Abstract
Xeroderma pigmentosum (XP) complementation group A (XPA) is an essential scaffolding protein in the multiprotein nucleotide excision repair (NER) machinery. The interaction of XPA with DNA is a core function of this protein; a number of mutations in the DNA-binding domain (DBD) are associated with XP disease. Although structures of the central globular domain of human XPA and data on binding of DNA substrates have been reported, the structural basis for XPA's DNA-binding activity remains unknown. X-ray crystal structures of the central globular domain of yeast XPA (Rad14) with lesion-containing DNA duplexes have provided valuable insights, but the DNA substrates used for this study do not correspond to the substrates of XPA as it functions within the NER machinery. To better understand the DNA-binding activity of human XPA in NER, we used NMR to investigate the interaction of its DBD with a range of DNA substrates. We found that XPA binds different single-stranded/double-stranded junction DNA substrates with a common surface. Comparisons of our NMR-based mapping of binding residues with the previously reported Rad14-DNA crystal structures revealed similarities and differences in substrate binding between XPA and Rad14. This includes direct evidence for DNA contacts to the residues extending C-terminally from the globular core, which are lacking in the Rad14 construct. Moreover, mutation of the XPA residue corresponding to Phe-262 in Rad14, previously reported as being critical for DNA binding, had only a moderate effect on the DNA-binding activity of XPA. The DNA-binding properties of several disease-associated mutations in the DBD were investigated. These results suggest that for XPA mutants exhibiting altered DNA-binding properties, a correlation exists between the extent of reduction in DNA-binding affinity and the severity of symptoms in XP patients.
Keywords: DNA-binding protein, DNA repair, nuclear magnetic resonance (NMR), nucleotide excision repair, protein-DNA interaction
Introduction
Nucleotide excision repair (NER)3 is a DNA damage repair pathway specialized for removing bulky lesions arising from exposure to various types of endogenous and exogenous toxic agents (1–5). Human NER is a multistep process involving coordinated action of over 30 proteins (4, 6). Two NER pathways exist, one repairs lesions in actively transcribed DNA (transcription-coupled repair), and the other processes lesions more generally throughout the genome (global genome repair). These differ only in the mechanism by which the presence of damage is recognized. The subsequent steps, destabilization and unwinding of the DNA by transcription factor II H (TFIIH) to create a DNA structure termed the NER bubble, excision of the damaged nucleotide, and gap-filling synthesis, are understood to be the same (6–22).
XPA is recruited to the damage site by TFIIH once the duplex is unwound (6, 7). Although XPA does not have any enzymatic activity, it acts in concert with replication protein A (RPA) as a critical scaffolding protein through its interactions with DNA and other NER proteins (23).
Defects in NER result in the genetic disorder xeroderma pigmentosum (XP), which is characterized by hypersensitivity to sunlight and increased incidence of skin cancer (24–27). In severe cases, neurological defects are also observed (24, 28–30). XP arises from mutations in eight genes, seven of which (XPA-G) are directly involved in NER (4, 31). Among these, XPA mutations are usually associated with most severe disease phenotypes, many of which map to the DNA-binding domain (DBD) (28–30). Different XPA mutations lead to different disease phenotypes (24, 29, 30, 32); complete loss, substantial truncation, and unfolding of the DBD are known to lead to severe XP disease phenotypes. However, genotype-phenotype correlations and mechanisms behind missense mutations remain poorly understood (23, 30, 33–35).
Two structures of the globular core of the Saccharomyces cerevisiae homolog of XPA, Rad14, have been determined for complexes with a DNA duplex containing a cisplatin 1,2-deoxydiguanosine intrastrand cross-link or a N-(deoxyguanosin-8-ly)-2-acetylaminofluorene (AAF) lesion (36). Rad14 has remarkably higher affinity for these two modified duplexes than for unmodified duplexes or duplexes containing other lesions (36). The structure of the Rad14 globular core is quite similar to the previously reported structures of the globular core of human XPA obtained in the absence of DNA (37, 38). However, we and others have shown that the globular core of human XPA (XPA98–219) does not bind NER model DNA substrates with appreciable affinity and that additional C-terminal residues are required for full binding affinity (39, 40). Consequently, the high binding affinity for the two lesion-containing duplexes of the Rad14 globular core that lacks additional C-terminal residues is confounding and suggests that in the two studies, the Rad14 and XPA DBDs are not functioning in an equivalent manner biochemically and structurally. Notably, evidence has accumulated suggesting that XPA may have roles outside of NER (23, 41–46). Nevertheless, the difference is puzzling in the context of NER because, although XPA was first thought to be involved in damage recognition (47–49), it is now understood that XPA is not recruited to the NER preincision complex until after the presence of damaged DNA is recognized and unwound (6, 7, 50).
Using the XPA98–239 construct (XPA DBD) that we previously reported to exhibit full DNA-binding affinity (39), we report here an investigation of the interaction of human XPA with model NER bubble substrates. We used NMR and DNA affinity measurements to define the binding site of XPA DBD for model NER substrates and compared our findings to the structures of Rad14-DNA complexes (36). To confirm our findings, we determined the effect on DNA binding of selected structure-based and disease-associated mutants. Our results show human XPA and yeast Rad14 use similar DNA-binding surfaces, but XPA interactions in the context of NER are different from those observed for Rad14 interactions with high affinity lesion-containing duplexes (36). This study also provides insight into the molecular basis for the disease association of certain XPA mutations.
Results
Human XPA binds a range of ss-dsDNA junction substrates
It had been established previously that XPA binds to a ss-dsDNA junction in the NER bubble (51), although controversy remains over whether it is the 5′ or 3′ junction. Moreover, the footprint of XPA on the junction remains unclear. To address these issues, we measured the affinities of the XPA DBD for different junction structures and lengths. We have previously used a DNA fluorescence anisotropy assay to characterize affinities, but turned instead here to microscale thermophoresis (MST) to avoid the tendency of XPA-DNA complexes to aggregate in the 384-well plates during the course of fluorescence anisotropy experiments. Comparisons of the values obtained by the two methods revealed the same trends among substrates but higher dissociation constants (Kd) for MST by ∼5-fold. The systematically weaker binding in the MST experiments is attributable to the higher ionic strength of the buffer.
We first confirmed that XPA Y binds to a previously characterized 8/12 splayed-arm substrate (Fig. 1, diagram 1, Y-shaped junction substrate with an 8-bp duplex and 12-nucleotide (nt) 5′ and 3′ overhangs) with high affinity (39). In NER incision complexes, RPA coats the undamaged ssDNA opposite the damage-containing strand, so Y-shaped junctions with overhangs on both sides are not likely to represent the relevant substrate in a NER complex. Therefore, we next sought to find a junction DNA substrate that is optimized in length and shape so that it interacts with XPA in a manner that avoids nonspecific secondary binding to the substrate. To systematically screen the types of junction and the length of overhangs, a series of DNA substrates were designed containing a GC-rich duplex (sealed with a 4-nt hairpin (HP) for stability) and oligo(dT) overhangs (Fig. 1, diagrams 2–9). We found a modest but clearly significant difference in affinity for different nucleotide sequences (supplemental Fig. S1), as has been observed for other sequence nonspecific DNA-binding proteins such as RPA (52).
