the purified proteins in vitro. As we anticipated, purified FLAG-ME1 protein exhibited high capability of converting NADP+ to NADPH in the presence of sufficient substrate malate and showed high enzymatic activity (supplemental Fig. 1, A–C). Similar results were observed with purified His-6PGD proteins (supplemental Fig. 1, D–F), suggesting that enzymatic activities of the recombinant proteins were sustained during the process of purification. Having these recombined proteins, we next investigated the effect of ME1 on 6PGD activity in vitro. ME1 incubation with rHis-6PGD increased NADPH production rates of 6PGD and enhanced the 6PGD activity in a dose-dependent manner (Fig. 3, A–C). To exclude the possibility that the increased 6PGD activity might be due to nonspecific conversion of 6PGD substrate (6PG) by ME1 or the ME1-associated cellular 6PGD during purification by protein A/G affinity chromatography, we examined whether ME1 possessed “6PGD enzymatic activity.” In the presence of 6PG and NADP+, ME1 failed to elevate NADPH production, in other words, no 6PGD activity exhibited by recombined ME1 was observed, indicating that ME1 increased 6PGD activity directly and specifically (Fig. 3D, supplemental Fig. 2A). Increased 6PGD enzymatic activity might be induced by higher affinity to its substrate. To confirm this, we assayed the effect of ME1 on 6PGD enzymatic activity in vitro with increasing amounts of substrate 6PG (Fig. 3E). Surprisingly, at each dose of 6PG, FLAG-ME1 supplementation increased 6PGD activity, suggesting that ME1 enhances the capability of 6PGD binding to it substrate. Additionally, no activity was detected when 6PG is absent (0), underscoring the specificity of the assay.
Figure 3.
ME1 directly enhanced 6PGD enzymatic activity by increasing the binding ability of 6PGD to its substrate, which is independent of ME1 activity. A–C, recombinant (r) His-6PGD and Falg-ME1 were purified by using nickel-nitrilotriacetic acid-agarose and FLAG-M2-agarose, respectively. Purified proteins were analyzed by Coomassie Blue staining (A). A constant amount of purified His-6PGD protein was incubated with a different amount of either purified FLAG-ME1 or FLAG peptide at a mole ratio of 1:3, 1:1, or 3:1. The ability of 6PGD-associated NADPH generation in the presence of 6PG (B) and 6PGD activity (C) is shown. The concentration of 6PG is 200 μm. Data are the means ± S.D. (n = 3 independent experiments). D, 6PGD activity of purified FLAG-ME1 incubated with a reaction buffer containing 6PGD substrate 6PG and NADP+. Data are the means ± S.D. (n = 3 independent experiments). E, purified His-6PGD was incubated with either FLAG-ME1 proteins or FLAG peptide for 30 min. The enzymatic activity of 6PGD was measured in the presence of increasing amounts of 6PG. Data are the means ± S.D. (n = 3 independent experiments). F, activity of purified FLAG-6PGD after incubation with FLAG peptide, wild-type, or mutant ME1 proteins at different mole ratios (1:3, 1:1 and 3:1) as indicated. The protein expression was visualized by Coomassie Blue staining. Data are the means ± S.D. (n = 3 independent experiments). G, cells lysates from 293T cells transfected with FLAG-ME1, FLAG-ME1mut1, or FLAG-ME1mut2 were analyzed for G6PD activity (top) and protein expression (bottom). Data are the means ± S.D. (n = 3 independent experiments). H, colony formation of A549 cells stably overexpressing RNAi-resistant wild-type ME1, ME1 mutants, or control vector in the presence of ME1 siRNA or control siRNA as indicated. Numbers of colonies with a diameter >20 μm were quantified (means ± S.D., n = 3) (top panel). Protein expression is shown below.