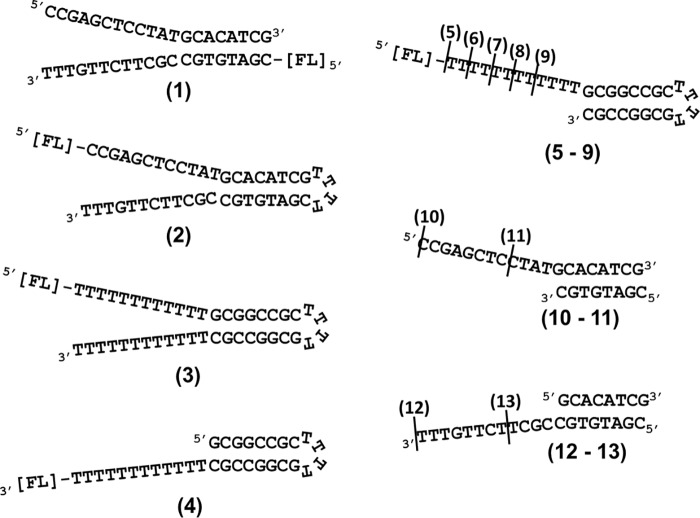
Figure 1.
DNA substrates. Structures and sequences of ss-dsDNA junction substrates used for binding assays and NMR analyses are shown. The names are based on the number of basepairs in the duplex region followed by the number of nucleotides in the overhang. Diagram 1, 8/12 splayed arm; diagram 2, 8/12 HP splayed arm with mixed sequence; diagram 3, 8/12 HP splayed arm; diagram 4, 8/12 HP 3′ overhang; diagram 5, 8/12 HP 5′ overhang; diagram 6, 8/10 HP 5′ overhang; diagram 7, 8/8 HP 5′ overhang; diagram 8, 8/6 HP 5′ overhang; diagram 9, 8/4 HP 5′ overhang; diagram 10, 8/12 5′ overhang; diagram 11, 8/4 5′ overhang; diagram 12, 8/12 3′ overhang; and diagram 13, 8/4 3′ overhang. All HPs are composed of four Ts. The positions of fluorescein tags are indicated by [FL].
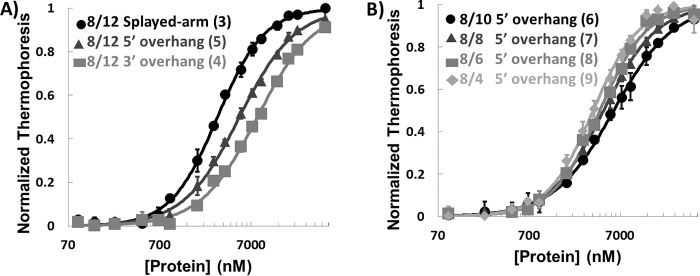
The data show that XPA DBD binds both 3′ and 5′ overhang substrates with approximately the same affinity as Y-shaped substrates (Fig. 2A and Table 1). It is interesting that the length of the overhang could be shortened to 4 nt without any significant effect on the affinity for substrate (Fig. 2B and Table 1). The shorter overhang was advantageous for NMR studies, so we selected an 8/4 5′ overhang substrate for detailed analysis.
Figure 2.
XPA binding to ss-ds junction DNA substrates. A, plot of MST data for XPA DBD binding 8/12 HP splayed arm (circle), 8/12 5′ HP overhang (triangle), and 8/12 3′ HP overhang (square) (substrates 3, 5, and 4 in Fig. 1, respectively). B, plot of MST data for XPA DBD binding DNA substrates with 8-nt duplex and different lengths of 5′ overhangs (substrates 6–9 in Fig. 1). All measurements were made at room temperature in a buffer containing 50 mm Tris-HCl, pH 7.8, 150 mm NaCl, 10 mm MgCl2, 0.05% Tween 20, and 1 mm DTT. The error bars indicate standard deviations from at least three measurements.
Table 1.
The affinity of XPA DBD for ss-ds junction DNA substrates
Dissociation constants were extracted from the MST data plotted in Fig. 2. The 8/12 HP splayed-arm substrate is composed of a Y-shaped ss-dsDNA junction with 8-nt duplex and 12-nt ssDNA overhangs on both 5′ and 3′ ends and a 4-nt HP on the opposite end. The nomenclature for other substrates follows the same system. The numbers in parentheses identify each substrate as shown in Fig. 1, which also includes the chemical structures. The errors are standard deviations from at least three measurements. The less than 3-fold difference between the Kd values indicates that the protein does not have substantial junction substrate specificity.
| Substrate | Kd |
|---|---|
| μm | |
| 8/12 HP splayed arm (3) | 3.0 ± 0.1 |
| 8/12 HP 5′ overhang (5) | 5.2 ± 0.2 |
| 8/12 HP 3′ overhang (4) | 8.2 ± 0.5 |
| 8/10 HP 5′ overhang (6) | 5.9 ± 0.2 |
| 8/8 HP 5′ overhang (7) | 4.6 ± 0.1 |
| 8/6 HP 5′ overhang (8) | 4.2 ± 0.2 |
| 8/4 HP 5′ overhang (9) | 3.5 ± 0.2 |
NMR analysis defines the DNA-binding sites of human XPA
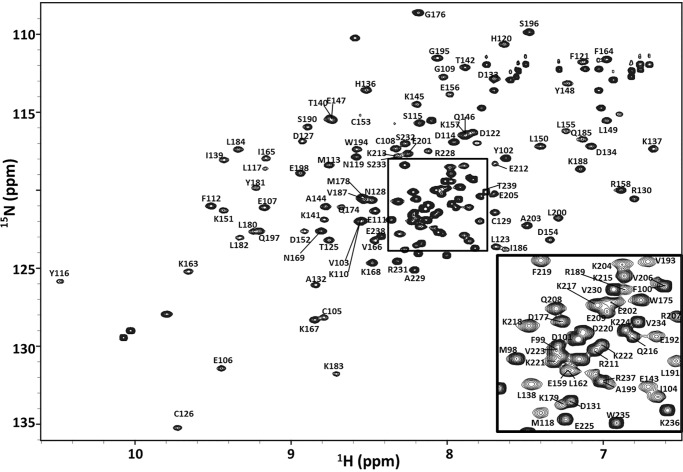
The binding of DNA substrates by XPA DBD (XPA98–239) was investigated using NMR spectroscopy. The first step in any detailed NMR analysis is the assignment of the signals (resonances) to specific atoms within the molecule. The NMR backbone resonance assignments for XPA DBD were obtained using a standard series of double and triple resonance 2D and 3D experiments (53). This analysis produced assignments for 98, 98, 94, 98, and 97% of the 15N, 1H, 13CO, 13Cα, and 13Cβ resonances, respectively (supplemental Table S1). Fig. 3 shows the 2D 15N-1H HSQC spectrum labeled with the corresponding backbone resonance assignments used for the titration analyses of the binding of DNA substrates. Because of the extensive overlap and some highly dynamic peak intensities in the spectrum, assignment of the stretch of residues between Gln-208 to Arg-228 was challenging. In this region, there are groups of residues whose connectivity is unambiguously established, however, there are breaks in between that create uncertainty in exactly how they fit to the sequence. The final assignments were made to maximize internal consistency, in conjunction with the NMR titrations, and the mutational analysis described below.
Figure 3.
NMR backbone resonance assignment of XPA DBD. The region shown is from the 600-MHz 15N-1H HSQC spectrum of XPA DBD acquired at 25 °C in a buffer containing 20 mm Tris, pH 7.0, 500 mm KCl, 1 mm TCEP, and 5% 2H2O. The inset is an expansion of the central region within the rectangle. Chemical shifts have been deposited at the Biological Magnetic Resonance Bank under the accession code 27131.
The results from NMR titrations of 15N-enriched XPA DBD with different DNA substrates were used to map the interaction surface of XPA DBD (supplemental Fig. S2). Overall, the trends in chemical shift perturbations (CSPs) were the same for the different substrates. Major perturbations were found in the globular core between residues 130 and 210 and in residues extending C-terminally from the core between residues 215 and 232 (supplemental Fig. S2). The N-terminal region of the globular core containing a zinc finger remained mostly unaffected. Although the overall patterns are similar, each substrate had some unique features. For example, perturbation of Glu-156 and Trp-175 was only observed for the Y-shaped junction, whereas Ala-229 was only perturbed by the 8/12 5′ overhang substrate. These observations reflect the adaptation of XPA DBD to the substrate.
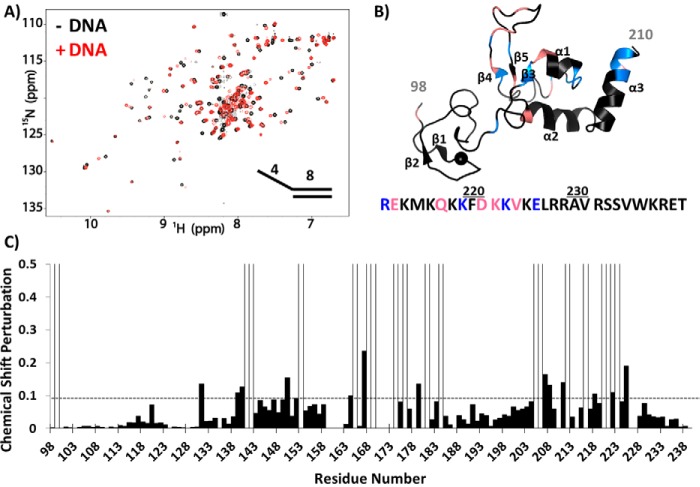
An in-depth analysis of NMR chemical shifts was performed for the titration of 15N-labeled XPA DBD with the 8/4 5′ overhang substrate. Significant CSPs were observed primarily in the C-terminal portion of XPA DBD including β3, α1, the hairpin between β4 and β5, the C-terminal end of α3, and a number of residues in the C-terminal extension (Fig. 4). Of the 34 residues perturbed by addition of the substrate, 16 exhibited fast exchange on the NMR time scale, whereas the 18 others exhibited intermediate exchange and were broadened beyond detection. Making the logical assumption that all CSPs arise from the same DNA-binding phenomenon, the residues with broadened signals have the largest chemical shift differences between the free and bound state and are presumed to be centered in the binding site. This group of residues is highlighted in Fig. 4B, which shows the CSPs mapped on the NMR structure of the human XPA globular core.
Figure 4.
NMR titration of XPA DBD with 8/4 5′ overhang DNA. A, overlay of the 900-MHz 15N-1H HSQC spectrum of XPA DBD in the presence (red) and absence (black) of 8/4 5′ overhang DNA (Fig. 1, substrate 11). The spectra were acquired in a buffer containing 20 mm Tris, pH 7.0, 150 mm KCl, 1 mm TCEP, and 5% 2H2O. B, CSPs from spectra shown in A mapped on the XPA NMR structure (PDB ID: 1XPA). Significant CSPs of C-terminal residues are mapped on the amino acid sequence below the structure. Blue indicates residues exhibited significant CSPs, whereas salmon indicates resonances exchange broadened upon DNA binding. C, plot of CSPs versus residue number from the spectra shown in A. Peaks exhibiting exchange broadening are shown as open bars. The threshold for significant CSP is indicated by the dashed line.
Comparative structural analysis reveals similarities and differences between the binding of DNA by XPA and Rad14
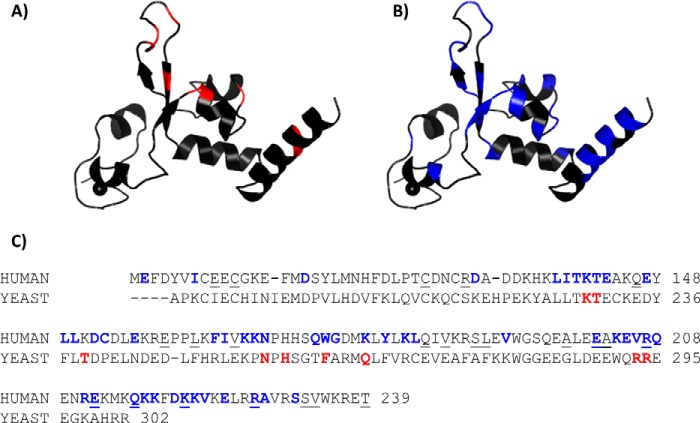
To obtain insight into whether the NMR analysis of DNA binding to the XPA DBD was consistent with the structure of the Rad14-DNA complexes, the XPA CSPs induced upon binding DNA were mapped onto the structure. Fig. 5 shows ribbon diagrams of the XPA homology model constructed from a Rad14 crystal structure (Protein Data Bank code 5A3D), with the residues highlighted that are involved in DNA binding as identified on the basis of the crystal structures of Rad14 (red, panel A) or by NMR (blue, panel B). Unlike Fig. 4, Fig. 5C shows the summary of all perturbed residues from all five ss-ds junction DNA substrates analyzed by NMR.
Figure 5.
Comparing DNA-binding residues identified in Rad14 crystal structures and NMR analyses of human XPA. A and B, mapping of DNA-binding residues identified in the Rad14 crystal structures (A) and NMR titrations (B) on the homology model of human XPA102–214. C, sequence alignment of the DNA binding construct of human XPA (top row) and S. cerevisiae Rad14 (bottom row). DNA-binding residues are colored (colors matched with A and B). The residues reported to be mutated in cancer patients are indicated by underlining (missense mutations only).
Most of the residues involved in contact with DNA in the Rad14 crystal structures exhibit CSPs in the NMR titrations of the human XPA DBD. However, the NMR study revealed many additional residues with significant CSPs induced by the binding of DNA. The most important differences were the CSPs observed in the extra 20 C-terminal residues extending beyond the globular core, which are not present in the Rad14 construct (Fig. 5C). These CSPs are fully consistent with our previous analysis showing that the C-terminal extension beyond the globular core is essential to recapitulating the DNA-binding affinity of full-length XPA (39). Two Rad14 residues, Thr-239 and His-258, contact the DNA, but the corresponding residues in XPA DBD (Lys-151 and His-171) did not exhibit a CSP in the DNA titrations. The absence of an effect on K151 is likely due to a structural adjustment in the flexible loop in which it is found; both neighboring residues around Lys-151, Leu-150, and Asp-152, showed significant CSPs (Fig. 5C), suggesting that one of these residues replaces the Thr-238 contact. As for His-171, involvement of this residue in DNA binding remains unclear because we were unable to assign its resonances.
The NMR analysis indicates four XPA lysine residues (Lys-168, Lys-179, Lys-221, and Lys-222) are involved in DNA binding (Fig. 5C). These data confirm the results of a previous mass spectrometry footprinting study that reported six XPA lysine residues (Lys-168, Lys-179, Lys-221, Lys-222, Lys-224, and Lys-236) are involved in the binding of junction DNA (40). Although Lys-224 did not exhibit a significant DNA binding–induced CSP, both adjacent residues Val-223 and Glu-225 did. Hence, biotinylation on Lys-224 presumably occurred not because this residue directly engages DNA but because the reagent was recruited to the vicinity via the interaction with the adjacent residues. The origin of biotinylation but lack of a CSP for Lys-236 is more puzzling. We have established that progressive truncation of the C-terminal extension from the XPA globular core results in progressively reduced DNA-binding affinity (see below), consistent with a general electrostatic contribution from basic residues such as Lys-236 in the C-terminal extension. Although the NMR chemical shift is very sensitive to even weak interaction, we cannot rule out the possibility that a very weak interaction with DNA was canceled out by an induced structural change, and therefore no NMR CSP was observed. This highlights the need for a high resolution structure of an XPA-DNA complex. In the Rad14 crystal structure, Lys-168 does not contact the DNA and Lys-179 does, but further comparison is limited because the construct is truncated and does not contain Lys-221, Lys-222, and Lys-224.