To rule out the possibility that different fusion tags may change 6PGD activity induced by ME1, we thus switched to a FLAG tag for 6PGD protein purification. Indeed, FLAG-6PGD showed higher activity than His-6PGD, which may be caused by fusing tag or activity loss during purification (Fig. 3, F versus C). Nevertheless, consistent with above findings, ME1 noticeably elevated 6PGD activity in a dose-dependent manner regardless of different tags used (Fig. 3F). By contrast, FLAG alone failed to affect 6PGD enzymatic activity (Fig. 3F). To determine whether these effects of ME1 are mediated by its own enzymatic activity, we generated two ME1 mutations (ME1mut1 and ME1mut2), each of which exhibited little or no enzymatic activity as described previously (10). Interestingly, both mutants were able to markedly increase 6PGD enzymatic activity and 6PGD-mediated NADPH generation in vitro (Fig. 3F, supplemental Fig. 2B). Similarly, these effects were also observed in 293T cells. Cells transfected with increasing amounts of either wild-type ME1 showed increased 6PGD enzymatic activities and NADPH generation ability compared with control cells (supplemental Fig. 2, C and D). Expression of these two mutant forms of ME1 led to augmentation of 6PGD activity, in which both ME1 mutants were still able to interact with 6PGD (Fig. 3G, supplemental Fig. 2E). Together, these data suggest that ME1 directly promotes 6PGD activation independent of its own activity.
Next, we wanted to investigate the effect of these ME1 mutants on cell proliferation. Consistent with our previous findings (10), ME1mut1 and ME1mut2 failed to remarkably promote cell proliferation under normal condition (supplemental Fig. 2, F and G, columns 1–4). This may due to the dominant negative effect of ME1 mutants on endogenous ME1. To test this possibility, we knocked down endogenous ME1 by introducing ME1 siRNA into the cells stably expressing RNAi-resistant ME1 or ME1 mutants. Notably, when endogenous ME1 was absent, expression of mutant ME1 led to increased cell proliferation to a similar extent as that of wild-type ME1 (supplemental Fig. 2, F and G, columns 5–8). Similar results were obtained with the anchorage-independent growth. Like wild-type ME1, these mutant forms of ME1 increased the number of colonies (Fig. 3H). Together, these findings suggest that ME1 promotes cell proliferation independent of its enzymatic activity, correlating with its effect on 6PGD (Fig. 3G). To extend our findings to other species, we tested the effect of ME1 on yeast 6PGD protein. Similarly, ME1 enhanced the yeast 6PGD activity and the ability of yeast 6PGD to generate NADPH in a dose-dependent manner (supplemental Fig. 3, A–C). Together, these results show that ME1 directly enhanced 6PGD enzymatic activity, which is independent of its own enzymatic activity.
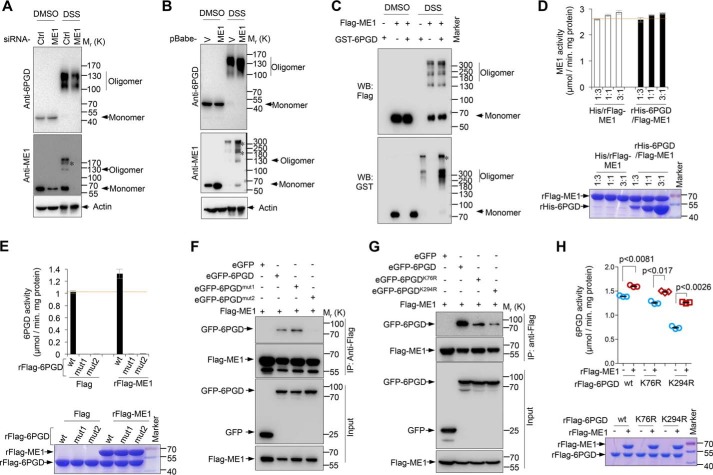
ME1 and 6PGD form an active hetero-oligomer to promote 6PGD enzymatic activity