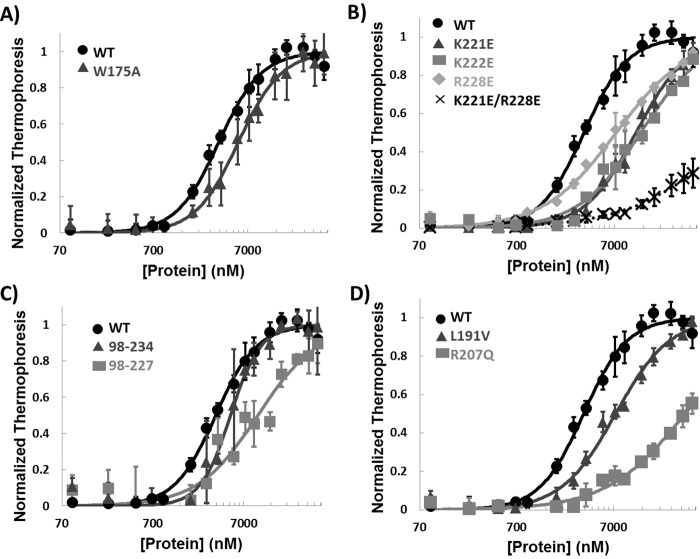
The F262A mutation in Rad14 was reported to cause complete loss of binding to damage-containing duplexes, suggesting the critical importance of this residue in DNA binding by yeast Rad14 (36). The equivalent residue in XPA, Trp-175, showed a significant CSP (Fig. 4), consistent with its involvement in binding DNA and the positioning of the side chain in the homology model. However, when the DNA binding of a W175A mutant was tested, only a slight reduction in DNA-binding affinity to the NER model substrates was observed (Fig. 6A and Table 2). In addition, among the five ss-ds junction DNA substrates tested by NMR, a Trp-175 CSP was observed only in the titration of 8/12 splayed-arm substrate.
Figure 6.
DNA binding of mutant XPA. A–D, MST analyses of DNA binding of WT XPA DBD and W175A mutant (A), mutations of residues in the C-terminal extension from the globular core (B), truncation mutants (C), and disease-associated missense mutants (D). All experiments used the 8/4 HP 5′ overhang DNA (Fig. 1, 9). The error bars indicate standard deviations from at least three measurements.
Table 2.
The affinity of XPA mutants for substrate 9
Dissociation constants (Kd) were extracted from the MST data plotted in Fig. 6. The errors are standard deviations from at least three measurements. Dashes indicate the binding was too weak to extract an accurate Kd value.
| XPA construct | Kd |
|---|---|
| μm | |
| XPA98–239 | 3.4 ± 0.2 |
| XPA98–239 W175A | 5.5 ± 0.34 |
| XPA98–239 K221E | 12.0 ± 1.0 |
| XPA98–239 K222E | 13.0 ± 3.1 |
| XPA98–239 R228E | 7.4 ± 0.6 |
| XPA98–239 K221E/R228E | – |
| XPA98–234 | 5.0 ± 0.3 |
| XPA98–227 | 10.0 ± 3.6 |
| XPA98–239 L191V | 7.4 ± 0.7 |
| XPA98–239 R207Q | – |
Mutation of residues in the C-terminal extension of XPA DBD inhibits binding of DNA
To investigate the contribution of residues 220–239 to DNA binding more closely, we prepared a series of point mutations of basic residues in this region. Three charge reversal, single-site mutations (K221E, K222E, and R228E) resulted in mild reduction in DNA-binding affinity, whereas the double mutation K221E/R228E had a much more dramatic effect (Fig. 6B and Table 2). Truncations of the DBD also caused significant reductions in DNA-binding affinity (Fig. 6C and Table 2). Although both truncation mutants retained some DNA-binding activity, the shorter construct XPA98–227 had weaker DNA-binding activity than the longer XPA98–234. These results confirm that the cluster of basic residues in the C-terminal of XPA DBD contribute significantly to DNA binding (36, 40).
Biophysical and structural studies enhance the general understanding of genotype–phenotype correlations for XPA mutations
The available data for generating genotype-phenotype correlations are limited, especially for many missense mutations discovered in cancer patients (23), simply because there have been few investigations of phenotypes or biochemical malfunctions of specific XPA mutants. The results reported here enable some genotype-phenotype correlations to be made.
Insights were obtained for three disease-associated mutants, V166A, L191V, and R207Q. These mutations were prepared in the XPA DBD construct, expressed, and purified, and their structural integrity and DNA-binding affinity were characterized. In WT XPA DBD, Val-166 did not have significant CSPs upon binding of DNA but is between residues that did (Ile-165 and Lys-167), Leu-191 is distant from residues affected by DNA binding, and Arg-207 has significant CSPs (Fig. 5). As anticipated, L191V did not cause any significant effect on DNA binding, whereas R207Q caused a dramatic decrease in DNA-binding affinity. Interestingly, we were unable to concentrate V166A sufficiently to conduct the DNA-binding assay, suggesting reduced stability of this mutant. We were, however, able to establish that the structural integrity of this mutant was maintained, because both the NMR and CD spectra were the same as WT (supplemental Fig. S3). To directly determine whether the mutation did indeed alter the stability of the domain, thermal denaturation was monitored by CD for the mutant and the WT protein (supplemental Fig. S3B). These data show that the V166A mutation results in a 4 °C lower apparent thermal denaturation midpoint than the WT. Hence, although the V166A mutation does not grossly alter folding, it does significantly reduce the stability of the globular core. In the Rad14 crystal structures and homology models of XPA, the Val-166 residue is a part of the β-hairpin that intercalates into the ss-dsDNA junction. The decrease in stability could be due to the loss of hydrophobic interactions that support the cross-strand interaction in the β-hairpin. Given its important role in XPA binding of DNA, destabilization of the β-hairpin may well prove to be the biochemical malfunction at the origin of the disease-association of the V166A mutant.
Discussion
XPA is central scaffold for human NER machinery, and although loss of DNA-binding activity of XPA is anticipated to lead to severe XP symptoms (35), XPA binding to the NER bubble and its effect on NER efficiency or correlation to XP symptoms have not been investigated. One contributing factor was that the DNA-binding domain had been incorrectly assigned to the XPA globular core, which has only weak affinity for DNA (39, 54). Important new insights were obtained recently when the first high-resolution structures in the presence of DNA were reported for the yeast homolog of XPA, Rad14 (36). However, whether or not these structures serve as accurate models for human XPA in the context of NER remains unclear because the DNA substrates in the crystal structures were 15- or 16-bp DNA duplexes modified at the center with specific lesions. These substrates are not representative of canonical NER, because XPA is not recruited to the site of damage until TFIIH unwinds the damaged DNA (6, 7). The specific lesion-containing duplexes appear to be very unique substrates because in general, XPA binds ss-ds junctions much more tightly than duplexes (51). Indeed, Rad14 binds unmodified duplexes and other lesion-containing duplexes with substantially lower affinity than duplexes modified specifically with either cisplatin and AAF (36). Although these observations are consistent with the observations that purified XPA preferentially binds to damage containing DNA in vitro (55–57), it is widely accepted that these lesions are first recognized by XPC in global genome repair or by stalling of RNA polymerase in transcription-coupled repair (2, 50, 58). XPA has been shown to interact with proteins not involved in NER (e.g. XPA binding proteins (41–44) and proliferating cell nuclear antigen (23, 45)), and it is conceivable that the Rad14 structures reveal the molecular basis of DNA interactions in the other pathways for cellular processing or bypass of damaged DNA. Regardless, structural information on human XPA in complex with NER substrates is required to better understand the molecular basis of XPA function in NER.
We have investigated the interaction of human XPA with DNA using a DBD construct that exhibits the same DNA-binding affinity as the full-length protein and with ss-dsDNA junction substrates that model the NER bubble created after the damaged duplex is unwound by TFIIH. NMR backbone resonance assignments allowed identification of the residues affected by binding to DNA substrates. Analysis of different DNA substrates resulted in similar CSPs, suggesting that the same set of residues is involved in DNA binding for variety of DNA structures with different binding affinities. For the most part, there was a good correlation between NMR CSPs of residues in the globular core and the corresponding DNA binding site in the yeast Rad14 crystal structures. Thus, the two proteins are found to utilize the same general mode of binding DNA. This result is consistent with the conservation of key DNA-binding residues in the globular core (23, 36).
Nevertheless, uncertainties remain as to whether the Rad14 structures provide an accurate model for DNA binding by XPA in the context of NER. In particular, the NMR analysis revealed significant DNA contacts with residues in the C-terminal residues that extend beyond the globular core and are missing from the yeast construct, consistent with our finding that the XPA globular core lacking these residues does not bind DNA with appreciable affinity (39). Our mutational analysis of basic residues at the C terminus of XPA DBD shows that these residues collectively contribute to the high affinity for DNA. It is possible that the absence of C-terminal residues in the Rad14 construct used for crystallization lead to differences in the mode of binding of differences substrates.
The observation that Rad14 binds with high affinity only to duplexes containing cisplatin or AAF lesions, and not to unmodified duplexes or duplexes with different lesions, further supports the idea that the crystal structure may reveal a unique mode of DNA binding (36). It is also conceivable that the yeast and human proteins do not completely correspond; for example, the F262A mutant in the Rad14 globular core completely abolished binding to DNA (36), whereas the corresponding mutation in human XPA DBD (W175A) did not significantly alter DNA-binding activity (Fig. 6A). However, it is not possible to rule out the possibility that this difference is the result of the differences in the constructs used in these two studies.
It is also difficult to rationalize that the crystal structures of Rad14 have two molecules bound, one on either end of the lesion-containing duplex (36). That said, it is conceivable that the end-on mode of interaction of Rad14 is in some way mimicked when XPA is engaged at the ss-dsDNA junction within the NER bubble. In fact, characterization of XPA in vitro revealed that it can form a dimer, although the nature of this dimer is not known (57, 59). Nevertheless, considering the extensive network of protein-protein interactions of XPA in NER complexes (60–65), it is difficult to imagine how two molecules of XPA could interact simultaneously within the preincision and incision complexes. Indeed, all current models of NER complexes incorporate only a single XPA molecule (23, 66).
Our studies also shed light on genotype-phenotype relationships for certain XPA mutants; in particular, we believe a correlation exists between the effect of mutation on DNA-binding affinity and severity of symptoms in XP patients. For example, XPA patients expressing truncation mutant XPA1–227 are known to exhibit mild neurological disorders, whereas those with XPA1–220 have more severe neurological symptoms (30). Our current and previous data show that XPA DBD truncated at Arg-227 retains weak DNA binding (Fig. 6C and Table 2), whereas truncation at Phe-219 barely retains any DNA-binding activity (39). These observations suggest a correlation exists between the degree of inhibition of XPA DNA-binding activity and XP disease phenotype. Hence, the observation of only very small reductions in DNA-binding affinity for the single-site charge reversal mutations of Lys-221 and Arg-228 (Fig. 6B and Table 2) suggests that the K221Q and R228Q mutations will result in relatively mild disease symptoms.
DNA-binding affinity is but one of several factors that can contribute to biochemical malfunction of disease-associated mutations. For example, we found that the V166A mutation is destabilized compared with the WT protein, which could affect not only DNA binding but also protein interactions and cellular turnover. Arg-207 presents another interesting example. We found that the disease-associated R207Q mutation has significantly weakened DNA-binding affinity, whereas a host reactivation assay showed that the R207E mutant is capable of the repair of UV lesions (67). The assay is not sensitive to effects on the rate of repair, which may be compromised when DNA-binding affinity is diminished. Moreover, this residue is also important for interactions with the NER factor XPE (68).
The remarkably rapid progress in recent years in structural analysis of large multiprotein complexes, such as those assembled along the trajectory of NER, holds promise for dissecting such complex relationships. Systematic functional analyses, for example of UV lesion repair efficiency, will also be required. The combination of mechanistic and functional data will be critical to attaining the ultimate objectives of a more complete understanding of the relationship of XPA DNA-binding affinity to NER activity and of the genotype-phenotype correlation of disease-associated mutations.
Experimental procedures
XPA DBD mutant construction
We previously reported the construction and purification of XPA DBD (XPA98–239) (39). Using the pBG100 XPA DBD plasmid as the template, single-site mutations were introduced using the Q5 site-directed mutagenesis kit (New England BioLabs, Inc.) following the manufacturer's protocols. The K221E/R228E double mutant was created using K221E primers and pBG100 XPA DBD R228E plasmid as the template. Primers for mutagenesis are listed in supplemental Table S2. For the truncation mutants R228X (XPA98–227) and W235X (XPA98–234), XPA constructs were amplified by PCR using the oligonucleotides indicated in supplemental Table S2 to introduce 5′ BamHI and 3′ NotI cleavage sites. All mutants were cloned into the pBG100 in-house expression vector (L. S. Mizoue, Center for Structural Biology, Vanderbilt University), which incorporates an N-terminal human rhinovirus 3C (HRV3C) protease cleavable His6 tag.
DNA substrate preparation
Fig. 1 shows the structures of DNA substrates used in this study. The position of the FITC tag is indicated if applicable. Desalted oligodeoxynucleotides were purchased from Sigma–Aldrich. Y-shaped ssDNA-dsDNA junctions, and duplexes were prepared by mixing an equimolar amount at 0.5–1 mm of each strand in the buffers listed below for NMR or DNA binding experiments. Then the mixture was heated in a boiling water bath and allowed to cool to room temperature for annealing. Hairpin-containing DNA substrates were dissolved in TNE buffer (10 mm Tris, pH 7.0, 50 mm NaCl, 1 mm EDTA) at 2 μm concentration and annealed by heating in a boiling water bath followed by immediate cooling on ice.
XPA DBD production
WT and mutant XPA DBD proteins were expressed and purified as described previously (39). The XPA DBD used for NMR titrations was expressed in minimal medium containing 0.5 g/liter of 15NH4Cl (Cambridge Isotope Laboratories, Inc.). The preparation of samples for backbone resonance assignments also contained 2 g/liter 13C6-glucose (CIL, Inc.).