6PGD is an equilibrium of inactive monomer and active oligomer. Studies mentioned above reveal that ME1 formed complexes with 6PGD (Fig. 2, G and H) and increased 6PGD activity (Fig. 3E). These findings led us to further investigate whether ME1 promotes the formation of ME1–6PGD hetero-oligomer, thereby increasing the binding affinity between 6PGD and its substrate 6PG. In U2OS cells, the lack of ME1 resulted in a decrease in 6PGD-associated oligomers (Fig. 4A). Consistently, stably overexpressing ME1 increased the formation of 6PGD-associated oligomers (Fig. 4B). Moreover, co-expression of GST-6PGD and FLAG-ME1 in 293T cells resulted in a significant increase in both 6PGD-associated oligomers and ME1-associated oligomers (Fig. 4C). Together with findings mentioned above (Fig. 2, G and H), these data indicate that ME1 and 6PGD may form a hetero-oligomer that harbors elevated 6PGD activity. Conversely, we sought to examine whether the formation of ME1/6PGD oligomer also exhibits increased enzymatic activity of ME1. Intriguingly, ME1 activity remained unaffected when 6PGD was present, indicating that formation of hetero-oligomers increased 6PGD activity unilaterally (Fig. 4D). Expectedly, ME1 failed to restore or increase the enzymatic activity of a mutant 6PGD, 6PGDmut1 (G10I, L11N, A12K, V13K, M14K, G15I), with a defect in binding to its cofactor NADP+ (Fig. 4E, supplemental Fig. 4, A–C), but even this mutant was still able to bind to ME1 (Fig. 4F, third lane). To further confirm this, we generated another 6PGD mutation, 6PGDmut2 (H187N, D188L), which exhibits no enzymatic activity (supplemental Fig. 4, A and B). Immunoprecipitation assays showed that this mutant 6PGD lost the ability to bind to ME1 (Fig. 4F, fourth lane), and consistently, no enhanced 6PGD enzymatic activity was observed when ME1 was present (Fig. 4E, supplemental Fig. 4C). These data suggest that direct protein-protein association is required for ME1-mediated 6PGD activation.
Figure 4.
ME1 and 6PGD formed hetero-oligomers to promote 6PGD enzymatic activity. A, lysates from U2OS cells transfected with ME1 siRNA or control siRNA were treated with DMSO or the cross-linker DSS. Cell lysates were then prepared and analyzed by Western blotting. B, lysates from U2OS cells stably expressing ME1 or control vector were treated with DMSO or DSS. The mixtures were subjected to Western blotting analysis. C, 293T cells were transfected with GST-6PGD and FLAG-ME1 as indicated. Cell lysates were treated with DMSO or DSS, and analyzed by Western blotting. D, malic enzyme activity of purified FLAG-ME1 after being incubated with His peptide or His-6PGD proteins at different mole ratio as indicated (top). The protein expression was visualized by Coomassie Blue staining (bottom). Data are the means ± S.D. (n = 3 independent experiments). E, activity of purified wild-type FLAG-6PGD, FLAG-6PGDmut1 (G10I, L11N, A12K, V13K, M14K, G15I), or FLAG-6PGDmut2 (H187N and D188L) protein in presence or absence of ME1 protein (top). Protein expression was analyzed by Coomassie Blue staining (bottom). Data are the means ± S.D. (n = 3 independent experiments). F, 293T cells were transfected with FLAG-ME1 and eGFP-6PGD, eGFP-6PGDmut1, or eGFP-6PGDmut2 as indicated. Cell lysates were immunoprecipitated with anti-FLAG antibody. Input and immunoprecipitates were analyzed by Western blotting. G, 293T cells were transfected with FLAG-ME1 and eGFP-6PGD, eGFP-6PGDK76R, or eGFP-6PGDK294R as indicated. Cell lysates were immunoprecipitated with anti-FLAG antibody and analyzed by Western blotting. H, purified FLAG-6PGD, FLAG-6PGDK76R, and FLAG-6PGDK294R proteins were incubated with either FLAG-ME1 or FLAG peptide as indicated. 6PGD activity was measured (top), and purified proteins were analyzed by Coomassie Blue staining (bottom). All results are representative of three independent experiments.