Measurement of DNA binding affinities
The protocol for the fluorescence anisotropy DNA-binding assay was described previously (39). For measurements using MST (69–72), proteins were dialyzed into MST buffer (50 mm Tris-HCl, pH 7.8, 150 mm NaCl, 10 mm MgCl2, 0.05% Tween 20, and 1 mm DTT). Fluorescein-labeled DNA stocks were also diluted in the MST buffer. All samples and buffer were passed through a 0.2-μm filter. For each experiment, 16 dilutions of the protein were prepared to varying concentrations. DNA was added to each of the sample to the final concentration of 40 nm in the tubes provided in the Monolith NT.115 series standard treated capillaries kit (NanoTemper, Inc.). All experiments were carried out at room temperature using the capillaries in a Monolith NT.115 Blue/Red instrument (NanoTemper, Inc.) at 20% LED power and 40% MST power. The data were analyzed using MO.Affinity software (NanoTemper, Inc.).
Circular dichroism
Samples of XPA DBD WT and the V166A mutant were dialyzed into a buffer containing 20 mm Tris, pH 7.0, 150 mm KCl, and 1 mm DTT. All samples and buffer were passed through a 0.2-μm filter before data collection. The concentration of protein was adjusted to 11 μm, and far-UV CD data were collected at 220 nm over the range 15–65 °C using a Jasco J-810 CD spectropolarimeter (Easton, MD).
Generation of the XPA DBD homology model
A homology model of human XPA102–214 was generated using the X-ray crystal structure of S. cerevisiae Rad14 (Protein Data Bank code 5A3D) as template for calculations using Modeler 9.14 (73).
1H 1D NMR analysis of XPA DBD V166A
XPA DBD WT and V166A mutant were concentrated to 30 μm in a buffer containing 20 mm Tris, pH 7.0, 75 mm KCl, and 1 mm TCEP to which 5% 2H2O was added prior to performing the experiment. The 1H 1D spectrum of each sample was acquired in a 3-mm tube at 25 °C using a Bruker Avance 600-MHz spectrometer equipped with a cryoprobe.
NMR backbone resonance assignments
13C,15N-enriched XPA DBD was concentrated to 460 μm in a buffer containing 20 mm Tris, pH 7.0, 500 mm KCl, 1 mm TCEP, and 5% 2H2O. 15N-1H HSQC and a series of standard heteronuclear triple resonance 3D experiments (HNCO, HNCA, HNCACB, HNCOCA, and CBCACONH) were acquired in a shaped tube at 25 °C using a Bruker Avance-III 600-MHz spectrometer equipped with a QCI cryoprobe. All 3D data sets were collected using flip-back pulses and a Watergate sequence to suppress the water signal (74, 75), as well as 25 and 50% non-uniform sampling (76–81) for the backbone and side chain experiments, respectively. Reconstruction and processing of the raw data were performed with the standard Topspin (Bruker) software, using compressed sensing, one time 0 filling, and shifted squared sine bell (60–90 degree) apodization. Further data processing and figure preparation were carried out using SPARKY (82). Resonance assignments were obtained and their reliability evaluated using NMRView software, utilizing the RunAbout tool (One Moon Scientific).
NMR titration of XPA DBD with DNA
All samples for DNA titrations were concentrated to 50 μm in the NMR buffer containing 20 mm Tris, pH 7.0, 150 mm KCl, 1 mm TCEP, and 5% 2H2O. Titrations were performed at 35 °C using a Bruker Avance-III 800- or 900-MHz spectrometer equipped with a TCI cryoprobe following the procedure described previously (39). A second titration with 8/4 5′ overhang substrate was performed in the NMR buffer with 250 mm instead of 150 mm KCl at 25 °C to enable transfer of the assignments from the conditions used for obtaining resonance assignments.
Author contributions
N. S. and W. J. C. designed research. N. S. conducted most of the experiments and analyzed data. M. W. V. performed resonance assignment and provided assistance for NMR experiments. M. S. R. subcloned, expressed, purified, and generated preliminary DNA-binding data for some of the disease-associated XPA DBD mutants. A. M. T.-W. assisted preparation of NMR samples, as well as some MST experiments. N. S. and W. J. C. wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Nicole Eckart from NanoTemper Technologies for the valuable input to optimize and interpret MST data, Dr. Jonathan Sheehan for assistance with construction of the homology model, and Dr. Sarah E. Soss for advice on backbone resonance assignment. NMR instrumentation was acquired with support from the National Science Foundation through Grant 0922862, from the National Institutes of Health through Grant S10 RR025677, and from Vanderbilt University. MST instrumentation was acquired from National Institutes of Health through Grant S10 OD021483.
This work was supported by National Institutes of Health Grants P01 CA092584, R35 GM118089, P30 CA068485, and P30 ES000267. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
- NER
- nucleotide excision repair
- DNA-binding affinity
- xeroderma pigmentosum
- XPA
- xeroderma pigmentosum complementation group A
- DBD
- DNA-binding domain
- RPA
- replication protein A
- CSP
- chemical shift perturbation
- MST
- microscale thermophoresis
- HP
- hairpin
- TCEP
- tris(2-carboxymethyl) phosphine
- nt
- nucleotide
- AAF
- N-(deoxyguanosin-8-ly)-2-acetylaminofluorene
- ds
- double-stranded
- ss
- single-stranded.
References
- 1. Schärer O. D. (2013) Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 5, a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gillet L. C., and Schärer O. D. (2006) Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 106, 253–276 [DOI] [PubMed] [Google Scholar]
- 3. Truglio J. J., Croteau D. L., Van Houten B., and Kisker C. (2006) Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 106, 233–252 [DOI] [PubMed] [Google Scholar]
- 4. Alekseev S., and Coin F. (2015) Orchestral maneuvers at the damaged sites in nucleotide excision repair. Cell. Mol. Life Sci. 72, 2177–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marteijn J. A., Lans H., Vermeulen W., and Hoeijmakers J. H. (2014) Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15, 465–481 [DOI] [PubMed] [Google Scholar]
- 6. Riedl T., Hanaoka F., and Egly J. (2003) The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22, 5293–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li C.-L., Golebiowski F. M., Onishi Y., Samara N. L., Sugasawa K., and Yang W. (2015) Tripartite DNA lesion recognition and verification by XPC, TFIIH, and XPA in nucleotide excision repair. Mol. Cell 59, 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shell S. M., Hawkins E. K., Tsai M. S., Hlaing A. S., Rizzo C. J., and Chazin W. J. (2013) Xeroderma pigmentosum complementation group C protein (XPC) serves as a general sensor of damaged DNA. DNA Repair (Amst.) 12, 947–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Batty D., Rapic'-Otrin V., Levine A. S., and Wood R. D. (2000) Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 300, 275–290 [DOI] [PubMed] [Google Scholar]
- 10. Uchida A., Sugasawa K., Masutani C., Dohmae N., Araki M., Yokoi M., Ohkuma Y., and Hanaoka F. (2002) The carboxy-terminal domain of the XPC protein plays a crucial role in nucleotide excision repair through interactions with transcription factor IIH. DNA Repair (Amst.) 1, 449–461 [DOI] [PubMed] [Google Scholar]
- 11. Min J. H., and Pavletich N. P. (2007) Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature 449, 570–575 [DOI] [PubMed] [Google Scholar]
- 12. Yokoi M., Masutani C., Maekawa T., Sugasawa K., Ohkuma Y., and Hanaoka F. (2000) The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 275, 9870–9875 [DOI] [PubMed] [Google Scholar]