6PGD activity can be modulated by post-translational modification such as acetylation (9). We, therefore, generated two acetyl-deficient 6PGD mutants (6PGD K76R and K294R), which exhibited decreased enzymatic activity compared to wild-type 6PGD (Fig. 4H). Mechanistically, K76R mutation reduced NADP+-binding ability of 6PGD, whereas K294R mutation impaired the formation of active 6PGD dimmers (9). Thus, we investigated whether ME1 could reverse the reduction of enzymatic activity of these mutants. First, we tested the interaction between ME1 and 6PGD (K76R) or 6PGD (K294R) by performing an immunoprecipitation assay. Although K76R or K294R mutation decrease the binding ability of 6PGD to ME1, a significant interaction between ME1 and 6PGD (K76R) or 6PGD (K294R) was observed (Fig. 4G). Interestingly, a substantial increase in the oligomerization of each of these mutants was detected related to wild-type 6PGD proteins in the presence of ME1 (supplemental Fig. 4D). Consistently, ME1 markedly restored the activity of both 6PGD (K294) and 6PGD (K76R) mutants (Fig. 4H). Together, these results suggest that ME1 forms hetero-oligomers with 6PGD to enhance 6PGD activity.
Discussion
As a key factor required for both antioxidant defense and reductive biosynthesis, NADPH can be produced from multiple pathways, of which PPP, malic enzymes, and folate metabolism are the three major contributors to cellular NADPH generation. Here we show that PPP and malic enzyme pathway are coordinated intrinsically to orchestrate NADPH metabolism. ME1 enhances PPP flux by forming hetero-complexes with 6PGD. Interestingly, although direct interaction between ME1 and 6PGD is required, the ME1 enzymatic activity is totally dispensable for the activation of 6PGD by ME1. Subsequent immunoprecipitation and cross-linking assays demonstrate that ME1/6PGD can form hetero-oligomers, which may mimic the active form of 6PGD homo-oligomer. Thus, this conformational simulation may lead to an increase in the binding ability of 6PGD to its substrate. To support this, we also generated the NADP+-binding defect 6PGDmut1 with no enzymatic activity. Similar to ME1, this mutant 6PGD could interact and form oligomers with the wild-type 6PGD (supplemental Fig. 5, A and B) and increase wild-type 6PGD activity (supplemental Fig. 5C). Hence, ME1 may act as a structural analogue of 6PGD that helps to activate 6PGD by forming ME1/6PGD hetero-oligomerization when cellular 6PGD level is low or insufficient. Consistent with hypothesis, 3D structure comparison revealed that 6PGD dimers and ME1 dimers shared similar conformation (15) (supplemental Fig. 5D). Besides this, hetero-oligomerization formation enhanced activity of 6PGD but not ME1. Therefore, further structural analysis might help us to gain insight into how the ME1/6PGD hetero-oligomers function.
Like 6PGD, the other NADPH-producing enzyme G6PD could also co-localized with ME1 (supplemental Fig. 6A) and directly bind ME1 with a Kd of ∼1.243 μm (supplemental Fig. 6, B–D). Consistently, ME1 stimulated G6PD activity and enhanced the capability of G6PD to bind its substrate glucose 6-phosphate (G6P) (supplemental Fig. 6, E and F). Likewise, mutant forms of ME1 dose-dependently activated G6PD (supplemental Fig. 6, E and F). Thus, given the existence of multiple NADPH-generating enzymes within the cells, it would be interesting to further determine the coordination network of these NADPH metabolic pathways. Moreover, some metabolic enzymes share the same coenzyme or substrate; therefore, the existence of the intrinsic coordination via conformational simulation between these metabolic enzymes might enable efficient metabolic adaptation to different stresses. Overall, our findings have highlighted the direct cross-talk between the malic enzyme pathway and the pentose phosphate pathway and may reveal an alternative mechanism for signaling transduction via conformation simulation among the metabolic enzymes.
Experimental procedures
Antibodies and reagents
The antibodies against the following proteins were purchased from the indicated sources: ME1 and phosphogluconate dehydrogenase (Santa Cruz Biotechnology); FLAG (Sigma); GFP (Seajet Scientific); GST (CMCTAG); actin (Proteintech). The following reagents were all purchased from Sigma: G6P, 6PG, β-nicotinamide adenine dinucleotide 2′-phosphate (NADP+), 6PGD from yeast, trichostatin A, FLAG peptide, and anti-FLAG M2 affinity gel except disuccinimidyl suberate (DSS) from Thermo Scientific and dimethyl sulfoxide (DMSO) from Amresco.
Cell culture, gene knockdown with siRNA, and stable lines
HCT116 cells and 293T cells were cultured in DMEM (Macgene). U2OS cells were maintained in McCoy's 5A Medium (HyClone). Cells were cultured in standard culture conditions without any antibiotic. siRNAs for ME1 and 6PGD were purchased from Invitrogen. siRNA sequences were 5′-AUAACAAUCAGGUAGAAUCUGGUCA-3′ (human ME1), 5′-GCCCAGGCCUUUGAGGAUUGGAAUA-3′ (human 6PGD), and 5′-GUGCGUUGCUAGUACCAACUU-3′ (scrambled siRNA). siRNAs were transfected into U2OS cells using Lipofectamine RNAiMAX Transfection Agent (Invitrogen). To generate cells with stable overexpression of ME1, U2OS and HCT116 cells were infected with pBabe vector or pBabe-ME1, and then cells were selected with puromycin (1 μg/ml) for 2 weeks.
6PGD and ME1 activity assay
For the 6PGD enzyme activity assay, cell lysates or purified proteins were measured by the conversion of NADP+ to NADPH in the presence of 6PG. For the ME1 activity assay, cell lysates or purified proteins were analyzed in the reaction buffer containing 67 mm triethanolamine, 3.3 mm l-malic acid, 0.3 mm β-NADP+, and 5.0 mm manganese chloride, and the increase in 341 nm absorbance (OD341) as a measure of NADPH production was obtained every 30 s for 30 min.
Primers for 6PGD mutation
The mutation primers for 6PGD were: mut1, 5′-GACATCGCGCTGATCATAAATAAGAAGAAGATCCAGAACTTAA-3′ and 5′-CAGAATTAAGTTCTGGATCTTCTTCTTATTTATGATCAGCGCG-3′; mut2, 5′-GAAGATGGTGGACCTCGGGATAGAGTATGG-3′ and 5′-CTCTATCCCGAGGTCCACCATCTTCACGAAG-3′; K76R, 5′-ATCATCCTCCTGGTGAGGGCTGGGCAAG-3′ and 5′-CACAGCTTGCCCAGCCCTCACCAGGAGG-3′; K294R, 5′-TGCTTATCATCTCTGAGGGATGAGAGAA-3′ and 5′-TTGAATTCTCTCATCCCTCAGAGATGAT-3′. Overlapping PCR was performed to construct 6PGD mutation.
Protein purification
FLAG-ME1, FLAG-ME1-mut1, FLAG-ME1-mut2, FLAG-6PGD, FLAG-6PGD-mut1, FLAG-6PGD-mut2, FLAG-6PGDK76R, and FLAG-6PGDK294R were expressed in HEK293T cells and purified by anti-FLAG M2 affinity gel. His-6PGD and His-ME1 were expressed in Escherichia coli and purified by nickel-nitrilotriacetic acid His bind resin (GE Healthcare). GST-tagged 6PGD, 6PGDK76R, and 6PGDK294R were expressed in HEK293T cells and purified by glutathione beads (GE Healthcare Life Sciences).