- 13. Fuss J. O., and Tainer J. A. (2011) XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst.) 10, 697–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hermanson-Miller I. L., and Turchi J. J. (2002) Strand-specific binding of RPA and XPA to damaged duplex DNA. Biochemistry 41, 2402–2408 [DOI] [PubMed] [Google Scholar]
- 15. Krasikova Y. S., Rechkunova N. I., Maltseva E. A., Petruseva I. O., and Lavrik O. I. (2010) Localization of xeroderma pigmentosum group A protein and replication protein A on damaged DNA in nucleotide excision repair. Nucleic Acids Res. 38, 8083–8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Donovan A., Davies A. A., Moggs J. G., West S. C., and Wood R. D. (1994) XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371, 432–435 [DOI] [PubMed] [Google Scholar]
- 17. Staresincic L., Fagbemi A. F., Enzlin J. H., Gourdin A. M., Wijgers N., Dunand-Sauthier I., Giglia-Mari G., Clarkson S. G., Vermeulen W., and Schärer O. D. (2009) Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 28, 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsodikov O. V., Ivanov D., Orelli B., Staresincic L., Shoshani I., Oberman R., Schärer O. D., Wagner G., and Ellenberger T. (2007) Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. EMBO J 26, 4768–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shivji M., Moggs J., Kuraoka I., and Wood R. (1999) Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol. Biol. 113, 373–392 [DOI] [PubMed] [Google Scholar]
- 20. Spivak G. (2015) Nucleotide excision repair in humans. DNA Repair (Amst.) 36, 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spivak G., and Ganesan A. K. (2014) The complex choreography of transcription-coupled repair. DNA Repair (Amst.) 19, 64–70 [DOI] [PubMed] [Google Scholar]
- 22. Vermeulen W., and Fousteri M. (2013) Mammalian transcription-coupled excision repair. Cold Spring Harb. Perspect. Biol. 5, a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sugitani N., Sivley R. M., Perry K. E., Capra J. A., and Chazin W. J. (2016) XPA: a key scaffold for human nucleotide excision repair. DNA Repair (Amst.) 44, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DiGiovanna J. J., and Kraemer K. H. (2012) Shining a light on xeroderma pigmentosum. J. Invest. Dermatol. 132, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoeijmakers J. H. (2009) DNA damage, aging, and cancer. N. Engl., J. Med. 361, 1475–1485 [DOI] [PubMed] [Google Scholar]
- 26. Lehmann A. R., McGibbon D., and Stefanini M. (2011) Xeroderma pigmentosum. Orphanet J. Rare Dis. 6, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lehmann A. R. (2012) DNA repair, DNA replication and human disorders: a personal journey. DNA Repair (Amst.) 11, 328–334 [DOI] [PubMed] [Google Scholar]
- 28. Bradford P. T., Goldstein A. M., Tamura D., Khan S. G., Ueda T., Boyle J., Oh K. S., Imoto K., Inui H., Moriwaki S., Emmert S., Pike K. M., Raziuddin A., Plona T. M., DiGiovanna J. J., et al. (2011) Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. J. Med. Genet. 48, 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cleaver J. E. (2005) Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer 5, 564–573 [DOI] [PubMed] [Google Scholar]
- 30. Cleaver J. E., and States J. C. (1997) The DNA damage-recognition problem in human and other eukaryotic cells: the XPA damage binding protein. Biochem. J. 328, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rouillon C., and White M. F. (2011) The evolution and mechanisms of nucleotide excision repair proteins. Res. Microbiol. 162, 19–26 [DOI] [PubMed] [Google Scholar]
- 32. Anttinen A., Koulu L., Nikoskelainen E., Portin R., Kurki T., Erkinjuntti M., Jaspers N. G., Raams A., Green M. H., Lehmann A. R., Wing J. F., Arlett C. F., and Marttila R. J. (2008) Neurological symptoms and natural course of xeroderma pigmentosum. Brain 131, 1979–1989 [DOI] [PubMed] [Google Scholar]
- 33. Asahina H., Kuraoka I., Shirakawa M., Morita E. H., Miura N., Miyamoto I., Ohtsuka E., Okada Y., and Tanaka K. (1994) The XPA protein is a zinc metalloprotein with an ability to recognize various kinds of DNA damage. Mutat Res. 315, 229–237 [DOI] [PubMed] [Google Scholar]
- 34. Morita E. H., Ohkubo T., Kuraoka I., Shirakawa M., Tanaka K., and Morikawa K. (1996) Implications of the zinc-finger motif found in the DNA-binding domain of the human XPA protein. Genes Cells 1, 437–442 [DOI] [PubMed] [Google Scholar]
- 35. States J. C., McDuffie E. R., Myrand S. P., McDowell M., and Cleaver J. E. (1998) Distribution of mutations in the human xeroderma pigmentosum group A gene and their relationships to the functional regions of the DNA damage recognition protein. Hum. Mutat. 12, 103–113 [DOI] [PubMed] [Google Scholar]
- 36. Koch S. C., Kuper J., Gasteiger K. L., Simon N., Strasser R., Eisen D., Geiger S., Schneider S., Kisker C., and Carell T. (2015) Structural insights into the recognition of cisplatin and AAF-dG lesion by Rad14 (XPA). Proc. Natl. Acad. Sci. U.S.A. 112, 8272–8277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ikegami T., Kuraoka I., Saijo M., Kodo N., Kyogoku Y., Morikawa K., Tanaka K., and Shirakawa M. (1998) Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA. Nat. Struct. Biol. 5, 701–706 [DOI] [PubMed] [Google Scholar]
- 38. Buchko G. W., Ni S., Thrall B. D., and Kennedy M. A. (1998) Structural features of the minimal DNA binding domain (M98–F219) of human nucleotide excision repair protein XPA. Nucleic Acids Res. 26, 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugitani N., Shell S. M., Soss S. E., and Chazin W. J. (2014) Redefining the DNA-binding domain of human XPA. J. Am. Chem. Soc. 136, 10830–10833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hilton B., Shkriabai N., Musich P. R., Kvaratskhelia M., Shell S., and Zou Y. (2014) A new structural insight into XPA-DNA interactions. Biosci. Rep. 34, e00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nitta M., Saijo M., Kodo N., Matsuda T., Nakatsu Y., Tamai H., and Tanaka K. (2000) A novel cytoplasmic GTPase XAB1 interacts with DNA repair protein XPA. Nucleic Acids Res. 28, 4212–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lembo F., Pero R., Angrisano T., Vitiello C., Iuliano R., Bruni C. B., and Chiariotti L. (2003) MBD in a novel MBD2-interacting protein relieves MBD2 repression potential and reactivates transcription from methylated promoters. Mol. Cell Biol. 23, 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yonemasu R., Minami M., Nakatsu Y., Takeuchi M., Kuraoka I., Matsuda Y., Higashi Y., Kondoh H., and Tanaka K. (2005) Disruption of mouse XAB2 gene involved in pre-mRNA splicing, transcription and transcription-coupled DNA repair results in preimplantation lethality. DNA Repair (Amst.) 4, 479–491 [DOI] [PubMed] [Google Scholar]
- 44. Nakatsu Y., Asahina H., Citterio E., Rademakers S., Vermeulen W., Kamiuchi S., Yeo J. P., Khaw M. C., Saijo M., Kodo N., Matsuda T., Hoeijmakers J. H., and Tanaka K. (2000) XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J. Biol. Chem. 275, 34931–34937 [DOI] [PubMed] [Google Scholar]
- 45. Gilljam K. M., Müller R., Liabakk N. B., and Otterlei M. (2012) Nucleotide excision repair is associated with the replisome and its efficiency depends on a direct interaction between XPA and PCNA. PLoS One 7, e49199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dusinská M., Dzupinková Z., Wsólová L., Harrington V., and Collins A. R. (2006) Possible involvement of XPA in repair of oxidative DNA damage deduced from analysis of damage, repair and genotype in a human population study. Mutagenesis 21, 205–211 [DOI] [PubMed] [Google Scholar]