Immunoprecipitation and indirect immunofluorescence
For immunoprecipitation, HEK293T cells were transfected with FLAG-ME1 and GFP-6PGD for 48 h, and then cells were lysed in IP lysis buffer containing 50 mm Tris, 150 mm NaCl (pH 7.4), 1 mm EDTA, 1% Triton-100, and protease inhibitors for 30 min. Anti-FLAG M2 affinity gels were adding to supernatants and incubated for 8 h. After incubation, beads were washed 3 times with lysis buffer and eluted by FLAG peptide. Protein samples were boiled in 5× loading buffer and resolved by SDS-PAGE. As for sequential immunoprecipitation, HEK293T cells were transfected with FLAG-ME1, GST-6PGD, and GFP-6PGD for 48 h. Anti-FLAG M2 affinity gels and glutathione beads were used in the first round IP and the second round IP, respectively. In addition, HEK293T cells were transfected with GST-6PGD, HA-ME1, and GFP-ME1 for 48 h. Glutathione beads and HA beads were used in the first round IP and the second round IP, respectively.
PPP flux and metabolites measurements
The flux of PPP was measured based on the rate of glucose consumption and the ratio of 13C incorporated into lactate determined by LC-MS (16). Briefly, cells were cultured in medium with or without 5 mm [1,2-13C2]glucose. After 12 h, medium was collected, and cells were treated with cold 80% methanol. Metabolites were extracted and analyzed by LC-MS. Flux analysis was performed on TSQ Quantiva Triple Quadrupole mass spectrometer (Thermo Fisher Scientific, San Jose, CA) with positive/negative ion switching. Multiple reaction monitoring (MRM) mode was used for data acquisition. Mobile phase A was prepared by adding 2.376 ml of tributylamine and 0.858 ml of acetic acid to HPLC-grade water, then HPLC-grade water to a 1-liter volume. Mobile phase B was HPLC-grade methanol. Synergi Hydro-RP 100A column was used for polar metabolites separation with a column temperature at 35 °C. The measured mass isotopomer distributions were corrected for natural enrichments.
Cell proliferation and soft agar assay
For cell proliferation assay cells were seeded in 6-well cell culture dishes in triplicate at a density of 20,000 cells as indicated per well in 2 ml of medium containing 10% FBS. After the indicated days of culture, cells were counted using a Bio-Rad cell counter.
For soft agar assay, cells were suspended in 1 ml of 10% FBS DMEM medium containing a 0.3% agarose and plated on a firm 0.6% agarose base in 6-well plates (5000 cells per well). Cells were then cultured in a 37 °C and 5% CO2 incubator for 2 weeks. Colonies were fixed and stained with 0.005% crystal violet (for 1 h) and visible colonies were counted.
Surface plasmon resonance analysis of interaction
Surface plasmon resonance analysis was carried out at 25 °C with a Biacore T200 biomolecular interaction analysis system (GE Healthcare) according to the manufacturer's instructions. FLAG-tagged 6PGD was immobilized on the surface of a CM5 sensor chip in 10 mm sodium acetate buffer (pH 4.5) and functioned as the stationary phase. As the mobile phase, purified His-ME1 protein flowed through the sensor chip surface at 8 different concentrations in a running buffer of PBS at a flow rate of 30 μl min−1. After each cycle of binding and dissociation, the surface was regenerated with 10 mm glycine (pH 2.0). The affinity parameters were analyzed by BIAcore T200 evaluation software.
Author contributions
P. Y. performed all the experiments. H. S., C. X., T. C., and B. Z. provided technical assistance. P.J. and W.D. designed and supervised the project. P. J. and W. D. wrote the manuscript with the help of P. Y.
Supplementary Material
Acknowledgments
We thank all members of Jiang laboratory and Du laboratory for discussion.
This work was supported by the 1000 Talents Program for Young Scholars, the Tsinghua-Peking Center for Life Sciences, and National Natural Science Foundation of China (31571470; to P. J.) and CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-4-002), National Natural Science Foundation of China (81672766), State Key Laboratory Special Fund (2060204), Peking Union College “Union Youth Fund” (3332016045), and the 1000 Talents Program for Young Scholars (to W. D.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S6.
- PPP
- pentose phosphate pathway
- G6PD
- glucose 6-phosphate (G6P) dehydrogenase
- 6PGD
- 6-phosphogluconate (6PG) dehydrogenase
- TCA
- tricarboxylic acid
- ME
- malic enzyme
- eGFP
- enhanced GFP
- rHis
- recombinant His
- DSS
- disuccinimidyl suberate
- IP
- immunoprecipitation.
References
- 1. Berg J. M., Tymoczko J. L., Gatto G. J., and Stryer L. (2006) Biochemistry, 6th Ed., pp. 577–589, W. H. Freeman & Co., a Macmillan Education Imprint, New York [Google Scholar]
- 2. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alving A. S., Carson P. E., Flanagan C. L., and Ickes C. E. (1956) Enzymatic deficiency in primaquine-sensitive erythrocytes. Science 124, 484–485 [DOI] [PubMed] [Google Scholar]
- 4. Patra K. C., and Hay N. (2014) The pentose phosphate pathway and cancer. Trends Biochem. Sci. 39, 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tian W. N., Braunstein L. D., Pang J., Stuhlmeier K. M., Xi Q. C., Tian X., and Stanton R. C. (1998) Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J. Biol. Chem. 273, 10609–10617 [DOI] [PubMed] [Google Scholar]
- 6. Du W., Jiang P., Mancuso A., Stonestrom A., Brewer M. D., Minn A. J., Mak T. W., Wu M., and Yang X. (2013) TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat. Cell Biol. 15, 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin R., Elf S., Shan C., Kang H. B., Ji Q., Zhou L., Hitosugi T., Zhang L., Zhang S., Seo J. H., Xie J., Tucker M., Gu T. L., Sudderth J., Jiang L., Mitsche M., et al. (2015) 6-Phosphogluconate dehydrogenase links oxidative PPP, lipogenesis and tumour growth by inhibiting LKB1-AMPK signalling. Nat. Cell Biol. 17, 1484–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang P., Du W., Wang X., Mancuso A., Gao X., Wu M., and Yang X. (2011) p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 13, 310–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shan C., Elf S., Ji Q., Kang H. B., Zhou L., Hitosugi T., Jin L., Lin R., Zhang L., Seo J. H., Xie J., Tucker M., Gu T. L., Sudderth J., Jiang L., et al. (2014) Lysine acetylation activates 6-phosphogluconate dehydrogenase to promote tumor growth. Mol. Cell 55, 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang P., Du W., Mancuso A., Wellen K. E., and Yang X. (2013) Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 493, 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fan J., Ye J., Kamphorst J. J., Shlomi T., Thompson C. B., and Rabinowitz J. D. (2014) Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu L., Shah S., Fan J., Park J. O., Wellen K. E., and Rabinowitz J. D. (2016) Malic enzyme tracers reveal hypoxia-induced switch in adipocyte NADPH pathway usage. Nat. Chem. Biol. 12, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lange K., and Proft E. R. (1970) Inhibition of the 6-phosphogluconate dehydrogenase in the rat kidney by 6-aminonicotinamide. Naunyn Schmiedebergs Arch. Pharmakol. 267, 177–180 [DOI] [PubMed] [Google Scholar]
- 14. Köhler E., Barrach H., and Neubert D. (1970) Inhibition of NADP dependent oxidoreductases by the 6-aminonicotinamide analogue of NADP. FEBS Lett. 6, 225–228 [DOI] [PubMed] [Google Scholar]
- 15. Hsieh J. Y., Li S. Y., Chen M. C., Yang P. C., Chen H. Y., Chan N. L., Liu J. H., and Hung H. C. (2014) Structural characteristics of the nonallosteric human cytosolic malic enzyme. Biochim. Biophys. Acta 1844, 1773–1783 [DOI] [PubMed] [Google Scholar]
- 16. Rao X., Duan X., Mao W., Li X., Li Z., Li Q., Zheng Z., Xu H., Chen M., Wang P. G., Wang Y., Shen B., and Yi W. (2015) O-GlcNAcylation of G6PD promotes the pentose phosphate pathway and tumor growth. Nat. Commun. 6, 8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.