- 47. Reardon J. T., and Sancar A. (2002) Molecular anatomy of the human excision nuclease assembled at sites of DNA damage. Mol. Cell. Biol. 22, 5938–5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wakasugi M., and Sancar A. (1999) Order of assembly of human DNA repair excision nuclease. J. Biol. Chem. 274, 18759–18768 [DOI] [PubMed] [Google Scholar]
- 49. Mu D., Wakasugi M., Hsu D. S., and Sancar A. (1997) Characterization of reaction intermediates of human excision repair nuclease. J. Biol. Chem. 272, 28971–28979 [DOI] [PubMed] [Google Scholar]
- 50. Sugasawa K. (2016) Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Repair (Amst.) 44, 110–117 [DOI] [PubMed] [Google Scholar]
- 51. Yang Z., Roginskaya M., Colis L. C., Basu A. K., Shell S. M., Liu Y., Musich P. R., Harris C. M., Harris T. M., and Zou Y. (2006) Specific and efficient binding of xeroderma pigmentosum complementation group A to double-strand/single-strand DNA junctions with 3′- and/or 5′-ssDNA branches. Biochemistry 45, 15921–15930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mou T. C., Shen M., Abdalla S., Delamora D., Bochkareva E., Bochkarev A., and Gray D. M. (2006) Effects of ssDNA sequence on non-sequence-specific protein binding. Chirality 18, 370–382 [DOI] [PubMed] [Google Scholar]
- 53. Sattler M., Schleucher J., and Griesinger C. (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 [Google Scholar]
- 54. Kuraoka I., Morita E. H., Saijo M., Matsuda T., Morikawa K., Shirakawa M., and Tanaka K. (1996) Identification of a damaged-DNA binding domain of the XPA protein. Mutat. Res. 362, 87–95 [DOI] [PubMed] [Google Scholar]
- 55. Jones C. J., and Wood R. D. (1993) Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry 32, 12096–12104 [DOI] [PubMed] [Google Scholar]
- 56. Missura M., Buterin T., Hindges R., Hübscher U., Kaspárková J., Brabec V., and Naegeli H. (2001) Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 20, 3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu Y., Yang Z., Utzat C., Wang G., Basu A. K., and Zou Y. (2005) Cooperative interaction of human XPA stabilizes and enhances specific binding of XPA to DNA damage. Biochemistry 44, 7361–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Spivak G. (2016) Transcription-coupled repair: an update. Arch. Toxicol. 90, 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang Z. G., Liu Y., Mao L. Y., Zhang J. T., and Zou Y. (2002) Dimerization of human XPA and formation of XPA2-RPA protein complex. Biochemistry 41, 13012–13020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Giglia-Mari G., Miquel C., Theil A. F., Mari P. O., Hoogstraten D., Ng J. M., Dinant C., Hoeijmakers J. H., and Vermeulen W. (2006) Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS Biol. 4, e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ziani S., Nagy Z., Alekseev S., Soutoglou E., Egly J. M., and Coin F. (2014) Sequential and ordered assembly of a large DNA repair complex on undamaged chromatin. J. Cell Biol. 206, 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mer G., Bochkarev A., Gupta R., Bochkareva E., Frappier L., Ingles C. J., Edwards A. M., and Chazin W. J. (2000) Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 103, 449–456 [DOI] [PubMed] [Google Scholar]
- 63. Lee S.-H., Kim D.-K., and Drissi R. (1995) Human xeroderma pigmentosum group A protein interacts with human replication protein A and inhibits DNA replication. J. Biol. Chem. 270, 21800–21805 [DOI] [PubMed] [Google Scholar]
- 64. Daughdrill G. W., Buchko G. W., Botuyan M. V., Arrowsmith C., Wold M.S., Kennedy M. A., and Lowry D. F. (2003) Chemical shift changes provide evidence for overlapping single-stranded DNA- and XPA-binding sites on the 70 kDa subunit of human replication protein A. Nucleic Acids Res. 31, 4176–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Buchko G. W., Daughdrill G. W., de Lorimier R., Rao B. K., Isern N. G., Lingbeck J. M., Taylor J. S., Wold M. S., Gochin M., Spicer L. D., Lowry D. F., and Kennedy M. A. (1999) Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies. Biochemistry 38, 15116–15128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fadda E. (2016) Role of the XPA protein in the NER pathway: a perspective on the function of structural disorder in macromolecular assembly. Comput. Struct. Biotechnol. J. 14, 78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Camenisch U., Dip R., Vitanescu M., and Naegeli H. (2007) Xeroderma pigmentosum complementation group A protein is driven to nucleotide excision repair sites by the electrostatic potential of distorted DNA. DNA Repair (Amst.) 6, 1819–1828 [DOI] [PubMed] [Google Scholar]
- 68. Wakasugi M., Kasashima H., Fukase Y., Imura M., Imai R., Yamada S., Cleaver J. E., and Matsunaga T. (2009) Physical and functional interaction between DDB and XPA in nucleotide excision repair. Nucleic Acids Res. 37, 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Duhr S., and Braun D. (2006) Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. U.S.A. 103, 19678–19682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jerabek-Willemsen M., André T., Wanner R., Roth H. M., Duhr S., Baaske P., and Breitsprecher D. (2014) Microscale thermophoresis: interaction analysis and beyond. J. Mol. Struct. 1077, 101–113 [Google Scholar]
- 71. Seidel S. A., Dijkman P. M., Lea W. A., van den Bogaart G., Jerabek-Willemsen M., Lazic A., Joseph J. S., Srinivasan P., Baaske P., Simeonov A., Katritch I., Melo F. A., Ladbury J. E., Schreiber G., Watts A., et al. (2013) Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Breitsprecher D., Schlinck N., Witte D., Duhr S., Baaske P., and Schubert T. (2016) Aptamer binding studies using microscale thermophoresis. Methods Mol. Biol. 1380, 99–111 [DOI] [PubMed] [Google Scholar]
- 73. Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., and Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 74. Grzesiek S., and Bax A. (1993) The importance of not saturating H2O in protein NMR: application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 115, 12593–12594 [Google Scholar]
- 75. Kay L. E., Xu G. Y., and Yamazaki T. (1994) Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J. Magn. Reson. 109, 129–133 [Google Scholar]
- 76. Hyberts S. G., Arthanari H., and Wagner G. (2012) Applications of non-uniform sampling and processing. Top. Curr. Chem. 316, 125–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hyberts S. G., Robson S. A., and Wagner G. (2013) Exploring signal-to-noise ratio and sensitivity in non-uniformly sampled multi-dimensional NMR spectra. J. Biomol. NMR 55, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hyberts S. G., Arthanari H., Robson S. A., and Wagner G. (2014) Perspectives in magnetic resonance: NMR in the post-FFT era. J. Magn. Reson. 241, 60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hyberts S. G., Frueh D. P., Arthanari H., and Wagner G. (2009) FM reconstruction of non-uniformly sampled protein NMR data at higher dimensions and optimization by distillation. J. Biomol. NMR 45, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hyberts S. G., Takeuchi K., and Wagner G. (2010) Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J. Am. Chem. Soc. 132, 2145–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hyberts S. G., Milbradt A. G., Wagner A. B., Arthanari H., and Wagner G. (2012) Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson gap scheduling. J. Biomol. NMR 52, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Goddard T. D., and Kneller D. G., SPARKY 3, University of California, San Francisco [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.